Abstract
Mammalian target-of-rapamycin (mTOR) triggers S6 kinase (S6K) activation to phosphorylate targets linked to translation in response to energy, nutrients, and hormones. Pathways of TOR activation in plants remain unknown. Here, we uncover the role of the phytohormone auxin in TOR signalling activation and reinitiation after upstream open reading frame (uORF) translation, which in plants is dependent on translation initiation factor eIF3h. We show that auxin triggers TOR activation followed by S6K1 phosphorylation at T449 and efficient loading of uORF-mRNAs onto polysomes in a manner sensitive to the TOR inhibitor Torin-1. Torin-1 mediates recruitment of inactive S6K1 to polysomes, while auxin triggers S6K1 dissociation and recruitment of activated TOR instead. A putative target of TOR/S6K1—eIF3h—is phosphorylated and detected in polysomes in response to auxin. In TOR-deficient plants, polysomes were prebound by inactive S6K1, and loading of uORF-mRNAs and eIF3h was impaired. Transient expression of eIF3h-S178D in plant protoplasts specifically upregulates uORF-mRNA translation. We propose that TOR functions in polysomes to maintain the active S6K1 (and thus eIF3h) phosphorylation status that is critical for translation reinitiation.
Keywords: eIF3 subunit h phosphorylation, gravitropic response, phytohormone auxin, polyribosomes, TOR
Introduction
Eukaryotic cells respond to changing environments by utilizing various signal transduction pathways. Regulation can occur at the transcriptional level as well as post-transcriptionally, for example, via regulation of translation of specific messages (Nilsson et al, 2004). Regulating translation via upstream open reading frames (uORFs) in the 5′-untranslated region (5′-UTR) of mRNA is now recognised as a means of controlling potent proteins such as growth factors, protein kinases, and transcription factors (Morris and Geballe, 2000). uORFs can reduce protein expression typically by 30–80%, but have only a modest impact on mRNA levels (Calvo et al, 2009). uORFs are especially common in Arabidopsis, being present in at least 30% of full-length mRNAs (Zhou et al, 2010).
Short uORF length—more importantly, the short time required for translation—and a sufficient intercistronic distance between two consecutive ORFs are the main parameters boosting the reinitiation capacity of ribosomes (Luukkonen et al, 1995; Kozak, 2001; Hinnebusch, 2005). Reinitiation potential is also regulated by specific RNA cis-acting elements and trans-acting factors (Sachs and Geballe, 2006; Rahmani et al, 2009; Medenbach et al, 2011).
The need for rapid uORF translation may be related to problems with de novo recruitment of initiator tRNA (tRNAiMet) in the ternary complex (TC; eIF2xGTPxMet-tRNAiMet), and the 60S ribosomal subunit (60S) required for the reinitiation event. Translation initiation factors (eIFs) needed for resumption of scanning and/or recruitment of TC and 60S were proposed to remain loosely associated with the elongating ribosome for a short time of a few cycles, and, after termination, to support a subsequent initiation event (Kozak, 2001; Pöyry et al, 2004), thus explaining why reinitiation is precluded after a long elongation event. Reinitiation-promoting factors (RPFs) include eukaryotic initiation factor 3 (eIF3) and eIF4F (Pöyry et al, 2004; Cuchalová et al, 2010; Roy et al, 2010; Munzarová et al, 2011). eIF3 is composed of 13 distinct subunits in humans and plants, and orchestrates assembly of the 43S pre-initiation complex (43S PIC) on mRNA (Browning et al, 2001; Hinnebusch, 2006).
In plants, eIF3 non-core subunit h (eIF3h) and the 60S protein RPL24 increase the reinitiation competence of uORF-containing mRNAs (uORF-RNAs) encoding two families of transcriptional factors—auxin response factors (ARFs) and basic zipper transcription factors (bZIPs)—via as yet unknown mechanisms (Kim et al, 2004; Nishimura et al, 2005; Zhou et al, 2010). Critically, translation reinitiation and auxin-mediated organogenesis are compromised severely by mutations in either eIF3h or RPL24. eIF3 is recruited to promote a special case of reinitiation after long ORF translation by the Cauliflower mosaic virus protein translational transactivator/viroplasmin (TAV; Park et al, 2001). TAV accomplishes reinitiation via retention in polyribosomes (polysomes) of eIF3 and a novel reinitiation supporting protein (RISP) during the long elongation event, and reuse of these factors for reinitiation (Park et al, 2001; Thiébeauld et al, 2009). More recent evidence has linked TAV to activation of target-of-rapamycin (TOR) in plants (Schepetilnikov et al, 2011). When activated by TAV, TOR associates with polysomes, ensuring the S6 kinase 1 (S6K1) and RISP phosphorylation that is essential for virus-activated reinitiation.
TOR—a critical sensor of nutritional and cellular energy and a regulator of cell growth (Gingras et al, 2001; Sengupta et al, 2010; Dobrenel et al, 2011)—is a large serine/threonine protein kinase. Mammalian TOR (mTOR) modulates the activity of two main substrate classes: 4E-binding proteins (4E-BPs) and protein kinases of RPS6 (S6Ks; Ma and Blenis, 2009). Recently, 4E-BPs were implicated in mTOR-dependent translation initiation control of mRNAs with 5′ terminal oligopyrimidine (TOP) motifs within the leader region (Thoreen et al, 2012). eIF3 works as a scaffold for mTOR and S6K1 binding (Holz et al, 2005). When inactive, S6K1, but not TOR, binds the non-polysome-associated eIF3 complex. Upon activation, TOR associates with eIF3 and phosphorylates S6K1, triggering its dissociation from eIF3 and further activation.
The Arabidopsis genome contains a single TOR gene (Menand et al, 2002), two RAPTOR genes (Menand et al, 2002; Mahfouz et al, 2006) and LST8 genes (Moreau et al, 2012) that encode components of the TOR complex. A TOR knockout mutant in Arabidopsis is embryo lethal, and altered TOR expression affects plant growth (Menand et al, 2002; Deprost et al, 2007). The Arabidopsis genome encodes two S6K1 homologues, S6K1 and S6K2; S6K1 is phosphorylated by TOR at hydrophobic motif residue T449 (Zhang et al, 1994; Schepetilnikov et al, 2011), while 4E-PBs remain illusive in plants.
An important question is what are the upstream effectors that trigger TOR activation in plants? Our data show that the phytohormone auxin can trigger TOR signalling pathway activation, thus providing a tool to study the role of TOR in plant translation control. Here, we uncover the role of the TOR signalling pathway in promoting reinitiation after uORF translation in Arabidopsis. Our data show that TOR and S6K1 contribute to assembly of reinitiation-competent ribosomes in response to auxin. We suggest that TOR functions in reinitiation via maintenance of eIF3h phosphorylation status in polysomes.
Results
Interaction of eIF3 with TOR or S6K1 is regulated by phytohormone auxin and TOR inhibitor, Torin-1
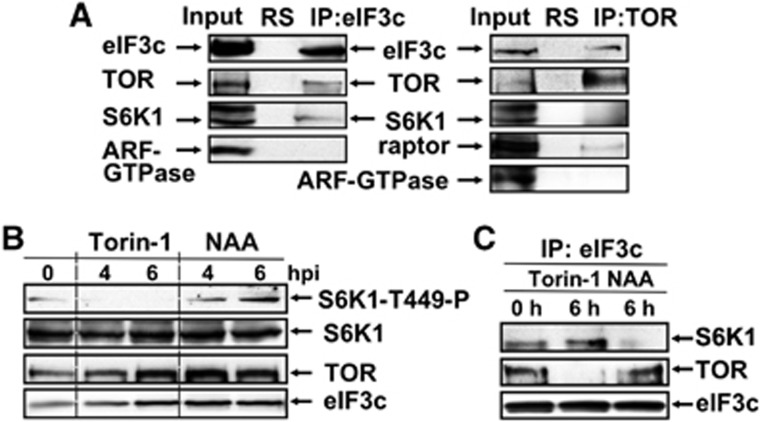
First, we asked if TOR and S6K1 form part of eIF3-containing complexes in Arabidopsis. Both TOR and S6K1 associate with eIF3-bound complexes immunoprecipitated with anti-eIF3c antibodies from Arabidopsis suspension cultures (Figure 1A). TOR immunoprecipitates contain an eIF3c marker, but not S6K1, suggesting that the TOR/eIF3 immunoprecipitation complex does not contain significant amounts of S6K1. Raptor—an important partner of TOR—was also found within TOR immunoprecipitates. In control experiments, the above proteins were not detected in non-immune RS complexes, and the control protein—ADP-ribosylation factor-GTPase (ARF GTPase)—did not co-immunoprecipitate with either TOR or eIF3c.
Figure 1.
Auxin-induced phosphorylation of AtS6K1 at TOR-specific residue T449 regulates interaction with eIF3. (A) eIF3c or TOR immunoprecipitated from WT extracts was assayed for complex formation between eIF3c, TOR, S6K1, raptor, and ARF-GTPase. Input, 5% of immunoprecipitation (IP) or normal rabbit serum (RS). (B) Suspension-culture cells treated with NAA or Torin-1 for 0, 4, and 6 h, lysed assayed by immunoblot analysis. (C) eIF3c immunoprecipitated from extracts in (B) and assayed for association with TOR and S6K1 by immunoblotting. Source data for this figure is available on the online supplementary information page.
Then, we examined the phytohormone auxin as an upstream TOR effector. To verify auxin activation of TOR, we used Torin-1, which inhibits TOR phosphorylation activity (Thoreen et al, 2009; Schepetilnikov et al, 2011). Torin-1 abolished phosphorylation of recombinant AtS6K1 at TOR-specific residue T449 in Arabidopsis extract, and triggered its rapid dephosphorylation (Supplementary Figure S1A). Next, the effect of the auxin analogue 1-naphthylacetic acid (NAA) or Torin-1 on S6K1 phosphorylation at T449, and TOR and S6K1 association with eIF3 was followed in stationary-phase suspension cultures. Phosphorylation of recombinant S6K1 at T449 was visualised by western blot using phospho-specific antibodies against mS6K1 phosphorylated at T389, which specifically recognises AtS6K1-T449-P (Schepetilnikov et al, 2011). Although expression of S6K1 did not change significantly upon NAA treatment, the basal level of S6K1 phosphorylation at T449 increased about seven-fold 6 h after treatment, while it fell below the limit of detection upon Torin-1 treatment (Figure 1B; for quantification, see Supplementary Figure S1B). Thus, auxin can induce phosphorylation of S6K1 at TOR-specific T449. TOR was present in eIF3c immunoprecipitates only when suspension cultures were treated with auxin (Figure 1C). In contrast, S6K1 accumulated in the eIF3c pellet after a 6h incubation with Torin-1, but no S6K1 precipitation was seen 6 h after NAA treatment.
