Abstract
Type IV secretion (T4S) systems are able to transport DNAs and/or proteins through the membranes of bacteria. They form large multiprotein complexes consisting of 12 proteins termed VirB1-11 and VirD4. VirB7, 9 and 10 assemble into a 1.07 MegaDalton membrane-spanning core complex (CC), around which all other components assemble. This complex is made of two parts, the O-layer inserted in the outer membrane and the I-layer inserted in the inner membrane. While the structure of the O-layer has been solved by X-ray crystallography, there is no detailed structural information on the I-layer. Using high-resolution cryo-electron microscopy and molecular modelling combined with biochemical approaches, we determined the I-layer structure and located its various components in the electron density. Our results provide new structural insights on the CC, from which the essential features of T4S system mechanisms can be derived.
Keywords: core complex, cryo electron microscopy, pKM101, structure, type 4 secretion system
Introduction
Type IV secretion (T4S) systems are multiprotein complexes present in both Gram-negative and Gram-positive bacteria, and also in some archea (Alvarez-Martinez and Christie, 2009). This versatile system can translocate various substrates ranging from single to multimeric proteins and nucleoprotein complexes (Alvarez-Martinez and Christie, 2009). T4S systems can be subdivided into three main groups based upon their function: (i) conjugation systems; (ii) DNA release or uptake systems; and (iii) effector translocation systems (Waksman and Fronzes, 2010). Conjugation systems mediate the transfer of DNA to recipient cells in a contact-dependent manner (Dreiseikelmann, 1994). These systems promote genome plasticity in bacteria and thus mediate rapid adaptive responses to changes in environment. They are also responsible for the spread of antibiotic-resistance genes among pathogenic bacteria (Wallden et al, 2010). T4S systems, mediating DNA release and uptake, also contribute to the expansion of genome diversity (Alvarez-Martinez and Christie, 2009). Finally, effector translocation systems deliver proteins into eukaryotic cells (Wallden et al, 2010). For example, Helicobacter pylori, the main causative agent of gastric ulcers and gastric adenocarcinoma, disrupts the homeostasis of the gastrointestinal tract via effectors secreted by the Cag T4S system (Terradot and Waksman, 2011); the plasmid pTi from Agrobacterium tumefaciens encodes the VirB/D T4S system, which is able to deliver an oncogenic DNA fragment named T-DNA into plant cells leading to crown gall disease (McCullen and Binns, 2006).
The most studied T4S systems are the VirB/D system from A. tumefaciens and the closely related systems from Escherichia coli encoded by the conjugative plasmids F, R388, pKM101 and RP4 (Alvarez-Martinez and Christie, 2009; Waksman and Fronzes, 2010). Protein components and the genetic organisation of the VirB/D T4S systems have high similarity across functionally diverse T4S systems, reflecting their evolutionary relationship (Lessl and Lanka, 1994). The VirB/D system consists of 12 proteins named VirB1–VirB11 and VirD4 that form a multiprotein envelope-spanning transport apparatus. Three cytoplasmic ATPases associated with the inner membrane (IM), VirB11, VirD4 and VirB4, power substrate translocation and the assembly of the secretion system (Gomis-Ruth et al, 2001; Savvides et al, 2003; Hare et al, 2006; Waksman and Fronzes, 2010; Wallden et al, 2012). Three proteins (VirB3, VirB6 and VirB8) are likely to form an IM subcomplex. The extracellular pilus is composed of major and minor pilins, termed VirB2 and VirB5, respectively. Three proteins (VirB7, VirB9 and VirB10) form the ‘core complex’ (CC), a large central structure of the T4S system. This complex is inserted in both the outer (OM) and IMs, respectively, and plays a fundamental role as a scaffold for the rest of the T4S system components. Because of its central position, the complex participates actively in T4S substrate transfer through the bacterial envelope (Llosa et al, 2003; Atmakuri et al, 2004; Cascales and Christie, 2004a; Alvarez-Martinez and Christie, 2009; Jakubowski et al, 2009). Therefore, detailed structural knowledge of the CC architecture and function is essential to understand the mechanism of secretion by T4S systems.
The T4S system CC encoded by the pKM101 plasmid comprises 14 copies each of the three proteins TraN/VirB7, TraO/VirB9 and TraF/VirB10. The first CC structure was solved by cryo-electron microscopy (cryo-EM) at 15 Å resolution (Fronzes et al, 2009). The CC forms a cylindrical pore that consists of two layers: an outer layer (OL) and an inner layer (IL). The OL is inserted in the OM and is composed of full-length TraN/VirB7 and the C-terminal domains of TraO/VirB9 and TraF/VirB10 (TraO/VirB9CT and TraF/VirB10CT, respectively). The atomic structure of a truncated form of the pKM101 CC corresponding to the entire OL was solved at 2.6 Å resolution (Chandran et al, 2009). This structure revealed that TraF/VirB10CT lines the interior of the OL while TraO/VirB9CT and TraN/VirB7 are at the periphery. The OL inserts in the OM by forming a helical barrel made of 14 copies of a two-helix bundle from TraF/VirB10CT (Chandran et al, 2009). The lipidated TraN/VirB7 anchors TraO/VirB9CT to the inner leaflet of the OM (Chandran et al, 2009; Fronzes et al, 2009). The IL is composed of the N-terminal domains of TraO/VirB9 and TraF/VirB10 (TraO/VirB9NT and TraF/VirB10NT, respectively). The latter domain contains the transmembrane (TM) region that anchors the CC to the IM.
Due to the low resolution of the first cryo-EM structure of the CC, it was not possible to accurately determine the structure and arrangement of TraO/VirB9NT and TraF/VirB10NT within the IL. We report here the structure of a truncated pKM101 CC that lacks the N-terminal part of TraF/VirB10 at a subnanometre resolution of 8.5 Å using cryo-EM and single-particle analysis. We have also refined the structure of the full-length CC to 12.4 Å resolution. A comparison of these two structures and the utilisation of hybrid methods based upon ternary structure prediction and flexible fitting provide key insights into the structure of the OL in its native state, into the position and structure of the N-terminal domains of TraO/VirB9 and TraF/VirB10 in the IL and their links with the OL.
