Abstract
Treatment options for patients with pancreatic ductal adenocarcinoma remain limited. Therapeutic targets of interest include mutated molecules that predispose to pancreatic cancer such as KRAS and TP53. Here we show that an element of the homologous recombination pathway of DNA repair, the PARP-binding protein C12orf48/PARI (PARPBP), is overexpressed specifically in pancreatic cancer cells where it is an appealing candidate for targeted therapy. PARI upregulation in pancreatic cancer cells or avian DT40 cells conferred DNA repair deficiency and genomic instability. Significantly, PARI silencing compromised cancer cell proliferation in vitro, leading to cell cycle alterations associated with S phase delay, perturbed DNA replication and activation of the DNA damage response pathway in the absence of DNA damage stimuli. Conversely, PARI overexpression produced tolerance to DNA damage by promoting replication of damaged DNA. In a mouse xenograft model of pancreatic cancer, PARI silencing was sufficient to reduce pancreatic tumor growth in vivo. Taken together, our findings offered a preclinical proof-of-concept for PARI as candidate therapeutic target to treat pancreatic ductal adenocarcinoma.
Keywords: Pancreatic cancer, Homologous Recombination, PARI, DNA damage response, DNA replication
Introduction
Pancreatic cancer is a devastatingly lethal disease that will be diagnosed in an estimated 43,920 men and women in the U.S.A. in 2012. Unfortunately pancreatic cancer patients have few viable treatment options, and it is predicted that 37,390 people will die from the disease this year (1). The most common form of pancreatic cancer, pancreatic ductal adenocarcinoma (PDAC), has the gravest prognosis of any major solid tumor, with a predicted 5-year survival rate of less than 5% (2). Only few patients present with localized potentially resectable cancers. Metastatic disease is intensely resistant to conventional chemo- and radiation therapy.
Progression of PDAC from pre-neoplastic legions has been closely studied. Adenocarcinomas of the pancreas progress along the PanIN (pancreatic intraepithelial neoplasia) cascade, with discrete histopathologies that define the three grades of precursor neoplasia: PanINI, PanINII, and PanINIII. Advancement into invasive carcinoma is concomitant with several distinct genetic alterations. Activating mutations in the KRas proto-oncogene occur in ~90% of PDACs and are usually accompanied by the loss of cardinal tumor suppressors TP53 (50–75%) and CDK2NA/Ink4a/p16 (95%) (2). Abnormalities in DNA repair mechanisms are also seen in PDAC. In particular, mutations in Homologous Recombination (HR) DNA repair factors were found to be associated with pancreatic cancer. Inactivation of the tumor suppressor BRCA2, an essential HR factor which catalyzes the loading of RAD51 at double strand breaks (DSBs) (3), has been implicated in both familial (4) and sporadic PDAC (5). Similarly, upstream pathways that regulate HR are important for suppressing pancreatic cancer. ATM is a kinase involved in recruiting repair factors to DNA strand breaks (6); ATM mutations have been associated with pancreatic cancer (7, 8). The Fanconi Anemia (FA) pathway co-ordinates the recognition and repair of DNA crosslinks by activating HR, and other repair mechanisms (9). BRCA2 itself is an FA factor, corresponding to the FANCD1 complementation group (10). Mutations in other FA genes including PALB2/FANCN, FANCC, and FANCG have been identified in familial clustering of pancreatic cancer (4, 11, 12). Finally, single nucleotide polymorphisms (SNPs) in HR factors XRCC2, XRCC3, RecQ1, and Rad54 have also been reported to contribute to pancreatic cancer risk (13, 14).
A number of murine models have been developed over the past decade, offering a valuable system for studying the biology of pancreatic cancer. Mice that harbor pancreas-specific activating mutations in Kras develop PanIN with 100% penetrance. Given the long latency for PDAC formation in these mice, additional genetic events are likely necessary for tumor progression. Indeed, conditional deletion of Trp53 in these mice promotes the rapid development of advanced murine PDAC with 100% penetrance (15). Strikingly, heterozygosity for a pathogenic germline truncation in BRCA2 can also promote Kras-driven carcinogenesis in the murine model (16).
The necessity for novel and effective therapies has underscored the search for crucial biomarkers in pancreatic cancer. Recently microarray expression analyses of human tumors have identified several genes trans-activated in PDAC, including KIAA0101/PAF (17), C2orf18/ANT2BP (18), and C12orf48/PARI (19). These factors have been proposed to be important for pancreatic cancer tumorigenesis.
We recently identified PARI as an inhibitor of HR in human cells (20). PARI is recruited to replication forks during S-phase, by interacting with SUMO-modified PCNA. PARI interferes with the formation of RAD51-DNA structures to suppress unwanted recombination events of replicating chromosomes.
Here we interrogated the role of PARI in PDAC. We show that PARI is overexpressed in PDAC cell lines, leading to HR inhibition, DNA damage hypersensitivity, and genomic instability. Furthermore, we show that PARI depletion from PARI-overexpressing pancreatic cell lines reduces their proliferation in vivo and in vitro by interfering with S-phase progression. We propose that PARI inhibitors can be used as targeted therapy for pancreatic cancer.
Materials and methods
Cell culture and protein techniques
8988T, CAPAN2, HeLa, U2OS, and 293T cells were grown in Dulbecco’s modified Eagle medium (Invitrogen), supplemented with 15% fetal bovine serum. PDAC cells were previously described (21). Cell lines were obtained from the American Type Culture Collection or the German Collection of Microorganisms and Cell Cultures. They are stored in our central cell bank and routinely checked for mycoplasma contamination and for changes in morphology. Whole cell extracts were prepared by lysing cells in RIPA buffer (50mM Tris, pH 7.3, 150mM NaCl, 1mM EDTA, 1% Triton X-100, 0.5% Na-deoxycholate, 0.1% SDS) with complete protease inhibitor, NaVO4, and NaF. RAD51 immunofluorescence was performed as previously described (20), using anti-RAD51 antibody (Santa Cruz Biotechnology). A list of antibodies used for Western blot, siRNA sequences, as well as additional Materials and Methods information, are available in Supplemental Material.
