Summary
In this issue of Cancer Cell, Weischenfeldt et al. report on whole genome sequencing of 11 early onset prostate cancers. Compared to elderly onset prostate cancer, these tumors demonstrate enrichment for androgen driven structural rearrangements involving ETS family genes. This study confirms observations that prostate cancer manifests discrete genomic subclasses.
Prostate cancer remains the most common type of cancer and a frequent cause of cancer-related mortality in men worldwide. Although prostate cancer predominantly affects older men, a subset of prostate cancer arises in men below the age of 50. Early onset cancers often display hereditary links to germline mutations, well-known examples include BRCA2 mutations in breast and ovarian cancers and p53 mutations in Li-Fraumeni syndrome. Among common solid tumors, prostate cancer has been shown to have the largest estimated effect of heritability (Lichtenstein et al., 2000). Despite suggestions that hereditary prostate cancer genes exist, most studies have failed to yield reproducible germline variants that account for early onset prostate cancer. Rare germline variants have been discovered, such as in HOXB13, but may only account for small percentage of early onset prostate cancer cases. Thus, no single germline variant accounts for a substantial proportion of early onset prostate cancer.
In the search for a genetic basis for early onset prostate cancer, Weischenfeldt and colleagues describe, in this issue of Cancer Cell, whole genome sequencing of 11 early onset prostate cancer (median age 47 years) selected from a German cohort (Weischenfeldt et al., 2013). They compared their results to 7 previously published whole genomes meeting their definition for elderly-onset prostate cancer (median age of 65 years) (Berger et al., 2011). A side-to-side evaluation revealed a statistically significant increase in structural rearrangements (SRs) in general, and balanced rearrangements in particular, in the 11 early onset-prostate cancer tumors as compared to the elderly-onset prostate cancer tumors. Interestingly, 91% (10/11) early onset-prostate cancer harbored an ETS gene rearrangement involving either ERG (n=9) or ETV1 (n=1), which is significantly higher than the estimated 50% for all prostate cancers (Rubin et al., 2011; Tomlins et al., 2005).
Androgen stimulation and genotoxic stress (e.g., radiation) have previously been shown to induce the TMPRSS2-ERG gene fusion, the most common prostate cancer ETS rearrangement (Mani et al., 2009). By exploiting a map of publicly available androgen receptor (AR) binding sites, Weischenfeldt et al. observed that the early onset-prostate cancer tumors exhibited SR break points situated nearer to AR binding sites than those in the 7 elderly onset prostate cancer from Berger et al. This finding raises the possibility that androgen stimulation preferentially induces certain types of SRs in early onset-prostate cancer, leading to a cascade of oncogenic molecular events such as recurrent ETS rearrangements. The authors conclude that early onset-prostate cancer may be more likely to harbor androgen-driven SRs, whereas elderly onset prostate cancers display a distinct rearrangement landscape.
The notion that androgen-triggered DNA damage might explain the majority of early onset-prostate cancer is certainly intriguing. However, comprehensive assessment of causal components should also account for how prostate cancer is often diagnosed. Today, most prostate cancer is detected through prostate specific antigen (PSA) screening. Widespread screening has increased the detection of low risk cancers. In fact, recent U.S. and European Screening studies have questioned whether the benefits of PSA screening outweigh the attendant morbidities and costs. Relevant to the Weischenfeldt et al. study, prostate cancer detected by PSA screening may be diagnosed up to 15 years earlier than may have been the case without PSA screening. Thus, a 50 year old man diagnosed with prostate cancer by PSA screening might not have presented clinically until he was 65 years old. In such a case, the “early onset” versus “elderly onset” comparison might represent a distinction without a difference.