Results
Cryo-EM structure of a truncated pKM101 CC at 8.5 Å resolution
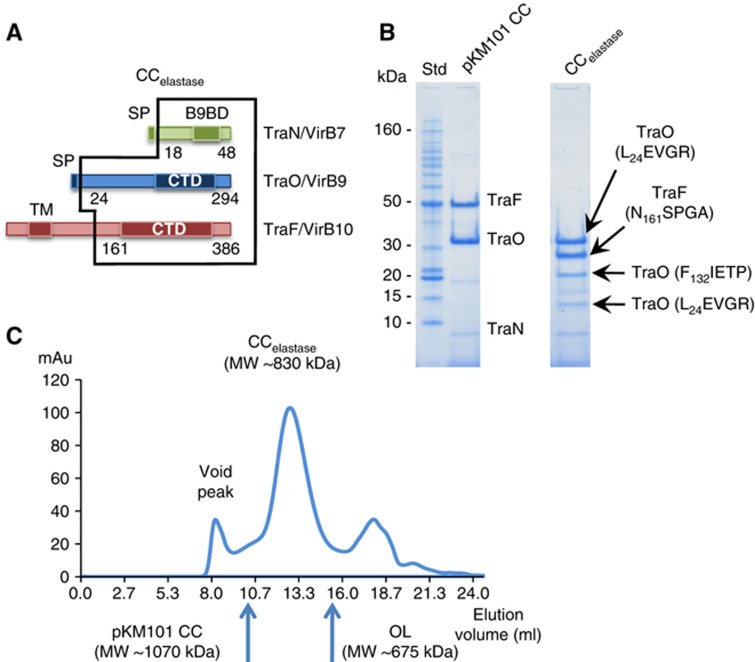
We have shown that the digestion of the full-length pKM101 CC (FLCC) (∼1070, kDa) yields stable complexes suitable for structural analysis (Chandran et al, 2009; Fronzes et al, 2009). TraF/VirB10NT contains a proline-rich region and most of this domain is predicted to be unstructured (Jakubowski et al, 2009). This domain is thus very susceptible to proteolysis. Digestion of the FLCC with elastase produced a stable truncated complex of around 880 kDa in molecular weight, termed the ‘CCelastase’ complex (Figure 1). The N-terminal sequencing of the two main bands in SDS-PAGE gels confirms that the complex is made of full-length TraO/VirB9 and the C-terminal domain of TraF/VirB10 starting at N161SPGA (the integrity of the C terminus of TraF/VirB10CT was confirmed by western blotting analysis of the Strep-tag present at the C terminus of the TraF/VirB10 construct). Therefore, the IL of this complex is only made of TraO/VirB9NT (Figure 1). The expected molecular weight of a 14-mer for this new truncated complex is ∼830 kDa, which is very close to the measured molecular weight indicated above.
Figure 1.
CCelastase purification. (A) Schematic representation of the regions of TraN/VirB7, TraO/VirB9 and TraF/VirB10 present in the CCelastase complex. Domains corresponding to the VirB9-binding domain (B9BD), the signal peptide (SP), the N-terminal TM helix and the C-terminal domains (CTD) are shown in darker colours. (B) SDS–PAGE of the CCelastase and the FLCC complexes. For the CCelastase complex, the first aa in the bands identified using Edman degradation are indicated. The CCelastase complex is composed of three major bands: full-length TraN/VirB7 and TraO/VirB9 and cleaved TraF/VirB10. In addition two minor byproducts are observed, both consisting of cleaved TraO/VirB9. Molecular weight standards (Std) are indicated. (C) Size exclusion chromatography of the CCelastase complex using a Superose 6 10/300 (GE Healthcare). The mobility of the pKM101 FLCC and the OL complex is also indicated by blue arrows in the size exclusion chromatography plot.
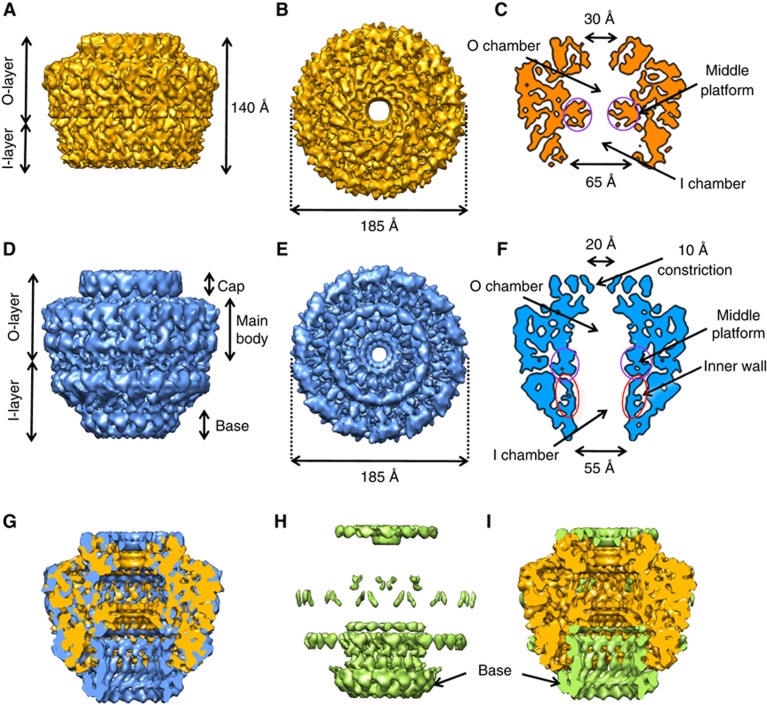
For structural analysis, samples of the CCelastase complex were vitrified and imaged using a FEG F20 microscope (FEI) (Supplementary Figure 1a and Supplementary Information). The structure of the CCelastase complex was solved at 8.5 Å resolution (Figure 2A, B and C, and Supplementary Figure 1, see Materials and methods). This complex is 185 Å in diameter and 140 Å in height. The characteristic separation into the OL and IL is clearly seen in the map. The OL and IL are connected by thin short linkers. The OL and IL contain chambers that are separated by large protrusions spanning inward from the main wall at the level of the linkers and forming a middle platform (indicated in purple circles in Figure 2C). These protrusions narrow the passage within the complex to ∼30 Å diameter. The O chamber is capped at the top by a ring of density with a small central opening of ∼20 Å in diameter while the IL chamber is widely open at the bottom with a diameter of ∼65 Å. Interestingly, the thickness of the chamber walls varies significantly. In the OL, the thickness of the wall is ∼50 Å, whereas in the IL it is ∼30 Å.