Functional cell-based assays
For survival assays, cells were transfected with siRNA and 48 hours later, seeded in 96-well plates and exposed to specified drugs. Viability was assessed after three days using CellTiterGlo (Promega). For clonogenic assays, cells were transfected with siRNA for 48 hours then seeded at low densities in 6-well plates and allowed to form colonies. The cells were fixed in Methanol/20% acetic acid and stained with 1% crystal violet. For FACS analysis, cells were fixed overnight at 4°C in 70% Ethanol, stained with PI for 1 hour, and analyzed for DNA content using a FACSCalibur (BD Biosciences) machine. SupF mutagenesis assay (22), and chromosomal aberration detection in mitotic spreads (10) were performed as previously described.
BrdU incorporation
For S-phase quantification by BrdU incorporation, 5×105 human 8988T cells were incubated with 20μM BrdU for 45 minutes, washed and fixed overnight at 4°C in 70% Ethanol. Cells were subsequently incubated with 2N HCl / 0.5% TritonX-100 for 30 minutes, and 0.1M sodium tetraborate pH 8.5 for 1 minute. Cells were washed and successively incubated with anti-BrdU antibodies (Pierce) and Alexa-Fluor 488 –conjugated anti-mouse secondary antibodies, (Invitrogen) for 30 minutes each, and analyzed using a FACSCalibur (BD Biosciences) machine.
Quantitative RT-PCR
For mRNA purification, the TRIZOL Reagent (Invitrogen) was used. Next, cDNA was amplified using the Transcriptor Reverse Transcriptaze kit (Roche), with oligo dT primers. Finally, mRNA quantification was done with QuantiTect SYBRGreen (Qiagen), using an iCycler machine (Bio-Rad). The cDNA of GAPDH gene was obtained and analyzed in parallel for normalization.
DT40 methods
Standard DT40 methods were used (23). DT40 PARI-knockout cells were previously described (20). For overexpression of human PARI in DT40 cells, human PARI cDNA was cloned with a Myc tag into pcDNA expression plasmid. DT40 cells were electroporated with 30 μg of linearized vector using Gene Pulser (BioRad) at 950V and 25 μF. Cell extracts were obtained by boiling cells in 100mM Tris, 4% SDS, 0.5M β-mercaptoethanol. For survival assays, chicken cells analyzed after 4 days incubation with the respective drug, using CellTiterGlo (Promega). For BrdU incorporation, 3×106 logarithmically growing DT40 cells were incubated with 20μM BrdU for 20 minutes and analyzed as described above.
In vivo xenograft studies
PARI and non-targeting (control) shRNA hairpins were transduced into 8988T cells by lentiviral transduction using pTripZ (Open Biosystems) and selected with puromycin. Cells were then amplified and re-selected for expression of RFP after doxycycline induction. For xenograft tumor formation, 1.5×106 sorted cells suspended in growth media and mixed 1:1 with Matrigel Basement Membrane (BD Biosciences) were then injected into both flanks of athymic nude mice (obtained from Charles River Laboratories). When tumors reached 200mm3 in volume, mice were administered 500 μg/ml doxycycline in their drinking water.
Results
PARI overexpression in pancreatic cancer cell lines
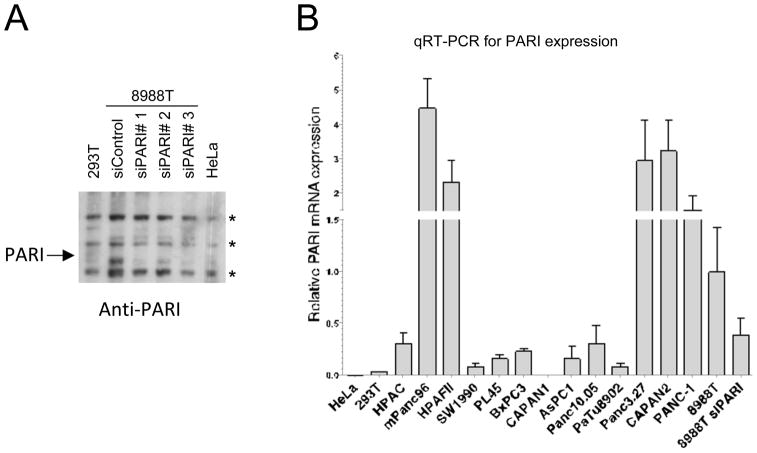
We recently identified PARI as a Homologous Recombination regulator in human cells. We hypothesized that PARI may be upregulated in PDAC resulting in a decrease in HR repair, a phenotype of many pancreatic cancer cells. To explore this possibility, we investigated PARI protein levels, using an antibody raised against PARI (20), in a panel of 40 human breast, ovarian and pancreatic cancer cell lines. As previously reported (20), human cell lines such as cervical cancer HeLa cells, transformed embryonic kidney 293T cells, and osteosarcoma U2OS cells, expressed PARI at very low levels; PARI protein is undetectable by Western blot. In our protein expression screen we found that, out of the 40 cell lines investigated, three of them exhibited a specific band at 72kDa when lysates were probed with anti-PARI antibodies in Western blot experiments. These three cell lines are: the ovarian cancer cell line PA-1 (24), and the pancreatic cancer cell lines 8988T (25) and CAPAN2 (26) (data not shown). Focusing on the pancreatic cell line 8988T, we showed that knockdown of PARI with three different siRNA oligonucleotides reduced the intensity of the 72kDa band, confirming the identity of this band as PARI (Figure 1A). Similar results were obtained when knocking down PARI in CAPAN2 and PA-1 cells (data not shown).
Figure 1. PARI is overexpressed in pancreatic adenocarcinoma (PDAC).
(A) Quantitative RT-PCR analysis for PARI mRNA expression showing that PARI is over-expressed in a number of PDAC cell lines, compared to control HeLa (cervical cancer) and 293T (transformed human embryonic kidney) cells. Expression is normalized to 8988T PDAC cells. The average of 3 independent experiments is shown. Error bars represent standard error of the mean. (B) Western blot using anti-PARI antibodies, showing that endogenous PARI can be detected in 8988T cells but not in 293T and HeLa cells. Three siRNA oligonucleotides can efficiently down-regulate PARI in 8988T cells. The arrow indicates the full-length protein; asterisks indicate crossreactive bands.
