Abstract
High resolution compressed sensing hyperpolarized 13C magnetic resonance spectroscopic imaging was applied in orthotopic human glioblastoma xenografts for quantitative assessment of spatial variations in 13C metabolic profiles and comparison with histopathology. A new compressed sensing sampling design with a factor of 3.72 acceleration was implemented to enable a factor of 4 increase in spatial resolution. Compressed sensing 3D 13C magnetic resonance spectroscopic imaging data were acquired from a phantom and 10 tumor-bearing rats following injection of hyperpolarized [1-13C]-pyruvate using a 3T scanner. The 13C metabolic profiles were compared with hematoxylin and eosin staining and carbonic anhydrase 9 staining. The high-resolution compressed sensing 13C magnetic resonance spectroscopic imaging data enabled the differentiation of distinct 13C metabolite patterns within abnormal tissues with high specificity in similar scan times compared to the fully sampled method. The results from pathology confirmed the different characteristics of 13C metabolic profiles between viable, non-necrotic, nonhypoxic tumor, and necrotic, hypoxic tissue.
Keywords: hyperpolarized 13C MRSI, compressed sensing, dynamic nuclear polarization, pyruvate, glioblastoma
Dynamic nuclear polarization and the recent development of a dissolution process that retains polarization in liquid state have allowed for the acquisition of 13C magnetic resonance spectroscopic imaging (MRSI) data with a substantial gain in sensitivity over conventional MR methods (1).
Recent studies using hyperpolarized [1-13C]-pyruvate as a substrate have demonstrated the promise of this technique for examining in vivo tumor metabolism in brain tumors (2–4). These preclinical studies have shown the feasibility of using this technique for differentiating tumor from normal brain tissue and detecting an early response to treatment in animal models of high-grade gliomas. The further refinement of 13C MRSI acquisition techniques with higher spatial resolution is of interest for assessing spatial variations in tissue metabolism due to the infiltrative nature and molecular heterogeneity of gliomas.
One of the major obstacles in acquiring hyperpolarized 13C signal is the time limitation imposed by the T1 decay of hyperpolarized compound. In order to more efficiently sample the rapidly decaying 13C signal, fast imaging techniques, such as compressed sensing, have been incorporated in the acquisition of hyperpolarized 13C data (5). By taking advantage of the sparsity in hyperpolarized 13C spectra and the undersampling of spectral k-space, the compressed sensing technique has enabled the acquisition of hyperpolarized 13C metabolites with high temporal and spatial resolution in preclinical studies involving transgenic prostate cancer and liver cancer murine models (5,6).
The purpose of this project was to implement a compressed sensing scheme tailored to the rat brain morphology and to demonstrate the utility of this method in obtaining hyperpolarized 13C MRSI data for evaluating heterogeneous 13C metabolic profiles within brain tumor tissue using an orthotopic human glioblastoma (GBM) xenograft model. The new compressed sensing sampling scheme was validated through phantom experiments and by comparing with fully sampled 13C 3D MRSI data. Our emphasis was to quantitatively assess the compressed sensing hyperpolarized 13C MRSI data and to compare 13C metabolic profiles from different regions of tumor with the results from histology and immunohistochemistry.
