Abstract
Purposes
To determine if the deregulation of genes relevant for normal thymus development can contribute to the biology of thymic epithelial tumors.
Experimental Design
Using array comparative genomic hybridization, we evaluated the copy number aberrations of genes regulating thymus development. The expression of genes most commonly involved in copy number aberrations was evaluated by immunohistochemistry and correlated with patients' outcome. Correlation between FOXC1 copy number loss and gene expression was determined in a confirmation cohort. Cell lines were used to test the role of FOXC1 in tumors.
Results
Among 31 thymus development-related genes, PBX1 copy number gain and FOXC1 copy number loss were presented in 43.0% and 39.5% of the tumors respectively. Immunohistochemistry on a series of 132 thymic epithelial tumors including those evaluated by comparative genomic hybridization, revealed a correlation between protein expression and copy number status only for FOXC1 but not for PBX1. Patients with FOXC1–negative tumors had a shorter time to progression and a trend for a shorter disease related survival. The correlation between FOXC1 copy number loss and mRNA expression was confirmed in a separate cohort of 27 thymic epithelial tumors. Ectopic FOXC1 expression attenuated anchorage-independent cell growth and cell migration in vitro.
Conclusion
Our data support a tumor suppressor role of FOXC1 in thymic epithelial tumors.
Keywords: thymic epithelial tumors, thymoma, thymic carcinoma, FOXC1, PBX1, thymus development
Introduction
Thymic epithelial tumors (TETs) are a group of rare neoplasms with heterogeneous histological features and clinical behavior. Thymic carcinomas are aggressive tumors, that microscopically remind the features of carcinomas of other organs(1). On the contrary, the histological appearance of thymomas resembles the structure of normal thymus and these tumors are grouped into five subcategories (A, AB, B1, B2, B3), depending on their cancer cell shape, degree of atypia and number of intratumoral thymocytes, according to the most recent WHO classification(1). Thymomas, but not thymic carcinomas, are frequently associated with paraneoplastic syndromes: myasthenia gravis being the most common (1). Surgery represents the mainstay of treatment for thymic malignancies, and prognosis is significantly influenced by pathological stage and by completeness of tumor resection (2). Metastatic and nonresectable TETs are candidates for systemic therapy. Although, combination chemotherapy is able to induce substantial tumor shrinkage of variable duration, it is not curative in patients with metastatic disease(2). There is very little understanding of the biology of these neoplasms and molecular prognostic markers and specific targets for therapy have so far not been identified.
Tumor cells share many stem cell-like properties with embryonic cells and the ectopic reactivation of embryo-restricted genes is observed during the neoplastic progression(3). Several cell lineage-specific transcription factors, implicated in organogenesis, have been found to be ectopically reactivated by copy number (CN) aberrations in cancers, including NKX2-1 (TITF) in lung cancer (4–8), ESR1 in breast cancer (9), GATA6 in pancreatic cancer (10) and MITF in melanoma (11).
The role of thymic developmental genes in TETs has not been explored to date. Thymic epithelial cells originate from the endoderm of the third pharyngeal pouch(12). Finely spatiotemporally regulated waves of proliferation and differentiation of thymic epithelial cell precursors are necessary for the normal maturation of the thymus as well as interactions with thymic septum cells of neural crest origin and lymphocyte precursors (thymocytes)(13). To efficiently sustain this process, genes controlling the cell proliferation need to be expressed or repressed at precise moments of the development of thymic epithelial cells (13).
We performed array Comparative Genomic Hybridization (CGH) in TETs to determine the CN status of genes involved in thymus development. Interestingly, FOXC1 and PBX1, two transcription factors that regulate TBX1 expression, were frequently included in regions of CN loss and CN gain, respectively. We suggest that FOXC1 presents tumor suppressor-like activity and loss of FOXC1 expression correlates with poorer time to progression in TET patients.
Materials and Methods
Our institution ethical review boards approved this research (ClinicalTrials.gov ID: NCT00965627). The study has been conducted in agreement with the Declaration of Helsinki.
Patients and samples
Cohort 1
Clinical data and Formalin Fixed Paraffin Embedded (FFPE) samples were collected from a series of 132 consecutive patients who underwent surgery for TET at the Istituto Clinico Humanitas (Rozzano-Milan, Italy) in the period from 1996 to 2008. One hundred and nine patients underwent surgery for primary tumors and 23 for a tumor relapse. In case of relapsed tumors, the date of the first surgery was considered, extending the observation to a period ranging from 1976 to 2008. This series has been described in detail elsewhere(14). Patients were staged according to the Masaoka staging system (15). The completeness of resection was defined as R0=complete resection, R1=microscopic residual disease infiltrating resection margins and R2=macroscopic residual disease (16). An experienced thoracic tumor pathologist (HSL) reviewed the retrieved material for the amount of tumor content, adequate storage and classified the TETs according to the 2004 WHO classification (1).
Confirmation cohort
Frozen TET samples from 27 patients were collected at NCI (Bethesda, MD). Patient characteristics are summarized in table 1.
Table 1.
FOXC1 copy number status and patients' characteristics of the confirmation cohort of 27 frozen thymic epithelial tumors.
| Number of Cases | Frequency of FOXC1 CN Loss | |
|---|---|---|
| Median age | 53 | |
| Range | 36–76 | |
| Sex | ||
| Male | 12 | 33.3% |
| Female | 15 | 60.0% |
| WHO histotype | ||
| A | 5 | 20.0% |
| AB | 2 | 100.0% |
| B2 | 5 | 40.0% |
| B3 | 6 | 83.3% |
| TC | 9 | 44.4% |
| Stage | ||
| IIA | 5 | 40.0% |
| IIB | 6 | 33.3% |
| III | 2 | 100.0% |
| IVA | 4 | 75.0% |
| IVB | 6 | 33.3% |
| Na | 4 | 50.0% |
Stage at the diagnosis was not available for 4 patients (Na), TC: thymic carcinoma.
Nucleic acid extraction and array CGH
Because of the heterogeneity of the histological features and the presence of non neoplastic intratumoral thymocytes only 59 tumor samples containing at least 80% of tumor cells were selected for array CGH, as assessed by Haematoxylin & Eosin (H&E) stained slides.