Figure 2.
Cryo-EM structures of the CCelastase and FLCC complexes. (A, B) Side view and top view, respectively, of the cryo-EM structure of the CCelastase complex. (C) Central section of the cryo-EM map shown in A and B. The density corresponding to the middle platform is indicated in purple circles. (D–E) Side view and top view, respectively, of the cryo-EM structure of the FLCC. (F) Central section of the FLCC cryo-EM map. The new density corresponding to the inner wall is outlined with red ellipses. (G) Cutaway view of the superposition of the cryo-EM maps of the FLCC (blue) and CCelastase (orange) complexes. (H) Difference map between the FLCC and CCelastase cryo-EM maps. (I) Cutaway view of the superposition of the difference map shown in H (green) and the cryo-EM structure of the CCelastase complex (orange). In H and I, arrows indicate the extra ring at the base of the FLCC (‘base’) not present in the CCelastase map.
Cryo-EM structure of the full-length pKM101 CC at 12.4 Å resolution
The previous structure of the FLCC was obtained at 15 Å resolution using cryo images (Fronzes et al, 2009). The resolution has been improved to 12.4 Å (Figure 2D, E and F; see Materials and methods and Supplementary Information). The overall dimensions of the refined structure are 185 Å in both height and diameter. The OL has an inner chamber that is open to the extracellular media through an opening of ∼20 Å in the cap (Figure 2F). Interestingly, the refined FLCC structure reveals new features that were not apparent in the previous one (Figure 2D, E and F). This structure shows the presence of an inner cylindrical wall of ∼50–55 Å in diameter formed by 14 columns of density separated by ∼7 Å (indicated in red ellipses in Figure 2F). This wall is separated from the outer wall by ∼7 Å. Each column has a diameter of ∼8 Å and is ∼70 Å in length. The top of each column joins the OL. The lower parts of these densities are connected to the base of the complex defining an opening of ∼55 Å in diameter on the cytoplasmic side of the complex.
Comparison of the full-length and the elastase-digested complexes
To investigate the differences between the higher-resolution FLCC structure and the structure of the CCelastase complex, we first low-pass filtered the CCelastase map to a resolution of 12.4 Å corresponding to the resolution of the FLCC. The two maps were then superimposed and a difference map was calculated (Figure 2G, H and I, Supplementary Figure 2).
This map reveals that the OL in the FLCC and CCelastase complexes are almost identical. They are both formed by a cap and the main body of the complex; the connection to the extracellular milieu takes place through an opening of ∼20 Å in diameter (Figure 2). Therefore, we propose that the 8.5 Å resolution structure of the OL found in the CCelastase complex reflects the native structure of the OL in the entire CC.
The two structures display significant differences in the IL. In the CCelastase complex, the opening on the cytoplasmic side has an increased diameter of 65 Å compared to ∼55 Å in the FLCC (Figure 2G and compare Figure 2C and F). The inner wall in the IL of the FLCC is absent in the CCelastase structure (Figure 2H and I). The FLCC inner wall consists of columns that run from the inner part of the OL to the FLCC bottom rim located on the cytoplasmic side of the complex that narrows the exit of the IL chamber (indicated as ‘Base’ in Figure 2H and I).
Modelling of the atomic structures of TraO and TraF N-termini
Since the structures of TraO/VirB9NT and TraF/VirB10NT are unknown, we used several structure-prediction packages (PSIPRED, Phyre2, HHpred and I-TASSER) to analyse the distribution of secondary structural elements within TraO/VirB9NT (residues 24–159) and TraF/VirB10NT (residues 1–160) and their possible folds (Jones, 1999; Soding, 2005; Zhang, 2008; Kelley and Sternberg, 2009; Roy et al, 2010). TraO/VirB9 has an approximate length of 300 amino acids (aa), and the structure of the C-terminal fragment containing the last 164 residues has been solved by X-ray crystallography (Chandran et al, 2009). The region corresponding to the first 23 aa of TraO/VirB9 is a signal peptide (Petersen et al, 2011): this part of the protein is removed during the translocation of TraO/VirB9 across the IM and it is therefore not present in the assembled CC. In silico analysis of the sequence of TraO/VirB9 between residues 24 and 159 reveals two regions, one comprising residues 136 and 159 where no secondary structures could be reliably predicted and another, between residues 24 and 135, which contains mainly β-strands and is likely to fold into a single domain. HHpred and PSIPRED identifies β-strands in this section of TraO/VirB9NT with high score and consistency. Phyre2 and I-TASSER predict a very compact organisation for this domain consisting of β-strands with a confidence score of ∼70%. (Supplementary Figure 3a and Supplementary Figure 4). In addition, I-TASSER suggests several slightly different folds that vary in the position and angles between the β-strands. The top five models were fitted as rigid bodies into the IL of the CCelastase complex (see below).
The sequence of TraF/VirB10NT (residues 1–160) was analysed using the same modelling tools (Jones, 1999; Soding, 2005; Zhang, 2008; Kelley and Sternberg, 2009; Roy et al, 2010). Since the EM structure of FLCC demonstrated the presence of inner columns, it was interesting to understand which part of the TraF/VirB10NT could correspond to these column-like features. The major part of TraF/VirB10NT was shown to be unstructured and the modelling of the complete N-terminal domain was not reliable (Supplementary Figure 3b). Thus, we have focused only on those secondary structural elements that were reliably identified by several algorithms. Three short helical peptides were found: one α-helix corresponding to the inner-membrane spanning region (αA: K40AF-LVF53) and two additional α-helical regions (αB: A107RA-QAA113; αC: P138EE-QRR146) (Supplementary Figure 3b). Their length is compatible with the diameter and the length of the observed columns in the FLCC cryo-EM map. Pseudo atomic models for αA, αB and αC were modelled by I-TASSER (Zhang, 2008; Roy et al, 2010) and used in the flexible fitting analysis.