METHODS
Design of Compressed Sensing Scheme
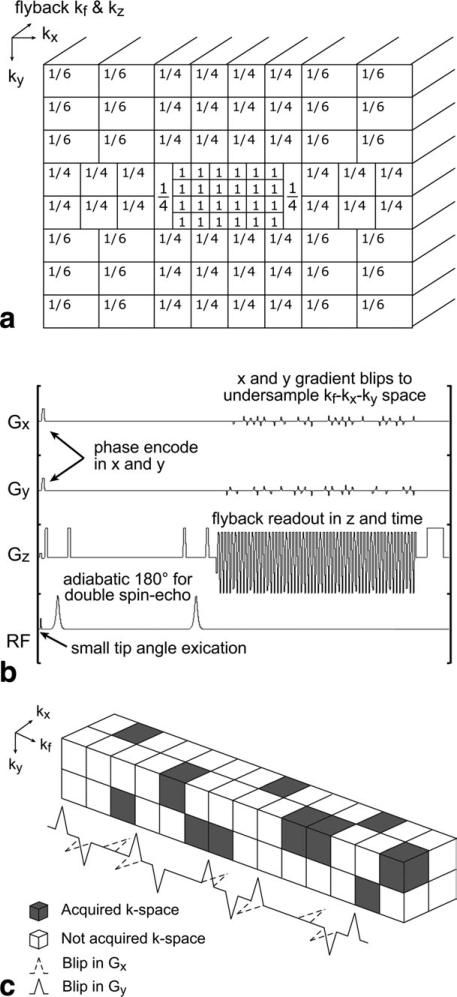
The compressed sensing scheme tailored to rat brain was based on a previously developed framework (5). First, an in-plane matrix size of 20 × 16 was selected based on the rat brain morphology. Then an undersampling pattern was generated in order to achieve an acceleration sufficient to cover this matrix within a reasonable acquisition time. Figure 1a shows the undersampling pattern design for the 20 × 16 in-plane matrix. Note that gradient blips were placed during the rewind portions of the flyback readout to undersample a 20 × 16 × 59 region of kx-ky-kf space and kz is fully sampled (Fig. 1b; 5). The central portion of k-space was fully sampled in order to enhance the stability of the compressed sensing reconstruction, which in this case, as shown in Figure 1a, is a 6 × 4 region of kx-ky space. The fractional values in Figure 1a show the sampling factors for different regions of kx-ky-kf space. For example, 1/6 corresponds to a 3 × 2 × 59 block of kx-ky-kf space undersampled by a factor of 6, i.e., covered after one excitation. Note that some of the 1/4 sampling regions are 2 × 2 × 59 kx-ky-kf space blocks and some are 1 × 4 × 59 kx-ky-kf space blocks in order to tile all the regions together into a 20 × 16 × 59 kx-ky-kf space whole. As shown in Figure 1a, there are 86 k-space blocks, thus 86 phase encodes are used to cover a 20 × 16 phase encoding matrix, corresponding to a factor of (20 × 16)/86 = 3.72 acceleration. Hu et al. has explored the limits of undersampling and the effect of noise on the compressed sensing nonlinear reconstruction (5). According to the simulated data from this study, the factor of 3.72 acceleration would produce practically perfect reconstruction of compressed sensing spectra. With this acceleration, spatial resolution can be doubled in x and y while keeping scan time approximately constant.
FIG. 1.
An undersampling pattern for the new compressed sensing scheme (a) and pulse sequence timing diagram for compressed sensing 13C MRSI (b). The numbers in (a) represent the fraction of samples collected in each kx-ky-kf block. c: Illustration of random undersampling in kx-ky-kf space. Random undersampling was achieved by implementing blips in the x and y gradients.
Phantom Data Acquisition
All experiments were performed using a 3T GE Signa™ scanner (GE Healthcare, Milwaukee, WI) equipped with the multinuclear spectroscopy hardware package and a dual-tuned 1H 13C coil (7). In order to validate the new compressed sensing sampling pattern, a spherical phantom containing enriched [1-13C]-acetate was scanned using the rat brain customized compressed sensing and fully sampled 13C 3D MRSI sequences in consecutive scans. A double spin echo (echo time/pulse repetition time = 140/1000 ms) and flyback echo-planar readout on the z-axis (Fig. 1b; 8) were also incorporated.
Animal Preparation
Two different human GBM cell lines, U87 (n = 5) and G55 (n = 5), were used for creating orthotopic GBM models in 10 six-week-old male athymic rats (rnu/rnu, homozygous) purchased from Harlan (Indianapolis, IN). The details of cell culture and intracerebral implantation have been described previously (2,3). Animal studies were approved by the Institutional Animal Care and Use Committee at our institution.
