Abstract
Dmrt1(doublesex and mab-3 related transcription factor 1) is a regulator of testis development in vertebrates that has been implicated in testicular germ cell tumors of mouse and human. In the fetal mouse testis Dmrt1 regulates germ cell pluripotency in a strain-dependent manner. Loss of Dmrt1 in 129Sv strain mice results in a >90% incidence of testicular teratomas, tumors consisting cells of multiple germ layers; by contrast, these tumors have never been observed in Dmrt1 mutants of C57BL/6J (B6) or mixed genetic backgrounds. To further investigate the interaction between Dmrt1 and genetic background we compared mRNA expression in wild type and Dmrt1 mutant fetal testes of 129Sv and B6 mice at embryonic day 15.5 (E15.5), prior to overt tumorigenesis. Loss of Dmrt1 caused misexpression of overlapping but distinct sets of mRNAs in the two strains. The mRNAs that were selectively affected included some that changed expression only in one strain or the other and some that changed in both strains but to a greater degree in one versus the other. In particular, loss of Dmrt1 in 129Sv testes caused a more severe failure to silence regulators of pluripotency than in B6 testes. A number of genes misregulated in 129Sv mutant testes also are misregulated in human testicular germ cell tumors (TGCTs), suggesting similar etiology between germ cell tumors in mouse and man. Expression profiling showed that DMRT1 also regulates pluripotency genes in the fetal ovary, although Dmrt1 mutant females do not develop teratomas. Pathway analysis indicated disruption of several signaling pathways in Dmrt1 mutant fetal testes, including Nodal, Notch, and GDNF. We used a Nanos3-cre knock-in allele to perform conditional gene targeting, testing the GDNF coreceptors Gfra1 and Ret for effects on teratoma susceptibility. Conditional deletion of Gfra1 but not Ret in fetal germ cells of animals outcrossed to 129Sv caused a modest but significant elevation in tumor incidence. Despite some variability in genetic background in these crosses, this result is consistent with previous genetic mapping of teratoma susceptibility loci to the region containing Gfra1. Using Nanos3-cre we also uncovered a strong genetic interaction between Dmrt1 and Nanos3, suggesting parallel functions for these two genes in fetal germ cells. Finally, we used chromatin immunoprecipitation (ChIP-seq) analysis to identify a number of potentially direct DMRT1 targets. This analysis suggested that DMRT1 controls pluripotency via transcriptional repression of Esrrb, Nr5a2/Lrh1, and Sox2. Given the strong evidence for involvement of DMRT1 in human TGCT, the downstream genes and pathways identified in this study provide potentially useful candidates for roles in the human disease.
Keywords: Dmrt1, Nanos3, teratoma, TGCT, pluripotency, germ cell
Introduction
Germ cells tread a narrow developmental path between pluripotency and lineage commitment. They are fated eventually to differentiate into one of two cell types — the spermatozoan or the oocyte — and they undergo an extended series of differentiation events that starts with germ lineage specification during early fetal development and concludes in adulthood with gamete differentiation. However, artificial manipulations or genetic defects can readily disrupt germ cell fate commitment, inducing germ cells to express their underlying pluripotency and adopt a variety of somatic cell fates (Durcova-Hills and Capel, 2008; Stevens, 1964).
Misregulation of pluripotency plays an important role in testicular germ cell tumors, the most common malignancy of young men (Oosterhuis and Looijenga, 2005). In the fetal mouse the result is formation of teratomas, tumors containing cells representing all three somatic germ layers (mesoderm, endoderm and ectoderm). Teratomas can be experimentally induced when embryonic genital ridges are grafted onto adult testes, but only if the donor tissue is transplanted prior to embryonic day 13.5 (E13.5) (Regenass et al., 1982; Stevens, 1964; Stevens, 1966), a timing that is consistent with the downregulation of pluripotency-associated mRNAs that begins during this period (Western et al., 2010; Yamaguchi et al., 2005). Testicular teratomas also can be induced genetically. Most mouse strains do not develop teratomas, but they occur spontaneously at a low incidence in mice of the 129Sv inbred strain (Stevens and Little, 1954) and can occur at higher incidence in mice mutant for germ cell developmental regulators including the RNA binding protein Dnd1 or the transcription factor Dmrt1, or mutant for the tumor suppressor Pten (Kimura et al., 2003; Krentz et al., 2009; Stevens, 1973; Youngren et al., 2005). These tumors involve inappropriate expression of a variety of pluripotency regulators, cell cycle regulators, and signaling pathway genes (Cook et al., 2011; Krentz et al., 2009), helping to highlight the pathways and regulators that normally constrain germ cell proliferation and pluripotency. Precisely how these pathways normally are controlled in the fetal germ line and why 129Sv mice are so sensitive to teratoma formation is poorly understood.
In humans, testicular germ cell tumors (TGCTs) also commonly express genes associated with pluripotency, such as OCT4 and NANOG, whereas normal adult germ cells do not (Clark et al., 2004; Hoei-Hansen et al., 2005; Jones et al., 2004; Rajpert-De Meyts et al., 2004). These differences in gene expression suggest that failure to regulate germ cell pluripotency also underlies human TGCT formation, and the persistence of early germ cell markers in germ cell tumors of mice and humans suggests that impaired developmental progression plays an important role. However, TGCT susceptibility is genetically complex and despite clear evidence that human TGCTs have a strong genetic component (Forman et al., 1992; Heimdal et al., 1996), familial linkage studies failed to identify predisposition loci. Linkage analysis in the mouse confirms the genetic component and suggests that 100 or more loci may significantly affect TGCT susceptibility (Lam and Nadeau, 2003). Recently human genome-wide association studies (GWAS) have identified six potential TGCT susceptibility loci (Kanetsky et al., 2009; Kanetsky et al., 2011; Rapley et al., 2009; Turnbull et al., 2010). Among these is DMRT1, a finding that strongly suggests a related etiology in formation of human and mouse TGCTs. In addition, in the mouse the availability of single gene mutations that dramatically elevate TGCT incidence, such as Dnd1 and Dmrt1, has provided an entry point to explore the role of genetic background and to define the genetic architecture involved in TGCT formation.
Dmrt1 is a member of a conserved gene family sharing the DM domain DNA binding motif and it functions in germ cells and somatic cells of the male gonad to transcriptionally regulate multiple aspects of gonadal development and function (Matson and Zarkower, 2012). We found previously that loss of Dmrt1 in mice of the 129Sv genetic background results in a very high incidence of teratoma formation, whereas Dmrt1−/− mice of the C57BL/6J or mixed strain backgrounds do not develop teratomas (Krentz et al., 2009). Mutations in Dnd1 also cause teratomas only in 129Sv mice, and this is due to a strain-dependent difference in apoptotic response; genetic suppression of apoptosis allows loss of Dnd1 to cause teratomas in B6/129Sv mixed background mice as well (Cook et al., 2009; Cook et al., 2011). By contrast, Dmrt1 mutant mice of B6, 129Sv or mixed background do not undergo elevated fetal germ cell apoptosis, but loss of Dmrt1 nevertheless causes teratomas only in 129Sv mice. In 129Sv mice Dmrt1 is required for repression of pluripotency regulators including the core regulators Sox2, Oct4, and Nanog, and for proper arrest of fetal germ cells in mitosis (Krentz et al., 2009).
Here have investigated Dmrt1-dependent TGCT formation by comparing how mRNA expression at E15.5 responds to loss of Dmrt1 in a teratoma-susceptible strain (129Sv) versus a teratoma-resistant strain (B6). We find that loss of Dmrt1 differentially affects mRNA expression in the two strains. In particular, loss of Dmrt1 more severely deregulates pluripotency gene repression in 129Sv than in B6 mice. Expression analysis also indicates that Nodal and Notch signaling are upregulated by loss of Dmrt1. The TGF family ligand GDNF and its tyrosine kinase coreceptors Gfra1 and Ret were underexpressed in Dmrt1 mutant testes, suggesting that GDNF signaling pathway might regulate fetal germ cell proliferation and/or pluripotency (this paper and (Krentz et al., 2009)). We conditionally deleted each GDNF coreceptor in fetal germ cells using a Nanos3-cre knockin allele and found that loss Gfra1 caused elevated teratoma susceptibility. We also found that Nanos3 functionally interacts with Dmrt1 to suppress TGCT formation, suggesting overlapping roles for translational and transcriptional regulation in the fetal germ line. Finally, we combined expression profiling with chromatin immunoprecipitation approaches (ChIP-chip, ChIP-seq, and qChIP) to identify genes whose transcription is likely to be regulated directly by DMRT1 in the fetal testis and ovary. We found that DMRT1 directly controls expression the pluripotency regulators Esrrb and Nr5a2/Lrh1 and that DMRT1 can apparently associate with DNA by two mechanisms in the fetal gonad, one involving its canonical DNA binding motif and one involving association with C-rich sequences, primarily in CpG islands.
Materials and Methods
Mouse breeding
Experiments involving Dmrt1flox/flox, Dmrt1+/− or Dmrt1−/− mice (Kim et al., 2007; Raymond et al., 2000) were performed on mice outcrossed to 129S6/SvEvTac (129S6) (Taconic Labs) at least seven times, or to C57BL/6J (B6) (Jackson Labs) at least ten times. Swiss Webster outbred mice (Charles River) were used for ChIP-seq experiments and mixed backgrounds (B6, 129Sv, and Swiss Webster) were used for ChIP-chip experiments. Nanos3Cre/+ mice on the MCH genetic background (Suzuki et al., 2008) were outcrossed 4x onto 129S1/SvImJ (129S1) (Jackson labs) and then crossed to 129S6 (N10) Dmrt1flox/flox animals for conditional targeting of Dmrt1 in embryonic germ cells. The mix of 129S1 and 129S6 substrains is presumed to cause the elevated teratoma background seen in Dmrt1fl/+ animals (Fig. 4B; TDK and DZ, unpublished results). Gfra1fl/fl (Uesaka et al., 2007) and Retfl/fl (Jain et al., 2006) mice were outcrossed 5x onto 129S1 and conditionally deleted in germ cells by crossing to Nanos3Cre/ mice previously outcrossed 4x onto 129S1. Presence of a copulation plug in the morning was recorded as E0.5. Genotyping of Dmrt1, Gfra1 and Ret was as described (Jain et al., 2006; Kim et al., 2007; Uesaka et al., 2007).