Frozen tumors were embedded in OCT and 8μm slides were cut in a −20°C cryostat. Slides were stained with Haematoxylin & Eosin (H&E) and sample regions with >80% of tumor cells were macro dissected for nucleic acid extraction.
From FFPE samples, DNA was extracted using DNeasy kit (Qiagen, Inc., Valencia, CA). Both RNA and DNA were extracted from cell lines and frozen tumors using AllPrep DNA/RNA Mini Kit (Qiagen). CGH was performed using Agilent platform according to Genomic DNA ULS labeling kit protocol (Agilent Technologies, Inc., Palo Alto, CA) as previously extensively described elsewhere(17). Twenty samples from FFPE were hybridized on Human Genome CGH Array 105A (Agilent) and the remaining on SurePrint G3 Human CGH Array 180K (Agilent). Frequency of CN aberrations was inferred using Nexus Copy Number 6 (Biodiscovery, Inc., El Segundo, CA, USA). Microarray data have been deposited in gene expression omnibus (GEO) repository with the number: GSE23540.
Copy Number and Real Time PCR
CN-PCR was adopted to confirm CGH results in a subset of tumors. RT-PCR was used for the evaluation of TBX1, PBX1 and FOXC1 gene expression in TET frozen tumors and cell lines. TaqMan gene expression assay (primers) were purchased from Applied Biosystems (Foster City, CA). The GAPDH and RPPH1 genes were used as endogenous controls for mRNA expression and gene copy number validation, respectively. Real-time PCRs were operated on ABI 7900HT fast real-time PCR system (Applied Biosystems). Fold change of mRNA expression was calculated by 2−ΔCt method and gene copy number was determined by CopyCaller software v1.0 (Applied Biosystems). For CN assay, normal male diploid genomic DNA (Promega) was used as a reference after 10 minutes heat fragmentation at 99°C. For CN-PCR, since FOXC1 contains only one exon, 2 couples of primers were designed to avoid possible issues related to residual RNA contamination: one covering the exonic sequence and the other one covering an intergenic region on the chromosome 6p.
FOXC1 sequencing
Primers used for sequencing of the FOXC1 gene are available upon request. Primers were tagged with M13 forward or M13 reverse sequences. DNA from FFPE samples were amplified by PCR using AmpliTaq Gold® PCR Master Mix (Applied Biosystems) and Veriti® 96-Well Thermal Cycler (Applied Biosystems). PCR products underwent ExoSAP-IT® (USB, Cleveland, OH) purification. The purified products were directly sequenced using a BigDye terminator v 3.1 cycle sequencing kit (Applied Biosystems) and 3730xl DNA Analyzer (Applied Biosystems). Data were analyzed using Mutation Surveyor v 3.23 (SoftGenetics LLC, State College, PA).
Immunohistochemistry
Immunohistochemistry was performed using a tissue microarray that included samples from 132 TET patients, as previously described (14, 18). Anti-FOXC1 (1:200, ab5079, Abcam, Cambridge, MA) and anti-PBX1 (1:150, HPA003881, Sigma, St. Louis, MO) antibodies were utilized. According to the human protein atlas website, normal human kidney and pancreas were used as controls for FOXC1 and PBX1, respectively. Positive controls were stained with anti-FOXC1 and anti-PBX1 antibodies and negative controls using respective isogenic serum instead of the primary antibodies. Both percentage and intensity of the stained cells were taken into consideration for immunohistochemistry scoring. Percentage of positive cells was ranked as 0 (0–5%), 1 (6–25%), 2 (26–50%), 3 (51–75%) or 4 (76–100%). For FOXC1, signal intensity 0–1 was considered negative and 2–3 positive; based on the rank of positive cells, samples were graded G0 (rank 0), G1 (rank 1), G2 (rank 2), or G3 (rank 3 and 4). For PBX1, signal intensity was multiplied by rank of positive cells and scores from 0–8 was considered negative (G0), scores of 9 were G1, 10–11 were G2, and 12 were G3.
Cell Lines and experiments
T1889 (thymic carcinoma) and T1682 (B1 thymoma) cell lines were kindly provided by Dr. Marco Breinig (19). TY82 thymic carcinoma cell line was purchased from Japan Health Science Foundation (Tokyo), whereas, NIH-H82, NIH-H69, NIH-H23, NIH-H460, NIH-H1355, HEK-293, NIH-3T3 and U2OS were obtained from ATCC (Manassas, VA). NIH-3T3 cells were cultured in DMEM and all other cell lines were cultured in RPMI 1640; media were supplemented with 200 mM L-Glutamine (Invitrogen, Grand Island, NY), 50 U/mL penicillin, 50 U/mL streptomycin (Invitrogen) and 10% heat-inactivated calf serum (Invitrogen) and grown in a 37°C incubator with humidified 5% CO2 atmosphere. For T1889 and T1682 medium was supplemented with 25nM Hepes.
Western blot
Protein extraction and western blot were performed as previously described (20, 21) using anti-N-terminal-FOXC1 (1:200, overnight 4°C incubation, Santa Cruz biotecgnology, Inc., Santa Cruz CA), anti-PBX1 (1:1000, overnight 4°C incubation, Cell Signaling Technology, Danvers, MA) anti-TBX1 (1:1000, overnight 4°C incubation, Epitomics, Burlingame, CA) and anti-α-Tubulin (1:2000, 1-hour room temperature incubation, Cell Signaling) antibodies, respectively.
Statistical analysis
Survival curves were generated using the Kaplan-Meier method. Disease related survival (DRS) was determined from the date of surgery on primary tumor to the date of death due to tumor progression. Time to progression (TTP) was calculated from date of surgery to relapse or progression. Differences between survival curves were, firstly, determined by a Log-rank test. Subsequently, those factors, which appeared to be at least modestly associated with outcome (p<0.10) in univariate analyses, were evaluated for their joint impact on TTP or DRS using a Cox proportional hazard model. All p-values are two tailed and have not been adjusted for multiple comparisons. Clinical and biological characteristics were compared using two-tail Fisher's exact test, or χ2-test or student's T-test or One-way ANOVA with Tukey's test for post-hoc comparisons, when appropriate. Pearson R coefficient described the correlation between FOXC1 and TBX1 expression. All tests were performed using the SPSS version 17 (SPSS, Inc., Chicago, IL), SAS version 8.2 (SAS Institute, Inc., Cary, NC.) and Prism 5 (GraphPad Software, La Jolla, CA).