Intrigued by the increased representation of pancreatic cancers among the cell lines we found to overexpress PARI, we collected fifteen more pancreatic tumor lines and analyzed them for PARI mRNA expression by quantitative RT-PCR. We found that most of them have significantly higher PARI mRNA levels than control HeLa and 293T cells; a few of them, including mPANC96, Panc3.27, CAPAN2, HPAF II, PANC-1 and 8988T had particularly high PARI expression (Figure 1B). Thus, we concluded that PARI is upregulated in pancreatic cancers. A recent study similarly found that PARI (also named C12orf48 and PARPBP1) mRNA is upregulated in a number of pancreatic cancer cell lines (19). Our results thus correlate with this study and raise the intriguing possibility that PARI upregulation is an important event in pancreatic tumor progression.
PARI inhibits Homologous Recombination and alters DNA repair in pancreatic cancer cell lines
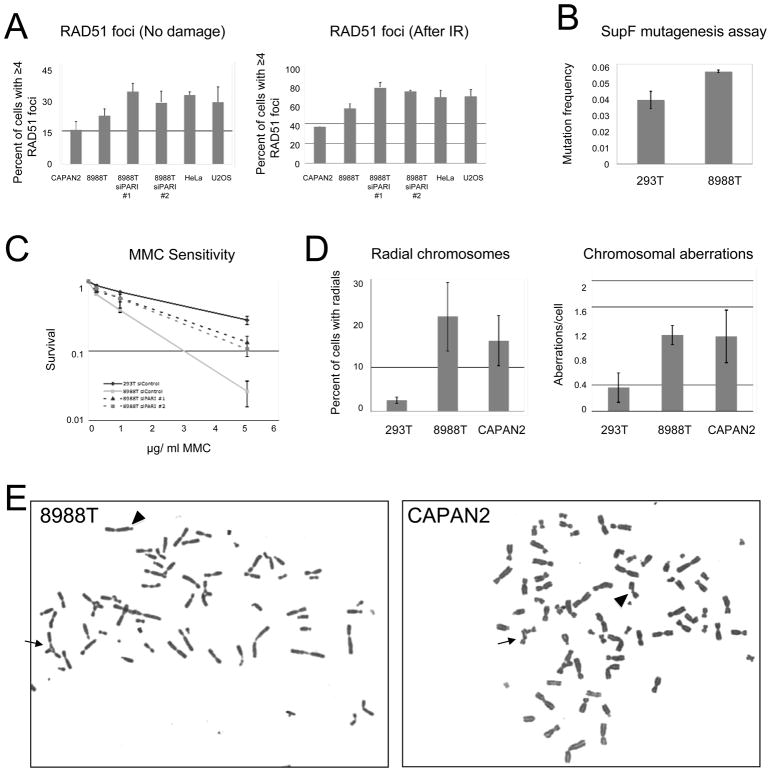
We previously showed that PARI inhibits Homologous Recombination in human cells. We next tested whether pancreatic cancer cell lines overexpressing PARI have reduced HR due to PARI upregulation. We examined the number of RAD51 foci –a surrogate marker of HR efficiency. As predicted, both spontaneous and IR-induced RAD51 foci were significantly reduced in PARI-overexpressing CAPAN2 and 8988T compared to control HeLa and U2OS cell lines; PARI knockdown in 8988T cells restored RAD51 foci to the levels found in non-overexpressing cells (Figure 2A, Supplementary Figure 1). RAD51 focus formation is not always a reliable indicator of HR proficiency (27); thus, we also investigated cellular sensitivity to PARP-1 inhibitors, a hallmark of Homologous Recombination deficiency (28, 29). 8988T cells were found to be hypersensitive to PARP-1 inhibitor ABT-888 (Supplementary Figure 2). We conclude that PARI overexpression results in reduced Homologous Recombination in pancreatic cancer cell lines.
Figure 2. PARI overexpression in PDAC cells is associated with genomic instability, reduced Homologous Recombination and altered DNA damage response.
(A) PARI-overexpressing PDAC cells 8988T and CAPAN2 show reduced Homologous Recombination as measured by RAD51 foci. PARI depletion restores RAD51 foci in 8988T cells. Shown is a quantification of spontaneous (left panel) and IR-induced (6h after treatment with 5Gray IR; right panel) RAD51 foci in pancreatic cancer cell lines CAPAN 2 and 8988T compared to control cells. The average of two to three independent experiments is shown. For each condition, 40–100 cells were counted. Error bars represent standard errors. Representative micrographs are shown in Supplementary Figure 1. (B) Translesion synthesis is increased in PARI-overexpressing PDAC cells. 8988T cells have higher UVC-induced mutation frequency than 293T cells as quantified using SupF mutagenesis assay. Data represents average of two independent experiments. Error bars show standard deviations. Overexpression of Myc-PARI in 293T cells also increased mutagenesis (Supplementary Figure 3). (C–E) PARI-overexpressing pancreatic cancer cell lines show hypersensitivity to DNA damage. (C) 8988T cells have increased cytotoxicity when exposed to MMC. Survival was measured after three days exposure to indicated concentrations of MMC. Average of two independent experiments is shown. Error bars represent standard deviations. (D) 8988T and CAPAN2 cells show elevated MMC-induced chromosomal damage in metaphase spreads. Quantifications of radial chromosomes (left panel) and total chromosomal aberrations (right panel) following treatment with 20ng/ml MMC for three days is shown. For each condition, 30–50 cells were analyzed. Data is shown as the average of two independent experiments. Error bars represent standard deviations. (E) Representative metaphase spreads of 8988T and CAPAN2 cells treated with 20 ng/ml MMC. Arrows indicate radial chromosomes and arrow heads indicate chromatid breaks.