Overall, these results suggest that eIF3 associates with S6K1 upon Torin-1 treatment, and with TOR in the presence of auxin. Thus, plant eIF3-containing complexes can interact with TOR and S6K1 in a defined temporal order and may serve as a platform for S6K1 phosphorylation by TOR, likely within non-polysomal PICs, as suggested in mammals (Holz et al, 2005).
Auxin stimulation of polysomal loading of uORF-containing mRNAs in planta
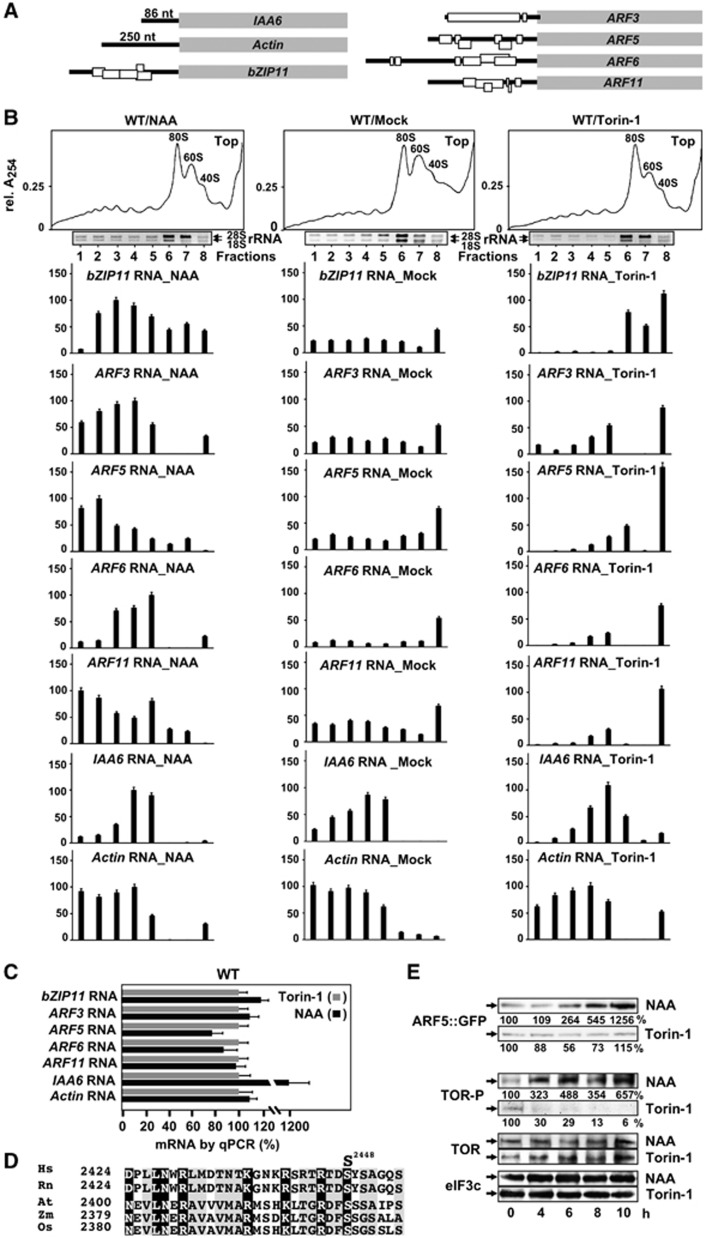
Our studies of CaMV TAV-mediated polycistronic translation in plants revealed that the TOR signalling pathway is essential for activation of reinitiation after long ORF translation (Schepetilnikov et al, 2011). Here, we asked whether TOR could modulate the reinitiation capacity of uORF-containing mRNAs such as ARF3, ARF5, ARF6, ARF11, and bZIP11 (Figure 2A). Translation of these messages requires reinitiation to translate the main ORF (Zhou et al, 2010). To determine the effects of TOR on initiation per se, we included uORF-less mRNAs encoding actin or an auxin/indole-3-acetic acid 6 (IAA6) transcriptional inhibitor. Notably, the mRNAs used do not contain TOP motifs (Thoreen et al, 2012).
Figure 2.
uORF-mRNA abundance in polysomes is regulated by auxin and Torin-1. (A) Schematic representation of mRNAs coding for ARFs, bZIP11, IAA6, and Actin (open rectangles uORFs). (B) Distribution of mRNAs—ARF3, ARF5, ARF6, ARF11, bZIP11, IAA6 and Actin analysed in polysome gradient fractions from extracts prepared from 7-dag seedlings treated (or not, Mock) with either auxin (NAA) or Torin-1 for 8 h. A set of graphs shows quantification of semi-quantitative RT–PCR (sqRT–PCR; Supplementary Figure S2A) corrected for polysome volume in non-, or NAA-, or Torin-1-treated ribosomal profiles. The highest value of each WT/NAA polysome-bound mRNA was set as 100%. Error bars indicate standard deviation of the mean of three replicates. (C) Quantitative RT–PCR (qRT–PCR) of each mRNA in total extracts as in (B). The RNA value in WT/Torin-1 extracts was set as 100%. Values, expressed in arbitrary units, are averages of two replicates, and error bars indicate s.d. (D) Alignment of phosphorylation site patterns from TOR homologues Human, Rattus norvegicus, Arabidopsis, Zea mays, and Oryza sativa. The human phosphorylation site S2448 within the motif is indicated. Similar residues are printed in reverse type and conserved residues are shaded in agreement with Blossom 62 and Jonson amino-acid substitution matrixes. (E) Time course of ARF5::GFP accumulation in 7-dag seedlings expressing GFP tag fused to ARF5 under the control of the natural promoter (ARF5:ARF5::GFP) before (0 h) and after transfer to medium with NAA or Torin-1 analysed by immunoblot with anti GFP ABs (see quantification below the blot line). TOR-P, TOR, and eIF3c protein values in above conditions assayed by immunoblot and ARF5::GFP were corrected for loading control (LC; TOR phosphorylation was quantified). The value at 0 h (no incubation) for each line was set as 100%. Data shown are the means of three independent blots. Source data for this figure is available on the online supplementary information page.
We quantified polysomal levels of selected endogenous uORF-mRNAs under conditions that affect TOR activation differentially in planta. mRNA mobilization was monitored by semi-quantitative RT–PCR (sqRT–PCR) of mRNA in polysome gradient fractions in extracts obtained from Arabidopsis seedlings treated with NAA or Torin-1 7 days after germination (dag). Possible translation repression mediated by uORF2b within the leader of bZIP11 mRNA in response to sucrose was suppressed by using low sucrose levels (Wiese et al, 2004).
In non-treated seedlings, accumulation in polysomes of bZIP11, ARF3, ARF5, ARF6, and ARF11 mRNAs was significantly lower than in ribosomal subunit fractions, while actin and IAA6 mRNAs accumulated exclusively in polysomes (Figure 2B; band density quantification of results shown in Supplementary Figure S2A; for sqRT–PCR specificity control, see Supplementary Figure S2B). This correlates with the inhibiting effect of uORFs diminishing ARF3 and ARF5 mRNA translation levels to 50 and 12–15% in Arabidopsis protoplasts, respectively (Nishimura et al, 2005). Surprisingly, all our uORF-containing mRNAs, including bZIP11 mRNA, accumulated to a high extent in polysome fractions in response to auxin (Figure 2B, WT/NAA). In contrast, TOR inactivation by Torin-1 reduced their accumulation in polysomes, and shifted uORF-mRNAs from polysomes to ribosomal fractions, mainly 40S (fractions 7 and 8). Note that Torin-1 did not abolish uORF-mRNA loading on 40S PICs in our conditions. Torin-1 did not affect the first initiation event significantly—heavy polysomal loading of both actin and IAA6 mRNAs was practically the same. In contrast, Torin-1 greatly impaired uORF-mRNA complex formation with heavy polysomes, while light polysomes—with probably 2–3 ribosomes sitting on the uORFs—still formed. We concluded that uORF-mRNA abundance in polysomes is regulated by auxin in a TOR-dependent manner, not only for auxin-related genes, but also for bZIP11, suggesting a role for TOR in translation of these mRNAs.
To verify that the effect of auxin on polysomal loading of uORF-mRNAs was mediated by TOR activation, TOR was inactivated by Torin-1 in seedlings treated with NAA. Supplementary Figure S3A and B demonstrates that auxin treatment failed to support polysomal loading of uORF-mRNAs in conditions of TOR inactivation (WT/NAA+Torin-1), as compared with NAA treatment (WT/NAA). Again, loading of Actin and IAA6 mRNAs was affected only slightly by NAA/Torin-1 application.
Comparative analysis of total mRNAs in extracts just prior to loading on sucrose gradients revealed no significant difference in mRNA levels 8 h after NAA or Torin-1-treatment (Figure 2C). In contrast, IAA6 mRNA levels were significantly higher after NAA treatment, but there was no impact on polysomal loading.
Next, we verified whether auxin can promote protein synthesis from uORF-containing ARF5::GFP mRNA in ARF5:ARF5::GFP transgenic seedlings. There was a significant increase in accumulation of ARF5::GFP fusion protein in response to auxin, but not in the presence of Torin-1 (Figure 2E). To monitor the phosphorylation status of endogenous TOR during 4, 6, 8, and 10 h of auxin or Torin-1 treatment, we used phospho-specific antibodies against mTOR phosphorylated at S2448 (see Schepetilnikov et al, 2011; S6K1 is the major protein kinase responsible for S2448 phosphorylation of mTOR (Chiang, 2005); S2448 appears to be conserved in Arabidopsis TOR (see alignment in Figure 2D)). The partial TOR phosphorylation RxxS/T site (TGRDFS) can be phosphorylated as well (Jastrzebski et al, 2011). Strikingly, auxin triggered TOR phosphorylation after 4–6 h of NAA application, without altering TOR or eIF3c protein levels, while Torin-1 triggered TOR dephosphorylation (Figure 2E, TOR-P). In contrast, there was no significant effect of auxin on Arabidopsis ERK1/ERK2-like MAPK phosphorylation upon auxin application during 10 h (Supplementary Figure S3C), as shown by western blot with phospho-specific antibodies raised against Arabidopsis MPK3/4/6-P (Asai et al, 2002).
An effect of ARF mRNA leaders on GFP protein levels was studied in transgenic plants, with GFP placed downstream of the authentic ARF promoter/5′-UTR (Rademacher et al, 2011). As expected, GFP levels were elevated after auxin treatment (Supplementary Figure S3D).