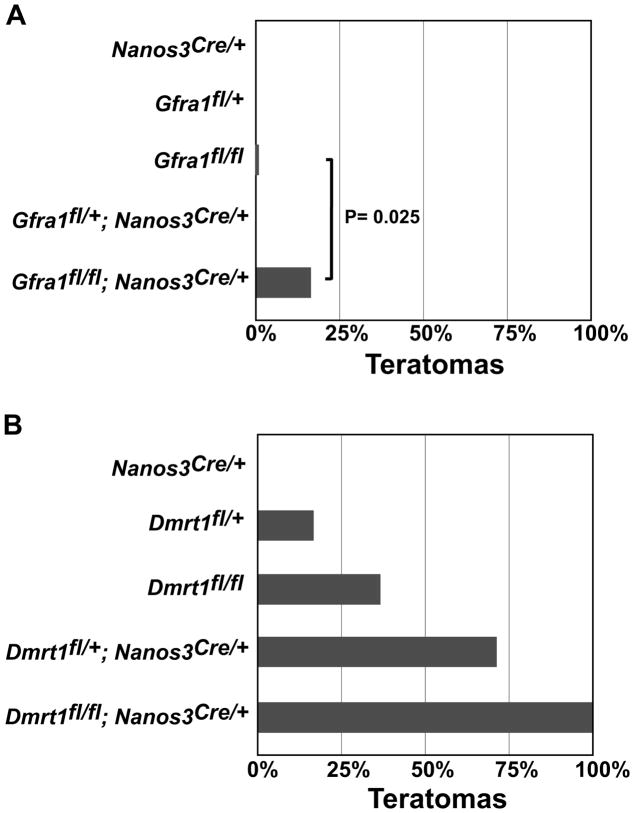
Figure 4. Gfra1 and Nanos3 help control proliferation/pluripotency in the fetal testis.
(A) Incidence of teratomas in 129Sv mice carrying a functional “floxed” allele of Gfra1 or conditionally deleted for Gfra1 using Nanos3Cre. Numbers of testes examined was: Nanos3Cre/+ (N = 12); Gfra1fl/+ (N=6); Gfra1fl/fl (N=49); Gfra1fl/+;Nanos3Cre/+ (N=10); Gfra1fl/fl; Nanos3Cre/+ (N=44). Indicated P value (0.025) is from two-tailed Fisher’s exact test. (B) Incidence of teratomas in 129Sv mice carrying a functional “floxed” allele of Dmrt1 or conditionally deleted with Nanos3Cre. Number of testes examined was: Nanos3Cre/+ (N=22); Dmrt1fl/+ (N=12); Dmrt1fl/fl (N=30); Dmrt1fl/+;Nanos3Cre/+ (N=29); Dmrt1fl/fl;Nanos3Cre/+ (N=34).
mRNA expression profiling
Testes and ovaries were dissected from E15.5 embryos and placed in RNAlater (Qiagen) until all genotypes were collected. RNA was prepared from gonad pairs and hybridized to Affymetrix 430 2.0 expression arrays (one gonad pair per array; each genotype analyzed in triplicate) and analyzed as previously described (Krentz et al., 2009). Expression profiling from wild type gonads from E11.5–E18.5 was downloaded from GEO (GSE6881) (Small et al., 2005). Temporally regulated mRNAs were defined as those with more than two-fold higher or lower expression between E11.5 and E18.5 (p > 0.05). RNA preparation and qRT-PCR were performed as previously described, using Hprt as a normalization control (Krentz et al., 2009). Primer sequences are in Table S1.
ChIP
Embryos were collected at E13.5 and sexed by gonadal morphology. Testes from 65–80 males were dissected and pooled for each biological replicate in ChIP-chip and qChIP. ChIP-seq was performed using 600 E13.5 gonad pairs per experiment. Chromatin was prepared as described (Krentz et al., 2009; Murphy et al., 2010). DNA from three independent DMRT1 ChIP samples was amplified by ligation-mediated PCR (Oberley et al., 2004) modified to use DNATerminator (Lucigen Corp.). Amplified ChIP and input material was labeled by Nimblegen and hybridized to Mouse ChIP 385K RefSeq Promoter Arrays (Nimblegen). Promoters with enrichment were identified as described (Murphy et al., 2010). For validation of ChIP data, qPCR was performed on unamplified immunoprecipitated DNA using gene specific primers that directly flank the ChIP peak (Table S1). qPCR was performed in an Eppendorf Mastercycler epgradient S thermocycler using SYBR green detection (Life Sciences) and the comparative C(T) method (Schmittgen and Livak, 2008).
ChIP-seq data analysis
Illumina FASTQ sequence files were converted to Sanger FASTQ file format using FASTQ Groomer (Blankenberg et al., 2010) and mapped to mouse genome (MM9) using Bowtie (Langmead et al., 2009). Areas of enrichment were identified using Model-based Analysis of ChIP-seq (MACS version 1.4.1) (Zhang et al., 2008). MACS was run using the command line: macs14 -t target.bam -c control.bam -n target_peaks -s 25 -g 1.87e9 -p 1e-20 --slocal 100 --llocal 1000. ChIP-seq peak calls are available in Table S9.
ChIP motif search
The nucleotide sequence under the area of the DMRT1 binding peak was used for motif scanning using W-ChIPMotif (Jin et al., 2009). The P-values used for motif discovery were from the 99.9% confidence interval.
Results
Strain-dependent responses to loss of Dmrt1 in the fetal testis
To establish baselines for examining the effects of Dmrt1 loss at E15.5, we first analyzed mRNA expression in wild type testes of teratoma-resistant B6 mice and teratoma-prone 129Sv mice by microarray hybridization (Fig. S1). We chose E15.5 as the developmental time for this analysis in part because previous mRNA profiling at E13.5 identified very few misexpressed genes in Dmrt1 mutant 129Sv testes (Krentz et al., 2009). At E15.5 wild type germ cells normally have finished downregulating expression of pluripotency genes and have undergone cell cycle arrest; misexpression of genes involved in these processes therefore should be more readily detectable by this stage of development.
Comparison of testicular mRNA expression between the two strains identified 827 genes that were differentially expressed more than 2-fold (Fig. S1, left; Table S2) (P<0.05). This result is consistent with the previous observation that the gonadal and germ cell transcriptomes of the two strains differ substantially at E11.5 and E14.5 (Cook et al., 2011; Munger et al., 2009). To ask whether this differential expression might reflect a difference in developmental timing between the two strains we used data from a published developmental profile of fetal testis mRNA expression in B6/129Sv hybrid mice (Small et al., 2005). This analysis showed that although some of the differentially expressed genes are normally temporally-regulated during fetal development, many are not (Fig. S1, right). In addition, based on examination of the temporally-regulated genes whose expression differed between the two strains, neither strain showed a unidirectional difference in the timing of testicular gene expression relative to the other at E15.5 (i.e., an overall developmental delay or acceleration) (Fig. S2). We conclude that the strain differences in testicular mRNA expression at E15.5 include differences in temporal gene regulation but they do not reflect a simple difference in the rate of testicular development between the two strains.
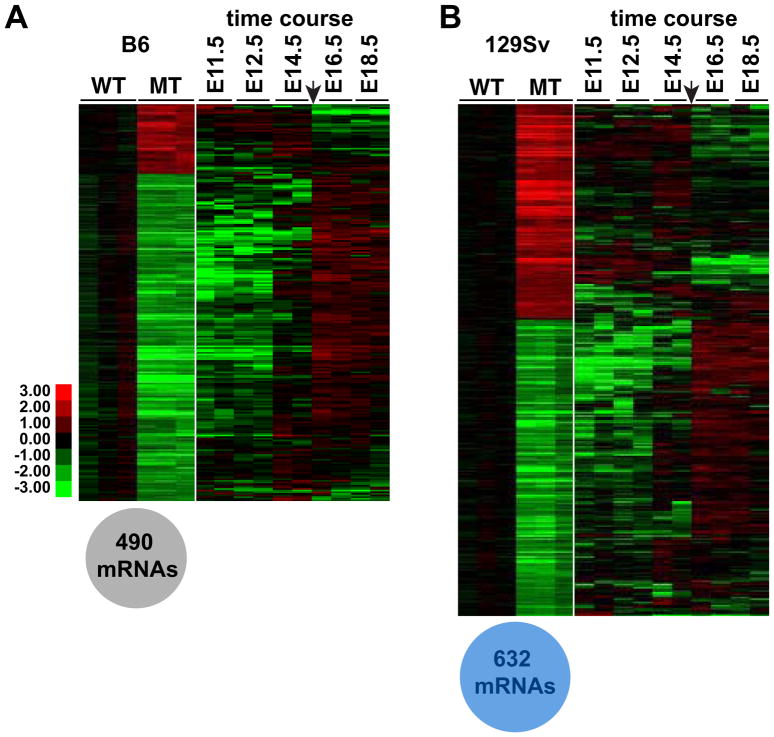
Next we compared the consequences of Dmrt1 loss in each strain. A Dmrt1 null mutation in B6 or mixed background mice causes postnatal defects in germ cell and Sertoli cell development and eventually leads to male-to-female transdifferentiation of Sertoli cells (Fahrioglu et al., 2007; Kim et al., 2007; Matson et al., 2011; Raymond et al., 2000). Although B6 Dmrt1 mutant testes appear normal until birth, loss of Dmrt1 in this strain caused misexpression of 490 mRNAs at E15.5, mainly involving reduced expression (Fig. 1A, left; Table S3) (>2-fold change; P < 0.05). Most of these affected mRNAs were developmentally regulated in the wild type fetal testis but failed to be up- or down-regulated in the mutant testis appropriately for this stage of development (Fig. 1A, right). From these results we conclude that loss of Dmrt1 in B6 mice impairs fetal testicular differentiation by E15.5. B6 mutant testes did not display a global disruption of testicular differentiation, however: only a small fraction of the 1787 mRNAs that normally change expression >2-fold from E11.5 to E18.5 in wild type testes were affected by loss of Dmrt1 (Fig. S2).
Figure 1. Misregulation of mRNA expression in B6 and 129Sv Dmrt1 mutant testes.
(A) Left: heat map showing relative expression of 490 mRNAs differing more than two-fold (P<0.05) between E15.5 B6 wild type and B6 Dmrt1 mutant testes. mRNA levels were normalized to wild type. Right: data from Small et al. (2005) showing relative expression of the same mRNAs from E11.5 to E18.5. Arrow indicates E15.5. (B) Left: heat map showing relative expression of 632 mRNAs that differed more than two-fold between E15.5 129Sv wild type and 129Sv Dmrt1 mutant testes. Right: data from Small et al. (2005) showing relative expression of the same mRNAs from E11.5 to E18.5 in B6/129 hybrid testes.
In 129Sv mice, loss of Dmrt1 disrupted expression of 632 mRNAs (>2-fold change; P < 0.05) (Fig. 1B; Table S4). As in B6 mutants, some of these mRNAs normally are developmentally regulated; however a greater proportion of affected mRNAs in 129Sv mutants were not developmentally regulated (Fig. 2B,D). In addition, while the affected mRNAs in mutants of either strain mainly showed reduced expression, the proportion with elevated expression was higher in 129Sv mutants (17% elevated in B6, versus 42% in 129Sv). We conclude that loss of Dmrt1 in the 129Sv testis compromises testicular differentiation as it does in B6, but also is likely to disrupt other functions, and these presumably contribute to the highly elevated incidence of teratoma formation in mutants of this strain.
