Abstract
Gaining insight in likely disease emergence scenarios is critical to preventing such events from happening. Recent focus has been on emerging zoonoses and on identifying common patterns and drivers of emerging diseases. However, no overarching framework exists to integrate knowledge on all emerging infectious disease events. Here, we propose such a conceptual framework based on changes in the interplay of pathogens, hosts and environment that lead to the formation of novel disease patterns and pathogen genetic adjustment. We categorize infectious disease emergence events into three groups: (i) pathogens showing up in a novel host, ranging from spill-over, including zoonoses, to complete species jumps; (ii) mutant pathogens displaying novel traits in the same host, including an increase in virulence, antimicrobial resistance and host immune escape; and (iii) disease complexes emerging in a new geographic area, either through range expansion or through long distance jumps. Each of these categories is characterized by a typical set of drivers of emergence, matching pathogen trait profiles, disease ecology and transmission dynamics. Our framework may assist in disentangling and structuring the rapidly growing amount of available information on infectious diseases. Moreover, it may contribute to a better understanding of how human action changes disease landscapes globally.
Keywords: drivers, geographic invasion, species jumps, virulence, zoonoses
Introduction
An emerging infectious disease (EID) can be defined as ‘an infectious disease whose incidence is increasing following its first introduction into a new host population or whose incidence is increasing in an existing host population as a result of long-term changes in its underlying epidemiology'.1 EID events may also be caused by a pathogen expanding into an area in which it has not previously been reported, or which has significantly changed its pathological or clinical presentation.2
Mostly, infectious disease emergence in humans is caused by pathogens of animal origin, so-called zoonoses.2,3,4,5 Likewise, cross-over events may occur between non-human species including between domestic animals and wildlife, and such events also involve transmission from a reservoir population into a novel host population (spill-over).5,6,7 Emergence in a novel host, which includes spill-over/zoonoses, has been extensively studied. An elaborate framework featuring the subsequent stages in the emergence process of a species jump has already been developed, describing how an established animal pathogen, through stages of spill-over and lengthening of the transmission chain in the novel host, may evolve all the way up to an established and genetically consolidated pathogenic agent.8,9,10 However, as implied by the above broader definition of EID, other categories can be distinguished in addition to emergence in a novel host, including disease outbreaks in an existing host or the emergence of a disease complex beyond the normal geographic range.
Here, we will argue that changes in host range, in pathogen traits displayed in the same host, and the geographic distribution of a disease complex, form three distinct sets of complementary and only slightly intersecting disease emergence scenarios. Together, these scenarios present the full picture and range of possible disease emergence dynamics. Hence, we categorize EIDs into three main groups, with emergence of (i) a pathogen in a novel host; (ii) a pathogen with novel traits within the same host; and (iii) a disease complex moving into a novel geographic area. Human actions that modulate the interplay between pathogens, hosts and environment are at the basis of almost all EID events, although the exact drivers and mechanisms differ. For each of the three groups, we will argue how the emergence process is driven by specific sets of causal factors, discuss the changes in disease ecology and transmission and elaborate on the invasion dynamics and on the characteristics of pathogens that are dominant in each group. Such structuring of the myriad of EID on the basis of the changes in the interplay between pathogens, hosts and environment will assist in better understanding of specific EID events and in designing tailored measures for prevention and prediction. Moreover, the framework contributes to understanding the effects of human actions that pave the way for the three distinct emergence scenarios. We propose that the resulting framework applies not just to pathogens affecting humans and animals in agriculture and natural ecosystems; it may be usefully applied also for pest and disease emergence in aquaculture, plant production and insect rearing.
Drivers of disease emergence
Drivers of EIDs can be defined as the underlying causal factors of emergence.2 In 1992, Lederberg et al.11 listed a set of ‘specific forces that shape infectious disease emergence' in humans. Subsequent authors adapted and/or expanded these lists of drivers and even proposed ranking of drivers according to incidence.2,4,11,13 Drivers shaping emergence in domestic animals and wildlife are analogous to the drivers for EIDs in humans.14 Notably, changes of anthropogenic nature are at the basis of virtually all EIDs in humans, domestic animals and wildlife.14,15,16,17,18,19 Human population growth and economic development have translated in a growing need for land, water and energy, and thus created a global set of more proximate drivers of disease emergence including deforestation and associated biodiversity loss, climate change, imbalances in agricultural and food supply systems, increases in travel, trade and traffic and a persistence of poor health systems and protection practices.