Fitting of the OL crystal structure in the CCelastase and FLCC cryo-EM maps
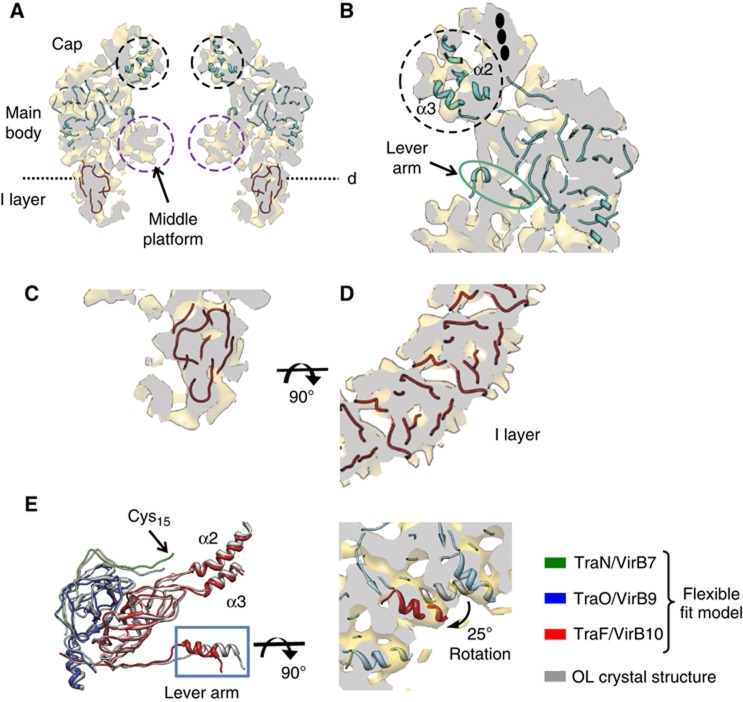
The position of proteins within the OL of both structures were localised by the flexible fitting of the OL crystal structure that contains TraO/VirB9CT, TraF/VirB10CT and TraN/VirB7 (Figure 3A and B; Supplementary Figure 5, PDB code 3JQO). The subnanometre resolution of the CCelastase map allowed us to identify densities corresponding to these three proteins within the OL. The correlation coefficient of the flexible fitting of the OL crystal structure (3JQO) into the FLCC and CCelastase complex maps is 0.63 and 0.71, respectively. Assessment of the RMSD of these fits was ∼1.21 Å indicating that at the level of resolution of 8.5 Å, these fits are nearly identical to the original OL crystal structure. Minor changes were observed in the lever arm area of the EM structure (Figure 3E).
Figure 3.
Modelling and fitting of atomic data in the cryo-EM map of the CCelastase complex. (A) Central vertical slice showing the fitting of the OL atomic structure and the atomic model obtained for TraO/VirB9NT. The densities corresponding to the middle platform and two α-helices α2 and α3 of TraF/VirB10CT are indicated with purple and black dashed circles, respectively. The dashed line corresponds to the equivalent cutting plane perpendicular to the vertical axis of the complex shown in panel D. (B) Detailed view of the crystal structure fitting in the OL. In the cap, there is clear extra density not accounted for by helices α2 and α3 (in a black dashed circle). The plausible location of the lipidic moiety covalently bound to TraN/VirB7 is indicated with black ovals. (C, D) Fitting of the model obtained for TraO/VirB9NT in the IL. In panel D, the cutting plane shown is in a perpendicular direction to the vertical axis of the CCelastase cryo-EM map shown in panel A. (E) Superposition of one subunit of the original atomic structure (grey) of the OL, and the corresponding subunit of the atomic model after flexible fitting (TraN/VirB7, TraO/VirB9 and TraF/VirB10 are shown in green, blue and red, respectively). The RMSD between the two models is 1.21 Å. On the right, the N-terminal α-helix present in the lever arm (red) is shown fitted in the equivalent density of the CCelastase cryo-EM map. The symmetry-related subunits are shown in blue. The same helix of the original atomic structure of the OL is shown in grey.
Under the lever arm area, in both complexes there is a substantial area of electron density called the middle platform, which is not accounted for by fitting of the OL crystal structure (purple circles in Figure 3A). Although there are an extra nine aa present at the N terminus of TraF/VirB10CT in the CCelastase complex compared to the OL crystal structure, the size of this peptide is not sufficient to account for the entire middle platform density. On the other hand, the middle platform could be formed by the region of the TraO/VirB9NT sequence between residues 136 and 159. This region could possibly act as a flexible linker connecting the TraO/VirB9 domains of the OL and IL and be involved in the interaction with TraF/VirB10 and a substrate within the central channel. We observe that the conformation of the middle platform is different in both structures, being more pronounced in the CCelastase complex than in the FLCC where it is more tubular. Clearly, removing the N-terminal domain of TraF/VirB10 leads to a conformational change in that region, the molecular basis of which remains unclear, given the current resolution of the maps.
In the region of the cap, the CCelastase map reveals densities for α-helices α2 and α3 of TraF/VirB10CT (Figure 3A and B, black circles). Fourteen of those form the OL channel through the OM. There is additional density that cannot be ascribed to the helices but might be attributed to the lipid moiety of TraN/VirB7 and detergent molecules associated to it (see black ovals in Figure 3B and also Discussion below).
The IL organisation: fitting of TraO/VirB9NT and TraF/VirB10NT
Several models for TraO/VirB9NT (residues 24–135) obtained from I-TASSER that had slightly different folds varying in positions and angles of the β-strands were fitted in the IL of the CCelastase map. To select the most appropriate model, we checked the correlation between the models and the EM density. The fitting of the pseudo atomic model of this β-domain has to satisfy two important constraints: the location of its C terminus has to be in close proximity to the linker region and the OL, and the resulting fit should not create clashes upon 14-fold symmetrisation. This allowed us to make a consistent assessment of the best five models and the final model was refined using flexible fitting into the experimentally derived densities of the EM map. The final atomic model (Figure 3C and D, and Supplementary Figure 4) was fitted to the CCelastase cryo-EM map with a cross-correlation coefficient of 0.82. The C terminus of this model is located close to the N terminus of TraO/VirB9CT in the OL crystal structure. The results of the docking suggest that the β-strands in the IL would run at an angle of 28° away from the central axis of the secretion pore (Figure 3C and D).