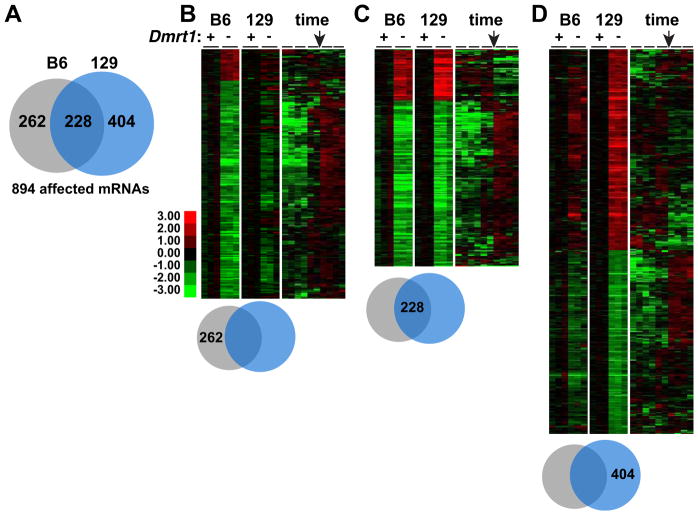
Figure 2. mRNA misexpression in Dmrt1 mutant testes is dependent on genetic background.
(A) Venn diagram indicating the proportion of 894 mRNAs misexpressed more than two-fold (P>0.05) in that were affected in Dmrt1 mutant testes of each genetic background. (B) Left: heat map showing relative expression of 262 mRNAs whose expression differed more than two-fold in B6 mutant testes and less than two-fold in 129Sv mutant testes at E15.5. Right: heat map using data of Small et al (2005) and showing relative expression of the same mRNAs from E11.5 to E18.5 in B6/129 hybrid testes. Arrow indicates E15.5. (C) As in panel B, but showing relative expression of 228 mRNAs affected more than two-fold in mutant testes of both strain backgrounds. (D) As in panel B but showing relative expression of 404 mRNAs affected more than two-fold in 129Sv mutant testes and less than two-fold in B6 mutant testes.
A potential concern in analyzing whole testes is that a large difference in the proportion of germ cells between strains or genotypes could introduce a global bias into the mRNA expression profiles. To address this question we examined expression of 361 mRNAs that are enriched in fetal male germ cells (Jameson et al., 2012), asking whether their expression levels varied systematically between the wild type testes of the two strains or between wild type and mutant testes of each strain. 167 germ line enriched mRNAs differed significantly (p < 0.05) between wild type testes of the two strains, with an average elevation of 1.33-fold in 129Sv versus B6. In B6 testes 93 mRNAs differed significantly between wild type and mutant, averaging 1.09-fold higher in the mutant. These data suggest that germ cell numbers are slightly higher in 129Sv than B6 testes and that B6 mutant testes have approximately normal germ cell numbers relative to wild type. In 129Sv testes 136 mRNAs differed significantly between wild type and mutant, averaging 1.66-fold higher in the mutant. This difference was strongly affected by a small number of highly elevated mRNAs (eg, >40-fold higher expression of L1td1 and Otx2) but it remained possible that 129Sv mutants also have an increase in germ cell numbers relative to wild type. We therefore examined E15.5 testes from 129Sv wild type and mutant animals by immunohistochemistry, staining germ cells for MHV (Fig. S3). We could not exclude a modest increase in germ cells in 129Sv mutant testes, but a major difference was not apparent at E15.5, consistent with the previous conclusion that germ cell numbers are normal in 129Sv mutant gonads at E13.5 and at birth (Krentz et al., 2009). From these analyses we conclude the threshold of two-fold change in germ cell gene expression that we used should be sufficient to identify misregulated mRNAs, whether upregulated or downregulated.
Comparison of which mRNAs were misregulated in Dmrt1 mutants of each strain showed that some were affected in both strains but, strikingly, about 75% were significantly misregulated only in one strain or the other (Fig. 2). In particular, loss of Dmrt1 in 129Sv testes affected many mRNAs that were not affected by loss in B6: of 894 mRNAs affected in the two strains, 262 were significantly affected only in testes of B6 mutants, 404 only in 129Sv mutants, and 228 in both (>2-fold change; P < 0.05). Most of the mRNAs that were affected in both strains normally are developmentally regulated but had failed to increase or decrease in expression appropriately, further indicating that loss of Dmrt1 disrupts testicular differentiation in both strains (Fig. 2B,C). However, among the mRNAs selectively affected in 129Sv mutant testes about half were overexpressed and were not normally subject to temporal regulation in the fetal testis (Fig. 2D). We suggest that these mRNAs are prime candidates to drive teratoma formation. Indeed, among mRNAs more strongly overexpressed in 129Sv than B6 mutant testis were a number already implicated in human testicular germ cell tumors, including the three most highly over-expressed (L1td1, Otx2, and Zic3; Table S5). For example, L1td1 regulates self-renewal and proliferation in human seminoma cells and ES cells (Narva et al., 2012).
Strain-dependent disruption of pluripotency regulation
Expression profiling indicated distinct transcriptional responses to loss of Dmrt1 in 129Sv and B6 fetal testes. To help identify functions and regulatory pathways disrupted by loss of Dmrt1, we used Ingenuity Pathway Analysis (IPA; www.ingenuity.com;) (Table S6). Function analysis indicated that B6 mutants were specifically enriched mainly for defects in spermatogenesis and gonadogenesis. This finding is consistent with our earlier conclusion that the main defect in B6 mutants appeared to be a disruption in testicular differentiation and with our previous studies showing that overt germ cell defects are apparent in Dmrt1 mutants shortly after birth (Fahrioglu et al., 2007). Disrupted functions specific to 129Sv mutants included organogenesis, cell morphology, infertility, cellular homeostasis, and morphology of gonadal cells. Pathway analysis highlighted pluripotency regulation as affected in both strains, but with more genes affected in 129Sv mutant testes (Table S6). We confirmed the differential effect on pluripotency gene expression in the two strains using quantitative RT-PCR (qRT-PCR), which showed that loss of Dmrt1 caused a more severe failure to silence pluripotency genes in 129Sv than in B6 (Fig. 3).
Figure 3. Misexpression of pluripotency regulators in Dmrt1 mutant testes is dependent on genetic background.
qRT-PCR comparing expression in E15.5 wild type and mutant testes of each background. For each mRNA, expression is relative to the level in wild type B6. Error bars indicate standard deviation (N=3 testes pairs for each genotype and background). *P<0.05, and **P<0.005, unpaired t-test, comparison of WT vs mutant.
We also used IPA transcription factor analysis to search for transcriptional networks affected by loss of Dmrt1 by assessing affected target mRNAs that function in these pathways (Table S6). Importantly, this tool can identify an affected transcription factor network even when the mRNA level of the transcription factor is normal, as long as a significant number of its predicted targets are misexpressed. Based on target gene expression, a larger number of transcriptional networks appeared to be misregulated in mutants of both strains including targets of CTNNB1/ -catenin, which was predicted to be activated; however, many more networks were specifically affected in 129Sv mutants (17 with p<0.001 in 129Sv, versus 3 in B6). These results suggest that while loss of Dmrt1 affects expression of similar numbers of mRNAs in the two strains (490 in B6 versus 632 in 129Sv), the consequences for transcriptional regulatory networks are substantially more severe in 129Sv mice.
Altered Nodal pathway gene expression in mutant testes
IPA analysis suggested that a number of other regulatory genes and pathways relevant to germ cell identity or cancer were affected in mutant testes, including some affected selectively in 129Sv mutants. Among these latter were the Nodal signaling pathway components Nodal, Tdgf1 and Lefty1 (Table S6) whose activity is important in embryonic development and stem cell maintenance and is linked to a variety of cancers (Bianco et al., 2010; Shen, 2007). Expression profiling detected elevated expression of mRNAs encoding the TGF -family signaling proteins NODAL (2.6-fold in B6, 6.2-fold in 129Sv) and GDF3, which potentiates NODAL signaling (unchanged in B6, 2.7-fold in 129Sv), as well as the NODAL receptor TDGF1 (Teratocarcinoma-derived growth factor 1)/CRIPTO (2.3-fold in B6, 11-fold in 129Sv) and the Nodal target LEFTY1 (3.4-fold in B6, 5.2-fold in 129Sv). Quantitative RT-PCR (qRT-PCR) for several of these components confirmed their elevated expression and stronger misregulation in 129Sv mutants (Fig. S4). These data suggest that the Nodal signaling pathway is likely to be overactive in Dmrt1 mutant fetal testes.
Gfra1 and Nanos3 as potential germ cell pluripotency regulators
One goal of profiling mRNA expression changes in Dmrt1 mutant testes is to identify regulatory pathways whose disruption might contribute to teratoma susceptibility. We previously identified GDNF signaling as a candidate for such a role. Our previous mRNA profiling at E13.5 identified 18 mRNAs misexpressed more than two-fold (p<0.05) in 129Sv Dmrt1 null mutant versus wild-type testes (Krentz et al., 2009). Among these was Ret, whose protein depletion in germ cells at E13.5 was confirmed by immunofluorescence (Krentz et al., 2009). RET and GFRA1 are co-receptors for GDNF, which is required postnatally for spermatogonial stem cell self-renewal (Hofmann, 2008; Jain et al., 2004; Naughton et al., 2006). In addition, loss of Ret, but not Gdnf, compromises germ cell survival in the fetal testis (Miles et al., 2012). At E15.5 expression profiling detected misregulation of both Ret (down 2.3-fold in B6 and 2.6-fold in 129Sv) and Gdnf (down 1.8-fold in B6 and 2.4-fold in 129Sv Dmrt1 mutants). We therefore tested the possibility that DMRT1 is required for proper GDNF signaling in the embryo and that loss of GDNF signaling may cause germ cells to differentiate into teratomas in 129Sv mice. Because Ret and Gfra1 null mutations are lethal, we bred conditional alleles of each gene onto the 129Sv genetic background and conditionally deleted each gene in fetal germ cells using a “knock-in” allele in which Cre is expressed from the Nanos3 locus (Nanos3Cre) (Suzuki et al., 2008).
Conditional deletion of Ret did not affect teratoma formation (0/18 Retfl/fl;Nanos3Cre/+ 129Sv testes had tumors), but loss of Gfra1 in germ cells resulted in teratoma formation in 16% of mutant testes (7/44; Fig. 4A). As controls we examined undeleted Gfra1fl/fl “floxed” testes (1/49 had tumors), Gfra1fl/+;Nanos3Cre/+ heterozygous conditional mutants (0/10 had tumors), and Nanos3cre/+ testes (0/12 had tumors). Comparison of tumor incidence in Gfra1fl/fl;Nanos3Cre/+ versus Gfra1fl/fl controls indicated that homozygous deletion of the conditional allele of Gfra1 causes a significant increase in tumor incidence (P = 0.025, Fisher’s exact test; Fig. 4A).