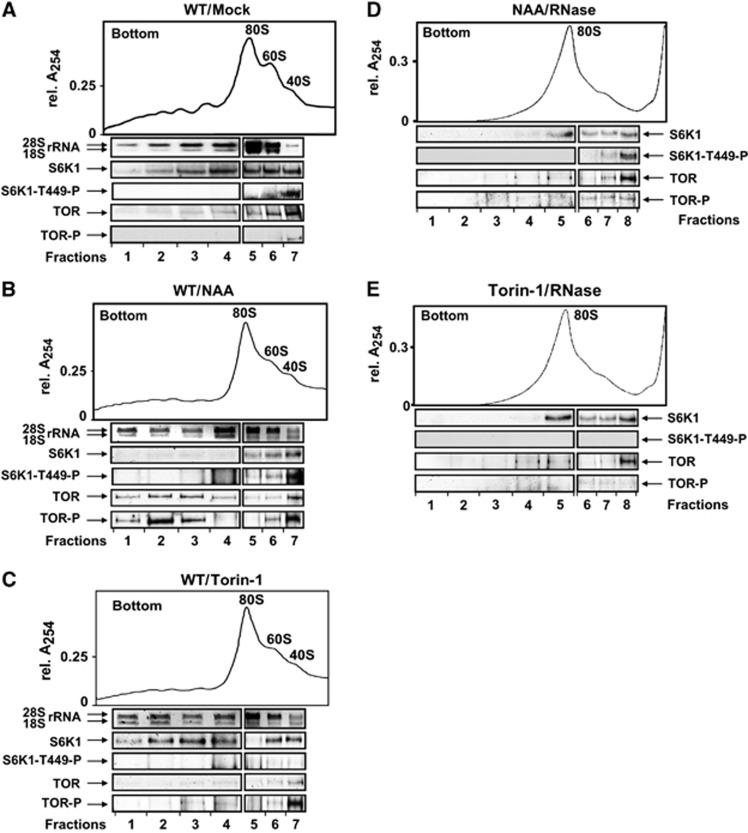
TOR is phosphorylated and recruited to polysomes in response to auxin, while Torin-1 triggers TOR and S6K1 dephosphorylation, resulting in replacement of TOR by S6K1 in polysomes
Although reinitiation efficiency depends on retention of RPFs in polysomes after the preceding initiation event, TOR pathway activation also seems important. We analysed the distribution of TOR and S6K1 between non-polysomal 48S PIC and polysomes in extracts of Arabidopsis seedlings treated with NAA or Torin-1 for 8 h (Figure 3). The initial state of seedlings without treatment showed that TOR and S6K1 are distributed between polysomal and ribosomal fractions (Figure 3A), with TOR and S6K1 detected in 40S ribosomal subunit fractions, while in 80S and light polysomes TOR levels are at the limit of detection, and S6K1 seems to preferentially occupy polysomes. Interestingly, in response to auxin, TOR is phosphorylated and associates not only with 80S and ribosomal subunit fractions as expected, but also with polysomes (Figure 3B). Although S6K1 association with polysomes was fully disrupted in NAA-treated seedlings, we noted that some S6K1-T449-P remained associated with ribosomal fractions. Torin-1 treatment triggers TOR dephosphorylation and dissociation from polysomes and binding of inactive S6K1 instead (Figure 3C). Inactive S6K1 associated with 40S as well. We noted that phosphorylated TOR could be detected in 40S fractions—most likely due to incomplete TOR inactivation by Torin-1, which may explain why Torin-1 had no major effect on the first initiation event. We conclude that reinitiation after uORF translation may require active TOR loading onto polysomes. In control experiments, RNase treatment resulted in the disruption of polysomal complexes and the concomitant redistribution of S6K1 and TOR to lighter fractions of sucrose gradients (Figure 3D and E).
Figure 3.
TOR and S6K1 are loaded on polysomes in an NAA- and Torin-1-sensitive manner. (A–C) Ribosomal profiles obtained from extracts prepared from 7-dag seedlings treated (or not, A) with either auxin (NAA, B) or Torin-1 (C) for 8 h. In all, 1 ml (1V, 80S/60S/40S) and 2 ml (2V, polysomes) aliquots were precipitated with 10% TCA; rRNA was analysed by agarose gel electrophoresis, and S6K1/TOR by immunoblot. Left (2V) and right (1V) panels below each profile are images from the same gel. Data shown are representative of three independent blots. (D, E) Ribosomal profiles of polyribosomes and ribosomal species from extracts prepared from auxin treated with RNase A (D) and Torin-1 treated with RNase A (E). Source data for this figure is available on the online supplementary information page.
Partial depletion of TOR in planta maintains inactive S6K1 bound to polysomes defective in uORF-mRNA recruitment
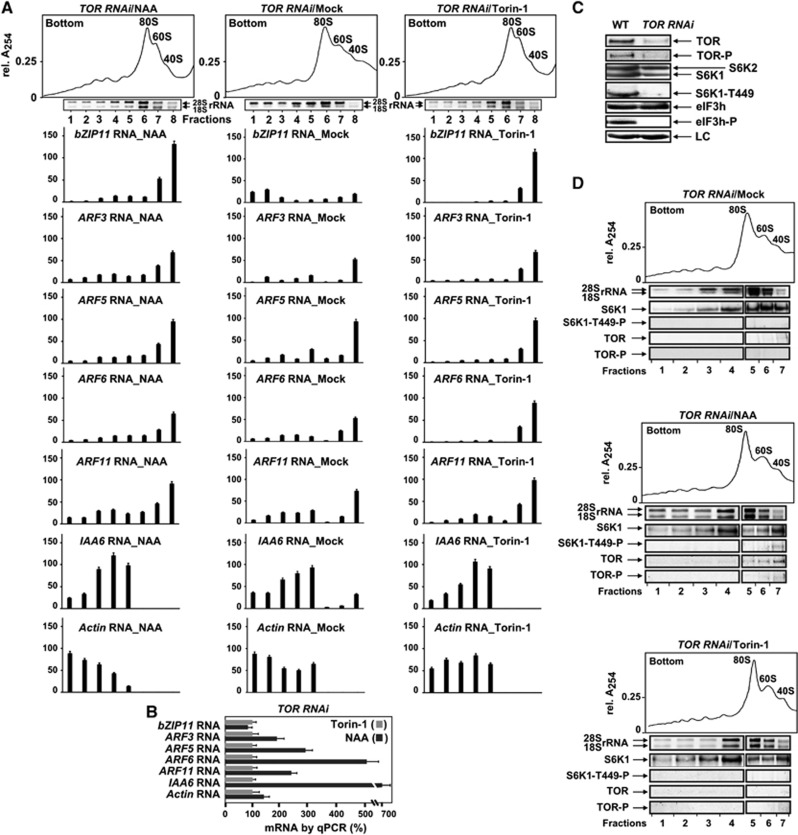
We next tested whether TOR inactivation in planta is able to impair reinitiation events specifically. As an assay system we employed plants ectopically expressing an RNAi construct targeting the TOR FRB domain (TOR-deficient RNAi line 35-7; Deprost et al, 2007; Schepetilnikov et al, 2011). Although TOR and TOR transcript accumulation are reduced by ∼10-fold, mesophyll protoplasts derived from these plants promote translation initiation of uORF-less reporters as efficiently as wild-type protoplasts (Schepetilnikov et al, 2011), suggesting that the remaining TOR levels are sufficient to promote initiation events.
Accordingly, under conditions of partial TOR depletion (TOR RNAi) polysomal loading of uORF-less mRNAs revealed no significant differences in either actin or IAA6 RNA with WT seedlings (Figure 2B), under any of the conditions applied (Mock/NAA/Torin-1 data shown in Figure 4A and Supplementary Figure S4A). In contrast, polysomal loading of uORF-mRNAs was reduced significantly in both non-treated (TOR RNAi/Mock) and NAA-treated conditions (cf Figures 2B and 4A). However, Torin-1 application reduced these levels even further.
Figure 4.
Polysomal loading of both uORF-mRNAs and TOR is impaired in TOR RNAi plants. (A) Comparison of uORF-RNA accumulation in polysomes in extracts prepared from 7-dag TOR RNAi seedlings treated (or not, Mock) with NAA or Torin-1 for 8 h. A set of graphs shows quantification of sqRT–PCR (Supplementary Figure S4A) corrected for polysome volume in non-, or NAA-, or Torin-1-treated ribosomal profiles. The highest value of each mRNA in polysomes from NAA/WT plants (Figure 2B) was set as 100%. Error bars indicate s.d. of the mean of three replicates. (B) qRT–PCR of mRNAs in total extracts prepared as in (A). The RNA value in TOR RNAi/Torin-1 extracts was set as 100%. Values are averages of three replicates. (C) Immunoblot analysis of TOR, S6K1, and eIF3h phosphorylation, as well as their accumulation levels in the control line (WT) and the TOR-deficient RNAi line. LC, loading control. (D) Ribosomal profiles from TOR RNAi were obtained as in (B). In all, 2 ml (2V, polysomes) and 1 ml (1V, 80S/60S/40S) were used to monitor distribution of rRNA on agarose gels and S6K1/TOR by immunoblot. Left (2V) and right (1V) panels of rRNA gel and immunoblot are from the same gel. Source data for this figure is available on the online supplementary information page.
In TOR RNAi seedlings, auxin treatment for 8 h augmented total ARF and IAA6 mRNA levels as compared with after Torin-1 application (Figure 4B). As shown in Supplementary Figure S4C, auxin can stimulate ARF, in contrast to bZIP11 mRNAs, while after about 8 h ARF mRNAs become unstable and their levels drop to those in Mock seedlings. Similarly, auxin upregulates ARF4 transiently in Arabidopsis plants (Marin et al, 2010). However, upon partial TOR depletion the auxin effect on ARF mRNA accumulation was delayed by about 2 h, resulting in elevated ARF mRNA levels at 8 h as compared with WT/NAA conditions (Supplementary Figure S4E). But the increase in ARF-mRNA levels in TOR RNAi seedlings failed to significantly shift uORF-RNA loading from 40S to polysomes (Figure 4A). Again, a seven-fold increase in IAA6 mRNA levels did not further improve polysomal loading of this mRNA.
Next, we examined TOR and S6K1 levels and their phosphorylation status in TOR RNAi seedlings. Phosphorylated TOR and S6K1 were both detected in WT plants, while TOR levels were at the limit of detection in TOR-deficient plants (Figure 4C). As for phosphorylation of the TOR-dependent substrate S6K1, phosphothreonine 449 was high in WT plants, and below the limit of detection in TOR RNAi (see also Schepetilnikov et al, 2011). Accordingly, TOR partial depletion revealed defects in polysomal loading of mRNAs that require reinitiation, but is TOR present within polysomes in TOR RNAi plants? Strikingly, regardless of the conditions applied—mock, NAA, and Torin-1 (Figure 4D)—polysomes were prebound exclusively by S6K1 in its inactive dephosphorylated state. TOR association with ribosomal subunits could be detected after NAA treatment, but was below the limit of detection in Torin-1 conditions. Accordingly, TOR RNAi extracts were not active in in vitro phosphorylation of recombinant S6K1 (Supplementary Figure S1A, right panel).