Inhibition or loss of one DNA repair pathway can cause a compensatory increase in the activity of other DNA repair pathways. To investigate if this is the case in PARI-overexpressing pancreatic cancer cell lines, we measured the efficiency of an alternative DNA repair pathway, Translesion Synthesis (TLS), using the SupF shuttle plasmid mutation assay (22). TLS is a damage tolerance mechanism which involves bypass of DNA lesions in S-phase by specific DNA polymerases that are able to replicate through damaged DNA, albeit frequently inserting the wrong nucleotides and thus leading to point mutations. TLS is an alternative to HR repair in S-phase and the two pathways can often compensate for each other (30). The pancreatic cancer cell line 8988T exhibited a specific increase in point mutation frequency, as measured by the SupF assay (Figure 2B, Supplementary Figure 3). Thus, HR deficiency in pancreatic cancers overexpressing PARI results in TLS hyperactivation. Another recent study found that some pancreatic cancer cell lines including 8988T and CAPAN2 show increased Non-Homologous End Joining (NHEJ) activity (21). Overall, these results suggest that PARI overexpression in pancreatic cancers results in reduced HR and a compensatory increase in other DNA repair pathways, including TLS and NHEJ.
Unlike HR, both TLS and NHEJ are error-prone pathways. Their activities often result in point mutations, or, in the case of NHEJ, deletions, translocations and chromosomal rearrangements. NHEJ is also involved in the formation of radial chromosomes in response to DNA crosslinks (9). 8988T cells were hypersensitive to the DNA crosslinking drug Mitomycin C (MMC; Figure 2C), as well as to other DNA damaging agents (data not shown), consistent with the cellular defect in HR. Moreover, cytological analysis of chromosomes in metaphase spreads showed an accumulation of chromosomal abnormalities, in particular radial chromosomes, in PARI-overexpressing pancreatic cancer cell lines 8988T and CAPAN2 treated with MMC (Figure 2D, E). PARI knockdown partially suppressed the MMC hypersensitivity and chromosomal aberrations of 8988T cells (Figure 2C, Supplementary Figure 4). Thus, we conclude that PARI-overexpressing pancreatic cancer cells have defective HR, causing genomic instability and chromosomal aberrations.
PARI downregulation blocks proliferation of pancreatic cancer cell lines in vitro
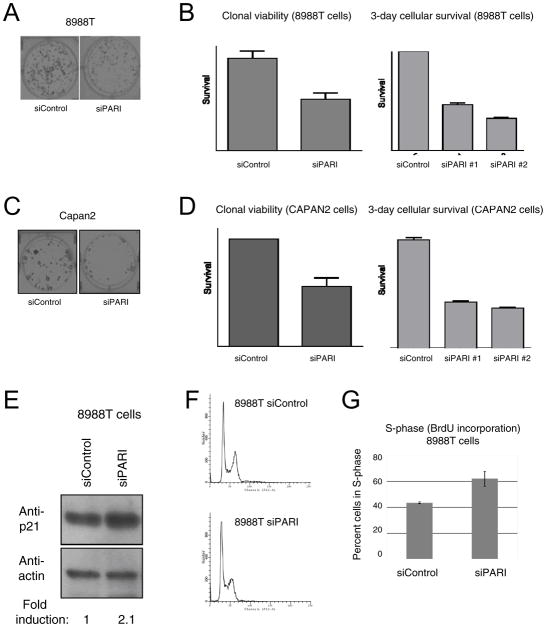
During our studies, we noticed that 8988T, CAPAN2 and other PDAC cells overexpressing PARI showed reduced viability when treated with PARI-targeting siRNA oligonucleotides. We next quantified clonal viability and cellular survival of these cells following PARI downregulation. Both 8988T (Figure 3A, B) and CAPAN2 (Figure 3C, D) cells showed a 50% reduction in survival in vitro, when PARI was suppressed. Importantly, cells that do not overexpress PARI, including pancreatic adenocarcinoma CAPAN1 cells, as well as HeLa and 293T cells, are not affected by PARI knockdown (Figure 1B, Supplementary Figure 5). Taken together, these results show that pancreatic cancers depend on PARI overexpression for their growth, and raise the possibility that targeting PARI may be a viable strategy in pancreatic cancer treatment.
Figure 3. PARI depletion impairs cellular proliferation and S-phase progression in PDAC cells.
(A, B) PARI knockdown in 8988T cells decreases cellular survival in culture. (A) Representative image of clonal viability of 8988T cells following PARI knockdown. (B) Depletion of PARI decreases both clonal viability and 3-day cellular survival in 8988T cells. Data represents average of two replicates (left panel) and six replicates (right panel). Error bars show standard error of the mean. (C, D) PARI knockdown in CAPAN2 cells decreases cellular survival. (C) Representative image of reduced clonal viability following PARI depletion in CAPAN2 cells. (D) Depletion of PARI significantly decreased both clonal viability and 3-day cellular survival in CAPAN2 cells. Data represents average of two replicates (left panel) and six replicates (right panel). Error bars represent standard error of the mean. (E) Western blot indicating that p21 is upregulated in 8988T cells upon PARI knockdown. The quantification was done normalizing against the actin signal, using ImageJ software. (F, G) PARI depletion from 8988T cells results in an accumulation of cells in S-phase. (F) FACS analysis of cycling 8988T cells showing that PARI depletion results in increased S-phase content. A quantification of this experiment is shown in Supplementary Figure 6. (G) Quantification of the percentage of 8988T cells in S-phase by BrdU incorporation. Bars represent the average of two independent experiments. Error bars represent standard deviations.
PARI is required for S-phase progression in pancreatic cancer cells
To further understand the mechanism of PARI-induced proliferation of pancreatic cancers, we investigated the effect of PARI depletion on cell cycle progression in PARI-overexpressing 8988T cells. 8988T cells, treated with siRNA targeting PARI, showed a specific induction of p21 protein (Figure 3E). Up-regulation of p21 is normally associated with S-phase arrest and replication defects (30). Indeed, we observed a specific accumulation of 8988T cells in S-phase upon PARI knockdown, both when analyzing the FACS profile (Figure 3F, Supplementary Figure 6) as well as in BrdU incorporation experiments (Figure 3G). These results suggest that PARI upregulation promotes S-phase progression of pancreatic cancer cells.