The Nanos3-cre allele causes loss of Nanos3 function and thus its potential role in teratoma formation also must be considered. Although Nanos-cre heterozygosity did not cause elevated teratoma formation either on its own (0/12) or when combined with either Retfl/+ or Retfl/fl (0/40), it remains possible that Nanos3 heterozygosity can enhance the phenotype of Gfra1 homozygosity. Indeed, in parallel experiments we found a strong genetic interaction between Nanos3 and Dmrt1. We previously found that heterozygosity of Dmrt1 alone did not significantly increase teratoma formation in 129Sv males (Krentz et al., 2009). By contrast, 129Sv males doubly heterozygous for Dmrt1fl and Nanos3cre had highly elevated teratoma formation (21/29 testes) relative to either Dmrt1fl/+ (2/12; P = 0.002, Fisher’s exact test) or Nanos3Cre/+ (0/22; P < 0.001, Fisher’s exact test) (Fig 4A,B). This synthetic phenotype suggests that Dmrt1 and Nanos3 have related functions in controlling pluripotency and proliferation. From this genetic analysis we conclude that both Gfra1 and Nanos3 can affect teratoma susceptibility and are good candidates to regulate fetal germ cell proliferation/pluripotency.
Direct transcriptional regulation of pluripotency by DMRT1 in the fetal testis
To investigate further which targets DMRT1 may regulate directly, we used genome-wide chromatin immunoprecipitation methods (ChIP-chip and ChIP-seq) to identify DMRT1 binding sites in the fetal testis. We chose E13.5, which is a key time for suppression of pluripotency based on testis transplantation experiments and mRNA expression studies (Durcova-Hills and Capel, 2008; Stevens, 1964; Stevens, 1966). ChIP-chip identified 245 promoter regions bound by DMRT1 in at least two out of three experiments at the stringency we employed (see Methods). Taking into account bidirectional promoters, DMRT1 bound to promoters associated with 362 genes. ChIP-seq identified 2430 regions bound by DMRT1 in E13.5 testis at the stringency we employed. Comparing the ChIP-chip and ChIP-seq data with mRNA expression data identified 63 genes adjacent to DMRT1 binding peaks that were misexpressed more than two-fold in Dmrt1 mutant testes at E15.5 (p<0.05) (Table S7). Of these potential direct target genes, 23 were misexpressed specifically in 129Sv Dmrt1−/− testes, 14 were misexpressed in B6 Dmrt1−/− testes, and 16 were misexpressed in mutants on both genetic backgrounds.
Among the putative direct DMRT1 transcriptional targets were a number with potential relevance to TGCT. Two putative direct targets that are upregulated in mutants are pluripotency regulators. One is Esrrb, a direct Nanog target gene that can substitute for Nanog (Festuccia et al., 2012) and the other is Nr5a2/Lrh1, which can function as a pluripotency regulator in iPS cell reprogramming (Heng et al., 2010; Wang et al., 2011). Two putative direct targets elevated in mutant testes are involved in Wnt signaling: one is Wnt inhibitory factor 1 (Wif1) and the other is Rnf43, a stem cell E3 ubiquitin ligase that can block Wnt responsiveness in a variety of cancer cells by inducing Wnt receptor endocytosis (Koo et al., 2012). Pmaip1/Noxa is a putative direct target elevated in mutant testes that is linked to cisplatin sensitivity in embryonal carcinoma cells (Grande et al., 2012). Among putative direct targets reduced in mutant testes, Rrad acts as a tumor suppressor in nasopharyngeal carcinoma, Prdm16 is an H3K9me1 histone methyltransferase involved in maintaining integrity of heterochromatin (Pinheiro et al., 2012), and Nedd4l is an E3 ubiquitin ligase that is reduced during development of prostate cancer and whose reduced expression is associated with aggressiveness and poor prognosis in other cancers (Gao et al., 2012; He et al., 2012; Hu et al., 2009). It will be important in the future to investigate genetically the roles of these genes in teratoma susceptibility.
Regulation of DNA methylases by DMRT1
Epigenetic reprogramming is a major germ cell-specific event that occurs around the time when teratomas form in Dmrt1 mutant mice (Hackett et al., 2012). Two important DNA methylases were misregulated in Dmrt1 mutant testes, suggesting that epigenetic defects may contribute to the mutant phenotype. One was Dnmt3l, a DNA methylase that is required for DNA methylation and normal proliferation of fetal germ cells (La Salle et al., 2007). Expression of Dnmt3l differed between B6 and 129Sv wild-type testes and was reduced by loss of Dmrt1 in both strains, with a much greater reduction in B6 mutants (Fig. S5). qChIP detected binding of DMRT1 to Dnmt3l in the fetal testis, suggesting possible direct regulation of DNA methylation by DMRT1. Another important germ cell DNA methylase, Dnmt3b, was elevated 4-fold in 129Sv mutant testes but unchanged in B6 mutants (Table S4) and appeared to be indirectly regulated. DNMT3L is expressed in human TGCTs and knockdown of DNMT3L in EC cell lines suppressed their growth and induced apoptosis, suggesting a role in human TGCT formation (Minami et al., 2010). We speculate that abnormal DNA methylation resulting from misregulated methylase expression may affect teratoma sensitivity. For example, the nearly complete elimination of Dnmt3l expression in B6 mutant germ cells may help block their ability to differentiate into teratomas, whereas overexpression of Dnmt3b may enhance teratoma formation in 129Sv mutant testes. Both of these possibilities can be tested in future genetic studies.
DMRT1 also regulates pluripotency in the fetal ovary
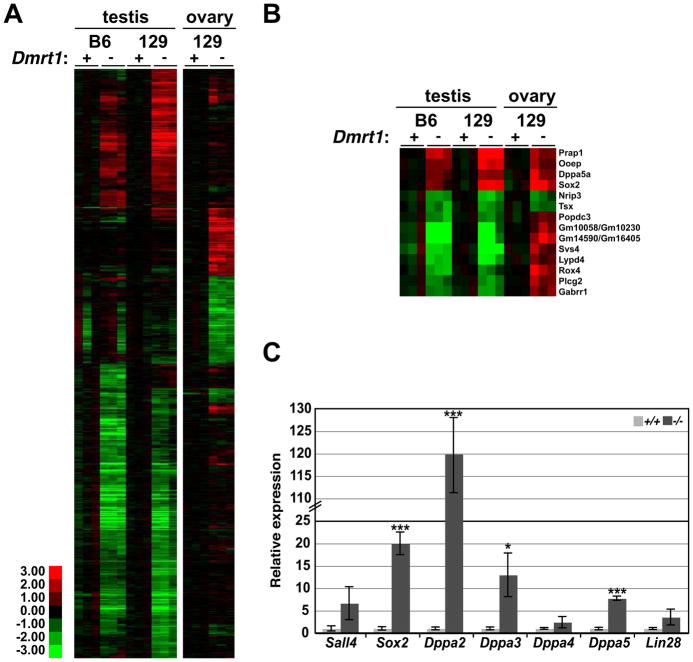
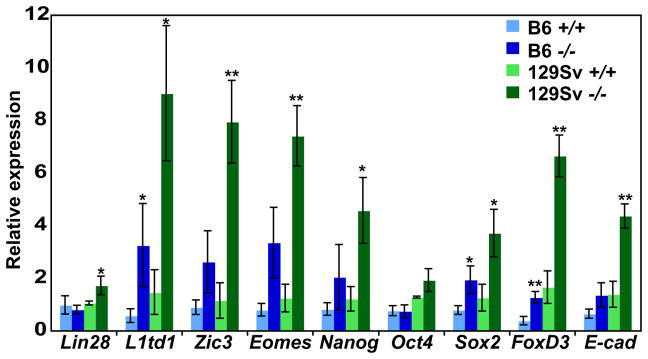
Dmrt1 is expressed transiently in ovarian fetal germ cells during the period when pluripotency genes normally are down-regulated and meiosis initiates (Krentz et al., 2011; Lei et al., 2007; Raymond et al., 1999) and we found previously that Dmrt1 mutant female germ cells undergo abnormal meiosis (Krentz et al., 2011). At E13.5, only eight mRNAs were misexpressed two-fold or more in mutant ovaries, including the meiotic regulator Stra8 (Krentz et al., 2011). However, since DMRT1 represses expression of pluripotency genes in the testis at E15.5, we used mRNA expression profiling to ask whether it also does so in the ovary. 326 genes were misexpressed more than two-fold (p<0.05) in mutant 129Sv ovaries at E15.5 (Fig. 5; Table S8). Most of the mRNAs regulated by Dmrt1 in the fetal ovary were different from those in the fetal testis (Fig. 5A; mRNAs misexpressed in both sexes are shown in Fig. 5B). However, as in males, the overexpressed mRNAs in Dmrt1 mutant females included a number that are associated with pluripotency (eg, Sall4, Sox2, Dppa2, Dppa3/Stella, Dppa4, Dppa5, and Lin28) (Bowles et al., 2003; Kim et al., 2005; Maldonado-Saldivia et al., 2007; Richards et al., 2004; Yang et al., 2010; Yuan et al., 1995). qRT-PCR confirmed the elevated expression of these pluripotency factors in Dmrt1−/− ovaries (Fig. 5C). We speculate that suppression of pluripotency is an important function of DMRT1 in the fetal gonad in both sexes and that this aberrant expression may contribute to the meiotic defects seen in Dmrt1 mutant oocytes (Krentz et al., 2011).
Figure 5. DMRT1 regulates distinct genes in testis and ovary, but regulates pluripotency in both.
(A) Heat map showing relative expression of 1192 mRNAs that differed more than twofold between B6 or 129Sv wild type and Dmrt1 mutant testes or wild type and Dmrt1 mutant 129Sv ovaries at E15.5. In each wild type versus mutant comparison transcripts were normalized to wild type. (B) Heat map showing mRNAs misexpressed both in testis and ovary. (C) qRT-PCR analysis confirming elevated mRNA expression of pluripotency regulators in mutant ovaries at E15.5. *P<0.05, and ***P<0.001, unpaired t-test, comparison of WT vs mutant.