Pathogen–host–environment interplay
There is growing awareness that drivers of disease emergence modulate the interplay between pathogens, hosts and environment.2,4,20,21,22,23 An EID event can be considered as a shift in the pathogen–host–environment interplay characteristics. Changes in the host–environment and the disease ecology are key to creating novel transmission patterns and selection of novel pathogens with fitter genetic traits. This process will finally result in a novel steady state pathogen–host–environment interplay. Yet, it is difficult to tell what this new pattern will look like until it materializes.
A key factor influencing both the likelihood and outcome of disease emergence is pathogen invasiveness, i.e., the ability of a pathogen to emerge. Such invasiveness is determined by the combination of pathogen traits, including opportunism and evolvability.20,24,25 Notably, RNA viruses with an inherent high mutational rate, bacteria capable of acquiring genetic material and pathogens infecting multiple hosts are more likely to turn into an emerging disease agent.2,4,8,24,26 On the other hand, emergence is influenced by the invasibility of the host–environment, in terms of the individual body, host population structure, host community composition and mosaic, and the associated landscape and its resilience towards pathogen invasion.25,27 The role of the environment extends also to the role of temperature and humidity in pathogen environmental survival and transmission, seasonality in abundance and distribution of arthropod vectors, and roles of geographic, physical or chemical barriers.
A comprehensive framework for disease emergence
The drivers of pathogen emergence change the overall pattern of the pathogen–host–environment interactions leading to either (i) a pathogen showing up in a novel host; (ii) a mutant pathogen with novel traits causing more frequent or more severe disease while remaining in the same host; or (iii) an invasion process involving a novel geographic area. As shown in Figure 1, disease emergence starts with an existing disease complex or pathogen–host–environment complex. While the three emergence categories are broadly speaking distinct, there are also grey areas, between existing and emerging disease events and at the interfaces of the three disease emergence categories. Taken together, these different sets of circumstances represent the full range of disease emergence. A brief introduction to the basics of this framework and the three distinct emergence categories is given in the present section. Details and examples further describing the three disease emergence categories are provided in subsequent sections.
Figure 1.
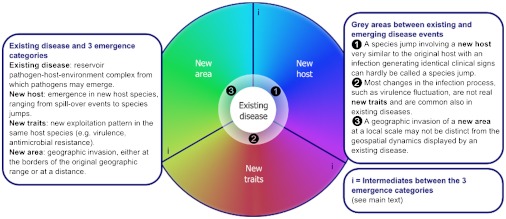
Schematic overview of the emerging infectious disease analysis framework. The rainbow spectrum reflects the full range of possible disease emergence scenarios that can be categorized as emergence in a new host, with new traits or in a new area.
As a first category, pathogens may enter in closer contact with novel host types, be it humans, domestic animals or wildlife. Increased pathogen spill-over may result from this mixing of species and eventually, given progressive exposure of the new host to the ‘new' pathogen, generate a ‘species jump' with sustained transmission in the novel host species. Examples of drivers comprise bush or wild meat hunting and consumption, deforestation and logging, other forms of human encroachment of forests and game reserves, and increased interspecies contacts at the wildlife/agriculture interface, between humans and their pet animals, and within food animal production systems.
Second, pathogens may develop novel traits while circulating in a given host. Key to these dynamics is that the novel trait allows the pathogen to unlock host resources that would otherwise remain unavailable. Mass rearing of animals and use of antimicrobials and vaccines may yield ‘virulence jumpers' with increased pathological or clinical presentation, pathogens acquiring antimicrobial resistance or escaping vaccine-acquired immunity.