Mutation of reinitiation factor eIF3h abolishes polysomal association of uORF-RNAs in the presence of a functional TOR signalling pathway
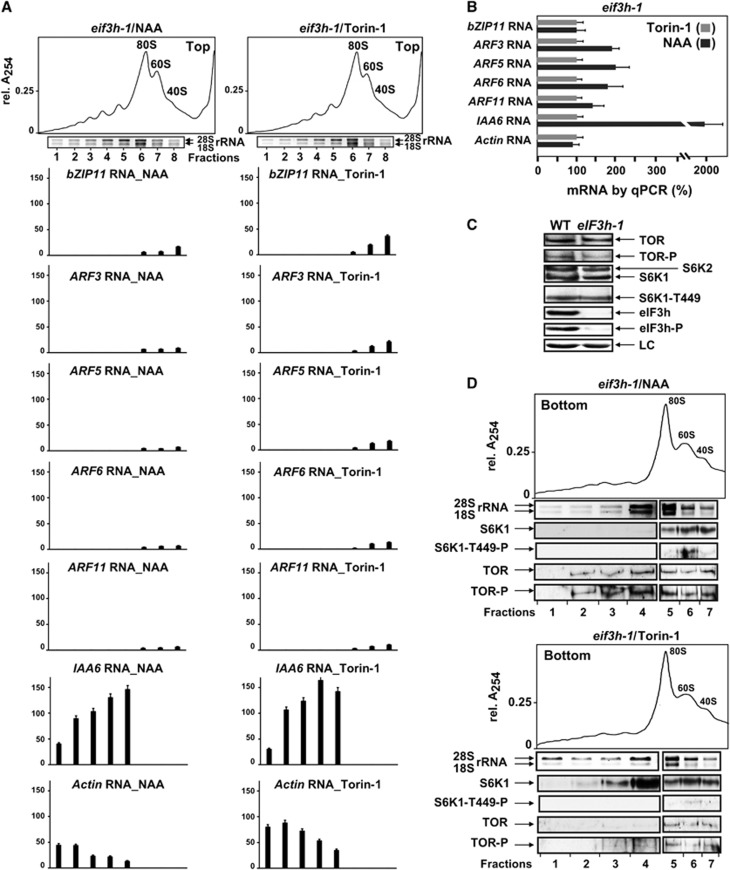
Translation initiation events are not reduced in eif3h-1 plants (carrying carboxyl-terminal truncation alleles of eIF3h), but there is a serious defect in translation of uORF-mRNAs (Roy et al, 2010). Accordingly, polysomal loading of ARF and bZIP11 mRNAs was fully abolished in non- (data not shown) and NAA/Torin-1-treated conditions (Figure 5A; band density quantification of results shown in Supplementary Figure S5A) in contrast to the recruitment of actin and IAA6 mRNAs into polysomes in all conditions. Note that total mRNA levels in the eif3h-1 mutant increased still further after 8 h of NAA treatment (Figure 5B), indicating that mRNA turnover is not responsible for the decrease in uORF-mRNA loading onto polysomes.
Figure 5.
Polysomal loading of uORF-mRNAs, but not TOR signalling, is impaired in eif3h-1 plants. (A) Comparison of uORF-RNA accumulation in polysomes in extracts prepared from 7-dag eif3h-1 seedlings treated for 8 h with NAA or Torin-1. Quantification of sqRT–PCR data (Supplementary Figure S5A) corrected for polysome volume in NAA- or Torin-1-treated ribosomal profiles. The highest value of each mRNA in NAA/WT polysomes (Figure 2B) was set as 100%. Error bars indicate s.d. of the mean of two replicates. (B) qRT–PCR of mRNAs in total eif3h-1 extracts prepared as in (A). The RNA value in eif3h-1/Torin-1 extracts was set as 100%. Each value is the average of two replicates. (C) Immunoblot analysis of TOR, S6K1, and eIF3h phosphorylation, as well as their accumulation levels in WT and the eif3h-1 line. LC, loading control. (D) Ribosomal profiles from eif3h-1 obtained as in (A). In all, 2 ml (2V, polysomes) and 1 ml (1V, 80S/60S/40S) were used to monitor rRNA and S6K1/TOR distribution by immunoblot. Left (2V) and right (1V) panels of rRNA gel and immunoblot are from the same gel. Source data for this figure is available on the online supplementary information page.
In eif3h-1 seedlings lacking full-length eIF3h, the level of TOR and S6K1 as well as their phosphorylation status was largely unaffected (Figure 5C). Neither TOR/S6K1 phosphorylation status nor their ordered binding to polysomes in eif3h-1 plants were significantly impaired: phosphorylated TOR was found in polysomes in response to NAA, and S6K1 was found after Torin-1 application (Figure 5D). Thus, while mRNA reinitiation capacity is severely affected in eif3h-1 mutants, normal TOR signalling pathway behaviour was unimpaired. This led us to suggest that eIF3h functions downstream of TOR and S6K1 in promoting reinitiation, raising the question of whether eIF3h is part of the TOR pathway.
Finally, we compared total polysomes from WT, TOR RNAi, and eif3h-1 treated with NAA or Torin-1 by superimposition of polysomal profiles (Supplementary Figure S5B). Interestingly, heavy polysomes in WT plants are often less pronounced in TOR RNAi or eif3h-1 mutant plants, and small, but reproducible decrease in polysomal levels in response to Torin-1 was detected.
Auxin treatment of Arabidopsis seedlings triggers eIF3h phosphorylation and binding to polysomes in Torin-1-sensitive manner
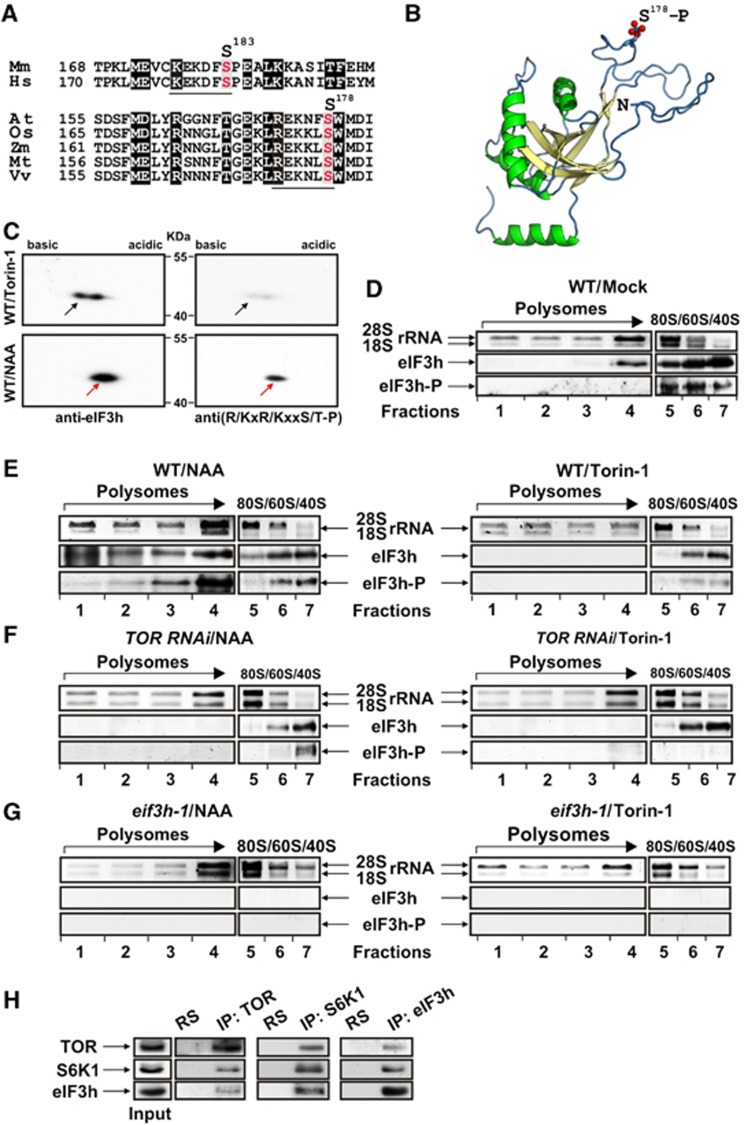
Comparing plant and mammalian eIF3h sequences revealed similar motifs (REKNFS178/KEKDFS183, Figure 6A)—a pattern found in many Akt or S6K1 substrates (R/KxR/KxxS/T)—suggesting eIF3h phosphorylation via TOR signalling. Using the crystallographic structure of the MPN domain of hMov34 as template, a 3D model of Arabidopsis eIF3h was generated (Sali et al, 1995); S178 is positioned within a specific loop spanning well-conserved residues in the N-terminal domain, suggesting high accessibility for phosphorylation (Figure 6B). In 2D western blots of extracts prepared from WT seedlings treated with either auxin or Torin-1 (Figure 6C), one major phosphoisoform was identified by eIF3h antibodies in addition to the eIF3h spot, which was present only upon Torin-1 application. Phospho-specific antibodies confirmed the opposing effects of NAA and Torin-1 on eIF3h phosphorylation by recognising an eIF3h-P spot mainly in extracts treated with NAA (Figure 6C). Strikingly, in TOR-deficient extract (TOR RNAi), where S6K1 phosphorylation was significantly reduced, phosphorylation of eIF3h was also impaired (Figure 4C), strongly suggesting that eIF3h is a novel target of the TOR signalling pathway.
Figure 6.
Auxin triggers eIF3h phosphorylation and association with polysomes in a Torin-1-sensitive manner. (A) Alignment of phosphorylation site patterns from eIF3h homologues Mus musculus, Homo sapiens, Arabidopsis, Oryza sativa, Zea mays, Medicago truncatula, and Vitis vinifera. Akt/S6K1 phosphorylation site consensus (R/KxR/KxxS/T) within the motif is underlined (putative phosphorylation residue in red). (B) The putative AteIF3h 3D structure was generated by PyMOL. The S178-P position within the conserved loop shown in (A) is highlighted in red, residues predicted to fold as helixes in green and β-sheets in yellow. (C) WT/NAA and WT/Torin-1 protein extracts were resolved in two dimensions and revealed by western blot with anti-eIF3h or anti-(R/KxR/KxxS/T-P) antibodies. Molecular masses are indicated. Arrow, eIF3h-P. (D–G) Ribosomal profiles from WT (D) not treated or WT, TOR RNAi, eif3h-1 treated with NAA or Torin-1 (E–G, respectively) were obtained as in (Figure 3) and indicated by distribution of rRNA. In all, 2 ml (2V, polysomes) and 1 ml (1V, 80S/60S/40S) were used to monitor distribution of rRNA by agarose gel and eIF3h/eIF3h-P by immunoblot with antibodies as in (C). Left (2V) and right (1V) panels of rRNA gel and immunoblot are from the same gel. (H) Extracts prepared from WT seedlings were used for immunoblotting of eIF3h, TOR, or S6K1 present in Input, normal rabbit serum (RS), and the entire immunoprecipitate (IP). Source data for this figure is available on the online supplementary information page.