Distinct modes of DMRT1 DNA association in fetal and postnatal gonads
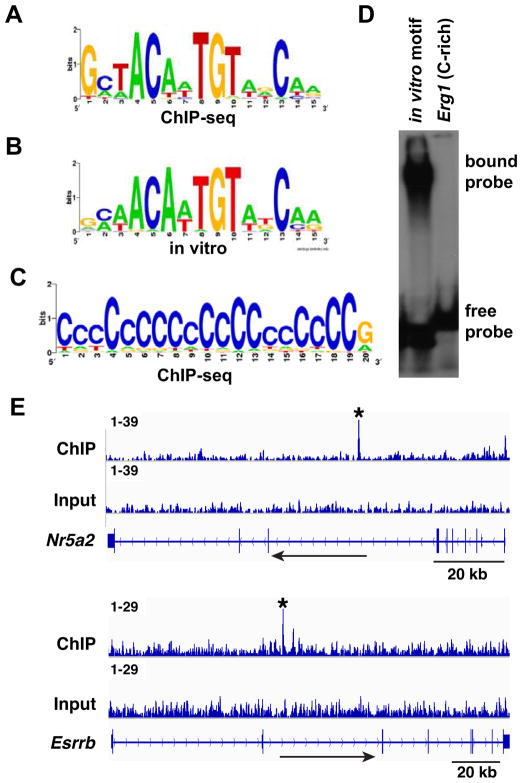
To identify potential DMRT1 recruitment elements we performed motif searches (Jin et al., 2009) of the DMRT1-bound regions. Analysis of the 100 top-ranked ChIP-seq binding peaks identified a motif almost exactly matching the previously identified in vitro DMRT1 binding motif (Fig. 6A; P = 3.6 × 10−10) (Murphy et al., 2007). We also detected enrichment of a polyC motif (Fig. 6B; P = 10−227). This sequence closely resembled the C-rich motif we previously found enriched in DMRT1 binding sites in the fetal ovary (Krentz et al., 2011) and appeared to reflect the association of DMRT1 with promoter-proximal CpG islands. The association of DMRT1 with polyC motifs suggests either that DMRT1 is able to bind this motif directly or that DMRT1 may be recruited to DNA via another transcription factor(s) in the fetal gonad. To test the first possibility we performed a gel shift assay using as the probe a C-rich motif found in the promoter of Egr1, a gene bound and transcriptionally regulated by DMRT1 at E15.5. DMRT1 was unable to bind this C-rich element in vitro (Figure 6C), suggesting either that its association with DMRT1 in vivo is indirect or that the association requires a modification of DMRT1 that is lacking in the in vitro translated protein.
Figure 6. DMRT1 DNA association in the fetal gonad.
(A) Enriched motif in top 100 peaks, closely resembling in vitro defined DMRT1 recognition sequence. (B) In vitro-defined DNA binding motif. (C) C-rich motif enriched in DMRT1 binding peaks. (D) Gel mobility shift assay showing that DMRT1 can bind to the in vitro defined DMRT1 DNA binding motif but not to a C-rich sequence (5′TCCTCCCCCTCCTACCCCCCCCCCCCACAC3′) derived from a prominent DMRT1 binding peak in the Erg1 gene. (E) ChIP-seq data showing binding of DMRT1 to Nr5a2/Lrh1 and to Esrrb in the E13.5 testis. Binding peaks called by MACS are indicated by asterisks. Each peak contains a close match to the DMRT1 consensus DNA binding motif.
Discussion
Here we have used mRNA expression profiling to compare wild type and Dmrt1−/− fetal testes on the 129Sv and B6 genetic backgrounds. We found loss of Dmrt1 in the two strains causes misregulation of overlapping but distinct sets of mRNAs. The combination of expression profiling and genetic analysis highlighted several signaling pathways, including NODAL and GDNF-related pathways, as well as RNA-based regulation by NANOS3, as strong candidates for involvement in germ cell tumor formation in the mouse. To better understand how DMRT1 controls gene expression in the fetal gonad we also used genome-wide ChIP-seq analysis to identify sites bound by DMRT1 in the E13.5 testis and ovary. This analysis suggested that DMRT1 controls pluripotency by direct repression of several key stem cell pluripotency genes.
Loss of Dmrt1 had very different consequences for mRNA expression in B6 versus 129Sv testes, with nearly half of affected mRNAs (404/894) significantly misexpressed only in 129Sv mutant testes. The most prominent group of genes misexpressed in Dmrt1 mutant testes were pluripotency regulators. Many were overexpressed in mutants of both strains, but the number of genes affected and the magnitude of overexpression both were higher in 129Sv mutants. There are many links between pluripotency regulators and human TGCT. Human TGCTs are thought to originate from fetal germ cells that transform into EC cells (pediatric tumors) or CIS cells (adult tumors including seminomas and nonseminomas) (Oosterhuis and Looijenga, 2005). Both of these progenitor cells express OCT4, NANOG and other key pluripotency genes and it is presumed that their expression plays an important role in tumor formation, especially in pediatric TGCTs and nonseminomas comprised of mixed somatic cell types. Previously we found by analysis of candidate genes that a number of pluripotency genes were upregulated in Dmrt1 mutant fetal testes and that DMRT1 binds to Sox2 in the fetal testis (Krentz et al., 2009). Here we used expression profiling to show that many pluripotency genes show elevated expression. In addition genome-wide ChIP analysis indicated that DMRT1 binds and negatively regulates two additional pluripotency genes, Esrrb and Nr5a2/Lrh1, suggesting that DMRT1 controls the pluripotency network by direct transcriptional repression. (ChIP-seq analysis in this study also detected binding of DMRT1 to Sox2, but below the significance cutoff that was used.)
The regulatory relationship between DMRT1 and the pluripotency network may be reciprocal. The Dmrt1 promoter is bound by NANOG, OCT4, SOX2, KLF4, and ZFX in mouse ES cells (Chen et al., 2008; Loh et al., 2006) and by NANOG, SOX2 and OCT4 in human ES cells (Boyer et al., 2005; Lister et al., 2009). Moreover, Dmrt1 expression is activated when Sox2 or Oct4 is depleted in ES cells (Masui et al., 2007; Sharov et al., 2008). Those results together with the data from this study suggest that the core pluripotency regulators and Dmrt1 may anchor antagonistic gene networks critical to proper developmental progression, with the pluripotency regulators silencing Dmrt1 in ES cells to help maintain “stemness” and Dmrt1 silencing pluripotency regulators in fetal germ cells to help restrict cell fates in the germ cell lineage. Dmrt1 is expressed only in the testis, so its repression of pluripotency represents an organ-specific mechanism. Germ cells differ from other cells in that they express pluripotency genes at several different stages of development, and thus they may need a specialized regulatory mechanism to allow the transient suppression of pluripotency.
Several of the pluripotency regulators misexpressed in Dmrt1 mutant testes (for example Nanog, Klf4, Nr5a2/Lrh1, Oct4) can help to reprogram differentiated somatic cells into pluripotent stem cells (Takahashi and Yamanaka, 2006). ChIP analysis suggests that DMRT1 controls pluripotency by transcriptional repression of Sox2, Esrrb, and Nr5a2/Lrh1 ((Krentz et al., 2009) and this paper). Lrh1 can replace Oct4 in iPS cell reprogramming (Heng et al., 2010). In postnatal Dmrt1 mutant testes, which undergo Sertoli-to-granulosa cell (male to female) reprogramming, Nr5a2/Lrh1 also is highly expressed (Matson et al., 2011). The involvement of Nr5a2/Lrh1 in both processes suggests a possible mechanistic link between teratoma formation and sexual transdifferentiation.
We found that loss of Dmrt1 also causes elevated pluripotency gene expression in fetal ovaries, and yet wild type and Dmrt1 mutant females of the 129Sv strain do not develop ovarian teratomas. This sex specificity is unlikely to derive from the sexual genotype of the germ cells, as XX germ cells developing in a male environment can generate EC clusters (Cook et al., 2009). One possibility is that the testicular but not the ovarian somatic microenvironment is conducive to teratoma formation and thus germ cells in a fetal testis must suppress pluripotency by E15.5 to prevent aberrant differentiation in response to that environment.
Recent studies have shown an association between ectopic meiotic initiation and teratoma formation in fetal germ cells, and mutation of the meiotic regulator Stra8 reduced teratoma incidence in 129Sv mice (Cook et al., 2011; Heaney et al., 2012). We previously found that DMRT1 directly represses Stra8 transcription in postnatal germ cells and activates Stra8 in the fetal ovary (Krentz et al., 2011; Matson et al., 2010). Although a difference in Stra8 expression was apparent in microarray experiments, the signal was extremely low. We confirmed the low expression by qRT-PCR (not shown), and antibody staining did not detect STRA8 in wild type or Dmrt1 mutant fetal male germ cells of either strain (Krentz et al., 2009) and unpublished results). These results suggest that teratoma formation in Dmrt1 mutants is likely independent of Stra8
In addition to pluripotency genes, a number of the mRNAs misregulated in Dmrt1 mutant 129Sv testes also are known to be associated with human TGCT formation (Table S5). Among the gene networks associated with human TGCT that stood out in this study was the Nodal signaling pathway. Loss of Dmrt1 caused elevated expression of several Nodal pathway components including the TGF family ligands Nodal and Gdf3, the receptor Tdgf1/Cripto, and the downstream target Lefty1, all with stronger misregulation in 129Sv than B6 mutant testes. Nodal signaling is clearly linked to human TGCT: NODAL, TDGF1/CRIPTO, and LEFTY1 are overexpressed in human TGCTS including embryonal carcinomas, yolk sac tumors, and non-seminomas (Almstrup et al., 2005; Baldassarre et al., 1997; Harrison et al., 2007; Looijenga et al., 2007; Spiller et al., 2012). Amplification of chromosome 12p is common in invasive TGCTs; GDF3 is found within the critical region of 12p and has elevated expression in invasive seminoma relative to normal testis (Almstrup et al., 2005; Caricasole et al., 1998; Ciccodicola et al., 1989; Harrison et al., 2007; Looijenga et al., 2007). Importantly, in a phase I clinical investigation, targeting of the Nodal pathway with an anti-CRIPTO immunoconjugate gave robust anti-tumor activity in human xenografts (Kelly et al., 2011). Nodal signaling also can affect pluripotency: overexpression of NODAL in human ES cells maintains the undifferentiated state and reduced expression causes reduced OCT4 and NANOG expression and decreased stem cell self-renewal (James et al., 2005; Vallier et al., 2005; Vallier et al., 2004). In mice reduced Nodal function reduces efficiency of embryonic germ cell (EG) colony derivation (Spiller et al., 2012). Nodal pathway components are transiently expressed in fetal germ cells between about E12.5 and E14.5 and reduced Nodal pathway activity can cause premature male germ cell differentiation (Spiller et al., 2012). Our data suggest that repression of Nodal signaling is important for silencing pluripotency and preventing TGCT formation in the mouse. It will be of interest to test whether activated Nodal signaling in fetal germ cells is sufficient to cause teratoma formation and whether loss of Nodal signaling is sufficient to suppress teratomas in 129Sv mice.
Many cancers have ES cell-like expression signatures (Kim and Orkin, 2011). ES cell pluripotency and self-renewal are maintained by three distinct gene regulatory networks, one controlled by MYC, one by Polycomb Group (PcG) repressors, and a third by the core pluripotency regulators including SOX2, NANOG and OCT4. In most cancers only the MYC signature is strongly enriched, but germ cell tumors appear to be an exception, showing activity of core pluripotency genes (Clark, 2007; Clark et al., 2004). Our results help elucidate the link between DMRT1 and the core pluripotency regulators in mouse germ cell tumors, a connection that could have therapeutic potential.
