Abstract
CD4+ helper and CD8+ cytotoxic T cells differentiate from common precursors in the thymus after T-cell receptor (TCR)-mediated selection. Commitment to the helper lineage depends on persistent TCR signals and expression of the ThPOK transcription factor, whereas a ThPOK cis-regulatory element, ThPOK silencer, represses Thpok gene expression during commitment to the cytotoxic lineage. Here, we show that silencer-mediated alterations of chromatin structures in cytotoxic-lineage thymocytes establish a repressive state that is epigenetically inherited in peripheral CD8+ T cells even after removal of the silencer. When silencer activity is enhanced in helper-lineage cells, by increasing its copy number, a similar heritable Thpok silencing occurs. Epigenetic locking of the Thpok locus may therefore be an independent event from commitment to the cytotoxic lineage. These findings imply that long-lasting TCR signals are needed to establish stable Thpok expression activity to commit to helper T-cell fate and that full commitment to the helper lineage requires persistent reversal of silencer activity during a particular time window.
Keywords: cell fate, epigenetics, gene silencing, T-cell development, ThPOK
Introduction
Differentiation of multi-potent precursors into a particular lineage upon exposure to stimuli from external environmental cues is accompanied by the expression of lineage-specific genes together with repression of alternative lineages, a process termed as lineage specification. As a consequence of sequential exposures to differentiation signals, specific gene expression signatures that confer unique cellular functions to differentiated cells are established (lineage commitment) (Cantor and Orkin, 2001; Murphy and Reiner, 2002; Rothenberg and Dionne, 2002). Mechanisms that guarantee the stable inheritance of established gene signatures even after multiple cell divisions enable differentiated cells to preserve their cell identity. Studies in the last decade have highlighted the crucial contributions of epigenetics in maintenance of gene expression status (Jenuwein and Allis, 2001; Probst et al, 2009; Bonasio et al, 2010). For instance, heterochromatin has been shown to be involved in heritable and stable gene repression (Henikoff, 2000), referred to as gene silencing, in part via relocating gene loci into specialized nuclear compartments such as the peri-nuclear lamina (Schneider and Grosschedl, 2007; Reddy et al, 2008). It is thus conceivable that lineage commitment would involve epigenetic changes towards heterochromatin-like structures at many developmentally regulated genes to assure their subsequent silent state. However, how the epigenetic machinery that delivers lineage-specific epigenetic marks is linked with lineage commitment remains uncharacterized. Specifically, it is not clear whether such epigenetic machinery becomes functional upon lineage commitment or acts in parallel with lineage specification. In order to better understand an epigenetic link between environmental cues and establishment of cell identity, it is important to unravel how specific epigenetic modifications are accumulated on the relevant genes.
There are two major lineages of T lymphocytes, the CD4+ helper and the CD8+ cytotoxic cells, and they differentiate from a common precursor, the CD4+CD8+ double-positive (DP) thymocyte, after positive selection. During this selection process, only DP thymocytes that receive optimal TCR-signal strength as a result of TCR engagement with peptide–MHC complexes are allowed to further differentiate (Germain, 2002; Starr et al, 2003; Singer et al, 2008). When CD4+CD8+ DP thymocytes are positively selected through MHC class II molecules (MHC class II-selected thymocytes), they develop into CD4+CD8− single-positive (SP) thymocytes that are committed to the helper lineage. On the contrary, DP thymocytes that undergo selection via MHC class I molecules with a help of CD8 co-receptors terminate CD4 expression and differentiate into CD4−CD8+ SP thymocytes committed to the cytotoxic lineage (Ellmeier et al, 1999). It has been proposed that differences in TCR-signal length during positive selection instruct distinct fates in post-selection thymocytes. Several lines of evidence have shown that persistent TCR signalling is essential for development of CD4+ helper T cells (Sarafova et al, 2005; Singer et al, 2008; Adoro et al, 2012). On the other hand, temporary termination of Cd8 gene expression in post-selection thymocytes (Brugnera et al, 2000) results in a disruption of TCR signals specifically in MHC class I-selected thymocytes, instructing them to become cytotoxic-lineage cells (Singer et al, 2008). However, it remains obscure why long-lasting TCR signals are necessary for commitment to the helper lineage.
Recently, it has been recognized that appropriate linkage of TCR specificity to MHC class during helper/cytotoxic lineage choice requires input from the zing finger transcription factor ThPOK (also known as cKrox), which is encoded by the Zbtb7b gene (hereafter referred to as the Thpok gene in this manuscript) (He et al, 2005, 2010; Sun et al, 2005). Gain and loss of function studies of ThPOK demonstrated a dominant role of ThPOK in acquiring a CD4+CD8− phenotype that is independent of TCR specificity to MHC class. Thus, both MHC class I-selected and class II-selected thymocytes are redirected to alternative CD4+CD8− helper and CD4−CD8+ cytotoxic lineages by ectopic induction or loss of ThPOK function, respectively (He et al, 2005; Sun et al, 2005). Because of its potent activity, expression of the Thpok gene from its two promoters, distal P1 and proximal P2, must be strictly regulated during thymocyte differentiation. During thymocyte maturation, ThPOK first appears after positive selection, increases in MHC class II-selected thymocytes, but then disappears in MHC class I-selected cells (He et al, 2005; Muroi et al, 2008). In sorting out mechanisms that control lineage- and stage-specific Thpok expression, previous studies have identified two essential cis-regulatory elements. A transcriptional silencer (referred to as the Thpok silencer in this manuscript) is essential to control helper lineage-specific expression of Thpok (He et al, 2008; Setoguchi et al, 2008). On the other hand, a transcriptional enhancer in the proximal region of the gene plays an essential role in increasing Thpok expression at later maturation stages in MHC class II-selected thymocytes (Muroi et al, 2008). Interestingly, conditional ablation of ThPOK function from peripheral CD4+ T cells indicated that extra-thymic expression of ThPOK is still necessary to maintain CD4+ helper T-cell identity (Wang et al, 2008). Furthermore, retroviral transduction of ThPOK into fully differentiated peripheral CD8+ T cells can activate helper lineage-related genes as well as repress cytotoxic-related genes, albeit only to limited extent (Jenkinson et al, 2007). These findings indicate that the helper lineage-specific Thpok expression established in the thymus must be sustained in the peripheral T cells to maintain their lineage identity. However, although roles of cis-regulatory elements in regulating Thpok expression during lineage commitment in thymus have begun to be characterized (Taniuchi, 2009), it remains unknown how the established state of the Thpok gene, either active or repressive, is stably maintained in differentiated T cells.
To address this point, we used genetic approaches that enabled us to modify Thpok silencer activity. We show that a silencer-dependent deposition of repressive epigenetic marks establishes a repressive state that is inherited independently of the silencer in peripheral CD8+ T cells via inhibiting the two promoters by different mechanisms. Furthermore, we provide evidence showing that such epigenetic Thpok silencing can also occur in CD4+ helper-lineage cells when silencer activity persists after positive selection. These findings suggest that epigenetic mechanisms that lock the Thpok locus can take place even in MHC class II signalled cells if the silencer activity is not properly terminated. Thus, our findings strongly suggest that, for appropriate helper-lineage choice by MHC class II-selected cells, reversal of the Thpok silencer upon receipt of TCR signals must persist for a certain time interval to avoid epigenetic sealing of the Thpok gene. We thus propose that such continuous counter-silencing by long-lasting TCR signals is necessary to establish stable Thpok expression, hence to fully commit to helper T-cell fate.