Results
Copy number aberrations of genes involved in thymus development
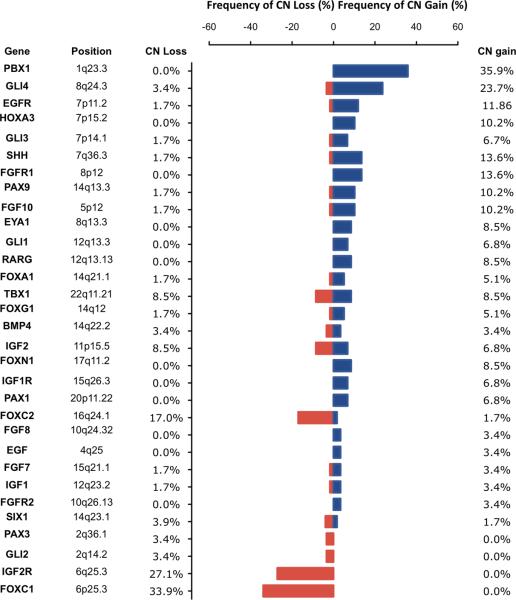
A group of 31 genes implicated in the thymus organogenesis was identified from an extensive literature review (12, 13, 22, 23). The CN aberrations of these 31 thymic developmental genes were determined using array CGH in a series of 59 FFPE TET samples. Frequencies of CN aberration are summarized in figure 1. The most frequent CN alterations were CN gains of PBX1 and GLI4, as well as CN losses of FOXC1 and IGF2R (p<0.05). Interestingly, both PBX1 and FOXC1 have been described to be able to regulate the expression of TBX1, a transcription factor indispensable for the thymus development.
Figure 1. Copy number aberrations of genes regulating normal thymus development.
Genomic CN aberrations of 59 FFPE samples of thymic epithelial tumors were evaluated using array CGH. The frequency of CN loss (red) and gain (blue) of genes implicated in normal thymus development are reported. 35.9% of the tumors presented CN gain of PBX1 locus and 33.9% CN loss of FOXC1.
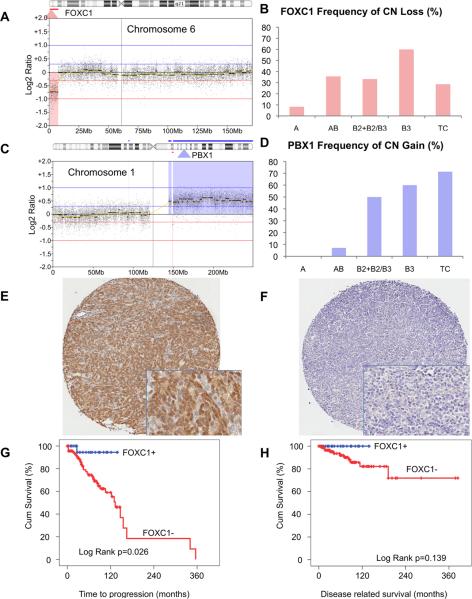
FOXC1 CN loss was found in a narrow region of CN loss affecting about 2Mb (1 996 266 nucleotides) of the short arm of chromosome 6 (Figure 2A). Based on our CGH data, chr6p25 CN loss, where FOXC1 resides, was observed in all histotypes, including type A thymomas that usually present infrequent CN aberrations (Figure 2B). The CN loss of chromosome 6p has also been described in another case of A thymoma by Zetel et al. (24). However, chr6p25 CN loss was less commonly observed in A thymomas (8%) than in other histotypes (40%; Fisher exact test; p=0.044; Figure 2B). FOXC1 CN loss was observed in 27% of stage I–II and in 35% of stage III–IV patients and the difference was not significant. Similarly, 30% of completely resected cases and 35% of cases with residual disease (R1+R2) carried FOXC1 CN loss. In contrast, PBX1 locus was included in a large region of CN gain involving the entire chromosome 1q (Figure 2C). PBX1 CN gain was a more frequent event in aggressive histotypes (B2+B2/B3, B3 and TC) than in A and AB (Fisher exact test p<0.001; Figure 2D), and in relapsed tumors than in primary tumors (χ2 test p=0.027).
Figure 2. CN loss of FOXC1 and CN gain of PBX1 in thymic epithelial tumors and prognostic value of immunohistochemistry results.
(A) An example of a focal CN loss of chromosome 6p affecting FOXC1 locus. The map of chromosome 6 is reported horizontally, the red highlighted area indicates the region of CN loss. (B) Frequency of FOXC1 CN loss according to WHO histotypes. (C) A representative PBX1 CN gain along with CN gain of approximately the entire long arm of chromosome 1. (D) Frequency of PBX1 CN gain according to WHO histotypes. Examples of (E) FOXC1 positive G3 and (F) negative G0 staining. Kaplan-Maier curves for (G) time to progression and (H) disease related survival of patients with FOXC1 positive and negative tumors.
CN aberrations of thymic developmental genes were confirmed using array CGH in an independent cohort of 27 frozen TET samples collected at NCI. Also in this series, PBX1 (59.3%) and GLI4 (37%) were the most frequent CN gains and FOXC1 (51.9%) and IGF2R (33.3%) were the most frequent CN losses (Supplementary Figure S1, Table 1). FOXC1 CN loss was present in 20% of type A thymoma and in 59.1% of the other histotypes (Fisher exact test p=0.165). Also in the confirmation cohort, PBX1 CN gain was more frequent in more aggressive histotypes (75%) than in A and AB tumors (14%, Fisher exact test p=0.009).
Across the two series (86 tumors) FOXC1 CN loss was observed in 34 tumors (39.5%) and PBX1 CN gain in 37 (43.0%; summary of combined data are reported in figure S2–S3A–B).
FOXC1 CN loss and PBX1 CN gain technical validation
Technical validation of CGH results was performed using CN-PCR. Chromosome 6p23.5 (FOXC1) deletion was evaluated in 6 tumors with and 5 without CN loss, according to CGH results. CN loss was confirmed in 5 out of 6 tumors that showed FOXC1 CN loss by CGH, whereas, in the control group without CN loss of FOXC1, CGH and CN-PCR data were 100% concordant (Fisher exact test p=0.015). CN gains of PBX1 locus were confirmed in all tested samples: 7 with and 7 without CGH gain (Fisher exact test p<0.001).