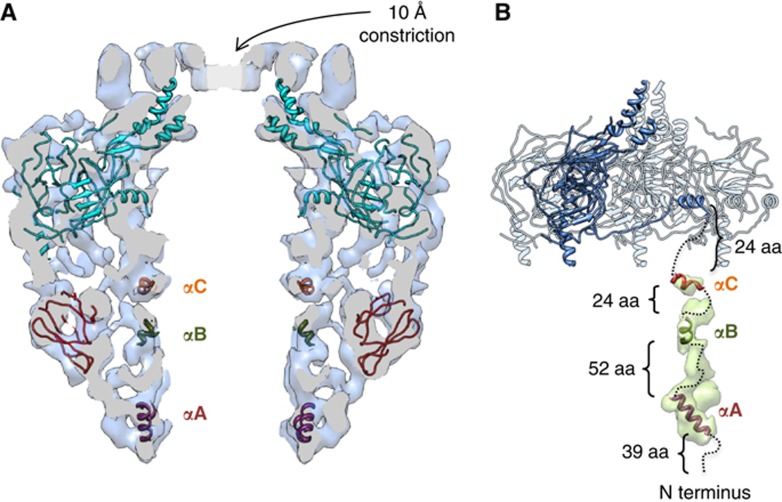
The superposition of the CCelastase and FLCC cryo-EM maps revealed extra densities in the IL of the FLCC (Figure 2G–I). Since TraF/VirB10NT is absent in the CCelastase structure, it is logical to assign the inner cylindrical wall of the FLCC IL to this domain of TraF/VirB10. Three α-helical regions in the N terminus of TraF/VirB10 were identified with high confidence (see above) with flexible linkers in between, and have been tentatively fitted in the column of densities that form the inner cylindrical wall in the IL of the FLCC (Figure 4). Helix αA, corresponding to the TM helix traversing the IM, was fitted at the base of the IL (Figure 4A and B). Detergents may also contribute to the density in the base of the IL. Helices αB and αC have been docked in stretches of density compatible with the size of these two helices, and helix αC has been located close to the N terminus of TraF/VirB10CT of the OL atomic structure (Figure 4B). Overall, the dimensions of the columns that form the inner wall in the IL are compatible with the sizes of the modelled helices. However, the remaining volume determined from the density at the 1σ threshold is insufficient to accommodate the linker polypeptide between the helices. It is likely that the linkers, which do not have a rigid organisation, were averaged out during the process of reconstruction.
Figure 4.
Modelling and fitting of atomic models in the cryo-EM map of the FLCC. (A) Central slice of the FLCC with fitted OL atomic structure and atomic models obtained for TraO/VirB9NT and the α-helices predicted in TraF/VirB10NT. (B) Schematic model for the organisation of TraF/VirB10 in the FLCC. Four TraN/VirB7+TraO/VirB9CT+TraF/VirB10CT subunits of the 14-mer present in the OL atomic structure are shown. One of the subunits is shown in dark blue. The tentative docking of the three TraF/VirB10NT α-helical regions modelled with high confidence together with a plausible connection with TraF/VirB10CT is shown in dashed lines. The density for one subunit in the difference map is shown in transparent green. This subunit is located immediately below the lever arm of TraF/VirB10CT shown in dark blue. The three α-helical regions of TraF/VirB10NT correspond to αA (purple, from K40 to F53), which transverses the IM, αB (green, from A107 to A113) and αC (red, from P138 to R146). The connections are shown in dashed lines together with the corresponding number of aa.
Localisation of TraO/VirB9NT in CC variants using domain-specific nanobodies
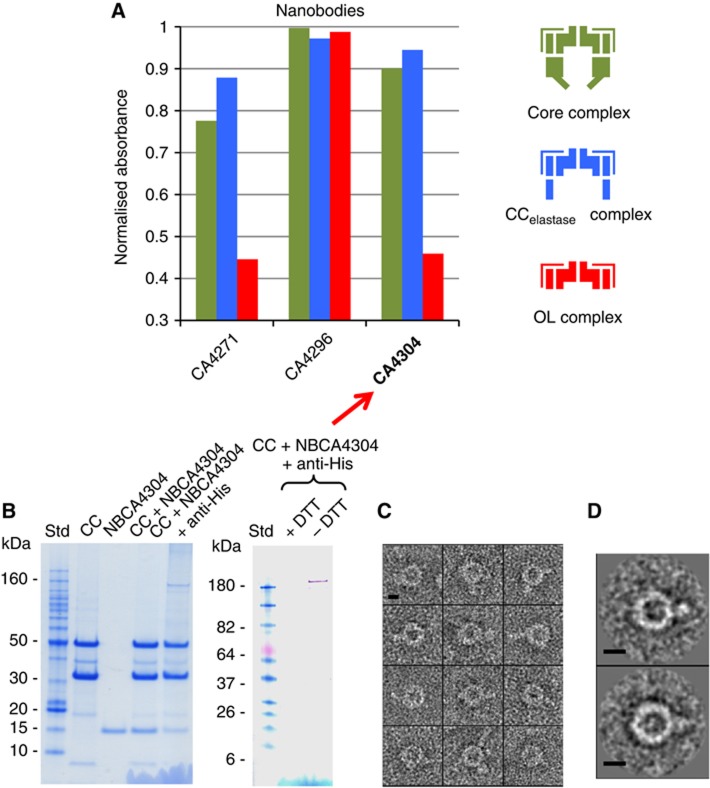
The cryo-EM map of the CCelastase complex and the fitting results for the atomic model of TraO/VirB9NT localise TraO/VirB9 at the periphery of the IL. To validate the domain localisation within the complex, we have employed another approach based on labelling experiments using nanobodies (NBs). ELISA experiments were carried out using the FLCC, the OL and CCelastase complexes. Among the 40 NBs that we tested, two NBs, NBCA4271 and NBCA4304, exhibited high specificity for TraO/VirB9NT, showing strong ELISA responses for CCelastase and FLCC and a low signal for the OL complex (Figure 5A; Supplementary Figure 6). Therefore, we expected that incubation of FLCC with NBCA4304 (or NBCA4271) would add an extra density that would be visible in the EM images, thereby allowing us to locate TraO/VirB9NT. In order to make the bound NBs more visible, we additionally labelled them using anti-His antibodies. Note that a NB/anti-His antibody complex would be too large to penetrate the CC through the 55 Å opening of the IL: thus labelling of TraO/VirB9NT might only occur if this domain is at the periphery of the IL.
Figure 5.