Results
Epigenetic modifications in the Thpok gene
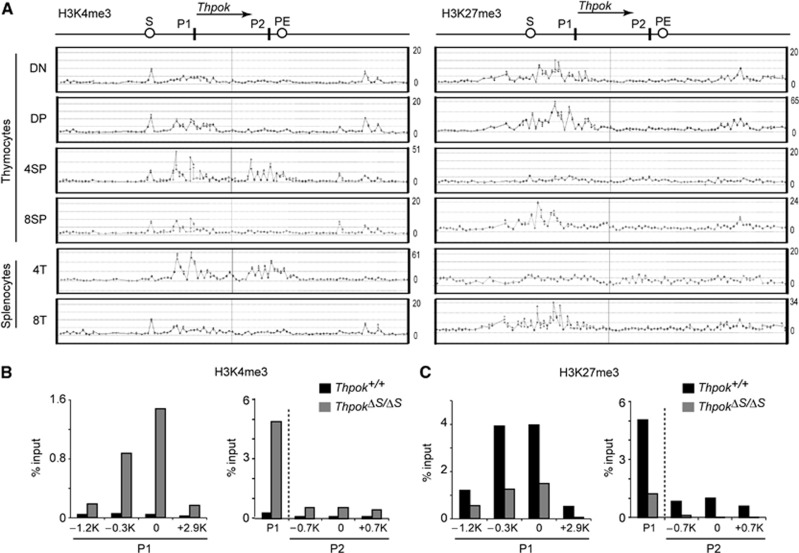
There is now compelling evidence that developmentally regulated genes receive dynamic modifications of local chromatin structure (Bonasio et al, 2010), and in particular, heterochromatin-like structures are known to be involved in stable gene repression (Henikoff, 2000). We therefore examined how the chromatin structure at the Thpok locus is altered during T-cell development. Since tri-methylation at two distinct histone H3 lysine residues, lysine 4 and lysine 27 (H3K4me3 and H3K27me3), has been shown to mark active and repressive states, respectively (Bernstein et al, 2005), of the target genes, we performed chromatin immunoprecipitation (ChIP)-on-chip analysis of distinct T-cell subsets. Accumulation of the repressive H3K27me3 mark at the distal P1 promoter and its upstream region was detected from immature CD4−CD8− DN thymocytes onward in cell subsets that do not express Thpok, while it was not present at a significant level at the proximal P2 promoter (Figure 1A and C). In contrast, H3K27me3 deposition at the distal P1 promoter was replaced by active H3K4me3 marks in cells expressing Thpok, such as CD4+CD8− SP thymocytes, along with accumulation of H3K4me3 at the proximal P2 promoter (Figure 1A). Interestingly, a comparison of the Thpok silencer-deficient and wild-type Thpok loci by analytical ChIP assays revealed that the loss of the Thpok silencer resulted in not only a decrease in H3K27me3 marks but also an increase in H3K4me3 marks around the P1 promoter in DP thymocytes (Figure 1B and C). Similar changes resulting from loss of the silencer were also observed at the P2 promoter, albeit to a lesser extent (Figure 1B and C). These results indicate that the silencer is involved in the initial deposition of H3K27me3 marks as well as in preventing premature H3K4me3 loading prior to positive selection.
Figure 1.
Changes of histone H3 modifications pattern in the Thpok gene during T-cell development. (A) Association of histone H3 K4 tri-methylation (H3K4me3) and histone H3 K27 tri-methylation (H3K27me3) in the Thpok locus. The signal intensities of individual oligonucleotide probe as revealed in a ChIP-on-chip assay in the indicated T-cell subsets are shown along with a schematic structure of the Thpok gene (top). Positions of the silencer (S), proximal enhancer (PE), distal P1 promoter (P1) and proximal P2 promoter (P2) are indicated. One representative result of two experiments is shown. (B, C) Comparison of H3K4me3 (B) and H3K27me3 (C) patterns at the region around the distal P1 and proximal P2 promoters between silencer-sufficient (Thpok+/+) and -deficient (ThpokΔS/ΔS) DP thymocytes determined by analytical ChIP assays. Positions of each region analysed are shown as distance from the transcriptional start site in the P1 or P2 promoter-derived Thpok mRNA. In the panel shown at the right, the P1 promoter is also included as a reference. Data are from one of two independent experiments.
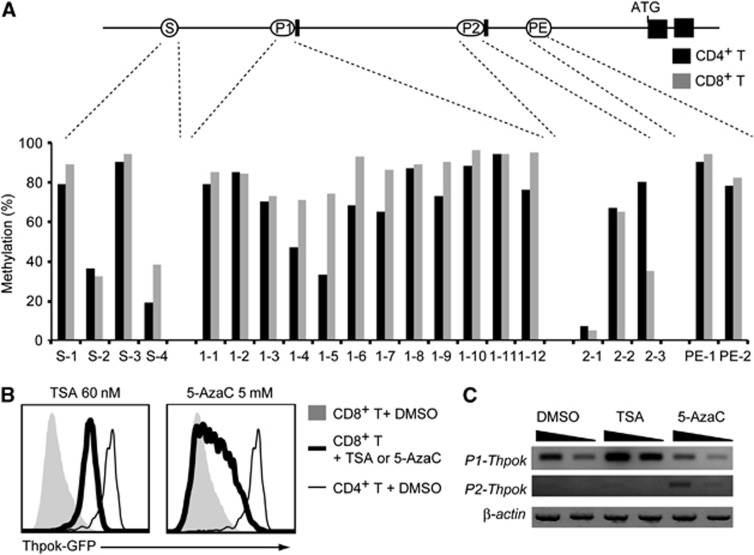
Another known epigenetic modification involved in gene silencing is DNA methylation (Jones, 2012). We therefore next compared the DNA methylation status of the Thpok gene in CD4+ and CD8+ peripheral T cells. We tested 21 CpG islands derived from the Thpok silencer, distal P1 promoter, proximal P2 promoter and proximal enhancer (PE) and only three regions (S-4, 1-4 and 1-5) exhibited slightly increased DNA methylation in CD8+ T cells compared to CD4+ T cells (Figure 2A). Thus, our analyses did not detect any clear differences in the DNA methylation status of active versus repressed Thpok genes, although they could not formally exclude involvement of DNA methylation at other sites in repression of the Thpok gene.
Figure 2.
Comparison of DNA methylation status of the Thpok locus in CD4+ and CD8+ T cells. (A) DNA methylation status in CD4+ and CD8+ T cells analysed by methylation-specific PCR at selected CpG islands located within the silencer (S), distal P1 promoter (P1), proximal P2 promoter (P2) and proximal enhancer (PE) are shown as percentages. (B) Effects of chemical inhibitors of histone deacetylases, Trichostatin A (TSA), and DNA methyltransferase, 5-aza-2′-cytidine (5-AzaC), on the Thpok repression. Histograms showing Thpok-GFP expression from the Thpokgfp allele in activated CD8+ T cells 3 days after treatment with either inhibitor in culture. Expression of GFP in control CD4+ T cells is shown as a thin line for reference. (C) Semi-quantitative RT–PCR for the P1 promoter- and the P2 promoter-derived Thpok mRNA after inhibitor treatment is shown with the β-actin mRNA as control. Wedges indicate 1:3 dilution of template. Data are from one of two independent experiments.
Source data for this figure is available on the online supplementary information page.
To further examine roles of epigenetic modifications in Thpok repression, we treated CD8+ T cells with Trichostatin A (TSA) or 5-aza-2′-deoxycytidine (5-AzaC), inhibitors of histone deacetylases (HDAC) and DNA methyltransferase, respectively. Three days after TSA treatment, Thpok expression in activated CD8+ T cells was significantly increased specifically from the P1 promoter, albeit at a level still well below that in CD4+ T cells. On the other hand, 5-AzaC induced a slight derepression of the Thpok gene mainly from the P2 promoter in a variegated manner (Figure 2B and C). Although it was not clear whether these inhibitors induced Thpok de-repression directly or indirectly, these results, along with the distinct H3K27me3 deposition pattern, suggested that different mechanisms are likely to be involved in preventing the activity of the two promoters for stable repression of the Thpok gene in CD8+ T cells.