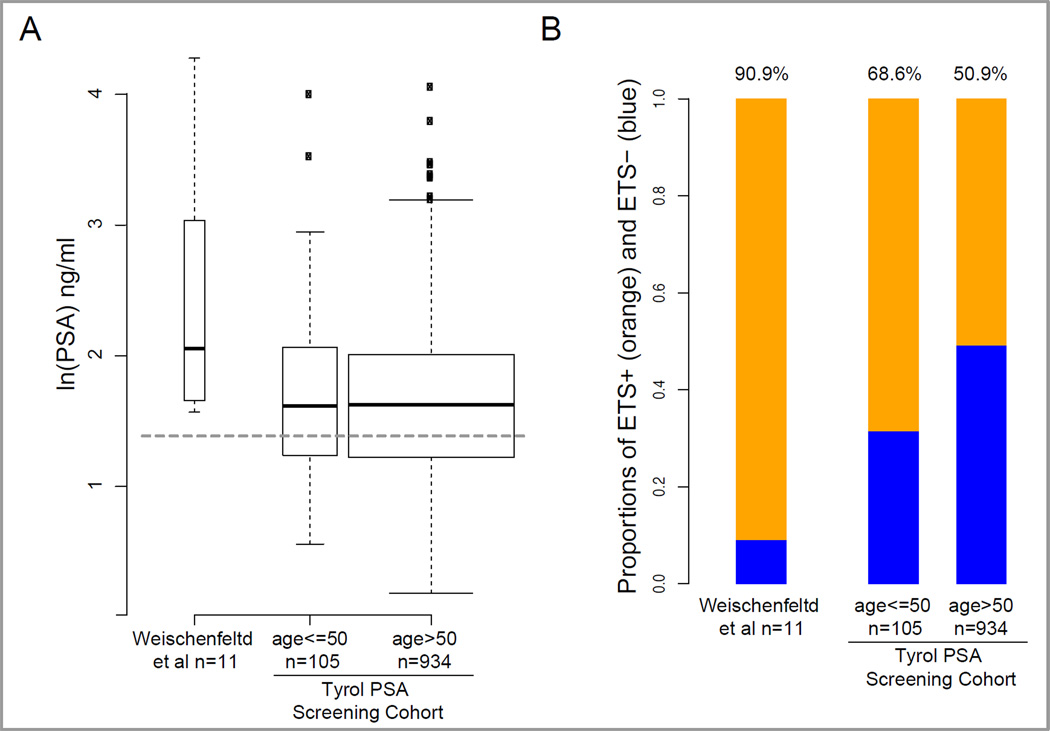
PSA vagaries aside, results of the Weischenfeldt et al. study also suggest that the constellation of genomic alterations (i.e., increased somatic androgen-related SRs and ETS rearrangements) influence earlier clinical detection. We recently completed a population-based study determining ERG overexpression, used as a surrogate for ERG rearrangements, in 1,039 radical prostatectomy tumor samples from the Tyrol PSA screening cohort (Schaefer et al., In Press). This study showed that early ERG rearranged tumors manifest clinically at lower PSA levels and their prevalence is age-dependent. Figure 1 shows the comparison of PSA levels and ETS/ERG rearrangement frequencies of the 11 cases from the Weischenfeldt et al. study with the Tyrol PSA screening population distinguishing between men of 50 years of age at diagnosis or younger versus patients above 50 using the cutoff defined by Weischenfeldt et al. Their 10 ETS rearrangement positive cases demonstrate significantly higher PSA levels than those observed in prostate cancer cases in the Tyrol PSA screening population. The higher PSA level, high tumor grade and stage might therefore reflect a particularly virulent subset of prostate cancer identified from a larger population of men with early–onset disease diagnosed by PSA screening. The question of whether a similar virulent, high-PSA subset may also exist amongst elderly prostate cancer patients —and at what prevalence— will be an important follow-up question.
Figure 1.
Comparison of PSA levels and ETS rearrangement frequencies of the 11 cases from the Weischenfeldt et al. study and data from the Tyrol PSA screening population. (A) Significantly higher PSA levels were found in the Weischenfeldt et al. cases than observed in the Tyrol PSA screening population of men with prostate cancer. The width of the boxes reflects differences in sample sizes; the dotted horizontal grey line corresponds to 4 ng/ml; age indicates years. (B) The percentage of ETS rearrangement positive prostate cancer in the 11 whole genomes from the Weischenfeldt et al. study shown compared to men from the Tyrol screening cohort stratified by age. The numbers at the top of each bar indicates the % of cases that are ETS positive.
A corollary of the Weischenfeldt et al. study findings posits that SRs arising from androgen-driven DNA damage also give rise to the TMPRSS2-ERG (and other ETS) rearrangements. This important study extends previously work that ETS-rearranged prostate cancer comprises a distinct molecular subclass. The TMPRSS2-ERG rearrangement can be observed in the prostate cancer precursor lesion (Perner et al., 2006) and is associated with distinct somatic copy number aberrations (Demichelis et al., 2009). TMPRSS2 is highly androgen regulated and drives ERG expression in the fusion gene. Berger et al. noted that ERG rearrangement-positive cases contained somatic SR breakpoints located near AR binding sites whereas ETS-negative prostate cancer harbored SR breakpoints significantly distant from AR binding sites. They also observed that these SRs in ERG rearranged tumors could exist as interwoven chains that involved cancer related genes, suggesting a possible selective growth advantage (Berger et al., 2011). More recent work suggests that ERG overexpression may mediate 3-dimentional DNA conformational changes through active transcription putatively facilitating SRs under genotoxic conditions (Rickman et al., 2012). The Weischenfeldt et al. study now suggests an alternative scenario where androgenic events drive SRs that lead to ETS rearrangements. This would imply the early onset “gene” might really represent a susceptibility to androgenic DNA damage.