Dmrt1 mutant 129Sv germ cells have reduced RET expression (this study and (Krentz et al., 2009), suggesting a possible role for GDNF signaling in teratoma formation. We tested this idea by conditional targeting of the coreceptors Ret and Gfra1. Deletion of Ret using Nanos3Cre had no affect on teratoma incidence, but deletion of Gfra1 caused a modest increase in teratomas from 2% to 16%. A caveat in these experiments is that the mice were not fully outbred to 129Sv and non-uniform strain backgrounds could affect teratoma incidence (see Methods for strain details). However, the numbers of animals analyzed in the experimental and control groups and the significant difference in teratoma incidence between these groups suggest a contribution of Gfra1 to teratoma susceptibility that could be relevant to pluripotency regulation in the mouse and to human germ cell tumor development. The relatively low incidence of teratomas in Gfra1 mutants compared to Dmrt1 mutants may indicate that other pathways act in parallel downstream of Dmrt1. There are a number of possible explanations for the lack of teratomas in conditional Ret mutants. The simplest is that Gfra1 functions in this context via a non-canonical signaling pathway that does not require Ret. However it also is possible that deletion of Ret by Nanos3Cre does not deplete RET protein early enough to reveal a function or that Ret mutant EC cells arrest or die before they can form teratomas. A role for Gfra1 also is supported by consomic strain analysis: introduction of a MOLF strain-derived segment of chromosome 19 containing Gfra1 into 129Sv mice causes elevated teratoma formation (Youngren et al., 2003).
The elevated tumor incidence in Dmrt1fl/+; Nanos3Cre/+ males relative to males heterozygous for either alone suggests that these two genes may function in parallel to regulate germ cell development. Here again a caveat is that the animals were not fully outbred to 129Sv (see Methods), but the significant differences in teratoma incidence between genotypes suggest role for Nanos3. There is evidence that Dmrt1 and Nanos3 are components of a gene network regulated by the transcription factor Tcfap2c. TCFAP2C/AP-2γ is a downstream target of the master germ cell regulator Blimp1 that is required for specification of germ cells during embryonic development (Schafer et al., 2011; Weber et al., 2010). Based on ChIP and gene expression studies, Tcfap2c is proposed to transcriptionally regulate Nanos3 and Dmrt1, as well as Dnmt3b (Kidder and Palmer, 2010; Weber et al., 2010; Woodfield et al., 2010). Dnmt3b expression in EC cells appears to convey high sensitivity to 5-aza-cytidine (Beyrouthy et al., 2009), suggesting the possibility that manipulation of this regulatory network might have a useful role in TGCT chemotherapy.
Nanos3 is expressed shortly after specification of PGCs in XX and XY germ cells and becomes male-specific shortly after sex determination (Tsuda et al., 2003). Nanos3 has not previously been implicated in pluripotency regulation or TGCT formation, likely because null mutant PGCs die during migration to the gonadal primordium (Suzuki et al., 2008; Tsuda et al., 2003). The teratoma phenotype in Dmrt1;Nanos3 doubly heterozygous males is an example how synthetic interactions can help reveal gene functions that are obscured by more severe null mutant phenotypes. Nanos genes are among the most deeply conserved of germ cells regulators (Tsuda et al., 2003). In Drosophila Nanos represses the expression of somatic cell markers and loss of Nanos causes some germ cells to adopt somatic fates (Hayashi et al., 2004). The genetic interaction of Nanos3Cre with Dmrt1 that we observed suggests that regulation of germ cell versus somatic cell identity may be among the conserved Nanos functions.
The data presented here have allowed us to identify genes and gene networks that differ between testes of TGCT-resistant and TGCT-prone mice as well as those that respond differently to loss of Dmrt1 in the two strains. In addition, in vivo DNA binding studies have implicated DMRT1 in the control of pluripotency in fetal germ cells via direct transcriptional repression of at least three key regulators. The 129Sv mouse provides a highly sensitive genetic model for the discovery and functional analysis of TGCT genes. Given the likely involvement in human TGCT of DMRT1 and many of the genes it regulates, we suggest that the Dmrt1-responsive mRNAs described here are good candidates for further investigation in human germ cell proliferation and TGCT formation.
Supplementary Material
Highlights.
Dmrt1 mutant mice develop testicular teratomas only on the 129Sv strain background.
mRNA profiling reveals strain-dependent transcriptional responses to loss of Dmrt1.
Dmrt1 regulates pluripotency and signaling pathways including Nodal, Notch, and Gdnf.
The GDNF coreceptor Gfra1 regulates teratoma susceptibility.
Dmrt1 genetically interacts with the germ cell RNA regulator Nanos3.
Acknowledgments
We thank members of the Zarkower and Bardwell laboratories for helpful discussions and critical reading of the manuscript, Chris Small, Katarina Chatzi, Ari Melnick, and Micah Gearhart for technical assistance, Yamiko Saga for providing Nanos3-Cre mice, and the Minnesota Supercomputing Institute for computational resources. This work was funded by the US National Institutes of Health (GM059152 and T32HD007480), by the University of Minnesota Masonic Cancer Center, and by a Doctoral Dissertation Fellowship from the University of Minnesota Graduate School.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almstrup K, Hoei-Hansen CE, Nielsen JE, Wirkner U, Ansorge W, Skakkebaek NE, Rajpert-De Meyts E, Leffers H. Genome-wide gene expression profiling of testicular carcinoma in situ progression into overt tumours. Br J Cancer. 2005;92:1934–41. doi: 10.1038/sj.bjc.6602560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldassarre G, Romano A, Armenante F, Rambaldi M, Paoletti I, Sandomenico C, Pepe S, Staibano S, Salvatore G, De Rosa G, Persico MG, Viglietto G. Expression of teratocarcinoma-derived growth factor-1 (TDGF-1) in testis germ cell tumors and its effects on growth and differentiation of embryonal carcinoma cell line NTERA2/D1. Oncogene. 1997;15:927–36. doi: 10.1038/sj.onc.1201260. [DOI] [PubMed] [Google Scholar]
- Beyrouthy MJ, Garner KM, Hever MP, Freemantle SJ, Eastman A, Dmitrovsky E, Spinella MJ. High DNA methyltransferase 3B expression mediates 5-aza-deoxycytidine hypersensitivity in testicular germ cell tumors. Cancer Res. 2009;69:9360–6. doi: 10.1158/0008-5472.CAN-09-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco C, Rangel MC, Castro NP, Nagaoka T, Rollman K, Gonzales M, Salomon DS. Role of Cripto-1 in stem cell maintenance and malignant progression. Am J Pathol. 2010;177:532–40. doi: 10.2353/ajpath.2010.100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D, Gordon A, Von Kuster G, Coraor N, Taylor J, Nekrutenko A. Manipulation of FASTQ data with Galaxy. Bioinformatics. 2010;26:1783–5. doi: 10.1093/bioinformatics/btq281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles J, Teasdale RP, James K, Koopman P. Dppa3 is a marker of pluripotency and has a human homologue that is expressed in germ cell tumours. Cytogenet Genome Res. 2003;101:261–5. doi: 10.1159/000074346. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–56. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caricasole AA, van Schaik RH, Zeinstra LM, Wierikx CD, van Gurp RJ, van den Pol M, Looijenga LH, Oosterhuis JW, Pera MF, Ward A, de Bruijn D, Kramer P, de Jong FH, van den Eijnden-van Raaij AJ. Human growth-differentiation factor 3 (hGDF3): developmental regulation in human teratocarcinoma cell lines and expression in primary testicular germ cell tumours. Oncogene. 1998;16:95–103. doi: 10.1038/sj.onc.1201515. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK, Clarke ND, Wei CL, Ng HH. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–17. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Ciccodicola A, Dono R, Obici S, Simeone A, Zollo M, Persico MG. Molecular characterization of a gene of the ‘EGF family’ expressed in undifferentiated human NTERA2 teratocarcinoma cells. EMBO J. 1989;8:1987–91. doi: 10.1002/j.1460-2075.1989.tb03605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AT. The stem cell identity of testicular cancer. Stem Cell Rev. 2007;3:49–59. doi: 10.1007/s12015-007-0002-x. [DOI] [PubMed] [Google Scholar]
- Clark AT, Rodriguez RT, Bodnar MS, Abeyta MJ, Cedars MI, Turek PJ, Firpo MT, Reijo Pera RA. Human STELLAR, NANOG, and GDF3 genes are expressed in pluripotent cells and map to chromosome 12p13, a hotspot for teratocarcinoma. Stem Cells. 2004;22:169–79. doi: 10.1634/stemcells.22-2-169. [DOI] [PubMed] [Google Scholar]
- Cook MS, Coveney D, Batchvarov I, Nadeau JH, Capel B. BAX-mediated cell death affects early germ cell loss and incidence of testicular teratomas in Dnd1(Ter/Ter) mice. Dev Biol. 2009;328:377–83. doi: 10.1016/j.ydbio.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook MS, Munger SC, Nadeau JH, Capel B. Regulation of male germ cell cycle arrest and differentiation by DND1 is modulated by genetic background. Development. 2011;138:23–32. doi: 10.1242/dev.057000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durcova-Hills G, Capel B. Development of germ cells in the mouse. Curr Top Dev Biol. 2008;83:185–212. doi: 10.1016/S0070-2153(08)00406-7. [DOI] [PubMed] [Google Scholar]
- Fahrioglu U, Murphy MW, Zarkower D, Bardwell VJ. mRNA expression analysis and the molecular basis of neonatal testis defects in Dmrt1 mutant mice. Sex Dev. 2007;1:42–58. doi: 10.1159/000096238. [DOI] [PubMed] [Google Scholar]
- Festuccia N, Osorno R, Halbritter F, Karwacki-Neisius V, Navarro P, Colby D, Wong F, Yates A, Tomlinson SR, Chambers I. Esrrb Is a Direct Nanog Target Gene that Can Substitute for Nanog Function in Pluripotent Cells. Cell Stem Cell. 2012;11:477–90. doi: 10.1016/j.stem.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman D, Oliver RT, Brett AR, Marsh SG, Moses JH, Bodmer JG, Chilvers CE, Pike MC. Familial testicular cancer: a report of the UK family register, estimation of risk and an HLA class 1 sib-pair analysis. Br J Cancer. 1992;65:255–62. doi: 10.1038/bjc.1992.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Pang L, Ren C, Ma T. Decreased expression of Nedd4L correlates with poor prognosis in gastric cancer patient. Med Oncol. 2012;29:1733–8. doi: 10.1007/s12032-011-0061-3. [DOI] [PubMed] [Google Scholar]