We next studied whether eIF3h, like TOR, is targeted to polysomes in response to auxin. Without treatment, eIF3h was found in sucrose gradient fractions of light polysomes and, in phosphorylated state (visualised by phospho-specific anti-(R/KxR/KxxS/T-P) antibodies), in 80S and ribosomal subunit fractions (Figure 6D). Strikingly, in response to auxin treatment, a significant fraction of phosphorylated eIF3h was found co-sedimenting with polysomes (Figure 6E). These signals are specific for eIF3h, and were not observed in extracts prepared from eif3h-1 plants (Figure 6G). Importantly, unlike with auxin, we detected no eIF3h in polysomes after Torin-1 treatment; however, phosphorylated eIF3h co-sedimented with ribosomal subunits, again suggesting that Torin-1 did not fully abolish TOR phosphorylation in Arabidopsis seedlings in our conditions (Figure 6E).
Next, we monitored eIF3h in polysomal and non-polysomal complexes from TOR RNAi seedlings separated by sucrose gradient sedimentation (Figure 6F). As with TOR in TOR RNAi polysomes (Figure 4D), we found no eIF3h in polysomes, further suggesting that TOR is required for both eIF3h phosphorylation and association with polysomes. Again, eIF3h was found in ribosomal subunit fractions and was phosphorylated after NAA application, or dephosphorylated if Torin-1 was applied. As expected, full-length eIF3h was not detected in polysomes or non-polysomal fractions of eif3h-1 mutant seedlings under any conditions tested (Figure 6G). To gain further evidence that eIF3h is the downstream target of TOR/S6K1, we show that eIF3h co-immunoprecipitates with S6K1 and/or TOR in Arabidopsis extracts (Figure 6H). In addition, S6K1 can interact physically with eIF3h as demonstrated by the yeast two-hybrid system (Supplementary Figure S6).
Reinitiation is upregulated by eIF3h phosphorylation in Arabidopsis protoplasts
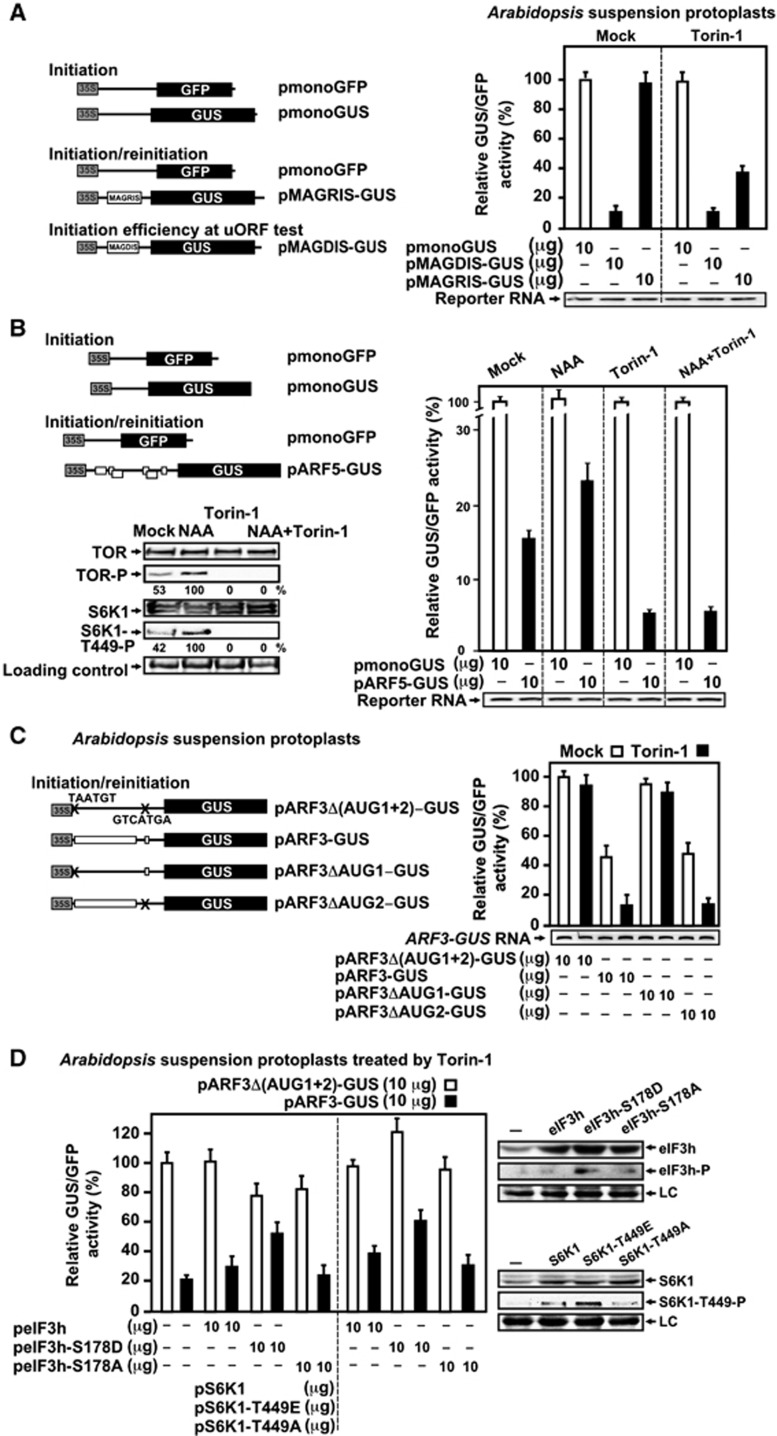
To study directly TOR and eIF3h involvement in translation reinitiation, we first tested whether transient expression of a reporter gene harbouring a single uORF within the leader region is sensitive to Torin-1. Protoplasts prepared from Arabidopsis suspension cultures were transformed with two reporter plasmids: pmonoGFP, containing a single GFP ORF (control for transformation efficiency), and either pMAGRIS-GUS with GUS (β-glucuronidase) reporting reinitiation after translation of the 6-amino-acid peptide MAGRIS, or pmonoGUS, with GUS indicating initiation efficiency (Figure 7A). The 7-codon uORF MAGRIS is highly permissive for reinitiation in protoplasts and its translation does not reduce reinitiation of translation of further downstream ORF in mock conditions (Ryabova and Hohn, 2000).
Figure 7.
Auxin and Torin-1 regulate reinitiation after uORF translation in Arabidopsis suspension protoplasts. (A) Transient expression experiments in Arabidopsis suspension protoplasts included the two reporter plasmids (left panel): pmonoGFP and pmonoGUS; and pmonoGFP and either pMAGRIS-GUS, or pMAGDIS-GUS in the amounts indicated below the graphs (right panel). After transfection, cells were incubated with Torin-1 or not (Mock) for 18 h, and GUS/GFP ratios are shown as open (pmonoGUS/pmonoGFP) and black (pMAGRIS/MAGDIS-GUS/pmonoGFP) bars. pmonoGUS expression in Mock protoplasts was set as 100% (197 800 RFU). Reporter mRNA levels were analysed by sqRT–PCR. (B) Arabidopsis suspension protoplasts were transformed with pmonoGFP and either pmonoGUS or pARF5-GUS (left panel). After transfection, cells were incubated or not (Mock) with Torin-1 or NAA or (NAA+Torin-1) for 18 h, and GUS/GFP ratios were calculated and shown as open (for pmonoGUS/pmonoGFP) and black bars (for pARF5-GUS/pmonoGFP). The GUS/GFP ratio found in Mock protoplasts (for pmonoGUS/pmonoGFP) was set as 100% (211 000 RFU). TOR, S6K1, and their phosphorylation status in protoplasts were assayed by immunoblotting (left panel). Densitometry was used to quantify western blot results from at least three independent replicates (NAA value set as 100%). (C) pmonoGFP and either pARF3-GUS or pARF3Δ(AUG1+2)-GUS or pARF3ΔAUG1-GUS or pARF3ΔAUG2-GUS (left panel) were used for transformation. GUS/GFP ratios were calculated and shown as open (Mock) and black bars (Torin-1). The GUS/GFP ratio found in Mock protoplasts with uORF-less ARF3 leader was set as 100% (150 000 RFU). (D) Protoplasts transformation with pmonoGFP and either pARF3Δ(AUG1+2)-GUS (open bars) or pARF3-GUS (black bars) with or without additional plasmids indicated below the graphs. The GUS/GFP ratio found in Mock protoplasts with uORF-less ARF3 leader was set as 100% (180 000 RFU). eIF3h, S6K1, and their phosphorylation status in protoplasts were assayed by immunoblotting (right panel). LC, loading control. Results in (A–D) represent the means of three independent experiments.
To investigate the percentage of scanning ribosomes that could initiate at the MAGRIS uORF start codon, and thus would need to reinitiate to translate GUS, we employed a specific uORF from the AdoMetDC gene (Ruan et al, 1996) encoding the peptide MAGDIS, which stalls ribosomes at the MAGDIS uORF stop codon, and no reinitiation occurs (Ryabova and Hohn, 2000). The stalling effect can be suppressed by a single amino-acid substitution (D4R) within the MAGDIS uORF coding region resulting in uORF MAGRIS. The MAGDIS uORF diminished GUS expression to 10% of that of pmonoGUS in mock conditions, indicating that most ribosomes initiated efficiently at MAGDIS uORF and thus are trapped at its stop codon (Figure 7A). Therefore, since the initiation context of the D4R mutant-coding uORF is identical, we expected to obtain similar high initiation efficiency for the MAGRIS ORF.
In control protoplasts reinitiation of GUS ORF from pMAGRIS-GUS as efficient as initiation from pmonoGUS as expected for uORF MAGRIS (Figure 7A). Adding Torin-1 to the protoplast incubation medium inhibits pMAGRIS-GUS expression by about 2.5-fold, but does not reduce pmonoGUS expression, suggesting that the reinitiation step is specifically affected. Note that levels of GUS-containing RNAs were unaltered during protoplast incubations (Figure 7A). Our results suggest strongly that reinitiation after translation of a 7-codon uORF is sensitive to TOR inactivation.