FOXC1 and PBX1 expression in TETs
We explored the correlation between CN status and protein expression, using immunohistochemistry performed on a tissue microarray containing 132 tumors that included the 59 samples evaluated by array CGH.
Out of 119 TET cases evaluable for FOXC1 immunostaining, 94 (79%) were negative (Table 2). In positive cells, staining was predominately nuclear according to the nature of FOXC1 that is a transcription factor (Figure 2E–F). Mainly tumor epithelial cells were stained but not thymocytes. Only epithelial cells were considered in scoring the intensity of FOXC1 staining. 81.8% of A thymoma were strongly positive for FOXC1 staining: all the positive cases were classified as G3. Overall 90% of the tumors strongly expressing FOXC1, with a score of G3, were A and AB. In B histotypes FOXC1 expression was uncommon and when present it was commonly with a reduced intensity. None of the 13 evaluable TCs showed positive FOXC1 staining. FOXC1 negative staining was associated with worse patient outcome and tumor characteristics: relapsed tumors, incomplete resections and more aggressive histotypes (Table 2). There was a significant correlation between FOXC1 CN loss and immunohistochemistry results (p= 0.008): 90% of tumors with FOXC1 CN loss did not express FOXC1 whereas only 55% of tumors with 2 copies of FOXC1 were negative for its expression. FOXC1-negative cases were associated with worse TTP than FOXC1-positive cases (10-year TTP 62% vs 93%, respectively; log rank p=0.026; Figure 2G). A similar trend was observed for DRS (log rank p=0.139; Figure 2H). However, multivariate models could not be constructed for DRS, since there were no death events observed in FOXC1-positive patients. FOXC1 was not an independent prognostic factor for TTP (p=0.15; HR: 0.23; 95% CI on HR: 0.03 to 1.73) after adjusting for WHO histotype (p=0.0047; HR: 4.02; 95% CI on HR: 1.53 to 10.55) in a Cox multivariate analysis.
Table 2.
Immunohistochemistry results and patients' characteristics
| Total | Frequency of FOXC1 IHC+ | p-value | Frequency of PBX1 IHC+ | p-value | |
|---|---|---|---|---|---|
| Total | 132 | 21.0% | 27.4% | ||
| Sex | |||||
| Female | 65 | 22.0% | 30.0% | ||
| Male | 67 | 20.0% | 24.6% | ||
| CGH results | |||||
| FOXC1 CN Loss | 20 | 10.0% | p=0.008* | ||
| FOXC1 2CN | 39 | 44.4% | |||
| PBX1 CN Gain | 21 | 24.6% | p=0.575 | ||
| PBX1 2CN | 38 | 24.6% | |||
| Tumor samples | |||||
| Primary | 108 | 24.7% | p=0.042* | 23.4% | p=0.53 |
| Relapse | 24 | 4.5% | 43.5% | ||
| Stage | |||||
| I | 35 | 36.4% | p=0.084 | 12.1% | p<0.001* |
| IIA | 26 | 25.0% | I–II/III–IV | 12.0% | I–II/III–IV |
| IIB | 17 | 21.4% | 14.3% | ||
| III | 19 | 6.3% | 40.0% | ||
| IVA | 5 | 25.0% | 75.0% | ||
| IVB | 12 | 18.2% | 55.6% | ||
| Na | 18 | ||||
| Completeness of resection | |||||
| R0 | 79 | 30.9% | p=0.023* | 23.5% | p=0.949 |
| R1 | 23 | 9.1% | R0/R1–2 | 13.6% | R0/R1–2 |
| R2 | 10 | 10.0% | 57.1% | ||
| Na | 20 | ||||
| WHO histotype | |||||
| A | 15 | 81.8% | p=0.001* | 36.4% | p=0.001* |
| AB | 28 | 38.5% | A-AB-B1/ | 15.4% | A-AB-B1/ |
| B1 | 24 | 0.0% | Remaining | 0.0% | Remaining |
| B1/B2 | 6 | 16.7% | 33.3% | ||
| B2 | 8 | 0.0% | 57.1% | ||
| B2/B3 | 11 | 10.0% | 55.6% | ||
| B3 | 24 | 13.0% | 50.0% | ||
| TC | 14 | 0.0% | 16.7% | ||
| Others | 2 | ||||
| Paraneoplastic syndromes | |||||
| No | 91 | 25.6% | p=0.205 | 21.0% | p=0.017* |
| Yes | 34 | 12.9% | 45.2% | ||
Total number of cases is reported to describe patients' characteristics but frequency of FOXC1 and PBX1 staining are calculated on the number of evaluable cases: 119 and 117, respectively. IHC+: positive staining determined by immunohistochemistry; FOXC1 2CN and PBX1 2CN represent tumors without FOXC1 CN loss and PBX1 CN gain respectively; Na: patients without available information at diagnosis; R0: complete resection, R1: microscopically incomplete resection, R2: macroscopically incomplete resection; TC: thymic carcinoma; Other: 1 micronodular and 1 cystic thymoma; Remaining: B1/B2, B2, B2/B3, B3 and TC;
significant differences calculated using Fisher exact test or χ2 test when appropriate.
No correlation was observed between copy number gain of the PBX1 locus and PBX1 protein expression by immunohistochemistry (p=0.575). Moreover, there was no statistically significant difference in DRS or TTP between PBX1-positive and -negative cases (log rank p=0.294 and p=0.229, respectively). PBX1 expression was higher in more advanced stages (III and IV) (50%; 14/28) than in stages I and II (12.7%; 9/72; Fisher exact test p<0.001). In advanced stages, PBX1-positive patients had a better outcome (10-year DRS 100%; 5-year TTP 100%) than PBX1-negative cases (10-year DRS 67%; log rank p=0.058; 5-year TTP 42%; log rank p<0.001). More aggressive histotypes (B1/B2, B2, B2/B3, B3 and TC) exhibited higher frequency of PBX1 expression (42.9%; 24/56) than less aggressive ones (A, AB, B1) (13.6%; 8/59; Fisher exact test p=0.001). In the subgroup of more aggressive histotypes, PBX1-positive cases had a better outcome (10-year DRS 94%; 10-year TTP 72%) than negative cases (10-year DRS 74%; log rank 0.069; 10-year TTP 36% log rank p=0.01).