1H MRI
During each imaging experiment, the rat was placed on a heated pad positioned in the radiofrequency coil in the MR scanner. Anesthesia was maintained with a constant delivery of isoflurane (1–2%). Prior to each 13C imaging study, high-resolution T2-weighted anatomical images were obtained in the axial plane using a fast spin-echo sequence (echo time/pulse repetition time = 60/4000 ms, 8 cm field of view, 192 × 192 matrix, 1.5 mm slice thickness, and 8 NEX). At the completion of 13C imaging, axial images were obtained using a T1-weighted spin-echo sequence (echo time/pulse repetition time = 10/700 ms, 8 cm field of view, 320 × 192 matrix, 1.2 mm slice thickness, and 6 NEX) after the injection of 0.1 mL Gadolinium (Gd)-diethylenetriaminepentaacetic acid (DTPA) (approximately 0.2 mmol/kg).
Polarization Procedure
A mixture of 35 μL (approximately 45 mg) [1-13C]-pyruvate (Isotec, Miamisburg, OH) and 15 mM OX63 trityl radical, along with 1.5 mM of Dotarem gadolinium was hyperpolarized using a HyperSense® dynamic nuclear polarization polarizer (Oxford Instruments, Abingdon, UK) at 3.35 T and 1.4°K by irradiation with 94.1 GHz microwaves using methods described previously (1). After approximately 60 min of microwave irradiation, the hyperpolarized pyruvic acid was rapidly dissolved in a saline solution with 5.96 g/L Tris (40 mM), 4.00 g/L NaOH (100 mM), and 0.1 mg/L Na2 ethylenediaminetetraacetic acid. The final dissolved solution had a concentration of 100 mM pyruvate and pH ~7.5.
Compressed Sensing and Fully Sampled 13C MRSI
Both compressed sensing and fully sampled 13C 3D MRSI data were acquired using a double spin echo sequence (echo time/pulse repetition time = 140/215 ms) with centric k-space encoding, a variable flip angle scheme and flyback echo-planar readout on the z-axis (Fig. 1b) (8,9) at 20 s from the start of the injection of approximately 2.5 mL hyperpolarized [1-13C]-pyruvate through the tail vein. The injection started ~10 s after dissolution and lasted 10 s. For the compressed sensing acquisition, 86 phase encodes were collected from a 20 × 16 matrix in 18.5 s, resulting in 2 × 2 × 5.4 mm resolution. For the fully sampled acquisition, 80 phase encodes were collected from a 10 × 8 matrix in 17.2 s, resulting in 4 × 4 × 5.4 mm resolution. Two rats with U87 tumor were scanned with both compressed sensing and fully sampled methods in consecutive experiments. Eight rats were scanned using the compressed sensing method only.
Data Processing and Analysis
The methods for processing 13C MRSI data have been described previously (10). The raw readout data were reordered into a 4D array. Only the k-space data from flat parts of the flyback trajectory were selected. The time domain signal was apodized by a 16-Hz Gaussian filter and zero-filled to 256 points. A 4D Fourier transform was used to produce a 3D spatial array of spectra. An additional linear phase correction was applied in the flyback dimension to correct for the offset of individual k-space points (8). For the compressed sensing acquisition, an additional step after reordering into a 4D array was needed to fill in the missing k-space data due to undersampling. The missing k-space points were filled in with an iterative L1-norm compressed sensing reconstruction. This reconstruction, described by Lustig et al. (11) and available online (http://www.eecs.berkeley.edu/~mlustig/), minimizes the L1-norm in the wavelet domain, enforces data consistency by constraining the variation of acquired (not undersampled) k-space points, and adds a total variation penalty to promote smoothness in the spatial domains. In other words, the reconstruction is related to a nonlinear conjugate gradient algorithm for the solution of the following optimization:
| [1] |
where three terms represent the data fidelity constraint, the L1-norm and a total variation penalty used to enforce some edge-preserving smoothness in the final solution, respectively. The detailed description of the Eq. 1 can be found in Ref. 5. The L1 reconstruction parameters were the same as those used in previous hyperpolarized studies, i.e., 0.0005 and 0.0001 for λ and α, respectively (5).