Conditional deletion of the Thpok silencer from CD8-lineage cells
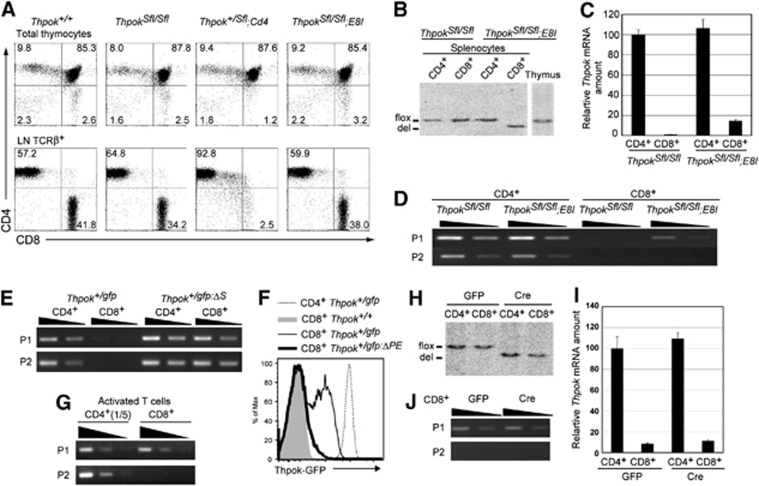
Our result showed that lineage-specific chromatin structures are established at the Thpok locus and that this correlates with the presence of the Thpok silencer. To further investigate the relevance of such epigenetic modifications for the regulation of Thpok expression, we wished to test whether the repressive state is maintained independently of the silencer in differentiated CD8+ T cells, as was observed at the Cd4 locus (Zou et al, 2001). To this end, we have generated a ThpokSfl allele in which an 800-bp region including the entire Thpok silencer sequence was flanked by two loxP sites (Supplementary Figure 1). The excision of the silencer in precursor DP thymocytes by Cd4-Cre transgene resulted in the loss of peripheral CD8+ T cells, as was previously observed in the case of germline deletion of the silencer (Figure 3A; Setoguchi et al, 2008). On the contrary, deletion of the Thpok silencer in developing CD4−CD8+ SP thymocytes by E8I-Cre transgene (Maekawa et al, 2008), which is expressed after the cells have downregulated the CD24 surface marker, did not impair development of CD8+ T cells (Figure 3A). Genotype analyses by Southern blot confirmed that the excision of the silencer by E8I-Cre was not at a detectable level in total thymocytes, but was almost complete in and restricted to CD8+ cells among splenic T cells (Figure 3B). The level of Thpok mRNA in these silencer-deficient CD8+ T cells was slightly elevated, but still <20% of that in CD4+ T cells (Figure 3C). Thus, the conditional removal of the Thpok silencer from post-selection thymocytes differentiating into the cytotoxic lineage did not cause a massive de-repression of the Thpok gene. Interestingly, only P1-derived Thpok transcripts were detected in the CD8+ T cells from ThpokSfl/Sfl;E8I mice (Figure 3D), whereas germline deletion of the silencer from a Thpokgfp reporter allele led to de-repression of the gfp gene from both the P1 and P2 promoters in CD8+ T cells (Figure 3E).
Figure 3.
Effects of conditional removal of the Thpok silencer in CD8-lineage cells on Thpok expression. (A) Dot plots showing CD4 and CD8 expression in total thymocytes and lymph node TCRβ+ T cells from mice of the indicated genotypes. Numbers in dot plots indicate the percentage of cells in each quadrant. (B–D) Genotype analyses by Southern blot (B), relative Thpok mRNA amounts (C) and semi-quantitative RT-PCR for the P1 promoter- and the P2 promoter-derived Thpok mRNA (D) in CD4+ and CD8+ T cells from the indicated mice. (E, G) Semi-quantitative RT–PCR for the P1 promoter- and the P2 promoter-derived gfp mRNA from the Thpokgfp and the silencer-deficient Thpokgfp:ΔS allele in CD4+ and CD8+ T cells (E), and for the P1- and P2-derived Thpok mRNA in activated CD4+ and CD8+ T cells (G). (F) Histogram showing Thpok-GFP expression in the indicated activated T cells of indicated genotypes. (H–J) Genotype analyses by Southern blot (H), relative Thpok mRNA amount (I) and semi-quantitative RT–PCR for the P1 promoter- and the P2 promoter-derived Thpok mRNA (J) in activated CD4+ and CD8+ T cells from ThpokSfl/Sfl mice transduced with control retroviral vector (GFP) or vector encoding Cre recombinase (Cre). Wedges in semi-quantitative RT–PCR assay indicate 1:3 dilution of template, except in (G) (1:5 dilutions). Data are from one of at least two independent experiments.
Source data for this figure is available on the online supplementary information page.
We next examined the effect of deleting the Thpok silencer from fully differentiated CD8+ T cells prepared from spleen. As previously reported (Setoguchi et al, 2009), Thpok-GFP expression was detected in activated CD8+ T cells from the Thpokgfp allele, but not the Thpokgfp:ΔPE allele lacking the PE, after in vitro TCR stimulation (Figure 3F), indicating that the PE is necessary for Thpok expression in activated CD8+ T cells. Both P1 and P2 promoter-derived Thpok mRNA was detected in activated CD4+ T cells, however, activated CD8+ T cells contained only the P1-derived Thpok mRNA (Figure 3G). When these CD4+ and CD8+ T cells from ThpokSfl/Sfl mice were transduced with retroviral vectors encoding Cre recombinase, the silencer region was efficiently excised within 48 h (Figure 3H). However, the amount of Thpok mRNA in activated CD8+ T cells was not elevated after removal of the silencer (Figure 3I) and the P2 promoter-derived Thpok mRNA remained undetectable (Figure 3J). These results demonstrate that sequential epigenetic modifications at the Thpok locus during commitment to the CD8+ cytotoxic lineage in the thymus established an epigenetically repressed state that is maintained in peripheral CD8+ T cells independently of the silencer.
Effect of enhanced Thpok silencer activity by increasing its copy number
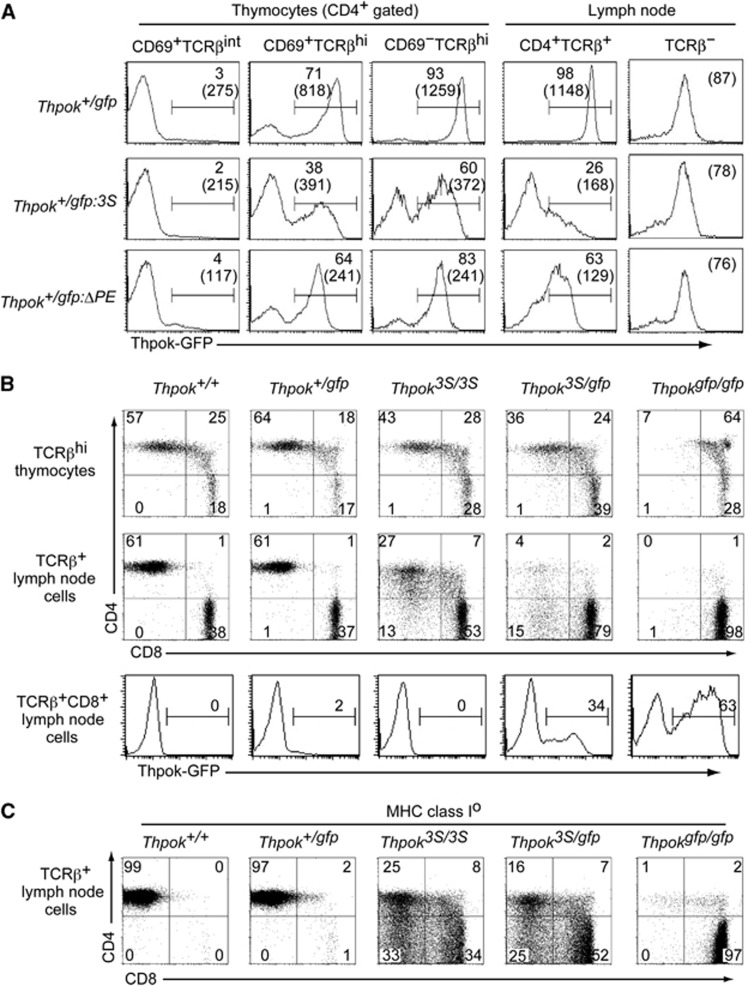
It is known that increasing the copy number of a cis-regulatory region can enhance its function (Herr and Gluzman, 1985). Indeed, three copies of the Thpok silencer in a reporter plasmid repressed luciferase reporter expression more efficiently than one copy in a transfection assay (Supplementary Figure 2). We then wished to examine whether and how an increase in silencer copy number at the endogenous Thpok gene would affect its expression. To this end, we have generated a Thpokgfp:3S allele, in which three tandem copies of 202–401 core silencer sequences were inserted into the Thpokgfp reporter allele by sequential gene targeting into ES cells of the Thpok+/gfp genotype (Supplementary Figure 3). Expression of GFP from the Thpokgfp:3S allele was analysed in Thpok+/gfp:3S mice, in which normal CD4+ T-cell development is supported by the half dosage of ThPOK produced from the wild-type allele. The induction of GFP in the CD69+ TCRβhi thymocyte subset from the Thpokgfp:3S allele was lower than from the control Thpokgfp allele with respect to both expression levels and the percentage of GFP+ cells (Figure 4A). Interestingly, GFP expression was lost in some CD4+ T cells but was only reduced in other CD4+ cells, thus exhibiting variegated repression rather than the uniform repression that was observed with the Thpokgfp:ΔPE allele (Figure 4A). Since reduced GFP expression was not observed in non-T cells (Figure 4A), the effect of increasing silencer copy number was likely to be CD4-lineage specific rather than a general phenomenon in any types of cells.