Third, diseases may become established in new areas and landscapes, as a result of passive and/or active redistribution of pathogens, vectors and hosts. Two distinct types of geographic invasion may be further considered: pathogens increasing the extent of their geographic range (‘geographic expansion') and pathogens that become dispersed over distances with saltation, across physical barriers in the landscape (‘geographic jumps'). A key factor in the first scenario, geographic expansion, is the suitability of the landscape for the disease complex to become introduced and established. Climate and weather and also land use changes may play a role as drivers. Geographic jumps are facilitated by international travel, trade and traffic, together enhancing the level of connectivity between distinct landscapes, and their respective host, vector and pathogen communities.
In addition, there are intermediates between the three disease emergence categories (indicated by ‘i' in Figure 1). Pathogens emerging in new host species will spread geographically when the species jump is successful, and host specificity adjustment may be an integral component of pathogen invasion of a new area. The expression of new traits of a pathogen in its original host species may lead to higher incidences and more spill-over events to new host species. And new traits may lead to geographic spread of a pathogen in a new area, and geographic spread of a pathogen may lead to adjustment of the infection course.
Distinctly different from the above three disease emergence categories is the disease behavior that that normally plays at the population level. All diseases feature a certain degree of plasticity in terms of their behavior in time and space, responding to host demographic cycles, host immune status, spatial population structure, seasonality or health protection measures (Figure 1, numbers 1, 2 and 3).
Emergence in a novel host
Disease emergence in a novel host includes all events ranging from incidental spill-over to emerging spill-over events (e.g., emerging zoonoses), and to full species jumps, the latter defined as successful infection and replication in a new host species leading to novel host-to-host transmission.28 It should be recalled that zoonoses and spill-over also occur in business-as-usual scenarios not associated with disease emergence such as certain food-borne diseases in humans. Species jumps have in the recent history brought multiple, devastating epidemics, including the influenza A and HIV-1 pandemics. Other potential pandemics were nipped in the bud, e.g., severe acute respiratory syndrome. Again other pathogens with apparent pandemic potential have so far never made it past spill-over events or short human-to-human transmission chains, e.g., Ebola, HEV and monkeypox viruses.
Dynamics of emergence in a novel host
The process of a pathogen that emerges in a novel host has been well described by Wolfe et al.10, subsequently adapted by others.8,9 In summary, different stages can be discerned starting from a pathogen that only infects reservoir hosts, to spill-over without subsequent transmission in the novel host (with R0 in novel host=0, where R0 is the expected number of secondary cases per primary case29), stuttering chains of infection (R0<130), and finally sustained novel host-to-novel host transmission (R0>1).
In some cases, pathogen adaptations in the novel host are not required for successful emergence in a novel host.31,32,33,34 Certain pathogens can shift from one host to the next via ecological fitting, using traits already present,31 or by pre-adaption in the reservoir host.34 One example is the 2009 pandemic H1N1 influenza A virus that showed evidence of pre-adaption to humans during its circulation in swine.35 However, adaptation in the novel host is often required for successful infection and subsequent sustained transmission between new hosts.
The probability of adaptation success is influenced by the number of primary infections, the initial R0, the number of genetic changes required, the pathogen evolvability and other factors.28 Occasionally, several rounds of stuttering chains of transmission in a novel host population are required for successful emergence. Even if the initial establishment of a pathogen fails, the novel host population can become partially immune and in this way primed for more prolonged epidemics upon reintroduction of the pathogen.36 For example, a recent study on the Nipah virus dynamics in Malaysia suggested that repeated introduction from bats into the same pig farm had created a conducive pig herd immunity status supporting on-farm virus circulation and spread to other pig farms.37
Pathogens prone to emerge in a novel host