For quantification of 13C metabolites, lactate over pyruvate ratio, lactate, pyruvate, and total carbon (tC: a sum of lactate, pyruvate-hydrate, alanine, and pyruvate) were calculated for each voxel using peak heights from magnitude spectra. The lactate, pyruvate, and tC levels were normalized to tC in blood vessels, which was obtained by taking the average of two maximum tC signals from a region outside the brain (Fig. 3). This region consistently exhibited a high level of pyruvate, which most likely originated from blood vessels, including the internal carotid artery and the vertebrobasilar arterial system that runs in the ventral surface of the brainstem. Regions of interests were manually contoured on T1-weighted post-Gd images for contrast enhancing (CE) lesions, necrotic regions, and normal appearing brain tissue, and the percentage of each regions of interest volume was calculated for each voxel. In order to evaluate the spatial variation of 13C metabolites, the median 13C parameters were compared between contrast enhancement (voxels with > 65% CE lesion), necrosis (voxels with > 65% necrotic region), and normal brain (voxels with 100% normal appearing brain tissue). Statistical significance was assessed using a Wilcoxon signed-rank test or a Mann-Whitney rank-sum test.
FIG. 3.
Compressed sensing (b) and fully sampled (c) 13C 3D MRSI data acquired in successive scans. T1 post-Gd images corresponding to the location of 13C spectra are shown in (a). The overall profiles of pyruvate and lactate were consistent between the two methods. The light grey voxels represent 13C spectra from tumor tissue. The dark grey voxels contained high pyruvate signal from blood vessels, which were used to normalize lactate, pyruvate, and total carbon levels.
Histopathology and Immunohistochemistry
At the end of the MR study, brains were removed and fixed in phosphate-buffered 4% formalin. Samples were then dehydrated by graded ethanols, and embedded in Paraplast Plus wax (McCormick Scientific). 5 μm sections were then examined following hematoxylin and eosin staining. Antibodies to carbonic anhydrase IX (CA9) were obtained from Novus Biologicals, Littleton Colorado and used at 1:500, 32 min/37°. Antigen retrieval for CA9 was performed for 8 min in Tris buffer (pH 8) at 90°. Immunohistochemistry was performed on the Benchmark XT (Ventana Medical Systems, Inc.) using the iView detection system. Histology and immunohistochemistry slices from the center of spectroscopic voxels were selected for comparison with 13C metabolites by correlating the morphological appearance of tumor with anatomical images.
RESULTS
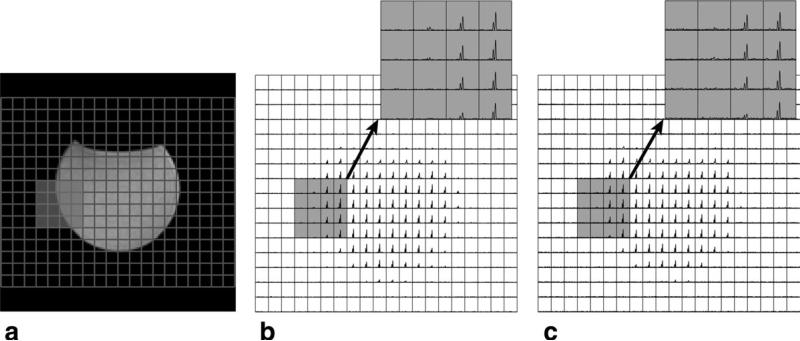
Figure 2 shows a T2-weighted image, compressed sensing, and fully sampled 13C MRSI data from the acetate phantom. The L1 reconstruction for the compressed sensing design produced 13C signal with excellent spectral quality. The compressed sensing data exhibited a similar spatial profile to the fully sampled data.
FIG. 2.
An anatomical image of a phantom (a) and 13C spectra acquired using the new compressed sensing design (b) and fully sampled method (c). The compressed sensing reconstructed data produced 13C acetate signal with high spectral quality and matched the fully sampled data.
Figure 3 illustrates a comparison between the compressed sensing and fully sampled 13C spectra acquired in consecutive scans from a rat with a brain tumor in 18.5 and 17.2 s, respectively. The factor of 3.72 acceleration in the compressed sensing scheme provided four 2 × 2 × 5.4 mm voxels (Figure 3b) for every 4 × 4 × 5.4 mm voxel in the fully sampled 13C 3D echo-planar spectroscopic imaging data (Fig. 3c) in a similar acquisition time. The undersampled compressed sensing reconstructed data were in excellent agreement with the fully sampled data in terms of the overall profiles of lactate and pyruvate. The compressed sensing data revealed heterogeneous metabolic profiles within a CE lesion (light grey voxels in Figure 3b). The four voxels, which corresponded to the location of tumor, had different levels of lactate and pyruvate. Partial voluming from normal appearing brain tissue at the lower part of the spectroscopic voxel (Fig. 3a, right) resulted in low metabolic levels in two of the four voxels comprising tumor.