NB labelling of the FLCC, CCelastase and OL complexes. (A) Selection of NBs preferentially binding the CCelastase (see also Supplementary Figure 6). The diagram reports the comparative affinity of three NBs for the FLCC (green), the CCelastase (blue) and the OL (red) complexes. NBCA4271 and NBCA4304 target the IL while NBCA4296 binds to the OL, a region present in the three complexes used for the ELISA experiment. NBCA4304 was chosen for the labelling experiments. The right panel represents the schematic organisation of the complexes used in the ELISA experiments coloured correspondingly. (B) SDS–PAGE analysis and western blot of the labelled FLCC. The different lanes correspond to the FLCC, the NBCA4304, the purified complex containing the FLCC and the NB (FLCC+NBCA4304), and the final purified complex containing the FLCC, the NB and the anti-His antibody (CC+NBCA4304+anti-His). This final complex was used to prepare negative stain grids. The right panel shows a western blot of the FLCC+NBCA4304+anti-His fraction in the presence or absence (+/− DTT) of reducing agent. The western blot was performed using only the secondary antibody HRP-conjugated. Molecular weight standards (Std) are indicated in both cases. (C) Gallery of negatively stained single particles with a clear extra density emerging from the CC. (D) Representative top view class averages of the FLCC+NBCA4304+anti-His sample. The scale bar corresponds to 10 nm.
Purified FLCC was incubated with NBCA4304 in a molar ratio of 1:7 FLCC:NB. The fractions containing NBs bound to the FLCC were pooled together, concentrated and further incubated with anti-His antibody targeted against the His-tag of the NB in a molar ratio anti-His:FLCC:NB of 5:1 (Figure 5B). Fractions containing FLCC:NB:anti-His complex were used to prepare negatively stained EM grids (Figure 5C and D, see Supplementary Information). Extra density was clearly seen on the end views of single complexes (Figure 5C). Tail-like density was protruding outward from the periphery of the CC. Figure 5D shows averaged images where extra density corresponding to the size of the antibody emerges radially from the outer part of the CC. This result adds to evidence that TraO/VirB9NT is located on the outer part of the IL.
Discussion
The CC is an essential structural feature of T4S systems. It is self-assembling and therefore is likely to be assembled first. It forms part of the IM channel and is a major component of the OM pore. It provides the central scaffold for assembly of all other components. Understanding its structure is thus crucial to the elucidation of T4S system transport mechanism. This report fills a major gap in our understanding of the CC structure: indeed, while the OL organisation and structure had been revealed previously by its X-ray crystal structure, very little was known about the organisation and structure of the IL.
In this report, we have re-analysed the FLCC at higher resolution and compared this more detailed structure to that of another complex where a part of the IL, TraF/VirB10NT, was selectively removed. Using immuno-labelling to selectively target the remaining component of the IL, TraO/VirB9NT, we unambiguously located and identified this part of the IL. Finally, our analysis was confirmed by fitting of predicted atomic structures into cryo-EM density maps. Given the high reliability of modern three-dimensional structure-prediction programmes, fitting of predictive atomic models (the only option in the absence of experimentally derived models) provides an important validation tool for the interpretation of cryo-EM maps. Conversely, the quality of the fit provides an important evaluation of the predicted models. With both issues, we were able to generate reliable models for most of the IL parts.
The difference map between the cryo-EM maps of the FLCC and the CCelastase complex (Figure 2H and I) clearly shows extra mass in the FLCC corresponding to the TraF/VirB10NT domain in the IL. This density is made up of 14 clearly defined columns that form the inner lining of the IL (Figure 2H and I). At the very bottom of the complex, these stretches join each other to form an extra ring at the base of the FLCC (indicated as ‘Base’ in Figure 2H and I). This ring might correspond to the 14 TM segments (helix αA) associated with detergent micelles. Fitting of the predicted helical regions (helices αA–C) of TraF/VirB10NT does not account for all the electron density in the observed columns. The unassigned electron density might be accounted for by the flexible linkers between the helices (Figure 4A and B).
Assessment of differences between the FLCC and the CCelastase structures, fitting of an atomic model for TraO/VirB9NT and labelling experiments with NB targeted against TraO/VirB9NT are all pointing towards an unambiguous conclusion that the TraO/VirB9 N-terminal part is located at the periphery of the IL. A recent EM structure of the pKM101 CC bound to the VirB4 ATPase also confirmed the outer location of TraO/VirB9 in the I-layer (Wallden et al, 2012). In this work, VirB4 is observed bound to the side of the CC, and the FLCC–VirB4 interaction is not disrupted in versions of the CC where TraF/VirB10 has been progressively truncated from the N-terminal end (Wallden et al, 2012).
Using a TrIP assay, Cascales and Christie (2004b) have demonstrated that VirB9 interacts with the T-DNA during the translocation of the substrate from the IM in A. tumefaciens. Since VirB9 traverses the FLCC from top to bottom except in the IM part of the complex, this would suggest that the T-DNA might access the CC from the outside. On the other hand, it could be that some of the non-assigned electron density such as the middle platform (Figure 3A) might also be formed by residues in VirB9 forming a structure that protrudes inwardly inside the CC. A part of the TraO/VirB9 sequence between the N-terminal and C-terminal domains was not assigned (residues P136WA to KAK159), but could account for this inwardly protruding region and thus contact DNA during transfer.
The structures of FLCC, CCelastase and OL demonstrate clearly that TraO/VirB9 forms the outer sheath of the CC while TraF/VirB10 is lining the inner surface of both chambers in the CC, and spans the whole length of the CC eventually forming the bottom ring of the IL. This location of TraF/VirB10 is in a full agreement with previous findings that suggest that TraF/VirB10 serves as a signal transmitter throughout the entire complex. By interacting not only with most of the system’s inner (VirB6 and VirB8) and outer (VirB9 and VirB7) membrane components, but also with the ATPases powering the system (VirB4, VirB11 and VirD4) (Llosa et al, 2003; Atmakuri et al, 2004; Alvarez-Martinez and Christie, 2009; Jakubowski et al, 2009), VirB10 acts as a cornerstone of the T4S system architecture. Its interactions with ATPases (Cascales and Christie, 2004a) are known to propagate ATP-driven conformational changes throughout the T4S system (Postle and Larsen, 2007; Jakubowski et al, 2009). The structural basis of these VirB10-mediated relay mechanisms can now be inferred from the structure presented here. Indeed, by assigning a continuous and extended density to TraF/VirB10NT within the IL, we can now map out the entire TraF/VirB10 structure and thus identify an uninterrupted path by which conformational changes would need to be relayed to reach out to the OM components.