A species jump requires utmost pathogen evolvability, and appears facilitated by a broad host range, frequent re-assortment or recombination events, a segmented genome structure, genetically conserved receptors, replication without nuclear entry and quasispecies formation.8,24,28,32,33,38 These features are especially common in single-stranded RNA viruses, dominant among species jumpers. These traits alone do not guarantee success in species cross-over.32,33 The collective pathogen trait profile and the ensuing transmission ecology specifics will have to be supportive. For example, pathogens with good environmental survival or transmitted by a range of arthropod vectors may stand a higher chance of infecting new host types than sexually transmitted pathogens.39
Certain pathogens are more prone to periodic species jumps and may again do so in the future. Influenza A viruses underwent multiple species jumps in recent history, including from avian host sources to horses, humans and pigs, from horses to dogs, from pigs to human, and from dogs to cats.40,41,42,43 Although the virus causing H5N1 highly pathogenic avian influenza (HPAI) has so far only shown rare stuttering transmission in humans, a mere five mutations would render this virus air-borne transmissible.44 For comprehensive inventories of pathogens in the different stages of emergence in a novel host, see elsewhere.8,9,45
Drivers of emergence in a novel host
Disease emergence in a novel host species depends on the contact rate between the reservoir host and the novel host, as well as on the suitability of the novel host for the concerned pathogens.9 In addition, major flare-up of disease in a reservoir host can increase spill over to other host types.7,9
In most cases, the main driver behind the emergence of a pathogen in a new host is increased contact between different host types. A set of global factors changes ecological landscapes worldwide and brings animals and humans in closer contact. Microbial reservoirs with potentially hazardous disease agents circulate in non-human primates, wild carnivores, birds, bats and rodents. Humans may become infected encroaching forest and game reserves, including hunting for and consumption of wild meat.45 Interspecies contact rates have gone up considerably also as a result of the steep recent and ongoing growth in human population and increases in food animal production. Wildlife migration can facilitate species jumps to domestic animals and humans.46 The pressures on the natural resource base forces wildlife into farming and urban landscapes, enhancing species mixing. One example is the emergence of Nipah virus in Malaysia in pigs and subsequently humans, and triggered by fruit bats foraging fruit trees near pig farms, using also pig manure as fertilizer.47
Emergence of a pathogen with novel traits in the same host
The second major component of the disease emergence framework concerns the emergence of a pathogen with novel characteristics bringing mostly sudden disease flare-up within the same host. This category comprises pathogens becoming hypervirulent (‘virulence jumpers'), certain pathogens acquiring antibiotic or antiviral resistance, and some pathogens circumventing the effects of vaccination. Common to these situations is a pathogen overcoming an obstacle that was precluding access to major host resources, creating a ‘winner takes all' scenario, often accompanied by severe clinical disease. Examples of pathogens with novel traits that inflict major host damage include recent H5 and H7 HPAI viruses in poultry, spilling back also to wild birds,48,49 and Escherichia (E.) coli O104:H4 with multiple antibiotic resistances that caused a major food safety and veterinary public health challenge in Germany in 2011.50
Dynamics of emergence of a pathogen with novel traits
A pathogen with novel traits can emerge as a result of mutation or because a latent trait with superior fitness becomes selected for, unlocking host resources that would otherwise remain out of reach. For virulence jumpers, the virulence increase may eventually turn less or even counter-productive, hindering transmission.51,52 Virulence-transmission tradeoff may take a variety of turns, depending also on additional factors such as the timing of transmission and associated virulence peak during the infection period,53 and the optimal virulence level may differ for each pathogen–host–environment configuration.54 For pathogens circumventing vaccines or antimicrobials, the superior fitness plays out as long as it lasts. It has to be noted that not all pathogens that gain resistance or escape vaccination count as EID; only the ones that bring sudden disease flare-up and/or that become dominant in a host community are truly emerging.
Pathogens prone to display novel traits in the same host
Pathogens capable of displaying novel traits while in the same host tend to feature a high mutation rate, and/or a capability of acquiring novel genetic material through re-assortment or recombination (viruses) or transfer of plasmids (bacteria). Enhanced virulence in RNA viruses is correlated with diversity of quasispecies, a collection of viruses with related sequences generated by mutation.55 Below, examples will be discussed, starting with viruses, followed by bacteria, fungi and parasites.