Figure 4.
Redirection of MHC class II-selected cells due to impaired Thpok expression by increase in the silencer copy number from one to three in the Thpok locus. (A) Histograms showing Thpok-GFP expression in the indicated T-cell subsets of Thpok+/gfp, Thpok+/gfp:3S and Thpok+/gfp:ΔPE mice. Numbers in the histogram indicate the percentage of GFP-positive cells, and numbers in parenthesis indicate the mean fluorescence intensity of GFP in GFP-positive cells. Notably, GFP expression in non-T cells of all three genotypes is same. (B) Dot plots showing CD4 and CD8 expression in mature (TCRβhi) thymocytes and lymph node (LN) TCRβ+ cells from mice of the indicated genotypes. Histograms show Thpok-GFP expression from the Thpokgfp allele in TCRβ+ CD8+ LN cells. Numbers in dot plots and histograms indicate the percentage of cells in each quadrant and the percentage of GFP-positive cells. (C) Dot plots showing CD4 and CD8 expression in lymph node TCRβ+ cells from mice of the indicated genotypes in the absence of cell surface expression of MHC class I molecules due to a genetic ablation of β2-microglobulin. Data are one representative of at least three independent experiments.
To examine how impaired ThPOK expression due to the insertion of three copies of the Thpok silencer affects T-cell development, we have generated a Thpok3S allele by targeting the same 3S mutation into the Thpok allele (Supplementary Figure 3). In Thpok3S/3S mice, the percentages of CD4+CD8– cells were reduced both in the thymus and in peripheral T-cell pools (Figure 4B). In Thpok3S/gfp mice in which ThPOK protein was produced only from the ThPOK3S allele, the decrease in the CD4+ T-cell subset became more evident with emergence of CD8+ T cells expressing GFP. Given that GFP expression from the Thpokgfp allele is specific to MHC class II-selected cells (Muroi et al, 2008), these CD8+ GFP+ cells are likely to be re-directed MHC class II-selected cells. Indeed, when we examined the differentiation of MHC class II-selected cells in mice with no surface expression of MHC class I molecules, cells with CD4−CD8−, CD4+CD8+ and CD4−CD8+ phenotype emerged in the peripheral lymphoid organs of Thpok3S/3S and Thpok3S/gfp mice whereas those cells were absent in control mice (Figure 4C). These observations indicated that enhanced Thpok silencer activity resulting from an increase in its copy number in the Thpok gene perturbed development of CD4+ T cells via inhibiting Thpok expression in a variegated manner.
Possible epigenetic Thpok silencing in CD4-lineage cells
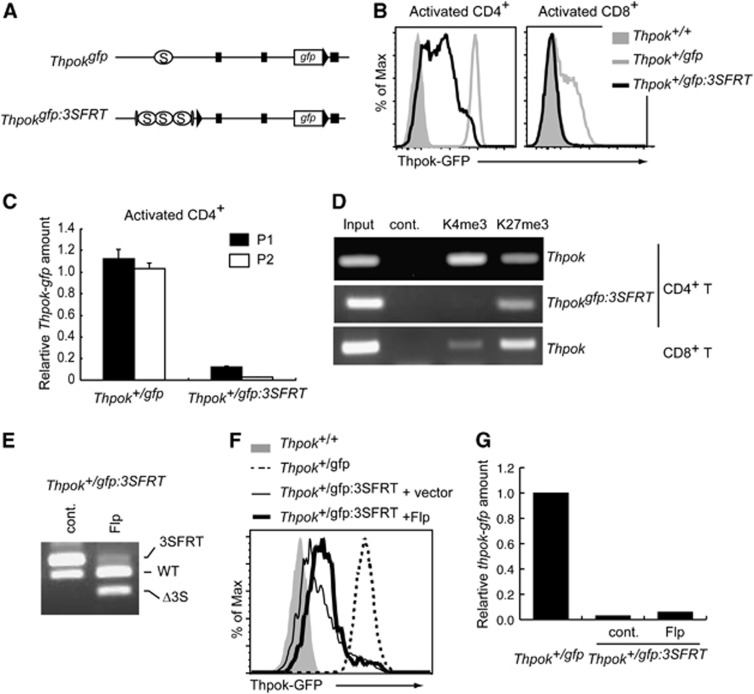
Variegated gene expression has been shown to be a characteristic of gene silencing mediated by heterochromatin-like structures (Weiler and Wakimoto, 1995). Given the variegated GFP expression from the Thpokgfp:3S allele in CD4+ T cells and the epigenetic control of the Thpok locus that results in a silencer-independent maintenance of Thpok silencing in CD8-lineage cells, we next addressed whether an epigenetic repressive state, which is maintained independently of the silencers, is similarly established at the Thpokgfp:3S allele in CD4+ helper-lineage cells. To this end, we have generated a Thpokgfp:3SFRT allele in which the three copies of the silencer can be excised upon expression of Flp recombinase (Figure 5A; Supplementary Figure 3). GFP expression from the Thpok+/gfp:3SFRT allele after in vitro TCR stimulation was still low in CD4+ T cells and was not detected in activated CD8+ T cells (Figure 5B). In addition, both P1 and P2 promoter-derived gfp mRNA was reduced in resting CD4+ T cells from Thpok+/gfp:3SFRT mice (Figure 5C). Consistent with these observations, ChIP assays demonstrated an increase and decrease in H3K27me3 and H3K4me3 marks, respectively, at the region nearby the silencer on the Thpokgfp:3SFRT allele, compared to those on the wild-type Thpok allele in the same CD4+ T cells of Thpok+/gfp:3SFRT mice (Figure 5D). Thus, chromatin structures at the Thpokgfp:3SFRT allele in CD4+ T cells were similar to those at the wild-type Thpok allele in CD8+ T cells. To examine whether GFP expression is restored if the three copies of the silencer are removed, we transduced CD4+ T cells with a retroviral vector encoding Flp recombinase to obtain a population of cells in which the FRT-flanked silencers were efficiently excised (Figure 5E). Interestingly, the mean fluorescent intensity of GFP and gfp mRNA levels were not elevated after excision of the 3 × silencer (Figure 5F and G), confirming that the silent state at the Thpokgfp:3SFRT allele in CD4+ helper-lineage cells could be maintained in the absence of the three Thpok silencer copies. These genetic results clearly demonstrate that epigenetic Thpok silencing could be established not only in cytotoxic-lineage cells but also in helper-lineage cells when the silencer activity is strengthened by increasing its copy number.
Figure 5.
Epigenetic Thpok silencing of the Thpokgfp:3SFRT allele in CD4+ helper T cells. (A) Schematic structure of Thpokgfp and Thpokgfp:3SFRT alleles. Black triangles, grey ovals, black boxes and open circles marked with S represent loxP sites, FRT sites, exons and the silencer region, respectively. (B) Histograms showing Thpok-GFP expression in the indicated activated T cells with indicated genotypes. (C) Relative amounts of the P1 promoter- and the P2 promoter-derived gfp mRNA from the Thpokgfp and Thpokgfp:3SFRT alleles in activated CD4+ T cells. (D) ChIP assay showing associations of histone H3K4 tri-methylation (K4me3) and histone H3K27 tri-methylation (K27me3) with the silencer region of the wild-type Thpok or Thpokgfp:3SFRT allele in CD4+ and CD8+ T cells from Thpok+/gfp:3SFRT mice. (E–G) Effect of removal of three copies of the silencers from the Thpokgfp:3SFRT allele in activated CD4+ T cells transduced with control vector (cont.) or vector encoding Flp recombinase (Flp). Genotyping analyses by DNA-PCR (E), histogram showing Thpok-GFP expression (F) and relative Thpok mRNA amounts (G) from Thpokgfp:3SFRT in the presence or absence of silencers and those from Thpokgfp or Thpok as controls (F). Data shown are one representative of at least two independent experiments.