- Grande L, Bretones G, Rosa-Garrido M, Garrido-Martin EM, Hernandez T, Fraile S, Botella L, de Alava E, Vidal A, Garcia del Muro X, Villanueva A, Delgado MD, Fernandez-Luna JL. Transcription factors Sp1 and p73 control the expression of the proapoptotic protein NOXA in the response of testicular embryonal carcinoma cells to cisplatin. J Biol Chem. 2012;287:26495–505. doi: 10.1074/jbc.M112.376319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Zylicz JJ, Surani MA. Parallel mechanisms of epigenetic reprogramming in the germline. Trends Genet. 2012;28:164–74. doi: 10.1016/j.tig.2012.01.005. [DOI] [PubMed] [Google Scholar]
- Harrison NJ, Baker D, Andrews PW. Culture adaptation of embryonic stem cells echoes germ cell malignancy. Int J Androl. 2007;30:275–81. doi: 10.1111/j.1365-2605.2007.00762.x. discussion 281. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Hayashi M, Kobayashi S. Nanos suppresses somatic cell fate in Drosophila germ line. Proc Natl Acad Sci U S A. 2004;101:10338–42. doi: 10.1073/pnas.0401647101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Deng J, Li G, Wang B, Cao Y, Tu Y. Down-regulation of Nedd4L is associated with the aggressive progression and worse prognosis of malignant glioma. Jpn J Clin Oncol. 2012;42:196–201. doi: 10.1093/jjco/hyr195. [DOI] [PubMed] [Google Scholar]
- Heaney JD, Anderson EL, Michelson MV, Zechel JL, Conrad PA, Page DC, Nadeau JH. Germ cell pluripotency, premature differentiation and susceptibility to testicular teratomas in mice. Development. 2012;139:1577–86. doi: 10.1242/dev.076851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimdal K, Olsson H, Tretli S, Flodgren P, Borresen AL, Fossa SD. Familial testicular cancer in Norway and southern Sweden. Br J Cancer. 1996;73:964–9. doi: 10.1038/bjc.1996.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng JC, Feng B, Han J, Jiang J, Kraus P, Ng JH, Orlov YL, Huss M, Yang L, Lufkin T, Lim B, Ng HH. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell. 2010;6:167–74. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Hoei-Hansen CE, Almstrup K, Nielsen JE, Brask Sonne S, Graem N, Skakkebaek NE, Leffers H, Rajpert-De Meyts E. Stem cell pluripotency factor NANOG is expressed in human fetal gonocytes, testicular carcinoma in situ and germ cell tumours. Histopathology. 2005;47:48–56. doi: 10.1111/j.1365-2559.2005.02182.x. [DOI] [PubMed] [Google Scholar]
- Hofmann MC. Gdnf signaling pathways within the mammalian spermatogonial stem cell niche. Mol Cell Endocrinol. 2008;288:95–103. doi: 10.1016/j.mce.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu XY, Xu YM, Fu Q, Yu JJ, Huang J. Nedd4L expression is downregulated in prostate cancer compared to benign prostatic hyperplasia. Eur J Surg Oncol. 2009;35:527–31. doi: 10.1016/j.ejso.2008.09.015. [DOI] [PubMed] [Google Scholar]
- Jain S, Golden JP, Wozniak D, Pehek E, Johnson EM, Jr, Milbrandt J. RET is dispensable for maintenance of midbrain dopaminergic neurons in adult mice. J Neurosci. 2006;26:11230–8. doi: 10.1523/JNEUROSCI.1876-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Naughton CK, Yang M, Strickland A, Vij K, Encinas M, Golden J, Gupta A, Heuckeroth R, Johnson EM, Jr, Milbrandt J. Mice expressing a dominant-negative Ret mutation phenocopy human Hirschsprung disease and delineate a direct role of Ret in spermatogenesis. Development. 2004;131:5503–13. doi: 10.1242/dev.01421. [DOI] [PubMed] [Google Scholar]
- James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132:1273–82. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- Jameson SA, Natarajan A, Cool J, DeFalco T, Maatouk DM, Mork L, Munger SC, Capel B. Temporal transcriptional profiling of somatic and germ cells reveals biased lineage priming of sexual fate in the fetal mouse gonad. PLoS Genet. 2012;8:e1002575. doi: 10.1371/journal.pgen.1002575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin VX, Apostolos J, Nagisetty NS, Farnham PJ. W-ChIPMotifs: a web application tool for de novo motif discovery from ChIP-based high-throughput data. Bioinformatics. 2009;25:3191–3. doi: 10.1093/bioinformatics/btp570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TD, Ulbright TM, Eble JN, Baldridge LA, Cheng L. OCT4 staining in testicular tumors: a sensitive and specific marker for seminoma and embryonal carcinoma. Am J Surg Pathol. 2004;28:935–40. doi: 10.1097/00000478-200407000-00014. [DOI] [PubMed] [Google Scholar]
- Kanetsky PA, Mitra N, Vardhanabhuti S, Li M, Vaughn DJ, Letrero R, Ciosek SL, Doody DR, Smith LM, Weaver J, Albano A, Chen C, Starr JR, Rader DJ, Godwin AK, Reilly MP, Hakonarson H, Schwartz SM, Nathanson KL. Common variation in KITLG and at 5q31.3 predisposes to testicular germ cell cancer. Nat Genet. 2009;41:811–5. doi: 10.1038/ng.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanetsky PA, Mitra N, Vardhanabhuti S, Vaughn DJ, Li M, Ciosek SL, Letrero R, D’Andrea K, Vaddi M, Doody DR, Weaver J, Chen C, Starr JR, Hakonarson H, Rader DJ, Godwin AK, Reilly MP, Schwartz SM, Nathanson KL. A Second Independent Locus within DMRT1 is Associated with Testicular Germ Cell Tumor Susceptibility. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly RK, Olson DL, Sun Y, Wen D, Wortham KA, Antognetti G, Cheung AE, Orozco OE, Yang L, Bailly V, Sanicola M. An antibody-cytotoxic conjugate, BIIB015, is a new targeted therapy for Cripto positive tumours. Eur J Cancer. 2011;47:1736–46. doi: 10.1016/j.ejca.2011.02.023. [DOI] [PubMed] [Google Scholar]
- Kidder BL, Palmer S. Examination of transcriptional networks reveals an important role for TCFAP2C, SMARCA4, and EOMES in trophoblast stem cell maintenance. Genome Res. 2010;20:458–72. doi: 10.1101/gr.101469.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Orkin SH. Embryonic stem cell-specific signatures in cancer: insights into genomic regulatory networks and implications for medicine. Genome Med. 2011;3:75. doi: 10.1186/gm291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Bardwell VJ, Zarkower D. Cell type-autonomous and non-autonomous requirements for Dmrt1 in postnatal testis differentiation. Dev Biol. 2007;307:314–27. doi: 10.1016/j.ydbio.2007.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Suh MR, Yoon HS, Lee JB, Oh SK, Moon SY, Moon SH, Lee JY, Hwang JH, Cho WJ, Kim KS. Identification of developmental pluripotency associated 5 expression in human pluripotent stem cells. Stem Cells. 2005;23:458–62. doi: 10.1634/stemcells.2004-0245. [DOI] [PubMed] [Google Scholar]
- Kimura T, Suzuki A, Fujita Y, Yomogida K, Lomeli H, Asada N, Ikeuchi M, Nagy A, Mak TW, Nakano T. Conditional loss of PTEN leads to testicular teratoma and enhances embryonic germ cell production. Development. 2003;130:1691–700. doi: 10.1242/dev.00392. [DOI] [PubMed] [Google Scholar]
- Koo BK, Spit M, Jordens I, Low TY, Stange DE, van de Wetering M, van Es JH, Mohammed S, Heck AJ, Maurice MM, Clevers H. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488:665–9. doi: 10.1038/nature11308. [DOI] [PubMed] [Google Scholar]
- Krentz AD, Murphy MW, Kim S, Cook MS, Capel B, Zhu R, Matin A, Sarver AL, Parker KL, Griswold MD, Looijenga LH, Bardwell VJ, Zarkower D. The DM domain protein DMRT1 is a dose-sensitive regulator of fetal germ cell proliferation and pluripotency. Proc Natl Acad Sci U S A. 2009;106:22323–8. doi: 10.1073/pnas.0905431106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krentz AD, Murphy MW, Sarver AL, Griswold MD, Bardwell VJ, Zarkower D. DMRT1 promotes oogenesis by transcriptional activation of Stra8 in the mammalian fetal ovary. Dev Biol. 2011;356:63–70. doi: 10.1016/j.ydbio.2011.05.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Salle S, Oakes CC, Neaga OR, Bourc’his D, Bestor TH, Trasler JM. Loss of spermatogonia and wide-spread DNA methylation defects in newborn male mice deficient in DNMT3L. BMC Dev Biol. 2007;7:104. doi: 10.1186/1471-213X-7-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MY, Nadeau JH. Genetic control of susceptibility to spontaneous testicular germ cell tumors in mice. APMIS. 2003;111:184–90. doi: 10.1034/j.1600-0463.2003.11101221.x. discussion 191. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei N, Hornbaker KI, Rice DA, Karpova T, Agbor VA, Heckert LL. Sex-specific differences in mouse DMRT1 expression are both cell type- and stage-dependent during gonad development. Biol Reprod. 2007;77:466–75. doi: 10.1095/biolreprod.106.058784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, Ruan Y, Lim B, Ng HH. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–40. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Looijenga LH, Gillis AJ, Stoop HJ, Hersmus R, Oosterhuis JW. Chromosomes and expression in human testicular germ-cell tumors: insight into their cell of origin and pathogenesis. Ann N Y Acad Sci. 2007;1120:187–214. doi: 10.1196/annals.1411.000. [DOI] [PubMed] [Google Scholar]
- Maldonado-Saldivia J, van den Bergen J, Krouskos M, Gilchrist M, Lee C, Li R, Sinclair AH, Surani MA, Western PS. Dppa2 and Dppa4 are closely linked SAP motif genes restricted to pluripotent cells and the germ line. Stem Cells. 2007;25:19–28. doi: 10.1634/stemcells.2006-0269. [DOI] [PubMed] [Google Scholar]
- Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, Ko MS, Niwa H. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–35. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- Matson CK, Murphy MW, Griswold MD, Yoshida S, Bardwell VJ, Zarkower D. The mammalian doublesex homolog DMRT1 is a transcriptional gatekeeper that controls the mitosis versus meiosis decision in male germ cells. Dev Cell. 2010;19:612–24. doi: 10.1016/j.devcel.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson CK, Murphy MW, Sarver AL, Griswold MD, Bardwell VJ, Zarkower D. DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature. 2011;476:101–4. doi: 10.1038/nature10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson CK, Zarkower D. Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nat Rev Genet. 2012;13:163–74. doi: 10.1038/nrg3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles DC, van den Bergen JA, Wakeling SI, Anderson RB, Sinclair AH, Western PS. The proto-oncogene Ret is required for male foetal germ cell survival. Dev Biol. 2012;365:101–9. doi: 10.1016/j.ydbio.2012.02.014. [DOI] [PubMed] [Google Scholar]