Compressed sensing hyperpolarized 13C MRSI provided a way to evaluate the heterogeneity of metabolic profiles within brain tumor tissue. Table 1 shows the summary of 13C metabolite quantification from the compressed sensing MRSI data at different radiographic regions. In all animals, lactate to pyruvate ratio, normalized lactate, pyruvate, and total carbon levels in contrast enhancement voxels (1.0 ± 0.36, 0.19 ± 0.12, 0.17 ± 0.05, and 0.40 ± 0.18, respectively) were significantly elevated compared to the respective values in normal brain voxels (0.29 ± 0.17, 0.03 ± 0.02, 0.12 ± 0.03, and 0.21 ± 0.05, respectively, P < 0.01). Four rats possessed necrotic voxels. Lactate to pyruvate ratio, normalized lactate and total carbon levels in these voxels (0.58 ± 0.23, 0.07 ± 0.06, and 0.21 ± 0.11, respectively) were significantly lower than those in contrast enhancement voxels (P < 0.05).
Table 1.
Summary of 13C metabolite Quantification at Different Radiographic Voxels
| Lac/Pyra,b | Laca,b | Pyra | tCa,b | |
|---|---|---|---|---|
| Normal brain (n = 10) | 0.29 ± 0.17 | 0.03 ± 0.02 | 0.12 ± 0.03 | 0.21 ± 0.05 |
| Contrast enhancement (n = 10) | 1.0 ± 0.36 | 0.19 ± 0.12 | 0.17 ± 0.05 | 0.40 ± 0.18 |
| Necrosis (n = 4) | 0.58 ± 0.23 | 0.07 ± 0.06 | 0.11 ± 0.05 | 0.21 ± 0.11 |
Lac/Pyr represents the lactate over pyruvate ratio. Lac, Pyr and tC represent the lactate, pyruvate, and total carbon levels normalized by vascular total carbon signal. All values are reported as median ± SD.
Significant difference between normal brain and contrast enhancement (P < 0.01, Wilcoxon signed-rank test).
Significant difference between contrast enhancement and necrosis (P < 0.05, Mann-Whitney rank-sum test).
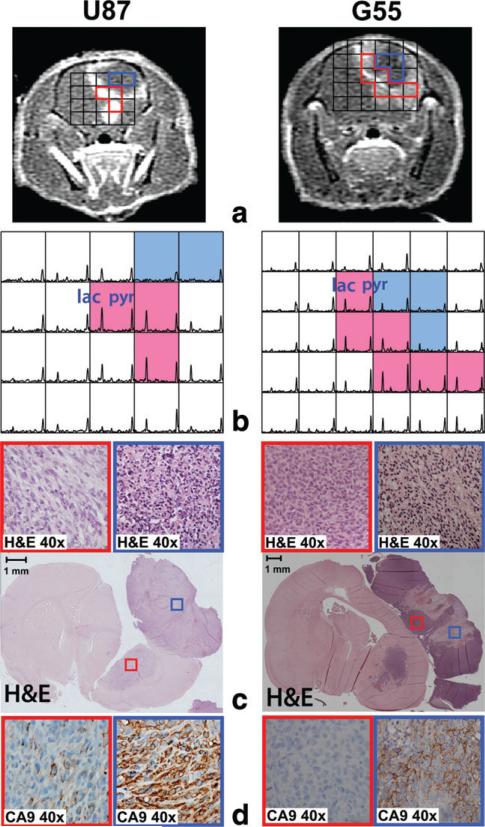
Figure 4 represents an example of two rats with heterogeneous tumor. Axial T1 post-Gd images showed areas with necrosis surrounded by CE lesion in the anterior part of the brain (Fig. 4a). The voxels that corresponded to necrosis (blue voxels in Figure 4b) exhibited decreased pyruvate (0.09 ± 0.03) and negligible lactate signal (0.05 ± 0.03) in the compressed sensing 13C spectra. In contrast, voxels associated with contrast enhancement (red voxels in Figure 4b) had relatively high levels of pyruvate (0.17 ± 0.05) and lactate (0.25 ± 0.25).