Source data for this figure is available on the online supplementary information page.
Distinct requirement for the Runx complex function in reversible Thpok repression versus establishment of irreversible Thpok silencing
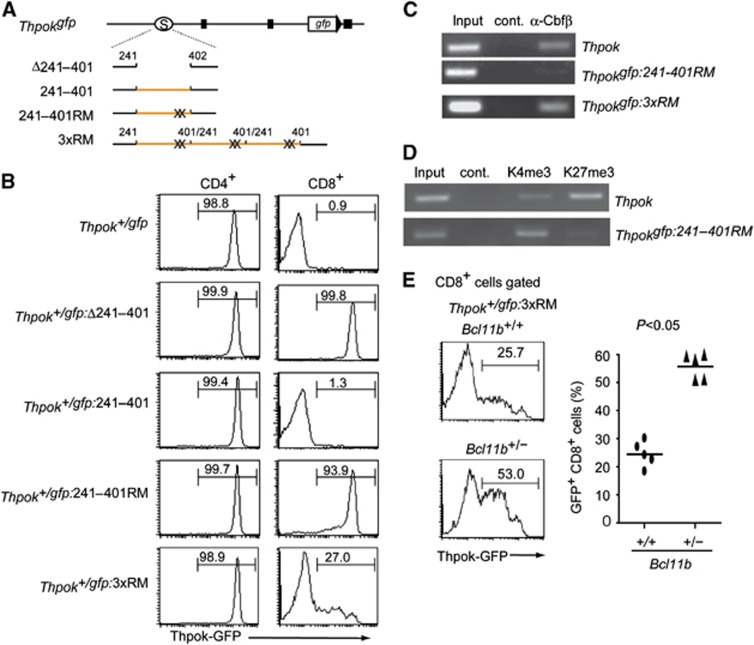
We next addressed which functional sites in the Thpok silencer are important to establish epigenetic Thpok silencing by a ‘knock-in’ mutagenesis assay (Figure 6A; Supplementary Figure 3). Whereas removal of the 242–401 core silencer sequences from the Thpokgfp allele led to full GFP de-repression in CD8+ T cells, repression was restored by reinsertion of wild-type 242–401 sequences (Figure 6B), confirming that junctional sequences created by ligation of the tested fragment did not affect silencer activity. Given the controversy about whether or not Runx sites are essential for Thpok silencer activity (He et al, 2008; Hedrick, 2008; Setoguchi et al, 2008), we introduced targeted mutations into the two Runx sites and generated a Thpokgfp:242–401RM reporter allele harbouring 242–401RM mutant sequences. Strikingly, upon loss of the two Runx sites, high GFP expression was induced in most of CD8+ T cells, although a small proportion of the cells expressed GFP at a low level (Figure 6B). ChIP assays with DP thymocytes from Thpok+/gfp:242–401RM mice showed that binding of Runx complexes to the Thpokgfp:242–401RM allele was severely reduced compared to the wild-type Thpok allele (Figure 6C). Furthermore, the ratio of H3K4me3 to H3K27me3 depositions at the Thpokgfp:242–401RM allele in CD8+ T cells of Thpok+/gfp:242–401RM mice was reversed compared to wild type (Figure 6D). This finding is inconsistent with previous results detecting substantial silencer activity in a mutant silencer fragment lacking Runx sites in a transgenic reporter expression assay (He et al, 2008). Given that silencer activity was enhanced by increasing its copy number, we next tested whether insertion of three copies of the 242–401RM mutant silencer restores Thpok silencing by generating a Thpokgfp:3xRM allele (Figure 6A). In the Thpok+/gfp:3xRM mice, GFP expression was lost in about three-quarter of CD8+ T cells (Figure 6B), along with restoration of Runx binding to the silencer to some extent (Figure 6C), although a low amount of GFP expression was still detected in about 25% of those cells. This result suggests that Runx complexes might be recruited to the Thpokgfp:3xRM allele through interaction with other silencer binding proteins. Indeed, the percentage of CD8+ T cells de-repressing GFP increased when the dosage of the Bcl11b gene, whose product, Bcl11b, binds to the Thpok silencer independently of Runx sites (Tanaka H, unpublished observation), was halved (Figure 6E). Taking into account the observed restoration of silencer activity by increasing copies of the mutant silencer lacking Runx sites from one to three, it is possible that integration of the transgene in tandem as multiple copies would influence the expression pattern of the reporter transgene and mask any effect of mutation in the cis-regulatory regions tested. In any cases, these genetic results clearly indicate that binding of Runx complexes to the Thpok silencer is essential for recruitment of nuclear protein complexes that catalyse epigenetic modifications to establish epigenetic Thpok silencing.
Figure 6.
Requirement for Runx complex binding to the silencer in establishment of epigenetic Thpok silencing. (A) Schematic structures of mutant Thpokgfp reporter alleles. Black boxes, open circles marked with S, orange lines and X represent exons, the silencer region, the 242–401 core silencer sequences and mutations at Runx sites, respectively. (B) Histograms showing Thpok-GFP expression in the indicated T-cell population from mice of the indicated genotypes. Numbers in the histogram indicate the percentage of GFP-positive cells. (C, D) ChIP assays showing binding of Runx/Cbfβ complexes to the silencer region on the Thpok, the Thpokgfp:242–401RM or the Thpokgfp:3xRM allele in DP thymocytes (C), histone H3K4 tri-methylation (K4me3) and histone H3K27 tri-methylation (K27me3) to the silencer region on the Thpok or Thpokgfp:242–401 RM allele in CD8+ T cells (D). Data are one representative of at least three individual experiments. (E) Histograms showing Thpok-GFP expression in CD8+ T cells of Thpok+/gfp:3xRM mice with full (Bcl11b+/+) or half dosage (Bcl11b+/−) of the Bcl11b gene. Numbers in the histogram indicate the percentage of GFP-positive cells. Graph at the right shows results with individual mice.
Source data for this figure is available on the online supplementary information page.
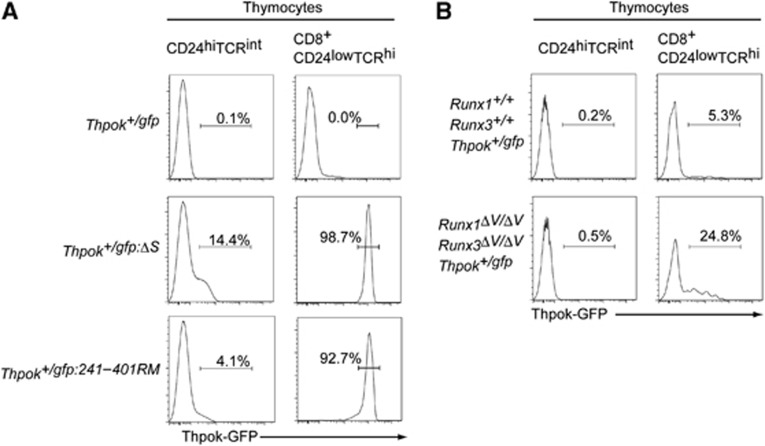
However, although Thpok repression in precursor DP thymocytes can be reversible, our analyses detected Runx complex binding to the silencer already in those cells (Figure 6C), raising the possibility of a distinct mode of action by Runx in repressing Thpok at two developmental stages. We therefore examined whether Runx complexes are functionally involved in Thpok repression by analysing Thpok-gfp reporter expression in precursor thymocytes. While low, but significant, GFP expression was induced in about 14% of CD24hi TCRint pre-selection thymocytes of Thpok+/gfp:ΔS mice, the percentage of GFP+ precursors was only ∼4% in Thpok+/gfp:242–401RM mice (Figure 7A). In contrast, in the cytotoxic-lineage committed CD8+CD24lowTCRhi mature thymocytes, GFP expression was quite similar between Thpok+/gfp:ΔS and Thpok+/gfp:242–401RM mice (Figure 7A), although lower GFP expression was observed in a small proportion of Thpok+/gfp:242–401RM cells, as was also observed in peripheral mature CD8+ T cells (Figure 6B). Thus, the requirement for Runx recognition sites for silencer activity could differ before and after positive selection.
Figure 7.