In contrast to a single uORF, multiple uORFs within the ARF5 leader fused to the GUS ORF in its authentic initiation context (pARF5-GUS) reduced GUS ORF translation by about 80% as compared with that of pmonoGUS (Mock, Figure 7B), suggesting that several uORFs become nearly reinitiation non-permissive in mock conditions. The low level of pARF5-GUS expression was decreased further by Torin-1, and increased in response to NAA. Importantly, the protoplast response to NAA was abolished by Torin-1 (NAA+Torin-1 versus Torin-1, Figure 7B). pmonoGUS expression was the same under all these conditions, indicating that normal initiation was unaffected.
Accordingly, TOR and S6K1, somewhat phosphorylated in protoplasts in mock conditions, were significantly dephosphoryated in the presence of Torin-1 with or without NAA (Figure 7B, left panel). In contrast, TOR and S6K1 phosphorylation increased by about two-fold in response to auxin. Thus, the GUS levels produced by ARF5-GUS mRNA correlate with TOR and S6K1 phosphorylation levels and all are determined by auxin or Torin-1 application.
Among uORF-containing leaders, ARF3 harbours the longest uORF that codes for a 92-aa peptide and a short one encoding a 5-aa peptide (Figure 2A). To dissect the effect of TOR on reinitiation after the first and/or second uORF translation, the AUG start of either one or both uORFs was replaced by a stop codon (Figure 7C). Although the ARF3 leader decreases the translation efficiency of the GUS ORF by 50% in mock and by 80% in Torin-1 conditions as compared with the uORF-less reporter, removal of uORF1 increases GUS up to the level of uORF-less or uORF2-containing reporters. Therefore, reinitiation of the GUS ORF after translation of uORF1, but not uORF2 is sensitive to Torin-1. However, the effect of Torin-1 on reinitiation is likely higher since uORF1/2 recognition can be decreased by a relatively weak initiation context of both uORF AUGs. Strikingly, GUS ORF translation from ARF3-GUS RNA in Torin-1 conditions can be increased by overexpression of eIF3h, or eIF3h-S178D (phosphorylation mimic), but not eIF3h-S178A (phosphorylation knock-out) or, similarly, by overexpression of S6K1 or S6K1-T449E, but less significantly by S6K1-S178A (Figure 7D). Here, uORF1/2-less ARF3-GUS RNA translation was somewhat improved also upon S6K1-T449E overexpression. In Torin-1 conditions, endogenous eIF3h/S6K1 phosphorylation levels were reduced significantly but, when overexpressed, these proteins display some phosphorylation albeit less than their phosphorylation mimics (Figure 7D, right panels).
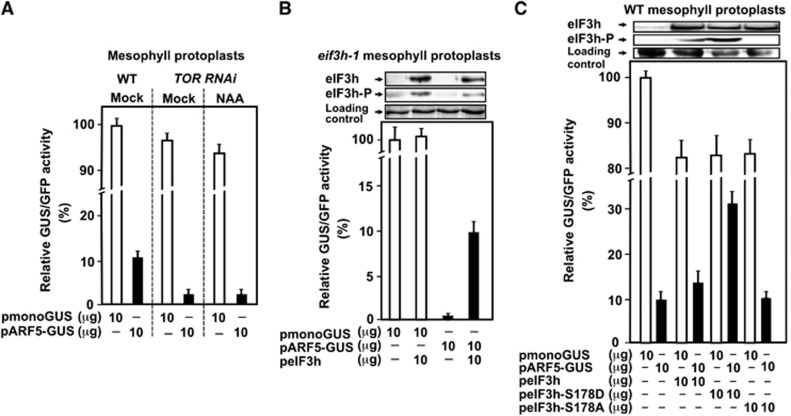
As expected, mesophyll protoplasts prepared from TOR RNAi seedlings support similar levels of pmonoGUS expression as WT protoplasts and significantly less pARF5-GUS transient expression in mock and NAA conditions (Figure 8A). Indeed, GUS expression levels from pARF5-GUS obtained in WT (11% from pmonoGUS) dropped to residual levels in TOR RNAi protoplasts. Furthermore, transient expression of pARF5-GUS, but not pmonoGUS in eif3h-1 mesophyll protoplasts failed, but was rescued by eIF3h overexpression (Figure 8B), suggesting strongly that protoplast reinitiation competence under the conditions used depends on eIF3h.
Figure 8.
TOR partial depletion or eIF3h C-terminal deletion impair reinitiation after uORF translation in mesophyll protoplasts. (A) WT or TOR RNAi protoplasts were co-transformed with reporter plasmids, which are shown in Figure 7B. Activity of GUS synthesized in protoplasts transfected with pmonoGUS was set as 100% (245 000 RFU). GUS/GFP ratios were calculated and shown as open (for pmonoGUS/pmonoGFP) and black bars (for pARF5-GUS/pmonoGFP). (B) eif3h-1 mesophyll protoplasts were co-transformed with reporters shown in Figure 7B with or without plasmid expressing eIF3h as indicated. Activity of GUS synthesized in protoplasts transfected with pmonoGUS was set as 100% (110 000 RFU). eIF3h and its phosphorylation status were assayed by immunoblotting (top panels). (C) WT mesophyll protoplasts were co-transfected with plasmids expressing eIF3h, or eIF3h-S178D, or S178A in addition to reporters shown in Figure 7B as indicated. The value of pmonoGUS expression was set as 100% (205 000 RFU). eIF3h and its phosphorylation status at S178 were assayed by immunoblotting (top panels). Results shown in (A–C) represent the means obtained in three independent experiments. Source data for this figure is available on the online supplementary information page.
To confirm further the significance of S178 phosphorylation for reinitiation, ribosome reinitiation capacity was compared in the presence of eIF3h, eIF3h-S178D, and eIF3h-S178A (Figure 8C). Strikingly, the eIF3h-S178D, when overexpressed, induces GUS functional activity by three-fold giving >30% of ARF5 leader-containing mRNA expression as compared with monoGUS mRNA. Transient overexpression of eIF3h (less phosphorylated than eIF3h-S178D, upper panel) slightly supported GUS activity in mock conditions, while eIF3h-S178A has no impact. GUS activity from pmonoGUS did not change significantly upon overexpression of eIF3h mutants. Since phosphorylation mimic of eIF3h is active in reinitiation, we expect to find it bound to polysomes, while eIF3h-S178A is not. Thus, we analysed polysomal association of eIF3h, eIF3h-S178D, or eIF3h-S178A mutants overexpressed in eIF3h-1 mesophyll protoplasts (Supplementary Figure S7). Only eIF3h and its phosphorylation mimic were found in polysomal fractions, again indicating that eIF3h phosphorylation is a prerequisite for polysomal association.
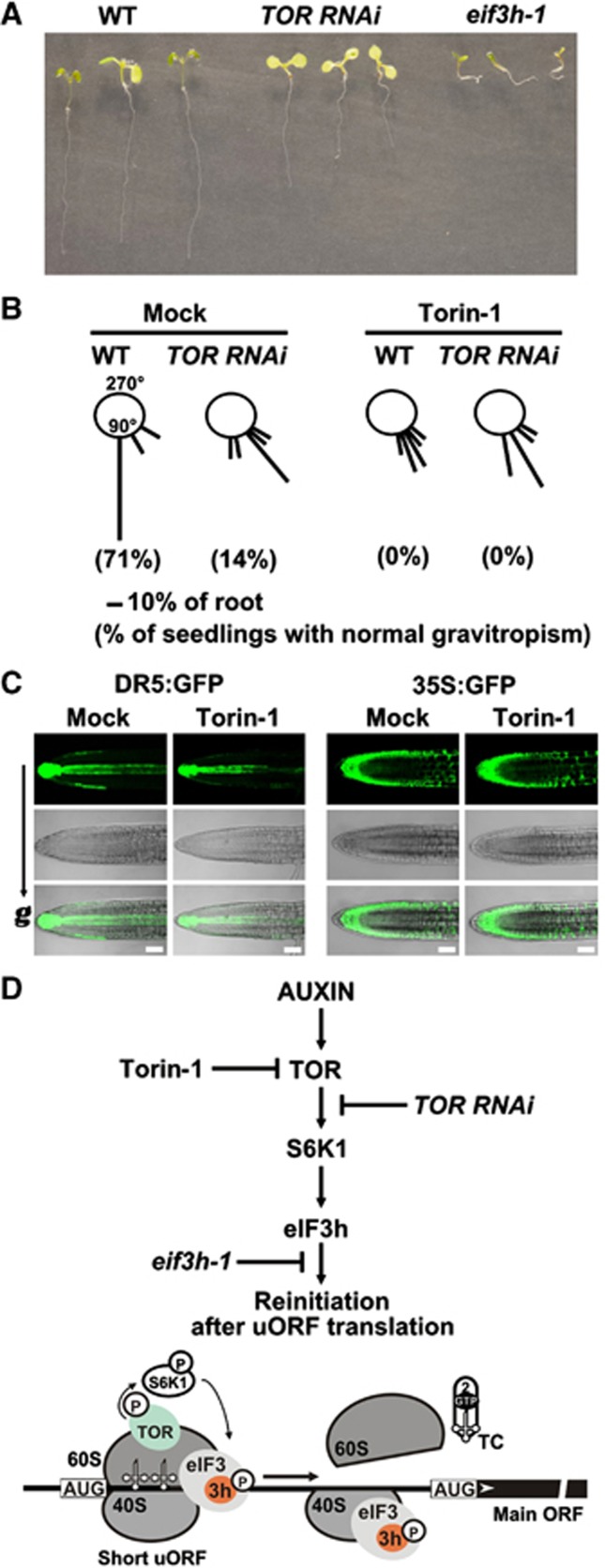
Auxin responses to gravity are impaired by Torin-1
The above results, together with the finding that eif3h-1 mutant plants display strong defects in root gravitropism (Figure 9A), raise the possibility that TOR may function in mediating the link between auxin signalling and root gravitropic response. Accordingly, a gravitropic defect was also seen in TOR RNAi seedlings after 24 h of 90° gravity stimulation (Figure 9B). We thus examined whether Torin-1 affects the slower phase of gravity root bending regulated by the asymmetric distribution of auxin (Wolverton et al, 2002). Strikingly, 4-dag WT and TOR RNAi seedlings grown for 48 h on agar plates with Torin-1 lost gravity perception after turning the seedlings through 90°, showing no or a smaller bending angle after Torin-1 application (drawn schematically in Figure 9B). These results indicate that TOR may exert its effects on gravity sensing by triggering translation reinitiation of mRNAs that encode members of the ARF family, and thus auxin-responsive gene expression. If true, then this would predict that Torin-1 will affect transcription from promoters regulated by ARFs.
Figure 9.