FIG. 4.
An example of two rats with heterogeneous tumors showing T1 post-Gd images (a), 13C spectra (b) and histopathology (c, d). The voxels representing necrosis (blue voxels in b) and contrast enhancement (red voxels in b) exhibited distinct 13C metabolic profiles from the compressed sensing data. Hematoxylin and eosin staining and CA9 immunostaining demonstrated substantial necrosis and hypoxia (blue boxes in c and d) in the area with small pyruvate and lactate (blue voxels in b) while the area with high 13C metabolites (red voxels in b) consisted of viable tumor with minimal or no necrosis and hypoxia (red boxes in c and d).
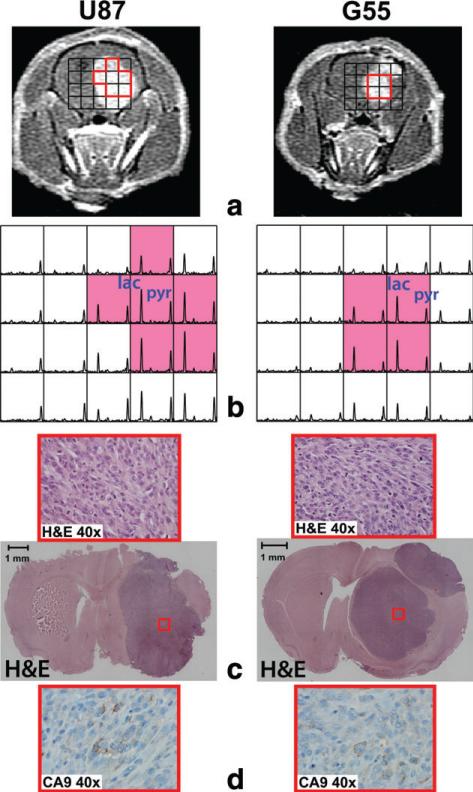
Figure 5 shows an example of rats with a uniform CE lesion. Axial T1 post-Gd images displayed the uniform level of contrast enhancement across tumor tissue (Fig. 5a). The compressed sensing spectra exhibited elevated pyruvate and lactate peaks in the CE lesion (Fig. 5b). In contrast to the previous example in Figure 4, all voxels with contrast enhancement (red voxels in Figure 5b) had relatively high levels of pyruvate (0.24 ± 0.05) and lactate (0.39 ± 0.17).
FIG. 5.
An example of two rats with a uniform CE lesion showing T1 post-Gd images (a), 13C spectra (b), and histopathology (c, d). Compressed sensing 13C spectra displayed highly elevated pyruvate and lactate consistently across the contrast enhancement voxel (red voxels in b). The corresponding hematoxylin and eosin staining and CA9 slides showed viable tumor without substantial necrosis and hypoxia (red boxes in c and d).
The distinct metabolic patterns also correlated with different histopathologic features. Rats with heterogeneous tumor (Fig. 4) displayed areas of necrosis (blue boxes in Figure 4c) and cellular hypoxia as determined by CA9 (blue boxes in Figure 4d). Interestingly, theses necrotic and hypoxic areas corresponded to the region of 13C spectra with relatively low 13C metabolites (blue voxels in Figure 4b). In contrast, the sections of tumor that exhibited high levels of pyruvate and lactate signal corresponded to the area of high contrast enhancement (red voxels in Figure 4b). These regions consisted of viable tumor with minimal or no necrosis and hypoxia (red boxes in Figure 4c and d). The rats with a homogeneous level of contrast enhancement exhibited viable tumor tissue without necrosis and with negligible hypoxia across the entire tumor volume in hematoxylin and eosin staining and CA9 slides (red boxes in Figure 5c and d). The area of viable tumor corresponded to the region of spectra with high pyruvate and lactate signal (red voxels in Figure 5b).