Runx complexes are nearly dispensable for Thpok repression in pre-selection thymocytes. (A) Histograms showing Thpok-GFP expression in pre-selection (CD24highTCRint) thymocytes and cytotoxic-lineage committed (CD8+CD24lowTCRhi) mature thymocytes from mice of the indicated genotypes. Numbers in the histogram indicate the percentage of GFP-positive cells. (B) Histograms showing Thpok-GFP expression in pre-selection (CD24highTCRint) thymocytes and mature CD8-lineage (CD8+CD24lowTCRhi) mature thymocytes from control mice (Runx1+/+:Runx3+/+:Thpok+/gfp) and mutant mice (Runx1ΔV/ΔV:Runx3ΔV/ΔV:Thpok+/gfp) that lack the VWRPY motif, a platform for recruiting TLE/Groucho co-repressors, from both Runx1 and Runx3 proteins. Numbers in the histogram indicate the percentage of GFP-positive cells. These results highlight a requirement for Runx proteins in establishment of epigenetic Thpok silencing that occurs specifically in post-selection thymocytes.
The VWRPY penta-peptide sequence is present in Runx proteins of all species examined and serves as a platform to recruit TLE/Groucho co-repressors to Runx proteins. Indeed, we recently reported that Thpok silencing in cytotoxic-lineage T cells was impaired in Runx1ΔV/ΔV:Runx3ΔV/ΔV mice in which the VWRPY motif was removed from both Runx1 and Runx3 proteins (Seo et al, 2012). We therefore next examined Thpok expression during differentiation of Runx1ΔV/ΔV:Runx3ΔV/ΔV thymocytes, and found that it remained repressed in precursor DP thymocytes whereas it was de-repressed in mature CD8-lineage thymocytes in a variegated manner (Figure 7B). These results suggest that regulation of the Thpok silencer function by Runx complexes may differ depending on developmental stage, with reversible repression in pre-selection precursors versus establishment of epigenetic silencing in post-selection thymocytes developing into the CD8 lineage.
Discussion
In this study, we examined how chromatin structure at the Thpok locus is modified during T-cell development. We observed that patterns of representative active and repressive histone modifications, H3K4me3 and H3K27me3, at the distal P1 promoter are dynamically altered and this correlated with promoter activity. Our genetic approach also uncovered a strong correlation between epigenetic modifications established during lineage commitment in the thymus and the maintenance of lineage-specific Thpok expression in the periphery. In particular, once silencer-dependent chromatin modifications were completed, the Thpok gene remained silenced independently of the Thpok silencer in CD8+ T cells, similar to the situation reported for the Cd4 locus (Zou et al, 2001). Furthermore, by generating mutant Thpok alleles harbouring three copies of the Thpok silencer, we showed that similar epigenetic Thpok silencing could be established in CD4+ T cells if the silencer activity is strengthened. Although the precise mechanism by which silencer activity is modified by increasing its copy number remains unclear, three silencers might potentially increase the rate of epigenetic modifications during a certain time period or extend the duration of a developmental window during which the silencer functions. Whatever the mechanisms, silencing by the three-copy silencers was not observed in B cells, strongly suggesting that this is not an artefact, and supporting its physiological relevance in the potency of epigenetic Thpok silencing in helper-lineage cells. Thus, even though this observation originated from an artificially modified Thpok locus, the potential for epigenetic Thpok silencing even in CD4+ T cells indicates that the nuclear machinery that establishes chromatin structures adequate for the silencer-independent maintenance of Thpok repression is not only present but also potentially functional even in cells differentiating towards the helper lineage. Thus, our findings indicate that epigenetic processes that stably silence the Thpok gene can occur independently of commitment to the cytotoxic lineage.
It has been proposed that proper development of MHC class II-selected thymocytes into the helper lineage requires persistence of TCR signals (Sarafova et al, 2005; Singer et al, 2008). However, it has been unclear how the length of TCR signalling can be linked with nuclear events controlling CD4/CD8 lineage choice, such as of Thpok expression, which clearly depends on TCR signals (Kappes, 2010). Our findings suggest that silencer-dependent chromatin modifications could potentially occur in all post-selection thymocytes unless the Thpok silencer is functionally inactivated. This silencer-mediated mode of regulation is likely to sequentially and gradually modify chromatin towards heterochromatin-like structures on the Thpok3S allele, even in cells developing into the helper lineage. The gradual nature of the epigenetic modification process presumably allows the Thpok locus to retain plasticity during a certain time window after the Thpok silencer is initially inactivated under TCR signals. It is therefore an intriguing possibility that the Thpok locus can undergo reverse chromatin modifications, leading towards the silenced state upon reactivation of the silencer. Physiologically, this could be induced by disruption of TCR signals, leading to a failure to produce enough ThPOK to support differentiation into CD4+ T cells. Along with previous finding that appropriate development of CD4+ helper T cells depends upon production of the appropriate amount of ThPOK protein (Muroi et al, 2008), our results suggest that reversal of the Thpok silencer activity must persist for a certain length of time to prevent accumulation of repressive epigenetic marks that convert the Thpok locus into a firmly repressed state. Thus, it is possible that chromatin alterations leading to stable silencing of the Thpok gene could serve as a mechanism that restricts a developmental time window to select the helper lineage by MHC class II-selected thymocytes. Interestingly, epigenetic regulatory mechanisms have been shown to be involved in determining stage-specific developmental potency in neural precursor cells (Hirabayashi and Gotoh, 2010), suggesting conserved features of epigenetic regulation in limiting the time available to multi-potent precursors for selecting a specific fate. In this case, sequential and temporally reversible epigenetic modifications at the Thpok locus would also serve as the mechanism underlying quantitative conversion of TCR-signal length into Thpok expression level. Precedent for such a mechanism exists in the Flowering Locus C gene in Arabidopsis, where the amount of H3K27me3 mirrors the duration of exposure to cold temperature (Huff and Zilberman, 2012). It is therefore important not only to further characterize the epigenetic machinery responsible for stable inheritance of Thpok silencing, but also to address its quantitative correlation with TCR signals.
Our results also indicated that the Thpok silencer-mediated regulation generates distinct repressive states at distinct developmental stages in terms of reversibility to an active state. In reporter transgene expression assays, Thpok silencer activity is detected from immature DN thymocyte to DP thymocyte stages (Muroi et al, 2013), consistent with the accumulation of H3K27me3 marks at the Thpok locus in those cells. Thus, the Thpok silencer already exerts its functions at the Thpok locus in uncommitted immature thymocytes to prevent its premature expression. However, the activation of the Thpok gene upon exposure to developmental cues that direct the cell towards the CD4 linegae indicates that the repressive state of the Thpok gene in such precursors retains the potential to readily revert into an active state. This concept is further supported with Thpok de-repression to the level sufficient to re-direct MHC class I-selected cells to CD4+ T cells by deletion of the silencer in precursors by Cd4-Cre. On the contrary, the conditional removal of the silencer in developing CD8 SP thymocytes by E8I-Cre transgene failed to induce high levels of Thpok expression. These findings suggest that alternation of chromatin structures towards stable and heritable epigenetic Thpok silencing might begin only after positive selection. Of note, the transcriptional silencer in the Cd4 locus can establish heritable epigenetic Cd4 silencing only in post-selection thymocytes developing into the cytotoxic lineage, even though repression of the Cd4 gene in immature DN thymocytes by the same silencer is reversible (Zou et al, 2001; Taniuchi et al, 2002b). Thus, epigenetic silencing at both Thpok and Cd4 loci commonly occurs only after positive selection. Given that the activity of both Thpok and Cd4 silencers requires Runx complex binding (Taniuchi et al, 2002a), loading of proteins specifically responsible for modifying chromatin leading to a heritable silent state might depend on both positive selection and Runx complexes. However, our results showed a distinct requirement for Runx complexes in reversible Thpok repression versus stable Thpok silencing. In contrast, Runx complexes are likely to be commonly essential for Cd4 repression in DN thymocytes and epigenetic Cd4 silencing in CD8-lineage cells (Taniuchi et al, 2002a). In addition, the VWRPY motif in Runx proteins is less necessary for epigenetic silencing at the Thpok than the Cd4 loci (Seo et al, 2012). Thus, there are both common and unique features of Thpok and Cd4 silencer-mediated gene repression. It will be important to understand the molecular basis that conveys commonality as well as uniqueness to these two silencers.