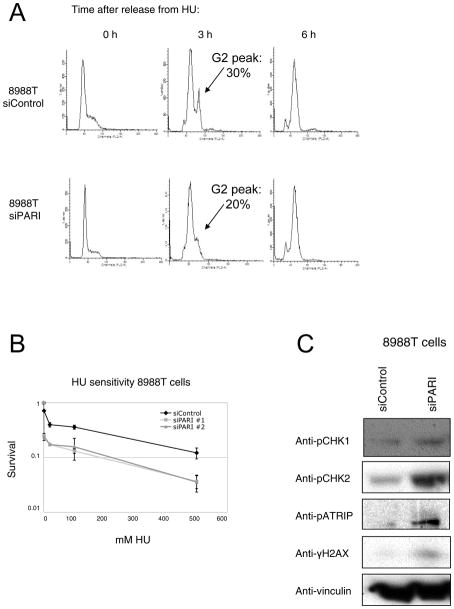
To test this model, we investigated the response of PARI-overexpressing cancer cell lines when DNA replication is hindered by addition of Hydroxyurea (HU). HU causes depletion of nucleotide pools and early S-phase arrest. We found that control and PARI-knocked-out 8988T cells similarly arrested in early S-phase upon HU treatment (Figure 4A, Supplementary Figure 7). We then released the cells in HU-free fresh media, and monitored cell cycle progression. Control cells readily reinitiated replication, and 3 hours later a significant proportion were already in G2; in contrast, PARI-depleted cells were slower to recover and enter G2 (Figure 4A). Accordingly, PARI-depleted pancreatic cancer cells were also hypersensitive to HU (Figure 4B, Supplementary Figure 8). These results suggest that PARI overexpression promotes S-phase progression following replication blockade.
Figure 4. PARI knockdown sensitizes pancreatic cancer cells to replication stress and activates the DNA damage checkpoint response.
(A) PARI-depleted 8988T cells recover slower from HU-arrest. Cells were arrested by treatment with 2mM HU for 24h, then washed and allowed to recover for indicated time points. PI-stained cells were then analyzed by FACS for DNA content. Arrows at 3h time point indicates emergence of the G2 peak; quantification of this peak was performed using Modfit software. (B) Depletion of PARI by two different siRNAs confers hypersensitivity to HU in 8988T cells. The average of two independent experiments is shown. Error bars represent standard deviations. (C) PARI knockdown results in strong activation of the DNA damage response, in the absence of exogenous DNA damage. The levels of phosphorylated CHK1, CHK2, ATRIP, and H2AX are shown.
To further investigate how PARI downregulation contributes to S-phase accumulation in pancreatic cancer cells, we studied the activation of the DNA damage response (DDR), a phosphorylation-based signaling cascade that triggers DNA repair and cell cycle arrest upon DNA damage (6). In the absence of exogenous DNA damage, PARI knockdown in 8988T cells causes a specific increase in phosphorylation of several DDR factors, including CHK1, CHK2, ATRIP and H2AX (Figure 4C). Thus, PARI depletion leads to DDR activation even in the absence of DNA damage. The activation of the DNA damage response accounts, at least in part, for the delayed S-phase progression of PARI-depleted 8988T pancreatic cancer cells.
DT40 cells overexpressing PARI show reduced DNA damage-induced replication arrest
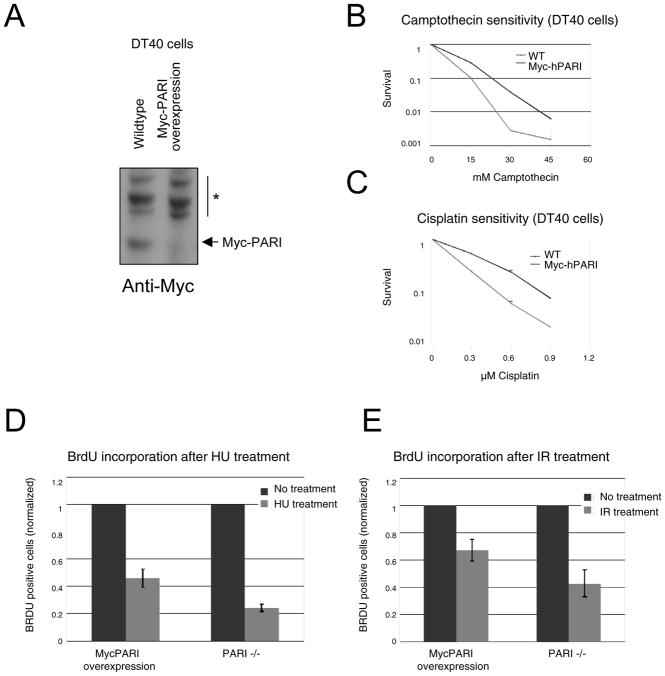
We previously generated a complete PARI knockout in DT40 cells and showed that PARI−/− cells are hyper-recombinogenic. We corrected this phenotype by complementing these cells with human PARI, showing that the human protein is functional in chicken cells (20). To test the model that PARI overexpression promotes S-phase progression following replication hindering, we attempted to mimic the situation in pancreatic cancer cell lines by overexpressing human PARI in wild-type chicken DT40 cells (Figure 5A). Similar to their human pancreatic cancer cell counterparts, DT40 cells overexpressing PARI showed hypersensitivity to DNA damaging agents including Camptothecin and Cisplatin (Figure 5B, C). These results confirm that PARI up-regulation affects DNA repair efficiency.
Figure 5. PARI overexpression in chicken DT40 promotes DNA replication.
(A) Western blot showing ectopic expression of human Myc-PARI in DT40 cells. Asterisk indicates cross-reactive bands. (B, C) Similar to their human counterparts, PARI-overexpressing DT40 cells are hypersensitive to DNA-damaging agents camptothecin (B) and cisplatin (C). Shown is 4-day cellular survival. The average of two independent experiments is present. Error bars represent standard deviations. (D, E) The suppression of DNA replication following exposure to DNA damaging agents HU (D) or IR (E) is milder in PARI-overexpressing DT40 cells compared to PARI-negative cells. Replicating cells were quantified by BrdU incorporation assays. Bars represent the average of two independent experiments. Error bars indicate standard deviations.