Torin-1 interferes with root gravitropic responses of wild type and abolishes that of TOR RNAi plants. (A) WT, TOR RNAi, and eif3h-1 seedlings grown vertically for 7 dag. (B) TOR RNAi/Torin-1 and WT/Torin-1 plants display agravitropic phenotype. Seedlings described in (A) were grown on medium without (Mock) or with 250 nM Torin-1 and analysed 24 h after gravity stimulation. The orientation of root growth of 12 seedlings was measured by assigning to 1 of 12 30° sectors; the length of each bar represents the percentage of seedlings showing this direction of root growth within the sector. (C) Seven-dag seedlings homozygous for the DR5:GFP construct were analysed 4 h after gravity (g) stimulation on control medium without (Mock) and with 250 nm Torin-1. Scale bars, 25 mm. (D) Proposed scheme of auxin-responsive TOR function in reinitiation after uORF translation (see text for details). Torin-1 application, or TOR deficiency, or eIF3h C-terminal deletion inhibit reinitiation at the steps indicated.
To test this hypothesis, we exploited an auxin-responsive promoter driving GFP expression (DR5:GFP) to follow in situ cellular activation of auxin-induced genes in gravity-stimulation experiments. Gravity determines auxin redistribution along the lower side of the root tip, creating supraoptimal auxin levels that inhibit cell elongation, causing the root to bend downwards (Takahashi et al, 2009). Four hours after gravity stimulation, the DR5:GFP signal was observed mainly in the columella, a stem-cell niche of the root meristem, and in a streak extending basipetally as expected (Figure 9C). As shown above, Torin-1 can block the gravity response, and here no asymmetry in DR5:GFP signal distribution was detected within cortical-endodermal cells in gravity-stimulated roots after 4 h. Torin-1 application had no effect on GFP redistribution along cortex/endodermal cells in response to gravity stimulation in a 35S:GFP Arabidopsis line.
Discussion
Protein synthesis is regulated intensively at the initiation stage by metabolic and signal transduction pathways in eukaryotes; however, this translational control is largely unexplored in plants. Here, we uncover a novel link between reinitiation of translation and the TOR signal transduction pathway. First, we found that the phytohormone auxin can specifically promote activation of TOR/S6K1 signalling in Arabidopsis. We demonstrated that reinitiation after uORF translation is controlled by the TOR signalling pathway in Arabidopsis plants. TOR can trigger translation reinitiation via phosphorylation of the plant reinitiation factor eIF3h—an RPF that, when phosphorylated, promotes translation of mRNAs harbouring uORFs within their leaders.
In plants, upstream TOR effectors are still unknown; however, auxin positively affects phosphorylation of S6 in maize (Beltrán-Peña et al, 2002) and S6K phosphorylation in Arabidopsis suspension culture (Turck et al, 2004). Here, we demonstrated that exogenous auxin can mediate TOR activation, triggering phosphorylation of S6K1 at the TOR-specific hydrophobic motif residue T449 followed by phosphorylation of eIF3h, which is critical for reinitiation in plants (Roy et al, 2010). Our data indicate that eIF3h functions in reinitiation under the control of TOR/S6K1.
Previous studies suggest that activated mTOR phosphorylates S6K1 at T389 in eIF3-PIC, triggering S6K1 dissociation (Holz et al, 2005). Examining TOR and S6K1 association with eIF3 revealed a similar scenario in Arabidopsis—auxin triggers both TOR association with eIF3-complexes and accumulation of phosphorylated S6K1 in a Torin-1-sensitive manner (Figure 1). Moreover, our results implicate polysomes as a second platform for S6K1 phosphorylation by TOR. When TOR is inactivated, S6K1 is dephosphorylated and occupies polysomes. Following an activating signal, such as auxin, TOR associates with polysomes, leading to phosphorylation of bound and inactive S6K1 at T449. Polysomal association of S6K1 is disrupted by its phosphorylation at T449. Concomitantly, eIF3h, when phosphorylated, occupies polysomes, where it can stimulate the reinitiation capacity of post-terminating ribosomes. According to our results, TOR and eIF3h recruitment to polysomes is a hallmark of efficient polysomal loading of uORF-containing mRNAs and thus translation reinitiation. The interaction of TOR with polysomes is dynamic, auxin-responsive, Torin-1 sensitive with or without auxin, and is blocked in TOR-deficient TOR RNAi plants. TOR activation improves polysomal loading not only for auxin-related ARFs, but also for auxin-unrelated bZIP11. All these data indicate a ubiquitous role for TOR in reinitiation after uORF translation.
Whether inactive S6K1 or activated TOR enters polysomes through direct interaction with ribosomes, or other polysomal components, remains unknown. In S. cerevisiae, TOR has the capacity to interact with 60S to accomplish TORC2 complex association with polysomes (Oh et al, 2010), and activation of TORC2 can also be triggered by the ribosome (Zinzalla et al, 2011). It is unlikely that eIF3h mediates TOR interaction with polysomes since, in eIF3h-1, activated TOR is recruited to polysomes (Figure 5D).
eIF3h seems to be a unique RPF that is critical for translation of uORF-mRNAs, but dispensable for translation of monocistronic messages in plants (Roy et al, 2010). In human cells, eIF3h is phosphorylated at S183 and, when overexpressed, triggers dysregulation of protein synthesis, where eIF3h phosphorylation at S183 is critical (Zhang et al, 2008). Phosphorylation of AteIF3h at S178 is also critical for uORF-mRNA reinitiation. S6K1 binds eIF3h in vivo and seems to be responsible for phosphorylation of eIF3h at S178—in TOR-deficient plants, S6K1 and eIF3h phosphorylation were nearly abolished. The effect of TOR on ribosomal reinitiation capacity is seen after a single (6-aa or 92-aa) uORF as well as multiple uORF translation. Since reinitiation competent uORF may become non-permissive if it has a secondary structure around uORF that would be expected to cause ribosome pausing or it is a further downstream uORF has to be initiated by the same ribosome, the effect of TOR may vary from case to case as well as modulated by the number, length, and nature of the uORF(s). It seems that inappropriately activated TOR/S6K1 signalling may trigger upregulation of uORF-mRNAs that encode proteins that are harmful when abundant.
The results obtained in this study allow us to postulate the following model for the role of TOR in translation of uORF-containing mRNAs (Figure 9D). During the rapid elongation event a set of RPFs recruited during the primary 5′-initiation event remain attached to the translating ribosome. One might expect that RPFs reinitiation competence while moving along the mRNA would require re-activation (eIF3h will undergo dephosphorylation). In response to auxin, TOR is activated, binds polysomes prebound by inactive S6K1 to phosphorylate S6K1 at T449. Activated S6K1 can maintain the phosphorylation state of eIF3h within polysomes.
How eIF3h contributes to translation reinitiation is unclear. Previous data suggest that the eIF3h C-terminal part is critical for eIF3h integration within eIF3, but not for eIF3 integrity—h-less eIF3 can still interact with 40S and participate in all initiation steps (Roy et al, 2010). Thus, we propose that eIF3h per se does not play an essential role in initiation steps. In contrast, it can provide a link between eIF3 and polysomes, thus ensuring polysomal retention of eIF3 via its N-terminus. Therefore, after termination, eIF3 bound to reinitiating ribosomes can promote scanning or TC/60S recruitment. Indeed, retention of eIF3 on translationally active ribosomes increases their reinitiation capacity (Park et al, 2001; Pöyry et al, 2007). Functional roles for several eIF3 subunits in reinitiation were proposed in yeast, where subunits a/Tif32, g/Tif35, i/Tif34 were shown to support resumption of scanning of post-terminating ribosomes in addition to their essential roles in translation initiation (Cuchalová et al, 2010; Munzarová et al, 2011).
The hormone auxin is essential for many aspects of growth, development, and pattern formation in plants (Vanneste and Friml, 2009). Based on our data, we predict that at least some auxin responses proceed via the TOR signalling pathway in a Torin-1-sensitive manner in Arabidopsis seedlings. Thus, auxin signalling can target the cell translation machinery by activating the TOR signalling pathway in the cytosol/ER. Note the suggestion of auxin trafficking in the cytoplasm (Friml and Jones, 2010), thus auxin functioning in translation control in the cytoplasm adds another layer of complexity to the well-known pathway in the nucleus, where auxin regulates activation of ARF transcriptional factors (Mockaitis and Estelle, 2008; Leyser, 2006). These ARF proteins regulate expression of multiple auxin-responsive genes and thus a multitude of functions in plant development, including root gravitropism (Blancaflor and Masson, 2003).
Strikingly, a root gravitropic response that is dependent on polar auxin transport and ARF-controlled auxin responsive factors (Palme et al, 2006; Takahashi et al, 2009) can be altered by TOR inactivation via Torin-1. TOR RNAi seedlings defective in reinitiation display defects in the gravity-sensing machinery, suggesting that auxin-responsive function of TOR is critical for plant development.
To summarise, our report describes a novel translational mechanism in which the translational efficiency of genes harbouring multiple uORFs in their leader regions is controlled at multiple levels by auxin, TOR signalling, and eIF3h. The details of how this regulation functions and the possible involvement of other players open up a fascinating area for further study.
Materials and methods
Plant material and growth conditions
Transgenic lines and vector constructions are described in Supplementary data. For seedling growth, cell culture details, extract preparations, production of recombinant S6K1, and the in vitro kinase phosphorylation assay, see Supplementary data and Schepetilnikov et al (2011). Fourteen-day-old Arabidopsis cultures, 7-dag seedlings were incubated in MS medium containing no, or 20 nM NAA, or 250 nM Torin-1 for the times indicated. For growth details, extract preparation, and analysis, see Supplementary methods.
Protein assays
For immunoprecipitation assays, we used 7-dag Arabidopsis Col-0 seedlings or 14-day-old Arabidopsis cultures that were transferred into fresh MS medium containing no or 20 nM NAA, or 250 nM Torin-1 and grown at 24°C. Total protein extracts, IP fractions, and 2D gel analysis were as described in Supplementary data.
Polyribosome analysis
Polysomes were isolated from 18 h mesophyll protoplasts, 7-dag Arabidopsis Col-0 WT, TOR RNAi, and eif3h-1 seedlings treated (or not) with NAA or Torin-1 for 8 h and analysed by density sucrose centrifugation (Supplementary data). ARF, bZIP11, IAA6, and ACTIN mRNA levels were analysed in ribosomal profiles by sqRT–PCR, and in total extracts by qRT–PCR, protein content by western blot.
Protoplast assays
Transient expression was analysed in protoplasts derived from an Arabidopsis suspension culture or 2-week eif3h-1 or TOR RNAi plantlets incubated with no or NAA, or Torin-1. For transfection protocol, see Supplementary data.
Assay for root gravitropism
Seedlings were germinated vertically in the dark at 22°C for 4 days. The plants were then placed on fresh MS agar plates with or without 250 nM Torin-1 maintaining the same orientation and growth conditions and allowed to grow for a further 2 days. The plates were then turned through 90 ° and grown for a further 4 h. GFP fluorescence in roots was analysed after 4 h of a 90° gravistimulation in the dark as described in Supplementary data.