Sequencing of FOXC1 gene
Since FOXC1 gene mutations have been shown in endometrial and ovarian cancers (25), we sequenced the entire FOXC1 coding region in the 59 FFPE TET samples analyzed by CGH and in the three TET cell lines. Three B3 samples showed an extra-glycine insertion in a poly-glycine region between amino acid 447 and 456. Normal DNA from one of the three patients was found to harbor this glycine insertion. One AB thymoma carried a glycine insertion in another poly-glycine region between amino acid 375 and 380. No insertion at the same position was found in normal DNA from the same patient.
FOXC1 and TBX1 mRNA expression do not correlate in TETs
FOXC1 has been described to regulate TBX1 expression during embryogenesis, therefore, we studied FOXC1 and TBX1 expression using RT-PCR in the confirmation cohort since good quality mRNA was available.
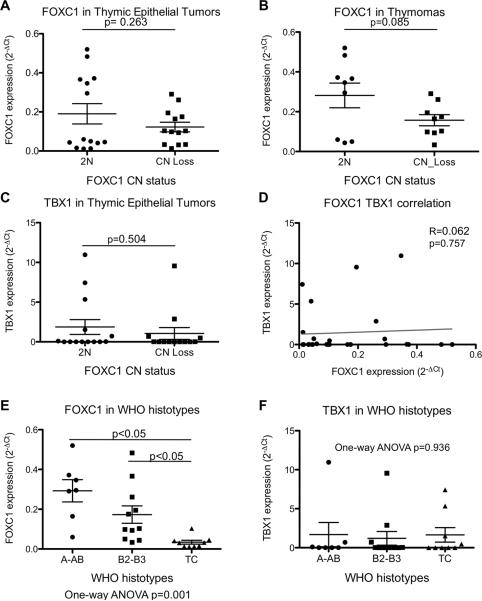
We observed a reduced expression of FOXC1 in tumors carrying FOXC1 CN loss, however the difference was not statistically significant (p=0.263; Figure 3A). This trend (p=0.085; Figure 3B) was more evident considering exclusively thymomas, and excluding thymic carcinomas. FOXC1 CN loss had no impact on the expression of TBX1 (p=0.504; Figure 3C), and also there was no correlation between FOXC1 and TBX1 expression (Pearson R=0.062 (95%CI −0.325; 0.432); p=0.757). Interestingly, FOXC1 expression was more significantly pronounced in type A and AB tumors, and progressively decreased from B2-B3 to TCs with the latter being almost completely negative for FOXC1 expression (One-way ANOVA p=0.001; B2-B3vsTC and TCvsA-AB; Turkey's test p<0.05). There was no difference of TBX1 expression in different WHO histotypes of TETs (One-way ANOVA p=0.936).
Figure 3. FOXC1 and TBX1 gene expression in the confirmation cohort of thymic epithelial tumors.
(A) FOXC1 relative gene expression (expressed as 2−ΔCt of FOXC1 and the housekeeping gene RPLO) according to FOXC1 CN status in thymic epithelial tumors (thymomas and thymic carcinomas). (B) Relative FOXC1 expression vs FOXC1 CN status in thymomas. (C) Relative TBX1 expression (expressed as 2−Ct of TBX1 and RPLO) vs FOXC1 CN status in thymic epithelial tumors. (D) No correlation between FOXC1 and TBX1 expression. (E) Relative FOXC1 expression vs WHO histotypes. (F) RelativeTBX1 expression vs WHO histotypes.
FOXC1, a candidate tumor suppressor gene in TETs
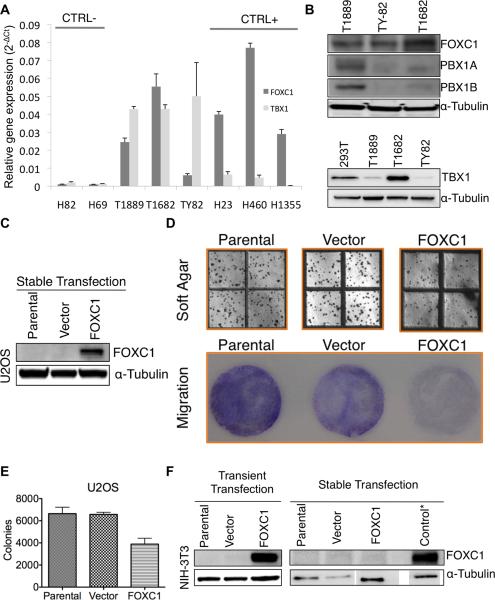
According to the cancer cell line encyclopedia microarray expression data (26), positive (H23, H460 and H1355) and negative control cell lines (H69 and H82) were selected based on FOXC1 expression. All TET cell lines, T1889 and TY82 thymic carcinoma and T1682 B1 thymoma, expressed FOXC1 (Figure 4A). Array CGH revealed that none of the TET cell lines had CN loss of FOXC1 locus. These observations were confirmed by protein expression using western blot (Figure 4B). Because none of the 3 TET lines was null for FOXC1 expression, the transfection of FOXC1 in these cells was not a suitable model to test its tumor suppressor properties. Therefore, we evaluated the effect of ectopic FOXC1 expression in U2OS (Figure 4C–E) and NIH-3T3 cells (Figure 4F). These cells were chosen over TET cells because they were negative for FOXC1 expression (Figure 4 C,F). If FOXC1 is a genuine tumor suppressor gene, reconstitution of FOXC1 into these cells should impede cell growth. Soft agar assay showed that constitutive expression of FOXC1 in U2OS cells significantly reduced the number of colony outgrowth in comparison to the vector-transfected clones and U2OS parental cells (Figure 4D–E). FOXC1 also attenuated the migration capacity of U2OS cells in trans-well migration assay (Figure 4D). Intriguingly, stable clones derived from FOXC1-transfected NIH3T3 cells were all negative for ectopic FOXC1 expression (Figure 4F) though FOXC1 was expressed in transiently-transfected NIH3T3 cells (Figure 4F), suggesting that FOXC1 reconstitution may be detrimental to the immortalized NIH3T3 cells and only the clones capable of evading FOXC1 expression could grow. Collectively, we postulate that FOXC1 may be a candidate tumor growth suppressor gene in TETs.
Figure 4. FOXC1, PBX1 and TBX1 expression in cell lines.