To further investigate the effects of PARI overexpression on DNA replication following DNA damage, we compared BrdU incorporation by PARI-knockout versus PARI-overexpressing cells after either HU or IR exposure. In both cases, PARI-knockout cells showed a severe reduction in BrdU incorporation after damage. In contrast, BrdU incorporation by PARI-overexpressing cells was less affected by both HU and IR (Figure 5D, E). Wild-type cells showed an intermediate phenotype (data not shown). These results show that PARI overexpression renders cells less sensitive to replication perturbations and suggests that PARI promotes replication of damaged DNA, perhaps resulting in further mutagenesis.
PARI depletion reduces pancreatic tumor growth in an in vivo xenograft model
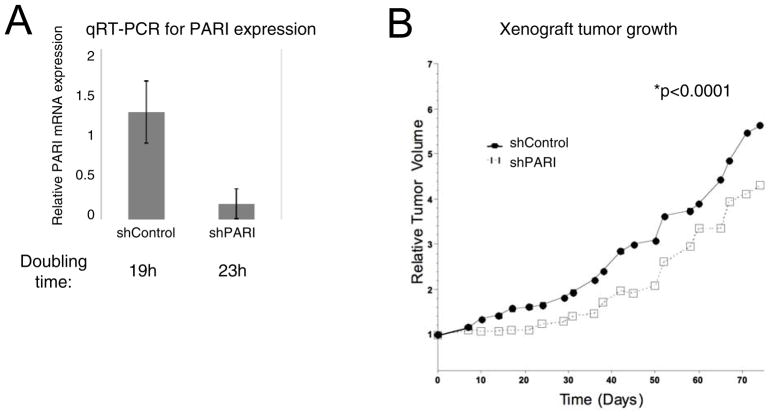
Since PARI knockdown reduces the in vitro proliferation of PARI-overexpressing pancreatic cancer cells, PARI may be a target for blocking pancreatic tumor growth in vivo. To test this hypothesis, we evaluated whether PARI overexpression is required for tumor formation in an in vivo model. To stably knockdown PARI, we created 8988T cells expressing a doxycycline-induced shRNA targeting PARI. A non-targeting shRNA was used as control. Quantitative RT-PCR analysis confirmed that PARI mRNA levels were down-regulated upon addition of doxycycline to the growth media (Figure 6A). As expected from the above-described experiments with siRNA, these cells also showed reduced proliferation in vitro, with a doubling time of 23h compared to 19h for control cells (Supplementary Figure 9).
Figure 6. PARI is required for in vivo growth in 8988T PDAC cells in a xenograft tumor model.
(A) Quantitative RT-PCR analysis shows that the doxycyline-induced shRNA hairpin system employed efficiently downregulates PARI mRNA when cells are grown in the presence of 20μg/ml doxycyline. A non-targeting hairpin is used as a control. Bars represent the average of two independent experiments. Error bars are standard deviations. The calculated doubling time is shown for each cell line (see also Supplementary Figure 9). (B) PARI depletion suppresses the growth of 8988T xenograft tumors in athymic nude mice. Tumor growth is shown as relative tumor volume after addition of doxycyline (N=11, 9 for shControl, shPARI, respectively). The results are statistically significant as measured by two-way paired t-test (p<0.0001).
8988T cells readily form xenograft tumors when injected subcutaneously in athymic nude mice (31). Upon implantation, the tumors were allowed to grow until the reached a volume of 200mm3. At this stage, mice were given doxycycline in the drinking water to induce shRNA expression and PARI depletion and monitored for up to 70 days. Compared to shControl tumors, shPARI tumors showed a statistically significant (p<0.0001) growth defect, as measured by tumor volume (Figure 6B). Analysis of biopsied tumors by hematoxylin and eosin staining showed significant levels of necrotic tissue in shPARI tumors, compared to control tumors (Supplementary Figure 10). These results show that PARI overexpression promotes the growth of tumors formed by pancreatic cancer cells in vivo.
Discussion
Pancreatic cancers and genomic instability
Homologous Recombination promotes error-free DNA repair of strand breaks in S-phase and G2 by using the sister chromatid as repair template (32). Thus, HR is essential for maintaining genomic stability. Our study reveals that PDAC cells overexpressing PARI are defective in the assembly of RAD51 foci (Figure 2A), indicating a deficiency in HR repair. Additionally these cells have hallmark features of genomic instability, as evidenced by elevated chromosomal breakage (Figure 2D, E). Interestingly, mutations in BRCA2, or other HR factors such as RAD54, XRCC2, XRCC3 and RECQ1 (4, 13, 14) have also been associated with pancreatic cancer. Similar to those mutations, PARI overexpression reduces HR. Thus, we propose that pancreatic cancers can employ two different mechanisms to suppress HR: inactivating mutations in HR pathway members, or overexpression of HR negative regulators. Upregulation of PARI is likely to constitute an alternate to BRCA2 inactivation in pancreatic cancers.
How a reduction in Homologous Recombination contributes to PDAC transformation is not clear. Genetic instability is a hallmark of most cancers (33), and it is likely to be important for generating a pool of mutations from which transforming mutations can be selected. NHEJ, which lacks a homologous template, is error-prone, as it involves end resection and subsequent ligation of DNA ends, a process that often results in loss or addition of nucleotides (34). It has recently been shown that PDAC cells have hyperactive NHEJ (21). Similarly, we show here that PDAC cells have increased TLS. TLS is also an error prone mechanism, involving mutagenic polymerases to bypass DNA lesions (35). Thus, it is possible that PARI overexpression in pancreatic cancers reduces HR, resulting in a compensatory increase in error-prone pathways like NHEJ and TLS, and thus favoring genomic instability and accumulation of mutations. This feature of PARI overexpression is reminiscent of the BRCA1-deficiency phenotype, which is also characterized by reduced HR and increased TLS (36). We predict that pancreatic cancer genome sequencing studies may reveal a hypermutation phenotype.