- Minami K, Chano T, Kawakami T, Ushida H, Kushima R, Okabe H, Okada Y, Okamoto K. DNMT3L is a novel marker and is essential for the growth of human embryonal carcinoma. Clin Cancer Res. 2010;16:2751–9. doi: 10.1158/1078-0432.CCR-09-3338. [DOI] [PubMed] [Google Scholar]
- Munger SC, Aylor DL, Syed HA, Magwene PM, Threadgill DW, Capel B. Elucidation of the transcription network governing mammalian sex determination by exploiting strain-specific susceptibility to sex reversal. Genes Dev. 2009;23:2521–36. doi: 10.1101/gad.1835809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MW, Sarver AL, Rice D, Hatzi K, Ye K, Melnick A, Heckert LL, Zarkower D, Bardwell VJ. Genome-wide analysis of DNA binding and transcriptional regulation by the mammalian Doublesex homolog DMRT1 in the juvenile testis. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1006243107. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MW, Zarkower D, Bardwell VJ. Vertebrate DM domain proteins bind similar DNA sequences and can heterodimerize on DNA. BMC Mol Biol. 2007;8:58. doi: 10.1186/1471-2199-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narva E, Rahkonen N, Emani MR, Lund R, Pursiheimo JP, Nasti J, Autio R, Rasool O, Denessiouk K, Lahdesmaki H, Rao A, Lahesmaa R. RNA-binding protein L1TD1 interacts with LIN28 via RNA and is required for human embryonic stem cell self-renewal and cancer cell proliferation. Stem Cells. 2012;30:452–60. doi: 10.1002/stem.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton CK, Jain S, Strickland AM, Gupta A, Milbrandt J. Glial cell-line derived neurotrophic factor-mediated RET signaling regulates spermatogonial stem cell fate. Biol Reprod. 2006;74:314–21. doi: 10.1095/biolreprod.105.047365. [DOI] [PubMed] [Google Scholar]
- Oberley MJ, Tsao J, Yau P, Farnham PJ. High-throughput screening of chromatin immunoprecipitates using CpG-island microarrays. Methods Enzymol. 2004;376:315–34. doi: 10.1016/S0076-6879(03)76021-2. [DOI] [PubMed] [Google Scholar]
- Oosterhuis JW, Looijenga LH. Testicular germ-cell tumours in a broader perspective. Nat Rev Cancer. 2005;5:210–22. doi: 10.1038/nrc1568. [DOI] [PubMed] [Google Scholar]
- Pinheiro I, Margueron R, Shukeir N, Eisold M, Fritzsch C, Richter FM, Mittler G, Genoud C, Goyama S, Kurokawa M, Son J, Reinberg D, Lachner M, Jenuwein T. Prdm3 and Prdm16 are H3K9me1 Methyltransferases Required for Mammalian Heterochromatin Integrity. Cell. 2012;150:948–60. doi: 10.1016/j.cell.2012.06.048. [DOI] [PubMed] [Google Scholar]
- Rajpert-De Meyts E, Hanstein R, Jorgensen N, Graem N, Vogt PH, Skakkebaek NE. Developmental expression of POU5F1 (OCT-3/4) in normal and dysgenetic human gonads. Hum Reprod. 2004;19:1338–44. doi: 10.1093/humrep/deh265. [DOI] [PubMed] [Google Scholar]
- Rapley EA, Turnbull C, Al Olama AA, Dermitzakis ET, Linger R, Huddart RA, Renwick A, Hughes D, Hines S, Seal S, Morrison J, Nsengimana J, Deloukas P, Rahman N, Bishop DT, Easton DF, Stratton MR. A genome-wide association study of testicular germ cell tumor. Nat Genet. 2009;41:807–10. doi: 10.1038/ng.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CS, Kettlewell JR, Hirsch B, Bardwell VJ, Zarkower D. Expression of Dmrt1 in the genital ridge of mouse and chicken embryos suggests a role in vertebrate sexual development. Dev Biol. 1999;215:208–20. doi: 10.1006/dbio.1999.9461. [DOI] [PubMed] [Google Scholar]
- Raymond CS, Murphy MW, O’Sullivan MG, Bardwell VJ, Zarkower D. Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 2000;14:2587–95. doi: 10.1101/gad.834100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regenass U, Friedrich TD, Stevens LC. Experimental induction of testicular teratomas in dissociated-reaggregated chimaeric gonads. J Embryol Exp Morphol. 1982;72:153–67. [PubMed] [Google Scholar]
- Richards M, Tan SP, Tan JH, Chan WK, Bongso A. The transcriptome profile of human embryonic stem cells as defined by SAGE. Stem Cells. 2004;22:51–64. doi: 10.1634/stemcells.22-1-51. [DOI] [PubMed] [Google Scholar]
- Schafer S, Anschlag J, Nettersheim D, Haas N, Pawig L, Schorle H. The role of BLIMP1 and its putative downstream target TFAP2C in germ cell development and germ cell tumours. Int J Androl. 2011 doi: 10.1111/j.1365-2605.2011.01167.x. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Sharov AA, Masui S, Sharova LV, Piao Y, Aiba K, Matoba R, Xin L, Niwa H, Ko MS. Identification of Pou5f1, Sox2, and Nanog downstream target genes with statistical confidence by applying a novel algorithm to time course microarray and genome-wide chromatin immunoprecipitation data. BMC Genomics. 2008;9:269. doi: 10.1186/1471-2164-9-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MM. Nodal signaling: developmental roles and regulation. Development. 2007;134:1023–34. doi: 10.1242/dev.000166. [DOI] [PubMed] [Google Scholar]
- Small CL, Shima JE, Uzumcu M, Skinner MK, Griswold MD. Profiling gene expression during the differentiation and development of the murine embryonic gonad. Biol Reprod. 2005;72:492–501. doi: 10.1095/biolreprod.104.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller CM, Feng CW, Jackson A, Gillis AJ, Rolland AD, Looijenga LH, Koopman P, Bowles J. Endogenous Nodal signaling regulates germ cell potency during mammalian testis development. Development. 2012 doi: 10.1242/dev.083006. [DOI] [PubMed] [Google Scholar]
- Stevens LC. Experimental Production of Testicular Teratomas in Mice. Proc Natl Acad Sci U S A. 1964;52:654–61. doi: 10.1073/pnas.52.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens LC. Development of resistance to teratocarcinogenesis by primordial germ cells in mice. J Natl Cancer Inst. 1966;37:859–67. [PubMed] [Google Scholar]
- Stevens LC. A new inbred subline of mice (129-terSv) with a high incidence of spontaneous congenital testicular teratomas. J Natl Cancer Inst. 1973;50:235–42. doi: 10.1093/jnci/50.1.235. [DOI] [PubMed] [Google Scholar]
- Stevens LC, Little CC. Spontaneous Testicular Teratomas in an Inbred Strain of Mice. Proc Natl Acad Sci U S A. 1954;40:1080–7. doi: 10.1073/pnas.40.11.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Tsuda M, Kiso M, Saga Y. Nanos3 maintains the germ cell lineage in the mouse by suppressing both Bax-dependent and -independent apoptotic pathways. Dev Biol. 2008;318:133–42. doi: 10.1016/j.ydbio.2008.03.020. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, Saga Y. Conserved role of nanos proteins in germ cell development. Science. 2003;301:1239–41. doi: 10.1126/science.1085222. [DOI] [PubMed] [Google Scholar]
- Turnbull C, Rapley EA, Seal S, Pernet D, Renwick A, Hughes D, Ricketts M, Linger R, Nsengimana J, Deloukas P, Huddart RA, Bishop DT, Easton DF, Stratton MR, Rahman N. Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat Genet. 2010;42:604–7. doi: 10.1038/ng.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesaka T, Jain S, Yonemura S, Uchiyama Y, Milbrandt J, Enomoto H. Conditional ablation of GFRalpha1 in postmigratory enteric neurons triggers unconventional neuronal death in the colon and causes a Hirschsprung’s disease phenotype. Development. 2007;134:2171–81. doi: 10.1242/dev.001388. [DOI] [PubMed] [Google Scholar]
- Vallier L, Alexander M, Pedersen RA. Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J Cell Sci. 2005;118:4495–509. doi: 10.1242/jcs.02553. [DOI] [PubMed] [Google Scholar]
- Vallier L, Reynolds D, Pedersen RA. Nodal inhibits differentiation of human embryonic stem cells along the neuroectodermal default pathway. Dev Biol. 2004;275:403–21. doi: 10.1016/j.ydbio.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Wang W, Yang J, Liu H, Lu D, Chen X, Zenonos Z, Campos LS, Rad R, Guo G, Zhang S, Bradley A, Liu P. Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor gamma and liver receptor homolog 1. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1100893108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S, Eckert D, Nettersheim D, Gillis AJ, Schafer S, Kuckenberg P, Ehlermann J, Werling U, Biermann K, Looijenga LH, Schorle H. Critical function of AP-2 gamma/TCFAP2C in mouse embryonic germ cell maintenance. Biol Reprod. 2010;82:214–23. doi: 10.1095/biolreprod.109.078717. [DOI] [PubMed] [Google Scholar]
- Western PS, van den Bergen JA, Miles DC, Sinclair AH. Male fetal germ cell differentiation involves complex repression of the regulatory network controlling pluripotency. FASEB J. 2010;24:3026–35. doi: 10.1096/fj.09-151555. [DOI] [PubMed] [Google Scholar]
- Woodfield GW, Chen Y, Bair TB, Domann FE, Weigel RJ. Identification of primary gene targets of TFAP2C in hormone responsive breast carcinoma cells. Genes Chromosomes Cancer. 2010;49:948–62. doi: 10.1002/gcc.20807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Kimura H, Tada M, Nakatsuji N, Tada T. Nanog expression in mouse germ cell development. Gene Expr Patterns. 2005;5:639–46. doi: 10.1016/j.modgep.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Yang J, Gao C, Chai L, Ma Y. A novel SALL4/OCT4 transcriptional feedback network for pluripotency of embryonic stem cells. PLoS One. 2010;5:e10766. doi: 10.1371/journal.pone.0010766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngren KK, Coveney D, Peng X, Bhattacharya C, Schmidt LS, Nickerson ML, Lamb BT, Deng JM, Behringer RR, Capel B, Rubin EM, Nadeau JH, Matin A. The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature. 2005;435:360–4. doi: 10.1038/nature03595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngren KK, Nadeau JH, Matin A. Testicular cancer susceptibility in the 129.MOLF-Chr19 mouse strain: additive effects, gene interactions and epigenetic modifications. Hum Mol Genet. 2003;12:389–98. doi: 10.1093/hmg/ddg036. [DOI] [PubMed] [Google Scholar]
- Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995;9:2635–45. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.