DISCUSSION
This study demonstrated the feasibility of evaluating heterogeneous 13C metabolic profiles within brain tumor tissue in human GBM orthotopic xenografts using compressed sensing hyperpolarized 3D 13C MRSI with [1-13C]-pyruvate as a substrate. The improved resolution achieved by the compressed sensing scheme enabled the acquisition of heterogeneous 13C metabolic profiles within tumor and made possible the differentiation between hypoxic/necrotic, nonhypoxic/necrotic tumor, and normal brain tissue (Table 1). The results from pathology confirmed that the absence or low level of pyruvate and lactate peaks was a characteristic of highly necrotic and hypoxic tissue, while tumor tissue with minimal levels of necrosis and hypoxia possessed relatively high levels of pyruvate and lactate (Figs. 4 and 5). The characterization of 13C metabolites in association with variable tumor hypoxia and necrosis would be difficult at best when using conventional fully sampled echo-planar 13C MRSI due to its lower resolution and the ensuing partial volume effect (Fig. 3).
Hyperpolarized 13C MRSI using dynamic nuclear polarization is an emerging technique that is capable of noninvasively probing carbon metabolism with dramatically increased sensitivity. A number of recent studies have used the 13C imaging of hyperpolarized [1-13C]-pyruvate to study carbon metabolism in the brain (12,13). Mayer et al.(12) acquired 2D spiral 13C chemical shift imaging data with in-plane resolution of 1.5 × 1.5 mm2 and 5 mm slice thickness from normal rat brain using a 3T clinical system combined with a high-performance gradient insert (600 mT/m, 3200 T/m/s). Day et al.(13) acquired 2D 13C chemical shift imaging data with 2.5 × 2.5 mm2 in-plane resolution and 6 mm slice thickness from tumor-bearing rat brain using a 4.7T magnet designed for animal studies. In the current study, a 3T clinical system without high-performance gradient inserts was used to acquire 3D 13C MRSI data with a 2 × 2 × 5.4 mm3 resolution in a brain tumor model. The use of a clinical system makes the compressed sensing method developed in this project easily translatable to future human studies.
The 13C 3D flyback MRSI sequence used in this study provides a fast method for acquiring hyperpolarized spectra and is less sensitive to timing errors, eddy currents, and B0 inhomogeneity than nonflyback versions (8). Compressed sensing in combination with a flyback readout was used to reduce acquisition time and improve spatial resolution. In order for compressed sensing to perform well, the underlying signal needs to possess sparsity and exhibit adequate signal-to-noise ratio (11). The parameters in Eq. 1 were chosen manually to balance the denoising and data fidelity constraints (6). The implementation of an automatic parameter selection scheme may be desirable in the future.
Noninvasive imaging of GBM is often complicated due to its heterogeneous and infiltrative nature (14). The ability to assess heterogeneous 13C metabolic profiles with high specificity and correlate them with histopathology is of great potential for monitoring GBM progression and response to therapy. The ability to characterize distinct 13C metabolic profiles using the compressed sensing method for higher spatial resolution than conventional 3D echo-planar spectroscopic imaging may assist in the noninvasive prediction of tumor tissue types and therefore, improve the management of patients with GBM.
In conclusion, we have shown that compressed sensing hyperpolarized 3D 13C MRSI can provide a unique tool for quantitative evaluation of tumor metabolism and characterization of 13C metabolite patterns between hypoxic/necrotic, nonhypoxic/necrotic tumor, and normal brain tissue. Future studies will validate the findings in a larger population of animals and examine the application of this technology to patients with brain tumors.
ACKNOWLEDGMENTS
The authors gratefully acknowledge the assistance of Dr. Michael Lustig for advice on implementing the compressed sensing methods, and Peter Shin and Llewelyn Jalbert for processing data and assisting in experiments.
Grant sponsor: American Brain Tumor Association (basic research fellowship to I. Park); Grant sponsor: NIH; Grant numbers: R01EB007588, P41EB13598; Grant sponsor: University of California and GE Healthcare; Grant number: ITL-BIO04-10148 (academic-industry partnership).