PARI overexpression suppresses replication arrest by DNA damage
PARI knockdown in the human pancreatic cancer cell line 8988T confers S-phase arrest and hypersensitivity to replication stalling, and activates the DNA damage response pathway even in the absence of exogenous DNA damage (Figure 4). Thus, PARI may function to promote replication through damaged DNA, perhaps resulting from endogenous stresses. This damage tolerance function ensures the suppression of the DDR when PARI is overexpressed in cancer cells. Following PARI knockdown, PDAC cells seem unable to progress through S-phase, perhaps because increased HR results in slower DNA damage processing, and/or in accumulation of a particular intermediate, eventually activating the DDR (Supplementary Figure 11). Indeed, TLS and NHEJ are simpler, and presumably faster mechanisms for repairing DNA lesions (37, 38). Interestingly, the yeast recombination inhibitor Srs2 (39), which shares significant structural and functional homology to PARI was similarly shown to terminate the DNA damage checkpoint (40). Although PARI-overexpressing cells replicate faster through damaged DNA, they do this by sacrificing mutation-suppressing mechanisms. This results in genomic instability, DNA damage sensitivity, and accumulation of mutations. It was recently shown that stalled replication forks are salvaged by a BRCA2-RAD51 dependent mechanism, rescuing them from degradation by the MRE11 nuclease (41, 42). It will be interesting to investigate if the PCNA-SUMO-PARI pathway plays any role in replication fork protection against MRE11 activity.
As detailed above, it is possible that PARI overexpression is essential in pancreatic cell transformation because of its ability to inhibit HR. Alternatively, it is possible that yet another, so far un-indentified function of PARI is involved in promoting S-phase progression and suppressing the DDR in pancreatic cancer cells. In this context, PARI was recently found as a substrate of the cell cycle kinase CDK2 in a proteomic approach (43). Perhaps CDK2 phosphorylation of PARI regulates its putative replication promoting function. A single protein that can regulate both DNA repair, and S-phase transition would ensure that cells have the means to quickly alter their cell cycle progression in response to DNA damage. Interestingly, yeast Srs2 and human BRCA2 were also shown to be phosphorylated by CDK kinases to coordinate HR with cell cycle progression (44, 45).
PARI overexpression and cancer therapy
In this study, we show that pancreatic cancer cells overexpressing PARI have DNA repair defects and are hypersensitive to DNA damaging agents (Figure 2). This raises the possibility that PARI expression is a potent biomarker for predicting the response of pancreatic and other tumors to genotoxic cancer treatment, including radiation and platinum-based chemotherapy. Consistent with this notion, PARI-overexpressing, HR-deficient pancreatic cancer cells are hypersensitive to PARP inhibitors (Supplementary Figure 2).
Importantly, we show that PARI knockdown greatly affects the proliferation of PARI-overexpressing pancreatic cancer cells in vitro (Figure 3). Furthermore we show that PARI overexpression in pancreatic cancer cells is required for tumor growth in a xenograft tumor mouse model (Figure 6). Thus, we propose that small molecule inhibitors of PARI could be used as novel targeted therapy for patients harboring PARI-overexpressing pancreatic tumors. Since PARI inhibition also restores DNA repair in pancreatic cancer cells, PARI inhibitors would be employed as single therapeutic agents, and not in combination with DNA damaging therapy. Our results strongly suggest an important function for PARI in pancreatic cancer; further research is required to understand exactly why pancreatic cancers become addicted to PARI overexpression.
Supplementary Material
Acknowledgments
Financial support: This work is supported by NIH grants R01DK43889, R37HL52725, P01HL048546, and P01CA092584 to A.D.D., and by Postdoctoral Fellowships from the Susan G. Komen Breast Cancer Foundation to G.L.M. and E.P.
Grant Support
We thank Benjamin Milne for early contributions to this project, Lisa Moreau for chromosome breakage analysis, Benjamin Primack for help with mouse work, Sofia Vidal-Cardenas for help with FACS experiments, and Erin Aho and Alexander Vlahos for technical assistance. G.L.M. and E.P. are supported by Postdoctoral Fellowships from the Susan G. Komen Breast Cancer Foundation. This work is supported by National Institutes of Health grants R01DK43889, R37HL52725, P01HL048546, and P01CA092584 to A.D.D.
Footnotes
Conflicts of interest: The authors disclose no potential conflicts of interest.
References
- 1.Siegel R, Desantis C, Virgo K, Stein K, Mariotto A, Smith T, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62:220–41. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 2.Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–17. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 3.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2012;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein AP. Genetic susceptibility to pancreatic cancer. Mol Carcinog. 2012;51:14–24. doi: 10.1002/mc.20855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goggins M, Schutte M, Lu J, Moskaluk CA, Weinstein CL, Petersen GM, et al. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res. 1996;56:5360–4. [PubMed] [Google Scholar]
- 6.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–94. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 7.Roberts NJ, Jiao Y, Yu J, Kopelovich L, Petersen GM, Bondy ML, et al. ATM mutations in patients with hereditary pancreatic cancer. Cancer Discov. 2012;2:41–6. doi: 10.1158/2159-8290.CD-11-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moldovan GL, D’Andrea AD. How the fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–49. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–9. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 11.Jones S, Hruban RH, Kamiyama M, Borges M, Zhang X, Parsons DW, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Heijden MS, Yeo CJ, Hruban RH, Kern SE. Fanconi anemia gene mutations in young-onset pancreatic cancer. Cancer Res. 2003;63:2585–8. [PubMed] [Google Scholar]