(A) Relative gene expression of FOXC1 (expressed as 2−Ct of FOXC1 and the housekeeping gene RPLO) and TBX1 (expressed as 2−Ct of TBX1 and RPLO) in thymic epithelial tumors cell lines. For FOXC1 gene expression, H82 and H69 small cell lung cancer cell lines were used as negative controls and H23, H460 and H1355 non-small cell lung cancer cell lines as positive controls, according to the cancer cell line encyclopedia(26). Results are the average of 4 independent replicate experiments with the relative standard deviation. (B) Western blots evaluating protein expression of FOXC1, PBX1 isoform-α, PBX1 isoform-β and TBX1 compared to α-Tubulin in TET cell lines. HEK-293T cells were used as a positive control for TBX1 expression. (C) Ectopic expression of FOXC1 in U2OS stable clones. (D) Soft agar (top panel) and migration (bottom panel) assays of parental, vector- and FOXC1-transfected U2OS cells. (E) Quantification of colony formation in soft agar assay shown in panel D. * denotes (p-value <0.05). (F) Ectopic expression of FOXC1 in NIH3T3. Note that FOXC1 stable clones stably expressing FOXC1 could not be established from NIH3T3 cells. * Control was NIH-3T3 cells transiently transfected with FOXC1.
Discussion
Recently, new technologies, such as array CGH, have enabled the evaluation of tumor specific events on a genome wide scale. Lineage-specific transcription factors, implicated in organogenesis, have been found deregulated by CN aberrations and they have been demonstrated to be able to drive the cancer phenotype. Examples include, NKX2-1 (TTF1) in lung cancer (4–8), ESR1 in breast cancer (9), GATA6 in pancreatic cancer (10) and MITF in melanoma (11). Using high-resolution array based CGH we have previously described the CN aberrations of a series of 59 TETs on a genome wide scale (17). Here we have focused on the CN aberrations of 31 genes implicated in thymus organogenesis (12, 13, 22, 23).
Subjects carrying the 22q11.2 deletion syndrome present variable pathologic phenotypes at birth including cardiac defects, abnormal faces, thymic hypoplasia, cleft palate and hypocalcaemia. The concomitant presence of some of these congenital abnormalities has been named as specific syndromes: for example DiGeorge, velocardiofacial and conotruncal anomaly face syndromes(27). Interestingly, several patients are immunocompromised because they were born with thymic aplasia or hypoplasia. TBX1, a member of the T-box containing family of transcription factors, is mapped on 22q11.2 and it is the more likely candidate responsible for the pharyngeal arch-derived defects observed in 22q11.2 deletion syndrome(27). TBX1 knockout mice demonstrated the importance of this gene in the congenital branchial arch pathologic development and consequently in the maturation of the epithelial compartment of the thymus that originates from the endoderm of the third pharyngeal pouch(28).
TBX1 exerts its activity integrated in a complex cascade of transcription factors that regulates thymus development. It has been shown that FOXC1 and PBX1 transcription factors regulate TBX1 expression in vitro and in animal models (29, 30), acting downstream of Sonic Hedgehog (SHH) pathway during thymus development (13, 30, 31). The TBX1 transcription is tightly spatiotemporally regulated by the SHH pathway and by HOX-EYA-PAX axis through FOXC1 and PBX1, respectively (30, 31). Upon the activation of SHH pathway, TBX1 is transcribed in order to regulate the expression of several genes, including FGF10 and FGF8 that support growth of surrounding cells and may also play a role in neural crest cells' migration (32). In vitro, TBX1 expression restores contact inhibition and reduces anchorage-independent cell growth in soft agar (33). In vivo, the ectopic expression of TBX1 suppresses the growth of skin carcinoma cells(33).
Among the 31 genes regulating thymus development, FOXC1 and PBX1 were those with the highest frequency of CN loss and gain in TETs, respectively. CN gain of PBX1 is frequently observed in Tumorscape, and has been described in 35% of all cancer analyzed, but it is often associated to a large region of CN aberration (34). FOXC1 CN loss is associated with a focal peak of deletion and less frequently observed in common cancers (12%, Tumorscape) than in TETs (39.5%). However, some types of tumors presented a remarkably high frequency of FOXC1 CN loss, the highest frequency being 89% in colorectal cancer (34). In contrast, CN gain of PBX1 is involved in a large region of CN gain of the whole chromosome 1q, being 1q CN gain a frequent event in TETs (17, 24). Therefore, it is difficult to link the deregulation of a single gene to this large region of CN gain (Figure 2C). On the contrary, in this study, we observed a frequent FOXC1 CN loss in a focal deletion including a limited number of genes. CN aberrations often affect the expression of the genes mapped to that region. Therefore, we correlated the protein expression of FOXC1 and PBX1, evaluated by immunohistochemistry, with their CN status. FOXC1 expression was reduced in TETs with the CN loss, but PBX1 expression was not increased in tumors with CN gain. Consistently, in the confirmation cohort, tumors with FOXC1 CN loss expressed less FOXC1 mRNA. Because there was no correlation between PBX1 expression and PBX1 CN gain, we suggest that FOXC1 CN loss but not PBX1 CN gain may play a role in TET development.
It is important to note that FOXC1 staining was negative in 90% of tumors carrying FOXC1 CN loss and 55% (20/36) of tumors with 2 FOXC1 alleles. Similarly, a very low expression of FOXC1 mRNA was observed in many tumors without FOXC1 CN loss (Figure 3A). Therefore, the CN status of FOXC1 is probably not the exclusive factor influencing FOXC1 expression. Probably, there are other mechanisms in thymic carcinomas and to some extent in B2 and B3 thymomas that inhibit FOXC1 expression. Indeed, FOXC1 was not an independent prognostic factor in the multivariate analysis after correction for WHO histotype. We excluded the possibility of FOXC1 mutation as cause of low expression by conventional sequencing in 59 cases. The extra-glycine insertions in polyglycine regions (of 6 and 10-glycine) were observed in three tumors and have been previously described as polymorphisms, thus unlikely affecting the protein function. Methylation of FOXC1 promoter is a candidate mechanism for repression of FOXC1 expression in tumors without FOXC1 CN loss, since FOXC1 promoter methylation has been described in other cancer types and correlated with FOXC1 silencing (35). A poor outcome of FOXC1-negative tumors has been observed in invasive ductal breast carcinoma and locally advanced breast cancer (36, 37), though contradictory results have also been reported (34).