REFERENCES
- 1.Ardenkjaer-Larsen JH, Fridlund B, Gram A, Hansson G, Hansson L, Lerche MH, Servin R, Thaning M, Golman K. Increase in signal-to-noise ratio of >10,000 times in liquid-state NMR. Proc Natl Acad Sci USA. 2003;100:10158–10163. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park I, Larson PE, Zierhut ML, et al. Hyperpolarized 13C magnetic resonance metabolic imaging: application to brain tumors. Neuro Oncol. 2010;12:133–144. doi: 10.1093/neuonc/nop043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park I, Bok R, Ozawa T, Phillips JJ, James CD, Vigneron DB, Ronen SM, Nelson SJ. Detection of early response to temozolomide treatment in brain tumors using hyperpolarized 13C MR metabolic imaging. J Magn Reson Imaging. 2011;33:1284–1290. doi: 10.1002/jmri.22563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaumeil MM, Ozawa T, Park I, Scott K, James CD, Nelson SJ, Ronen SM. Hyperpolarized (13)C MR spectroscopic imaging can be used to monitor Everolimus treatment in vivo in an orthotopic rodent model of glioblastoma. Neuroimage. 2012;59:193–201. doi: 10.1016/j.neuroimage.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu S, Lustig M, Balakrishnan A, et al. 3D compressed sensing for highly accelerated hyperpolarized 13C MRSI with in vivo applications to transgenic mouse models of cancer. Magn Reson Med. 2010;63:312–321. doi: 10.1002/mrm.22233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu S, Lustig M, Chen AP, et al. Compressed sensing for resolution enhancement of hyperpolarized 13C flyback 3D-MRSI. J Magn Reson. 2008;192:258–264. doi: 10.1016/j.jmr.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derby K, Tropp J, Hawryszko C. Design and evaluation of a novel dual-tuned resonator for spectroscopic imaging. J Magn Reson. 1990;86:645–651. [Google Scholar]
- 8.Cunningham CH, Vigneron DB, Chen AP, Xu D, Nelson SJ, Hurd RE, Kelley DA, Pauly JM. Design of flyback echo-planar readout gradients for magnetic resonance spectroscopic imaging. Magn Reson Med. 2005;54:1286–1289. doi: 10.1002/mrm.20663. [DOI] [PubMed] [Google Scholar]
- 9.Cunningham CH, Chen AP, Albers MJ, Kurhanewicz J, Hurd RE, Yen YF, Pauly JM, Nelson SJ, Vigneron DB. Double spin-echo sequence for rapid spectroscopic imaging of hyperpolarized 13C. J Magn Reson. 2007;187:357–362. doi: 10.1016/j.jmr.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 10.Nelson SJ. Analysis of volume MRI and MR spectroscopic imaging data for the evaluation of patients with brain tumors. Magn Reson Med. 2001;46:228–239. doi: 10.1002/mrm.1183. [DOI] [PubMed] [Google Scholar]
- 11.Lustig M, Donoho D, Pauly JM. Sparse MRI: the application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58:1182–1195. doi: 10.1002/mrm.21391. [DOI] [PubMed] [Google Scholar]
- 12.Mayer D, Yen YF, Takahashi A, Josan S, Tropp J, Rutt BK, Hurd RE, Spielman DM. Dynamic and high-resolution metabolic imaging of hyperpolarized [1–13C]-pyruvate in the rat brain using a high-performance gradient insert. Magn Reson Med. 2011;65:1228–1233. doi: 10.1002/mrm.22707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Day SE, Kettunen MI, Cherukuri MK, Mitchell JB, Lizak MJ, Morris HD, Matsumoto S, Koretsky AP, Brindle KM. Detecting response of rat C6 glioma tumors to radiotherapy using hyperpolarized [1-13C]pyruvate and 13C magnetic resonance spectroscopic imaging. Magn Reson Med. 2011;65:557–563. doi: 10.1002/mrm.22698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bohman LE, Swanson KR, Moore JL, Rockne R, Mandigo C, Hankinson T, Assanah M, Canoll P, Bruce JN. Magnetic resonance imaging characteristics of glioblastoma multiforme: implications for understanding glioma ontogeny. Neurosurgery. 2010;67:1319–1327. doi: 10.1227/NEU.0b013e3181f556ab. [DOI] [PMC free article] [PubMed] [Google Scholar]