- 13.Jiao L, Hassan MM, Bondy ML, Wolff RA, Evans DB, Abbruzzese JL, et al. XRCC2 and XRCC3 gene polymorphism and risk of pancreatic cancer. Am J Gastroenterol. 2008;103:360–7. doi: 10.1111/j.1572-0241.2007.01615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li D, Liu H, Jiao L, Chang DZ, Beinart G, Wolff RA, et al. Significant effect of homologous recombination DNA repair gene polymorphisms on pancreatic cancer survival. Cancer Res. 2006;66:3323–30. doi: 10.1158/0008-5472.CAN-05-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olive KP, Tuveson DA. The use of targeted mouse models for preclinical testing of novel cancer therapeutics. Clin Cancer Res. 2006;12:5277–87. doi: 10.1158/1078-0432.CCR-06-0436. [DOI] [PubMed] [Google Scholar]
- 16.Skoulidis F, Cassidy LD, Pisupati V, Jonasson JG, Bjarnason H, Eyfjord JE, et al. Germline Brca2 heterozygosity promotes Kras(G12D) -driven carcinogenesis in a murine model of familial pancreatic cancer. Cancer Cell. 2010;18:499–509. doi: 10.1016/j.ccr.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 17.Hosokawa M, Takehara A, Matsuda K, Eguchi H, Ohigashi H, Ishikawa O, et al. Oncogenic role of KIAA0101 interacting with proliferating cell nuclear antigen in pancreatic cancer. Cancer Res. 2007;67:2568–76. doi: 10.1158/0008-5472.CAN-06-4356. [DOI] [PubMed] [Google Scholar]
- 18.Kashiwaya K, Hosokawa M, Eguchi H, Ohigashi H, Ishikawa O, Shinomura Y, et al. Identification of C2orf18, termed ANT2BP (ANT2-binding protein), as one of the key molecules involved in pancreatic carcinogenesis. Cancer Sci. 2009;100:457–64. doi: 10.1111/j.1349-7006.2008.01058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piao L, Nakagawa H, Ueda K, Chung S, Kashiwaya K, Eguchi H, et al. C12orf48, termed PARP-1 binding protein, enhances poly(ADP-ribose) polymerase-1 (PARP-1) activity and protects pancreatic cancer cells from DNA damage. Genes Chromosomes Cancer. 2011;50:13–24. doi: 10.1002/gcc.20828. [DOI] [PubMed] [Google Scholar]
- 20.Moldovan GL, Dejsuphong D, Petalcorin MI, Hofmann K, Takeda S, Boulton SJ, et al. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol Cell. 2012;45:75–86. doi: 10.1016/j.molcel.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li YH, Wang X, Pan Y, Lee DH, Chowdhury D, Kimmelman AC. Inhibition of non-homologous end joining repair impairs pancreatic cancer growth and enhances radiation response. PLoS One. 2012;7:e39588. doi: 10.1371/journal.pone.0039588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mirchandani KD, McCaffrey RM, D’Andrea AD. The Fanconi anemia core complex is required for efficient point mutagenesis and Rev1 foci assembly. DNA Repair (Amst) 2008;7:902–11. doi: 10.1016/j.dnarep.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hochegger H, Dejsuphong D, Fukushima T, Morrison C, Sonoda E, Schreiber V, et al. Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. EMBO J. 2006;25:1305–14. doi: 10.1038/sj.emboj.7601015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tainsky MA, Krizman DB, Chiao PJ, Yim SO, Giovanella BC. PA-1, a human cell model for multistage carcinogenesis: oncogenes and other factors. Anticancer Res. 1988;8:899–913. [PubMed] [Google Scholar]
- 25.Elsasser HP, Lehr U, Agricola B, Kern HF. Establishment and characterisation of two cell lines with different grade of differentiation derived from one primary human pancreatic adenocarcinoma. Virchows Arch B Cell Pathol Incl Mol Pathol. 1992;61:295–306. doi: 10.1007/BF02890431. [DOI] [PubMed] [Google Scholar]
- 26.Sumi S, Beauchamp RD, Townsend CM, Jr, Uchida T, Murakami M, Rajaraman S, et al. Inhibition of pancreatic adenocarcinoma cell growth by lovastatin. Gastroenterology. 1992;103:982–9. doi: 10.1016/0016-5085(92)90032-t. [DOI] [PubMed] [Google Scholar]
- 27.Oplustilova L, Wolanin K, Mistrik M, Korinkova G, Simkova D, Bouchal J, et al. Evaluation of candidate biomarkers to predict cancer cell sensitivity or resistance to PARP-1 inhibitor treatment. Cell Cycle. 2012;11:3837–50. doi: 10.4161/cc.22026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 29.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 30.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–79. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 31.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–29. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu Rev Genet. 2010;44:113–39. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bakhoum SF, Compton DA. Chromosomal instability and cancer: a complex relationship with therapeutic potential. J Clin Invest. 2012;122:1138–43. doi: 10.1172/JCI59954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mladenov E, Iliakis G. Induction and repair of DNA double strand breaks: the increasing spectrum of non-homologous end joining pathways. Mutat Res. 2011;711:61–72. doi: 10.1016/j.mrfmmm.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 35.Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol. 2012;13:141–52. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pathania S, Nguyen J, Hill SJ, Scully R, Adelmant GO, Marto JA, et al. BRCA1 is required for postreplication repair after UV-induced DNA damage. Mol Cell. 2011;44:235–51. doi: 10.1016/j.molcel.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Helleday T. Pathways for mitotic homologous recombination in mammalian cells. Mutat Res. 2003;532:103–15. doi: 10.1016/j.mrfmmm.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 38.Mao Z, Bozzella M, Seluanov A, Gorbunova V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst) 2008;7:1765–71. doi: 10.1016/j.dnarep.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marini V, Krejci L. Srs2: the “Odd-Job Man” in DNA repair. DNA Repair (Amst) 2010;9:268–75. doi: 10.1016/j.dnarep.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaze MB, Pellicioli A, Lee SE, Ira G, Liberi G, Arbel-Eden A, et al. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol Cell. 2002;10:373–85. doi: 10.1016/s1097-2765(02)00593-2. [DOI] [PubMed] [Google Scholar]
- 41.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–42. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Errico A, Deshmukh K, Tanaka Y, Pozniakovsky A, Hunt T. Identification of substrates for cyclin dependent kinases. Adv Enzyme Regul. 2010;50:375–99. doi: 10.1016/j.advenzreg.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 44.Saponaro M, Callahan D, Zheng X, Krejci L, Haber JE, Klein HL, et al. Cdk1 targets Srs2 to complete synthesis-dependent strand annealing and to promote recombinational repair. PLoS Genet. 2010;6:e1000858. doi: 10.1371/journal.pgen.1000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Esashi F, Christ N, Gannon J, Liu Y, Hunt T, Jasin M, et al. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.