Materials and methods
Expression and purification of the pKM101 CC
The CC from IBA3c:traN-traO-traF/C-ST plasmid was purified as described previously (Fronzes et al, 2009).
Elastase digestion of the pKM101 CC
The pKM101 CC eluted from the StrepTrap HP column (GE Healthcare) was concentrated down to 500 μl at a final concentration between 5 and 10 mg/ml depending on the preparation. Ten microlitres of elastase (Sigma) at 1 mg/ml in 50 mM Tris-HCl pH 8.5, 200 mM NaCl were added to the 500 μl and the mix was incubated for 40 min at room temperature (RT). After the incubation, the solution was gel filtered as described previously (Fronzes et al, 2009). The N-terminal sequence of the main bands was determined using Edman degradation.
Induction of a humoral immune response in llama and NB identification
Llama immunisation and NB repertoire cloning and characterisation were as described in Supplementary Information. Periplasmic extracts containing different NBs were produced and purified from E. coli WK6 su− strain following standard procedures in order to screen the epitope of each NB (Rasmussen et al, 2011).
To perform the ELISA experiments presented in Figure 5 and Supplementary Figure 6, ELISA plates (flat-bottom Maxisorp 96-well plates, Nunc) were coated either with FLCC, OL or CCelastase complexes diluted to 5 μg/ml in sodium bicarbonate buffer pH 8.2 at 4°C overnight. The binding step was followed by washing (five times) with 1 × PBS containing 0.1% Tween-20 (PBST) at RT. Remaining binding sites were blocked with 2% skimmed milk powder in 1 × PBS for 2 h at RT followed by a washing step in PBST. The periplasmic extracts of each of the forty NBs were then applied diluted 1:5 in 1 × PBS for 1 h at RT followed by a washing step in PBST. To detect if the NBs were bound, the ELISA plates were incubated with a mouse monoclonal anti-His antibody coupled to HRP (Invitrogen) diluted 1:5000 in 1 × PBS containing 0.1% milk powder. The binding reaction was for 1 h at RT followed by a washing step. Then a peroxidase substrate (TMB, Sigma) was added and left incubating for 30 min at RT. The reaction was stopped adding 1 M H2SO4 and the absorption was measured at 450 nm. The ELISA screening was performed three times with duplicates for each NB in every experiment, and appropriate controls were included in each ELISA plate.
NBCA4304 purification for electron microscopy-labelling experiments
NBCA4304 was purified as described in Rasmussen et al (2011). The purity of the sample was assessed by SDS–PAGE and then concentrated using a centrifugal filter of 3 kDa cutoff (Millipore). Glycerol was added at a final concentration of 20% and the sample was frozen for latter use.
Labelling of the N terminus of TraO using NBs
The pKM101 CC was incubated with purified NBCA4304. The mix was then loaded onto Superose 6 10/300 (GE Healthcare) and the fractions corresponding to the peak containing the CC and NBCA4304 were incubated with anti-His antibody (GE Healthcare). A new top band that corresponds to the IgG bound to the His-tag of NBCA4304 in the CC:NB complex was apparent on the SDS–PAGE (Figure 5B). The fraction corresponding to the purified complex made of CC, NBCA4304 and anti-His antibody was used to prepare negative stain grids (see Supplementary Information).
Electron microscopy and image analysis of the FLCC and CCelastase complexes
Samples were vitrified and visualised on a FEI Tecnai F20 FEG microscope. Image frames were recorded on a 4k × 4k Gatan CCD camera under low-dose conditions. The pixel size at the specimen level was 2.22 for the FLCC and 2.08 for the CCelastase complex (Supplementary Information). The defocus on images was assessed using CTFFIND3 (Mindell and Grigorieff, 2003), and CTF correction was done by phase flipping using Bshow1.6 (Heymann, 2001; Heymann and Belnap, 2007). Corrected images were centred and subjected to multivariate statistical analysis (MSA). Alignment was improved by iterative procedure of multi-reference alignment (MRA) and MSA. Reconstructions for both CCelastase and FLCC samples were obtained independently (van Heel et al, 2000). The final 3D map of the CCelastase and FLCC complexes were calculated from 5430 particles and 3805 particles, respectively. The resolution was estimated by Fourier Shell Correlation (FSC) as 12.4 and 8.5 Å for the FLCC and CCelastase structures, according to the 0.5 criterion level (Supplementary Figure 1c; Supplementary Information). To compare the two structures, the CCelastase structure was low-pass filtered to a resolution of 12.4 Å. The two structures were aligned and normalised to the same sigma, then the difference map was calculated between the FLCC and the CCelastase complexes (see Supplementary Information).
Modelling of domain structures for the FLCC and CCelastase complexes
The sequences of TraO/VirB9NT (Q46704 UniProtKB) and TraF/VirB10NT (Q46705 UniProtKB) were analysed using the structure prediction servers Phyre2, HHpred and I-TASSER (Jones, 1999; Soding, 2005; Zhang, 2008; Kelley and Sternberg, 2009; Roy et al, 2010). Models for the first 160 aa of TraF/VirB10 were generated using I-TASSER (Zhang, 2008; Roy et al, 2010). This region is predicted to be mostly disordered; however, three areas have helical organisation predicted with high confidence by all servers used: helix αA (aa K40AF–LVF53), helix αB (aa A107RA–AA113) and helix αC (aa P138EE–QRR146).
Fitting of atomic data into the cryo-EM maps of the FLCC and CCelastase complexes
The initial fitting was done manually in Chimera (Pettersen et al, 2004). The crystal structure of the OL complex (PDB 3JQO (Chandran et al, 2009)) was fitted in the OL of the CCelastase cryo-EM map using a rigid body approach and then refined using the flexible fitting software Sculptor (Birmanns et al, 2011) and Flex-EM (Topf et al, 2008; Pandurangan and Topf, 2012). The N-terminal lever arm of TraF/VirB10 (Chandran et al, 2009) was manually adjusted to optimise the fitting in the corresponding density of the cryo-EM map (Supplementary Information).