Molecular modelling
The 3D structure of Arabidopsis eIF3h was created using Modeller (Sali et al, 1995) and represented graphically by PyMOL (http://www.pymol.org). Support structure for modelling the central region (1–200 aa) of eIF3h was found with NIH BLAST.
Supplementary Material
Acknowledgments
We are grateful to C Robaglia for TOR RNAi and A von Arnim for eif3h-1/anti eIF3h antibodies, D Weijers for providing ARF-encoding plasmids, M Yusupov and S Melnikov for help with molecular modelling, N Baumberger and P Hammann for helpful assistance in protein analysis. We thank A von Arnim and C Robaglia for comments on the manuscript. This work was supported by French Agence Nationale de la Recherche—BLAN06-2_135889 and BLAN-2011_BSV6 010 03, France—for funding LR.
Footnotes
The authors declare that they have no conflict of interest.
References
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu W-L, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. EMBO J 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Beltrán-Peña E, Aguilar R, Ortíz-López A, Dinkova TD, De Jiménez ES (2002) Auxin stimulates S6 ribosomal protein phosphorylation in maize thereby affecting protein synthesis regulation. Physiol Plant 115: 291–297 [DOI] [PubMed] [Google Scholar]
- Blancaflor EB, Masson PH (2003) Plant gravitropism. Unraveling the ups and downs of a complex process. Plant Physiol 133: 1677–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning KS, Gallie DR, Hershey JWB, Hinnebusch AG, Maitra U, Merrick WC, Norbury C (2001) Unified nomenclature for the subunits of eukaryotic initiation factor 3. Trends Biochem Sci 26: 284. [DOI] [PubMed] [Google Scholar]
- Calvo SE, Pagliarini DJ, Mootha VK (2009) Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci USA 106: 7507–7512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang GG (2005) Phosphorylation of mammalian target of rapamycin (mTOR) at Ser-2448 is mediated by p70S6 kinase. J Biol Chem 280: 25485–25490 [DOI] [PubMed] [Google Scholar]
- Cuchalová L, Kouba T, Herrmannová A, Dányi I, Chiu WL, Valášek L (2010) The RNA recognition motif of eukaryotic translation initiation factor 3g (eIF3g) is required for resumption of scanning of posttermination ribosomes for reinitiation on GCN4 and together with eIF3i stimulates linear scanning. Mol Cell Biol 30: 4671–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolai M, Bedu M, Robaglia C, Meyer C (2007) The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep 8: 864–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrenel T, Marchive C, Sormani R, Moreau M, Mozzo M, Montané MH, Menand B, Robaglia C, Meyer C (2011) Regulation of plant growth and metabolism by the TOR kinase. Biochem Soc Trans 39: 477–481 [DOI] [PubMed] [Google Scholar]
- Friml J, Jones AR (2010) Endoplasmic reticulum: the rising compartment in auxin biology. Plant Physiol 154: 458–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N (2001) Regulation of translation initiation by FRAP/mTOR. Genes Dev 15: 807–826 [DOI] [PubMed] [Google Scholar]
- Hinnebusch A (2006) eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci 31: 553–562 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59: 407–450 [DOI] [PubMed] [Google Scholar]
- Holz MK, Ballif BA, Gygi SP, Blenis J (2005) mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 123: 569–580 [DOI] [PubMed] [Google Scholar]
- Jastrzebski K, Hannan KM, House CM, Hung SSC, Pearson RB, Hannan RD (2011) A phospho-proteomic screen identifies novel S6K1 and mTORC1 substrates revealing additional complexity in the signaling network regulating cell growth. Cell Signal 23: 1338–1347 [DOI] [PubMed] [Google Scholar]
- Kim TH, Kim BH, Yahalom A, Chamovitz DA, Arnim von AG (2004) Translational regulation via 5' mRNA leader sequences revealed by mutational analysis of the Arabidopsis translation initiation factor subunit eIF3h. Plant Cell 16: 3341–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (2001) Constraints on reinitiation of translation in mammals. Nucleic Acids Res 29: 5226–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser O (2006) Dynamic integration of auxin transport and signaling. Curr Biol 16: 424–433 [DOI] [PubMed] [Google Scholar]
- Luukkonen BG, Tan W, Schwartz S (1995) Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. J Virol 69: 4086–4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Blenis J (2009) Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 10: 307–318 [DOI] [PubMed] [Google Scholar]
- Mahfouz MM, Kim S, Delauney AJ, Verma DP (2006) Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell 18: 477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, Vaucheret H, Nussaume L, Crespi MD, Maizel A (2010) miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22: 1104–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medenbach J, Seiler M, Hentze MW (2011) Translational control via protein-regulated upstream open reading frames. Cell 145: 902–913 [DOI] [PubMed] [Google Scholar]
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C (2002) Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci USA 99: 6422–6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockaitis K, Estelle M (2008) Auxin receptors and plant development: a new signaling paradigm. Annu Rev Cell Dev Biol 24: 55–80 [DOI] [PubMed] [Google Scholar]
- Moreau M, Azzopardi M, Clément G, Dobrenel T, Marchive C, Renne C, Martin-Magniette ML, Taconnat L, Renou JP, Robaglia C, Meyer C (2012) Mutations in the Arabidopsis homolog of LST8/GβL, a partner of the target of rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell 2: 463–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DR, Geballe AP (2000) Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol 20: 8635–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munzarová V, Pánek J, Gunišová S, Dányi I, Szamecz B, Valášek LS (2011) Translation reinitiation relies on the interaction between eIF3a/TIF32 and progressively folded cis-acting mRNA elements preceding short uORFs. PLoS Genet 7: e1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J, Sengupta J, Frank J, Nissen P (2004) Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep 5: 1137–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Wada T, Yamamoto KT, Okada K (2005) The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning. Plant Cell 17: 2940–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh WJ, Wu CC, Kim SJ, Facchinetti V, Julien LA, Finlan M, Roux PP, Su B, Jacinto E (2010) mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. EMBO J 29: 3939–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palme K, Dovzhenko A, Ditengou FA (2006) Auxin transport and gravitational research: perspectives. Protoplasma 229: 175–181 [DOI] [PubMed] [Google Scholar]
- Park HS, Himmelbach A, Browning KS, Hohn T, Ryabova LA (2001) A plant viral “reinitiation” factor interacts with the host translational machinery. Cell 106: 723–733 [DOI] [PubMed] [Google Scholar]
- Pöyry TA, Kaminski A, Connell EJ, Fraser CS, Jackson RJ (2007) The mechanism of an exceptional case of reinitiation after translation of a long ORF reveals why such events do not generally occur in mammalian mRNA translation. Genes Dev 21: 3149–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöyry TA, Kaminski A, Jackson RJ (2004) What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes Dev 18: 62–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher E, Möller B, Lokerse AS, Llavata-Peris CI, van den Berg W, Weijers D (2011) A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. Plant J 68: 597–606 [DOI] [PubMed] [Google Scholar]
- Rahmani F, Hummel M, Schuurmans J, Wiese-Klinkenberg A, Smeekens S, Hanson J (2009) Sucrose control of translation mediated by an upstream open reading frame-encoded peptide. Plant Physiol 150: 1356–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Vaughn JN, Kim BH, Zhou F, Gilchrist MA, von Arnim AG (2010) The h subunit of eIF3 promotes reinitiation competence during translation of mRNAs harboring upstream open reading frames. RNA 16: 748–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan H, Shantz LM, Pegg AE, Morris DR (1996) The upstream open reading frame of the mRNA encoding S-adenosylmethionine decarboxylase is a polyamine-responsive translational control element. J Biol Chem 271: 29576–29582 [DOI] [PubMed] [Google Scholar]
- Ryabova LA, Hohn T (2000) Ribosome shunting in the cauliflower mosaic virus 35S RNA leader is a special case of reinitiation of translation functioning in plant and animal systems. Genes Dev 14: 817–829 [PMC free article] [PubMed] [Google Scholar]
- Sachs MS, Geballe AP (2006) Downstream control of upstream open reading frames. Genes Dev 20: 915–921 [DOI] [PubMed] [Google Scholar]
- Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M (1995) Evaluation of comparative protein modeling by MODELLER. Proteins 23: 318–326 [DOI] [PubMed] [Google Scholar]
- Schepetilnikov M, Kobayashi K, Geldreich A, Caranta C, Robaglia C, Keller M, Ryabova LA (2011) Viral factor TAV recruits TOR/S6K1 signalling to activate reinitiation after long ORF translation. EMBO J 30: 1343–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Sabatini DM (2010) Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell 40: 310–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Miyazawa Y, Fujii N (2009) Hormonal interactions during root tropic growth: hydrotropism versus gravitropism. Plant Mol Biol 69: 489–502 [DOI] [PubMed] [Google Scholar]
- Thiébeauld O, Schepetilnikov M, Park HS, Geldreich A, Kobayashi K, Keller M, Hohn T, Ryabova LA (2009) A new plant protein interacts with eIF3 and 60S to enhance virus-activated translation re-initiation. EMBO J 28: 3171–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM (2012) A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485: 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS (2009) An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 284: 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Zilbermann F, Kozma SC, Thomas G, Nagy F (2004) Phytohormones participate in an S6 kinase signal transduction pathway in Arabidopsis. Plant Physiol 134: 1527–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, Friml J (2009) Auxin: a trigger for change in plant development. Cell 136: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Wiese A, Elzinga N, Wobbes B, Smeekens S (2004) A conserved upstream open reading frame mediates sucrose-induced repression of translation. Plant Cell 16: 1717–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolverton C, Ishikawa H, Evans ML (2002) The kinetics of root gravitropism: dual motors and sensors. J Plant Growth Regul 21: 102–112 [DOI] [PubMed] [Google Scholar]
- Zhang L, Smit-McBride Z, Pan X, Rheinhardt J, Hershey JWB (2008) An oncogenic role for the phosphorylated h-subunit of human translation initiation factor eIF3. J Biol Chem 283: 24047–24060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SH, Lawton MA, Hunter T, Lamb CJ (1994) Atpk1, a novel ribosomal protein kinase gene from Arabidopsis. I. Isolation, characterization, and expression. J Biol Chem 269: 17586–17592 [PubMed] [Google Scholar]
- Zhou F, Roy B, von Arnim AG (2010) Translation reinitiation and development are compromised in similar ways by mutations in translation initiation factor eIF3h and the ribosomal protein RPL24. BMC Plant Biol 10: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzalla V, Stracka D, Oppliger W, Hall MN (2011) Activation of mTORC2 by association with the ribosome. Cell 144: 757–768 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.