Another interesting finding was that the Thpok silencer regulates two promoters in the Thpok gene through different mechanisms. The presence of H3K27me3 repressive marks and a higher DNA methylation frequency in Thpok non-expressing cells were obvious only at the distal P1 promoter. However, despite the presence of such well-established repressive epigenetic marks, the P1 promoter retains the potential for reactivation in CD8+ T cells upon TCR stimulation, a process enhanced by HDAC inhibitor treatment. On the contrary, the proximal P2 promoter is stably silenced after activation of CD8+ T cells despite limited deposition of such repressive epigenetic marks. Given a slight increase in P2-derived Thpok mRNA upon treatment with 5-AzaC, the regulation of the P2 promoter is likely to involve uncharacterized epigenetic mechanisms that might cooperate with DNA methylation. Furthermore, leaky expression of P1 promoter-, but not the P2 promoter-, derived Thpok mRNA in resting CD8+ T cells of ThpokSfl/Sfl:E8I mice indicates that the processes that inactivate promoter function might be completed earlier at the P2 than at the P1 promoter. It will be important to address how a single silencer can inactivate two promoters located within an ∼6 kb interval by distinct manners.
Collectively, our results shed light on the nuclear regulatory event that would link TCR signals with the epigenetic regulation of Thpok expression. Nevertheless, uncovering properties of the molecular switch for Thpok silencer activity under TCR signals is extremely crucial to understand the underlying mechanisms of the link between signal length and lineage choice.
Materials and methods
Mice
β2-microglobulin-deficient mice (Koller et al, 1990), Thpokgfp mice (Muroi et al, 2008), E8I-Cre transgenic mice (Maekawa et al, 2008), Bcl11b-deficient mice (Wakabayashi et al, 2003) and Runx1ΔV/ΔV:Runx3ΔV/ΔV mice (Seo et al, 2012) have been described. All mice were bred and housed in the animal facility at RIKEN RCAI, and all experiments were performed in accordance with institutional guidelines for animal care.
Generation of mice harbouring mutations in the Thpok locus
Construction of each targeting vector for generating mutant Thpok alleles is described in Supplementary Methods. The targeting vectors that were targeted to the Thpokgfp allele were transfected into an ES cell clone harbouring the Thpok+/gfp genotype (Muroi et al, 2008), otherwise a wild-type ES cell line was used. Culture of ES cells and removal of the neor gene in ES cells by transient transfection of a Cre recombinase expression vector were performed as previously described (Muroi et al, 2008). Homologous recombination in ES cells was first screened by PCR or Southern blot. In order to determine whether the wild-type Thpok or Thpokgfp allele underwent homologous recombination with the targeting vector, ES clones were transduced with the retroviral vector encoding Cre recombinase and then screened by PCR that detected recombination between the loxP site downstream of the gfp gene and the loxP sites upstream of the neor gene (Supplementary Figure 3). Detection of the above recombination event was interpreted as confirmation of homologous recombination on the Thpokgfp allele. To establish a mouse strain containing the ThpokPEfl mutant allele, we used ES clone harbouring ThpokPEfl/gfp genotype that was obtained during generation of the ThpokΔPE mouse strain (Muroi et al, 2008).
Flow cytometry and cell sorting
Thymus, spleen and lymph nodes were removed from mice at 4–8 weeks of age and were used to make single-cell suspensions. Cells were stained with the following FITC, PE, PerCP or APC-conjugated antibodies purchased from BD Biosciences: CD4 (RM4-5), CD8 (53-6.7), CD69 (H1.2F3) and TCRβ (H57-597). For staining of intracellular Gata3, cells were fixed and permeabilized with Cytofix/perm solution (BD Biosciences) and stained with AlexaFluor 647-conjugated anti-Gata3 antibody (BD Biosciences). Multi-colour flow cytometry data were collected with a FACSCalibur or FACSCanto II (BD-Biosciences) and were analysed with BD CellQuest (BD Biosciences) or Flowjo (Tree Star) software. T-cell subsets were sorted using a FACSAria (BD Biosciences).
Cell culture with TSA and 5-AzaC
Sorted CD8+ T cells were stimulated with 2 μg/ml of plate-bound CD3ε antibody (BD Biosciences) and 2 μg/ml of soluble anti-CD28 antibody (BD Biosciences) in optimized DMEM (KOHJIN BIO) supplemented with 10% FBS for 48 h and were then maintained in medium supplemented with rIL-2 (20 U/ml). Twenty-four hours after initial stimulation, cells were treated with 60 nM of TSA (T8552, Sigma), or 5 mM 5-AzaC (014-20943, Wako Chemical) for 3 days and then analysed.
RNA isolation and RT–PCR
Total cellular RNA was extracted using Trizol Reagent (Invitrogen), and treated with RNase-free DNase I (Invitrogen) prior to reverse transcription in order to eliminate contaminating genomic DNA. cDNA was synthesized from total RNA using the SuperScript®III First Strand Synthesis System (Invitrogen). Quantitative RT–PCR was performed using the ABI/PRISM 7000 sequence detection system (Applied Biosystems) with an internal fluorescent TaqMan probe for the Thpok transcript and SYBR Green (Takara) for the gfp transcript. The amounts of Thpok and gfp mRNA were normalized against Hprt1 mRNA and β-actin mRNA expression, respectively. Primers and a probe for quantitative RT–PCR for Thpok were previously described (He et al, 2005). Sequences of all other primers are shown in Supplementary Table.
ChIP and tiling array
For a ChIP-on-chip experiment, chromatin DNA from 10 million cells was immune-precipitated with anti-H3K4me3 (ab1012, ABCOM) or anti-H3K27me3 (07-449, Millipore) antibody, and then amplified twice by LM-PCR according to manufacturer’s protocol (Agilent). We used custom microarrays generated by Agilent that tiled through the Thpok locus as previously described (Muroi et al, 2008). Probe hybridization and scanning of oligonucleotide array data were performed according to manufacturers’ protocol (Agilent). Feature Extraction software and Genomic Workbench (Agilent) were used for data analysis. For analytical ChIP assay, 1–10 million cells were immune-precipitated with anti-Cbfβ (Naoe et al, 2007) antibody.
DNA methylation assay
Methylation-specific PCR (Herman et al, 1996) was performed to analyse DNA methylation status by outsourcing (Towa Environment Science Co. Ltd). In brief, the MethylSEQr Bisulfite Conversion Kit (Applied Biosystems) was used for bisulfite conversion assay. Prediction of DNA methylation sites such as CpG islands within target regions and design of methylated primer sets and unmethylated primer sets were performed using MethylPrimer Express (Applied Biosystems). DNA methylation status was quantitatively analysed by real-time PCR, and was calculated as percentage of methylated Cytidine in methylated Cytidine+unmethylated Cytidine.
Retroviral transduction of T cells
A retroviral vector encoding Flp recombinase was constructed by insertion of a Flp encoding cDNA, which was PCR amplified from the pCAGGS-FLPe vector (GENE BRIDGES), into pMXs-IRES-hNGFR vector (a gift from Dr Yamashita at Chiba University). Transduction of retroviral products into T cells was performed as previously described (Naoe et al, 2007). Due to low recombination efficiency of the vector encoded Flp recombinase, we repeated Flp transduction three times over 2 days, sorted hNGFR-positive cells, then reactivated the cells, repeated the transductions, and sorted hNGFR-expressing cells 2 days after the last transduction.
Supplementary Material
Acknowledgments
We are grateful to C Yoshida and T Ishikura for aggregation of ES cells, to H Fujimoto and Y Hachiman for cell sorting and to Dr. Wilfried Ellmeier for critical reading of the manuscript. This work was supported by grants from Uehara Foundation and by the Grant-in-Aid for Scientific Research (S) and for Scientific Research on Priority Areas (IT).