Because of the FOXC1 role in regulation of TBX1, we studied the association between FOXC1 and TBX1 mRNA expression. There was no significant correlation in patients' tumors (Figure 3D) or TET cell lines. Moreover, we failed to find any correlation between PBX1 CN gain and TBX1 expression (data not shown). Therefore, TBX1 seems to be regulated in a more complex manner than solely by FOXC1. To evaluate the functional significance of FOXC1 deletion in TETs, we reconstituted FOXC1-negative U2OS cells, and demonstrated that ectopic FOXC1 attenuated anchorage-independent growth and motility of U2OS cells in vitro, suggesting that FOXC1 may act as a tumor growth suppressor gene. In line with these findings, we were not able to establish a stable clone of NIH3T3 cells transfected with FOXC1. Because of the small number of TET cell lines generated to date, only 3 were available to us. Unfortunately, all TET cell lines expressed FOXC1, thus, tumor suppressor activity of FOXC1 in TET cell lines cannot be tested.
Our FOXC1 results in TETs and in U2OS cells are discrepant to what has been described in basal-like breast cancer, where FOXC1 acts as a candidate oncogene. FOXC1 overexpression has been described in basal-like breast cancers where overexpression was a poor prognostic factor (38). Ectopic FOXC1 overexpression in breast cancer cell lines increases cell proliferation, invasion and migration (38). It has been suggested that FOXC1 may induce epithelial-mesenchymal transition (39), and consequently may increase the metastatic potential (39). Since FOXC1 is a transcription factor, epigenetic context of FOXC1 target genes may determine FOXC1 accessibility to their regulatory binding sites in the promoter sequences. Because thymic epithelial cells and precursors of basal like breast cancer undergo unique differentiating programs, chromatin conformation is expected to be different for thymic epithelial cells of endodermic origin and mammary glands cells of ectodermic origin. Therefore, different promoters will be accessible to FOXC1 in these 2 different contexts with possibly different effects on cancer cell growth. Frequently, the altered expression of transcription factors, usually restricted to normal cell precursors, requires a specific intracellular context to restore their function, and to induce proliferation. It is possible that such intracellular context is present exclusively in the committed line of descendant cells, indicating a state of “lineage-dependency” (11).
In conclusion, we identified frequent CN loss of FOXC1 in TETs, especially in more aggressive histotypes. A reduction of FOXC1 expression was observed in tumors with FOXC1 CN loss and was associated with poor prognosis. In vitro data support the candidate tumor suppressor activity of FOXC1 on cell growth.
Supplementary Material
Translational Relevance.
Thymic epithelial tumors are a group of neoplasms with heterogeneous histological features and clinical behavior. The identification of markers useful to predict patient prognosis and molecular targets for therapies is limited by a very little understanding of the biology of these neoplasms. We evaluated the copy number (CN) aberrations of genes involved in normal thymus development in thymic epithelial tumors, following the intriguing idea that the ectopic deregulation of genes relevant for proliferation and differentiation of embryonic cells, can contribute to tumor growth. Frequent CN losses of FOXC1 were observed in more aggressive tumors and correlated with a reduced protein expression; tumors negative for FOXC1 expression were associated with a shorter time to progression. In addition, FOXC1 showed tumor suppressor activity in in-vitro models. Our data indicate that FOXC1 loss can identify a group of thymic epithelial tumors with poor prognosis, possibly because its tumor suppressor properties.
Acknowledgments
Grant Support National Cancer Institute intramural program funded this research
Footnotes
Financial disclosures: the authors have no conflicts of interest to disclose.
References
- 1.Travis WD, Brambilla E, Muller-Hermelink HK, Harris CC. Pathology and genetics: Tumors of the lung, pleura, thymus and heart. IARC Press; Lyon, France: 2004. [Google Scholar]
- 2.Kelly RJ, Petrini I, Rajan A, Wang Y, Giaccone G. Thymic malignancies: from clinical management to targeted therapies. J Clin Oncol. 2011;29:4820–7. doi: 10.1200/JCO.2011.36.0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris PJ, Speranza G, Dansky Ullmann C. Targeting embryonic signaling pathways in cancer therapy. Expert Opin Ther Targets. 2012;16:131–45. doi: 10.1517/14728222.2011.645808. [DOI] [PubMed] [Google Scholar]
- 4.Winslow MM, Dayton TL, Verhaak RG, Kim-Kiselak C, Snyder EL, Feldser DM, et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature. 2011;473:101–4. doi: 10.1038/nature09881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwei KA, Kim YH, Girard L, Kao J, Pacyna-Gengelbach M, Salari K, et al. Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene. 2008;27:3635–40. doi: 10.1038/sj.onc.1211012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka H, Yanagisawa K, Shinjo K, Taguchi A, Maeno K, Tomida S, et al. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res. 2007;67:6007–11. doi: 10.1158/0008-5472.CAN-06-4774. [DOI] [PubMed] [Google Scholar]
- 7.Kendall J, Liu Q, Bakleh A, Krasnitz A, Nguyen KC, Lakshmi B, et al. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci U S A. 2007;104:16663–8. doi: 10.1073/pnas.0708286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–8. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holst F, Stahl PR, Ruiz C, Hellwinkel O, Jehan Z, Wendland M, et al. Estrogen receptor alpha (ESR1) gene amplification is frequent in breast cancer. Nat Genet. 2007;39:655–60. doi: 10.1038/ng2006. [DOI] [PubMed] [Google Scholar]
- 10.Kwei KA, Bashyam MD, Kao J, Ratheesh R, Reddy EC, Kim YH, et al. Genomic profiling identifies GATA6 as a candidate oncogene amplified in pancreatobiliary cancer. PLoS Genet. 2008;4:e1000081. doi: 10.1371/journal.pgen.1000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–22. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 12.Blackburn CC, Manley NR. Developing a new paradigm for thymus organogenesis. Nat Rev Immunol. 2004;4:278–89. doi: 10.1038/nri1331. [DOI] [PubMed] [Google Scholar]