Disentangling the effect of cancer screening from age may prove challenging for the foreseeable future given widespread PSA testing. Regardless, Weischenfeldt et al. confirm observations that prostate cancer manifests discrete genomic subclasses. The telltale molecular fingerprints are emerging through advances in whole genome sequencing that encompass important non-coding regulatory regions not captured by exome sequencing and innovations in large data analysis. Future studies should help elucidate the genomic events predisposing to androgen-driven SR breakpoints, genomic events that may trigger a cascade of prostate cancer alterations including the recurrent TMPRSS2-ERG rearrangements and the development of balanced chained loop rearrangements. One can imagine that germline polymorphisms (i.e., SNPs or Copy Number Variants) could deleteriously hinder DNA repair mechanisms, thus phenocopying BRCA2-deficiency. Epigenetic or environmental effects may play some modifying role in this process. Knowledge of somatic SRs may also have important implications with regards to diagnosis and response to targeted treatment after disease progression. Overall, the Weischenfeldt et al. study extends a growing paradigm regarding the links between complex rearrangements and prostate carcinogenesis, while also considering the age dimension as a possible player in the spectrum of clinical features that contribute to disease biology.
ACKNOWLEDGEMENTS
Work cited in this preview was supported by the Starr Cancer Consortium (M.A.R., F.D., and L.A.G), the Prostate Cancer Foundation (M.A.R.), US Department of Defense Synergy Awards (PC101020 to F.D., L.A.G. and M.A.R.) and the Early Detection Research Network (U01CA111275 and NCI EDRN to F.D. and M.A.R.). L.A.G. is an equity holder and consultant in Foundation Medicine, a consultant to Novartis and Millenium/Takeda, and a recipient of a grant from Novartis. F.D. and M.A.R. are listed as co-inventors of the patent on the detection of gene fusions in prostate cancer, filed by The University of Michigan and the Brigham and Women’s Hospital. The diagnostic field of use for ETS gene fusions has been licensed to Hologic Gen-Probe. The authors would like to thank Helmut A. Klocker and Scott M. Tomlins for their thoughtful comments.
Abbreviations
- PSA
prostate specific antigen
- ICGC
International Cancer Genome Consortium
- AR
androgen receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demichelis F, Setlur SR, Beroukhim R, Perner S, Korbel JO, Lafargue CJ, Pflueger D, Pina C, Hofer MD, Sboner A, et al. Distinct genomic aberrations associated with ERG rearranged prostate cancer. Genes Chromosomes Cancer. 2009;48:366–380. doi: 10.1002/gcc.20647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- Mani RS, Tomlins SA, Callahan K, Ghosh A, Nyati MK, Varambally S, Palanisamy N, Chinnaiyan AM. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perner S, Demichelis F, Beroukhim R, Schmidt FH, Mosquera JM, Setlur S, Tchinda J, Tomlins SA, Hofer MD, Pienta KG, et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- Rickman DS, Soong TD, Moss B, Mosquera JM, Dlabal J, Terry S, MacDonald TY, Tripodi J, Bunting K, Najfeld V, et al. Oncogene-mediated alterations in chromatin conformation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:9083–9088. doi: 10.1073/pnas.1112570109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin MA, Maher CA, Chinnaiyan AM. Common gene rearrangements in prostate cancer. J Clin Oncol. 2011;29:3659–3668. doi: 10.1200/JCO.2011.35.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer G, Mosquera JM, Ramoner R, Park K, Romanel A, Steiner E, Hominger W, Bektic J, Ladurner-Rennau M, Rubin MA, et al. Distinct prevalence of ERG overexpression in prostate cancer – higher frequency in young age and in low PSA prostate cancer. Prostate Cancer and Prostatic Diseases. 2013 doi: 10.1038/pcan.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Weischenfeldt J, Simon R, Feuerbach L, Schlangen K, Weichenhan D, Minner S, Wuttig D, Warnatz H-J. Integrative genomic analyses reveal androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer Cell. 2013 doi: 10.1016/j.ccr.2013.01.002. [DOI] [PubMed] [Google Scholar]