The model of the N terminus (L24EVG to FIET135) of TraO/VirB9 was fitted first manually and then it was refined using the Flex-EM software (Topf et al, 2008; Pandurangan and Topf, 2012). The models generated for helix αA, helix αB and helix αC located in TraF/VirB10NT were fitted manually in one monomer of the difference map and then symmetrised in Chimera (Pettersen et al, 2004). Supplementary Table I shows the cross-correlation coefficients for the different fittings.
Access codes
The EM maps of the FLCC and CCelastase complexes are deposited in the EMD data bank (http://www.ebi.ac.uk/pdbe/) under the accession codes EMD-2232 and EMD-2233.
Supplementary Material
Acknowledgments
This work was funded by grant 082227 from the Wellcome Trust to GW. Rémi Fronzes is supported by the Centre National de la Recherche Scientifique and Institut Pasteur. We would also like to thank the Wellcome Trust for the EM equipment, Drs D Houldershaw and R Westlake for computer support, and Drs M Topf and AP Pandurangan for their advice during the flexible fitting analysis.
Author contributions: AR-C, RF, EVO and GW designed the research; AR-C purified and characterised the CCelastase complex; AR-C and CGS collected data for the CCelastase complex; AR-C and EVO solved the structure of the CCelastase complex; RF and EVO solved the structure of the FLCC; AR-C and EVO performed the modelling of atomic structures and fitted them into EM maps; AR-C, VC and PWL were involved in the NB screening; AR-C performed the biochemistry and the EM of the labelling of the CCelastase complex using NBs; TL, EP, JS and HR were involved in NBs production and cloning; and AR-C, RF, EVO and GW wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alvarez-Martinez CE, Christie PJ (2009) Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev 73: 775–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmakuri K, Cascales E, Christie PJ (2004) Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol Microbiol 54: 1199–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmanns S, Rusu M, Wriggers W (2011) Using Sculptor and Situs for simultaneous assembly of atomic components into low-resolution shapes. J Struct Biol 173: 428–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Christie PJ (2004a) Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc Natl Acad Sci USA 101: 17228–17233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Christie PJ (2004b) Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304: 1170–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran V, Fronzes R, Duquerroy S, Cronin N, Navaza J, Waksman G (2009) Structure of the outer membrane complex of a type IV secretion system. Nature 462: 1011–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreiseikelmann B (1994) Translocation of DNA across bacterial membranes. Microbiol Rev 58: 293–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fronzes R, Schafer E, Wang L, Saibil HR, Orlova EV, Waksman G (2009) Structure of a type IV secretion system core complex. Science 323: 266–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis-Ruth FX, Moncalian G, Perez-Luque R, Gonzalez A, Cabezon E, de la Cruz F, Coll M (2001) The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409: 637–641 [DOI] [PubMed] [Google Scholar]
- Hare S, Bayliss R, Baron C, Waksman G (2006) A large domain swap in the VirB11 ATPase of Brucella suis leaves the hexameric assembly intact. J Mol Biol 360: 56–66 [DOI] [PubMed] [Google Scholar]
- Heymann JB (2001) Bsoft: image and molecular processing in electron microscopy. J Struct Biol 133: 156–169 [DOI] [PubMed] [Google Scholar]
- Heymann JB, Belnap DM (2007) Bsoft: image processing and molecular modeling for electron microscopy. J Struct Biol 157: 3–18 [DOI] [PubMed] [Google Scholar]
- Jakubowski SJ, Kerr JE, Garza I, Krishnamoorthy V, Bayliss R, Waksman G, Christie PJ (2009) Agrobacterium VirB10 domain requirements for type IV secretion and T pilus biogenesis. Mol Microbiol 71: 779–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT (1999) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292: 195–202 [DOI] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4: 363–371 [DOI] [PubMed] [Google Scholar]
- Lessl M, Lanka E (1994) Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell 77: 321–324 [DOI] [PubMed] [Google Scholar]
- Llosa M, Zunzunegui S, de la Cruz F (2003) Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc Natl Acad Sci USA 100: 10465–10470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullen CA, Binns AN (2006) Agrobacterium tumefaciens and plant cell interactions and activities required for interkingdom macromolecular transfer. Annu Rev Cell Dev Biol 22: 101–127 [DOI] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N (2003) Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142: 334–347 [DOI] [PubMed] [Google Scholar]
- Pandurangan AP, Topf M (2012) Finding rigid bodies in protein structures: application to flexible fitting into cryoEM maps. J Struct Biol 177: 520–531 [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Postle K, Larsen RA (2007) TonB-dependent energy transduction between outer and cytoplasmic membranes. Biometals 20: 453–465 [DOI] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, Devree BT, Rosenbaum DM, Thian FS, Kobilka TS, Schnapp A, Konetzki I, Sunahara RK, Gellman H, Pautsch A, Steyaert J, Weis WI, Kobilka BK (2011) Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature 469: 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5: 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savvides SN, Yeo HJ, Beck MR, Blaesing F, Lurz R, Lanka E, Buhrdorf R, Fischer W, Haas R, Waksman G (2003) VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J 22: 1969–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J (2005) Protein homology detection by HMM-HMM comparison. Bioinformatics 21: 951–960 [DOI] [PubMed] [Google Scholar]
- Terradot L, Waksman G (2011) Architecture of the Helicobacter pylori Cag-type IV secretion system. FEBS J 278: 1213–1222 [DOI] [PubMed] [Google Scholar]
- Topf M, Lasker K, Webb B, Wolfson H, Chiu W, Sali A (2008) Protein structure fitting and refinement guided by cryo-EM density. Structure 16: 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M, Gowen B, Matadeen R, Orlova EV, Finn R, Pape T, Cohen D, Stark H, Schmidt R, Schatz M, Patwardhan A (2000) Single-particle electron cryo-microscopy: towards atomic resolution. Q Rev Biophys 33: 307–369 [DOI] [PubMed] [Google Scholar]
- Waksman G, Fronzes R (2010) Molecular architecture of bacterial type IV secretion systems. Trends Biochem Sci 35: 691–698 [DOI] [PubMed] [Google Scholar]
- Wallden K, Rivera-Calzada A, Waksman G (2010) Type IV secretion systems: versatility and diversity in function. Cell Microbiol 12: 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallden K, Williams R, Yan J, Lian PW, Wang L, Thalassinos K, Orlova EV, Waksman G (2012) Structure of the VirB4 ATPase, alone and bound to the core complex of a type IV secretion system. Proc Natl Acad Sci USA 109: 11348–11353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.