Author contributions: HT and TN performed phenotypic analyses of mice and molecular biology experiments with the help of RC and CM. SM generated all mutant mouse strains. WS analysed Runx1ΔV/ΔV:Runx3ΔV/ΔV mice. KR provided important experimental material. IT designed the study and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adoro S, McCaughtry T, Erman B, Alag A, Van Laethem F, Park JH, Tai X, Kimura M, Wang L, Grinberg A, Kubo M, Bosselut R, Love P, Singer A (2012) Coreceptor gene imprinting governs thymocyte lineage fate. EMBO J 31: 366–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ 3rd, Gingeras TR, Schreiber SL, Lander ES (2005) Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120: 169–181 [DOI] [PubMed] [Google Scholar]
- Bonasio R, Tu S, Reinberg D (2010) Molecular signals of epigenetic states. Science 330: 612–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugnera E, Bhandoola A, Cibotti R, Yu Q, Guinter TI, Yamashita Y, Sharrow SO, Singer A (2000) Coreceptor reversal in the thymus: signaled CD4+8+ thymocytes initially terminate CD8 transcription even when differentiating into CD8+ T cells. Immunity 13: 59–71 [DOI] [PubMed] [Google Scholar]
- Cantor AB, Orkin SH (2001) Hematopoietic development: a balancing act. Curr Opin Genet Dev 11: 513–519 [DOI] [PubMed] [Google Scholar]
- Ellmeier W, Sawada S, Littman DR (1999) The regulation of CD4 and CD8 coreceptor gene expression during T cell development. Annu Rev Immunol 17: 523–554 [DOI] [PubMed] [Google Scholar]
- Germain RN (2002) T-cell development and the CD4-CD8 lineage decision. Nat Rev Immunol 2: 309–322 [DOI] [PubMed] [Google Scholar]
- He X, Dave VP, Zhang Y, Hua X, Nicolas E, Xu W, Roe BA, Kappes DJ (2005) The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature 433: 826–833 [DOI] [PubMed] [Google Scholar]
- He X, Park K, Kappes DJ (2010) The role of ThPOK in control of CD4/CD8 lineage commitment. Annu Rev Immunol 28: 295–320 [DOI] [PubMed] [Google Scholar]
- He X, Park K, Wang H, He X, Zhang Y, Hua X, Li Y, Kappes DJ (2008) CD4-CD8 lineage commitment is regulated by a silencer element at the ThPOK transcription-factor locus. Immunity 28: 346–358 [DOI] [PubMed] [Google Scholar]
- Hedrick SM (2008) Thymus lineage commitment: a single switch. Immunity 28: 297–299 [DOI] [PubMed] [Google Scholar]
- Henikoff S (2000) Heterochromatin function in complex genomes. Biochim Biophys Acta 1470: O1–O8 [DOI] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB (1996) Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 93: 9821–9826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr W, Gluzman Y (1985) Duplications of a mutated simian virus 40 enhancer restore its activity. Nature 313: 711–714 [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, Gotoh Y (2010) Epigenetic control of neural precursor cell fate during development. Nat Rev Neurosci 11: 377–388 [DOI] [PubMed] [Google Scholar]
- Huff JT, Zilberman D (2012) Regulation of biological accuracy, precision, and memory by plant chromatin organization. Curr Opin Genet Dev 22: 132–138 [DOI] [PubMed] [Google Scholar]
- Jenkinson SR, Intlekofer AM, Sun G, Feigenbaum L, Reiner SL, Bosselut R (2007) Expression of the transcription factor cKrox in peripheral CD8 T cells reveals substantial postthymic plasticity in CD4-CD8 lineage differentiation. J Exp Med 204: 267–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Jones PA (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13: 484–492 [DOI] [PubMed] [Google Scholar]
- Kappes DJ (2010) Expanding roles for ThPOK in thymic development. Immunol Rev 238: 182–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller BH, Marrack P, Kappler JW, Smithies O (1990) Normal development of mice deficient in beta 2M, MHC class I proteins, and CD8+ T cells. Science 248: 1227–1230 [DOI] [PubMed] [Google Scholar]
- Maekawa Y, Minato Y, Ishifune C, Kurihara T, Kitamura A, Kojima H, Yagita H, Sakata-Yanagimoto M, Saito T, Taniuchi I, Chiba S, Sone S, Yasutomo K (2008) Notch2 integrates signaling by the transcription factors RBP-J and CREB1 to promote T cell cytotoxicity. Nat Immunol 9: 1140–1147 [DOI] [PubMed] [Google Scholar]
- Muroi S, Naoe Y, Miyamoto C, Akiyama K, Ikawa T, Masuda K, Kawamoto H, Taniuchi I (2008) Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nat Immunol 9: 1113–1121 [DOI] [PubMed] [Google Scholar]
- Muroi S, Tanaka H, Miyamoto C, Taniuchi I (2013) Fine-tuning of Thpok gene activation by an enhancer in close proximity to its own silencer. J Immunol 190: 1397–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KM, Reiner SL (2002) The lineage decisions of helper T cells. Nat Rev Immunol 2: 933–944 [DOI] [PubMed] [Google Scholar]
- Naoe Y, Setoguchi R, Akiyama K, Muroi S, Kuroda M, Hatam F, Littman DR, Taniuchi I (2007) Repression of interleukin-4 in T helper type 1 cells by Runx/Cbfβ binding to the Il4 silencer. J Exp Med, 204: 1749–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, Dunleavy E, Almouzni G (2009) Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol 10: 192–206 [DOI] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H (2008) Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452: 243–247 [DOI] [PubMed] [Google Scholar]
- Rothenberg EV, Dionne CJ (2002) Lineage plasticity and commitment in T-cell development. Immunol Rev 187: 96–115 [DOI] [PubMed] [Google Scholar]
- Sarafova SD, Erman B, Yu Q, Van Laethem F, Guinter T, Sharrow SO, Feigenbaum L, Wildt KF, Ellmeier W, Singer A (2005) Modulation of coreceptor transcription during positive selection dictates lineage fate independently of TCR/coreceptor specificity. Immunity 23: 75–87 [DOI] [PubMed] [Google Scholar]
- Schneider R, Grosschedl R (2007) Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev 21: 3027–3043 [DOI] [PubMed] [Google Scholar]
- Seo W, Tanaka H, Miyamoto C, Levanon D, Groner Y, Taniuchi I (2012) Roles of VWRPY motif-mediated gene repression by Runx proteins during T-cell development. Immunol Cell Biol 90: 827–830 [DOI] [PubMed] [Google Scholar]
- Setoguchi R, Tachibana M, Naoe Y, Muroi S, Akiyama K, Tezuka C, Okuda T, Taniuchi I (2008) Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science 319: 822–825 [DOI] [PubMed] [Google Scholar]
- Setoguchi R, Taniuchi I, Bevan MJ (2009) ThPOK derepression is required for robust CD8 T cell responses to viral infection. J Immunol 183: 4467–4474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer A, Adoro S, Park JH (2008) Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat Rev Immunol 8: 788–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr TK, Jameson SC, Hogquist KA (2003) Positive and negative selection of T cells. Annu Rev Immunol 21: 139–176 [DOI] [PubMed] [Google Scholar]
- Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, Galera P, Bosselut R (2005) The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol 6: 373–381 [DOI] [PubMed] [Google Scholar]
- Taniuchi I (2009) Transcriptional regulation in helper versus cytotoxic-lineage decision. Curr Opin Immunol 21: 127–132 [DOI] [PubMed] [Google Scholar]
- Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, Ito Y, Littman DR (2002a) Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111: 621–633 [DOI] [PubMed] [Google Scholar]
- Taniuchi I, Sunshine MJ, Festenstein R, Littman DR (2002b) Evidence for distinct CD4 silencer functions at different stages of thymocyte differentiation. Mol Cell 10: 1083–1096 [DOI] [PubMed] [Google Scholar]
- Wakabayashi Y, Watanabe H, Inoue J, Takeda N, Sakata J, Mishima Y, Hitomi J, Yamamoto T, Utsuyama M, Niwa O, Aizawa S, Kominami R (2003) Bcl11b is required for differentiation and survival of alphabeta T lymphocytes. Nat Immunol 4: 533–539 [DOI] [PubMed] [Google Scholar]
- Wang L, Wildt KF, Castro E, Xiong Y, Feigenbaum L, Tessarollo L, Bosselut R (2008) The zinc finger transcription factor Zbtb7b represses CD8-lineage gene expression in peripheral CD4+ T cells. Immunity 29: 876–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler KS, Wakimoto BT (1995) Heterochromatin and gene expression in Drosophila. Annu Rev Genet 29: 577–605 [DOI] [PubMed] [Google Scholar]
- Zou YR, Sunshine MJ, Taniuchi I, Hatam F, Killeen N, Littman DR (2001) Epigenetic silencing of CD4 in T cells committed to the cytotoxic lineage. Nat Genet 29: 332–336 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.