- 13.Hollander G, Gill J, Zuklys S, Iwanami N, Liu C, Takahama Y. Cellular and molecular events during early thymus development. Immunol Rev. 2006;209:28–46. doi: 10.1111/j.0105-2896.2006.00357.x. [DOI] [PubMed] [Google Scholar]
- 14.Zucali PA, Petrini I, Lorenzi E, Merino M, Cao L, Di Tommaso L, et al. Insulin-like growth factor-1 receptor and phosphorylated AKT-serine 473 expression in 132 resected thymomas and thymic carcinomas. Cancer. 2010 doi: 10.1002/cncr.25367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masaoka A, Yamakawa Y, Niwa H, Fukai I, Saito Y, Tokudome S, et al. Thymectomy and malignancy. Eur J Cardiothorac Surg. 1994;8:251–3. doi: 10.1016/1010-7940(94)90155-4. [DOI] [PubMed] [Google Scholar]
- 16.Strobel P, Bauer A, Puppe B, Kraushaar T, Krein A, Toyka K, et al. Tumor recurrence and survival in patients treated for thymomas and thymic squamous cell carcinomas: a retrospective analysis. J Clin Oncol. 2004;22:1501–9. doi: 10.1200/JCO.2004.10.113. [DOI] [PubMed] [Google Scholar]
- 17.Petrini I, Meltzer PS, Zucali PA, Luo J, Lee C, Santoro A, et al. Copy number aberrations of BCL2 and CDKN2A/B identified by array-CGH in thymic epithelial tumors. Cell Death Dis. 2012;3:e351. doi: 10.1038/cddis.2012.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petrini I, Zucali PA, Lee HS, Pineda MA, Meltzer PS, Walter-Rodriguez B, et al. Expression and mutational status of c-kit in thymic epithelial tumors. J Thorac Oncol. 2010;5:1447–53. doi: 10.1097/JTO.0b013e3181e96e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ehemann V, Kern MA, Breinig M, Schnabel PA, Gunawan B, Schulten HJ, et al. Establishment, characterization and drug sensitivity testing in primary cultures of human thymoma and thymic carcinoma. Int J Cancer. 2008;122:2719–25. doi: 10.1002/ijc.23335. [DOI] [PubMed] [Google Scholar]
- 20.Chiarenza A, Lazarovici P, Lempereur L, Cantarella G, Bianchi A, Bernardini R. Tamoxifen inhibits nerve growth factor-induced proliferation of the human breast cancerous cell line MCF-7. Cancer Res. 2001;61:3002–8. [PubMed] [Google Scholar]
- 21.Harada T, Lopez-Chavez A, Xi L, Raffeld M, Wang Y, Giaccone G. Characterization of epidermal growth factor receptor mutations in non-small-cell lung cancer patients of African-American ancestry. Oncogene. 2011 doi: 10.1038/onc.2010.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordon J, Manley NR. Mechanisms of thymus organogenesis and morphogenesis. Development. 2011;138:3865–78. doi: 10.1242/dev.059998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodewald HR. Thymus organogenesis. Annu Rev Immunol. 2008;26:355–88. doi: 10.1146/annurev.immunol.26.021607.090408. [DOI] [PubMed] [Google Scholar]
- 24.Zettl A, Strobel P, Wagner K, Katzenberger T, Ott G, Rosenwald A, et al. Recurrent genetic aberrations in thymoma and thymic carcinoma. Am J Pathol. 2000;157:257–66. doi: 10.1016/S0002-9440(10)64536-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Y, Kato H, Asanoma K, Kondo H, Arima T, Kato K, et al. Identification of FOXC1 as a TGF-beta1 responsive gene and its involvement in negative regulation of cell growth. Genomics. 2002;80:465–72. [PubMed] [Google Scholar]
- 26.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindsay EA. Chromosomal microdeletions: dissecting del22q11 syndrome. Nat Rev Genet. 2001;2:858–68. doi: 10.1038/35098574. [DOI] [PubMed] [Google Scholar]
- 28.Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27:286–91. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- 29.Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–89. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 30.Manley NR, Selleri L, Brendolan A, Gordon J, Cleary ML. Abnormalities of caudal pharyngeal pouch development in Pbx1 knockout mice mimic loss of Hox3 paralogs. Dev Biol. 2004;276:301–12. doi: 10.1016/j.ydbio.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 31.Yamagishi H, Maeda J, Hu T, McAnally J, Conway SJ, Kume T, et al. Tbx1 is regulated by tissue-specific forkhead proteins through a common Sonic hedgehog-responsive enhancer. Genes Dev. 2003;17:269–81. doi: 10.1101/gad.1048903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDonald-McGinn DM, Sullivan KE. Chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome) Medicine (Baltimore) 2011;90:1–18. doi: 10.1097/MD.0b013e3182060469. [DOI] [PubMed] [Google Scholar]
- 33.Trempus CS, Wei SJ, Humble MM, Dang H, Bortner CD, Sifre MI, et al. A novel role for the T-box transcription factor Tbx1 as a negative regulator of tumor cell growth in mice. Mol Carcinog. 2011;50:981–91. doi: 10.1002/mc.20768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bloushtain-Qimron N, Yao J, Snyder EL, Shipitsin M, Campbell LL, Mani SA, et al. Cell type-specific DNA methylation patterns in the human breast. Proc Natl Acad Sci U S A. 2008;105:14076–81. doi: 10.1073/pnas.0805206105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muggerud AA, Ronneberg JA, Warnberg F, Botling J, Busato F, Jovanovic J, et al. Frequent aberrant DNA methylation of ABCB1, FOXC1, PPP2R2B and PTEN in ductal carcinoma in situ and early invasive breast cancer. Breast Cancer Res. 2010;12:R3. doi: 10.1186/bcr2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dejeux E, Ronneberg JA, Solvang H, Bukholm I, Geisler S, Aas T, et al. DNA methylation profiling in doxorubicin treated primary locally advanced breast tumours identifies novel genes associated with survival and treatment response. Mol Cancer. 2010;9:68. doi: 10.1186/1476-4598-9-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ray PS, Wang J, Qu Y, Sim MS, Shamonki J, Bagaria SP, et al. FOXC1 is a potential prognostic biomarker with functional significance in basal-like breast cancer. Cancer Res. 2010;70:3870–6. doi: 10.1158/0008-5472.CAN-09-4120. [DOI] [PubMed] [Google Scholar]
- 39.Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, et al. Core epithelial-tomesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci U S A. 2010;107:15449–54. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.