An important example of a virus capable to develop novel traits is influenza A virus, with a high mutation rate and formation of quasispecies, supporting the formation of a highly diverse gene pool across host reservoirs.40 Human influenza viruses resistant to antiviral drugs have emerged globally, while H5 and H7 avian influenza subtypes are seen to acquire increased virulence through mutation.40,56 One more example of a virulence jump is given by the emergence of a highly pathogenic form of porcine reproductive and respiratory syndrome virus in China in 2006, based on an accumulation of point mutations and deletions.57 Virulence jumps are also seen among plant viruses; the recombinant cassava mosaic virus UgV caused a very severe disease in cassava in the late 1980s in Sub-Saharan Africa.58
Bacteria capable of acquiring novel genetic material through horizontal gene transfer can obtain virulence factors, toxins and/or acquire antimicrobial resistance.59,60 For example, E. coli O157:H7 obtained the large virulence plasmid pO157 and a bacteriophage expressing Shiga toxin leading to an emerging disease challenge in the food chain.61 The plasmid containing New Delhi metallo-beta-lactamase forms another example; this plasmid is easily transferable by horizontal gene transfer and has conferred carbapenem resistance to many different Entero-bacteriaceae species.62 In fact, genes conferring antimicrobial resistance are ancient and known to circulate also in places out of reach of human and veterinary medicine.63 Thus, the presence of antimicrobial resistance genes in microbial communities is not new, but in response to the presence of antimicrobials, the frequencies of these genes may increase through horizontal gene transfer and natural selection.
Horizontal gene transfer is not restricted to just bacteria but also the fungus Pyrenophora tritici-repentis that causes tan spot in wheat acquired a virulence factor from another plant fungus through gene transfer.64
Protozoa and helminths may also acquire drug resistance. For example, Plasmodium falciparum with resistance to the malaria drug artemisin recently emerged in southeast Asia and is spreading.65
Pathogen characteristics pertaining to the transmission ecology play a main role in disease emergence through novel traits within the same host species. Food-, water- and vector-borne transmission, enhanced environmental robustness may assist in the emergence of new disease complexes. For instance, pathogen characteristics that optimize transmission in specific food production chains or host meta-populations may be selected for, and a ‘winner takes all' scenario may occur if a new pathogen is much fitter than its progenitors. In each situation, the full trait profile of a pathogen determines what the new disease complex will look like. Sometimes, the more host aggressive pathogens gain fitness through enhanced environmental survival, as transmission from immobilized or dead hosts may still be efficient.66,67,68 Examples of aggressive pathogens featuring environmental robustness include the H5N1 HPAI virus, E. coli bacteria including E. coli O157:H7 and the infectious bursal disease virus.61,69,70,71,72
Drivers of emergence of a pathogen with novel traits
Drivers of the evolution of pathogens emerging with novel traits comprise the rise in food animal populations, the process of agricultural intensification and global food supply dynamics as well as the extensive use of antimicrobials and vaccines.27,59,66,73
Food animal production, processing, marketing and distribution have intensified progressively in most industrial countries since the 1950s.16,59,73 Confined animal feeding operations for cattle, and large-scale rearing units with poultry and pigs form patches of ‘mono-cultures' in the host landscape mosaic. In these settings, there is a premium going to pathogens circumventing bio-exclusion and other health protection regimens. Also, mass rearing of food animals entails large numbers of genetically-similar animals of the same age (young) and sex, kept in high densities. Rapid population turnover and live animal transport support onward transmission once a pathogen with novel traits has emerged. For example, flocks of free-grazing ducks rotating in rice paddy fields and ending up in live bird markets played an important role in the transmission of H5N1 HPAI.49,74,75 Pathogens with novel traits may find ready access to human hosts via the food chain, through the handling of live animals, via aerosols emitted by animals in the intensive production units, in wet markets, through ingestion of manure contaminated food commodities, or related to waste disposal.59 One example forms a novel methicillin-resistant Staphylococcus aureus strain that emerged in humans in the Netherlands in 2003 and could be traced back to an origin in pig farming.76
Monoculture fruit, vegetable and crop production provide identical scenarios, generating bulk quantities of food, featuring very little genetic diversity and giving rise to the emergence of pathogens with novel traits.77 One example is the fungus Cochliobolus miyabeanus that destroyed rice crops resulting in the Great Bengal Famine of 1943. Also global fisheries production is undergoing a rapid scaling-up, triggering emergence, spread and persistence of pathogens with novel traits.73 Examples include the emergence of virulent strains of Flavobacterium columnare, Yersina ruckeri and infectious salmon anemia virus in intensively reared salmon.66,78,79
Ironically, the use of (partially effective) vaccines and antimicrobials to control pathogens in intensive systems (including antibiotics as feed additives for growth purposes) and in human medicine may drive selection of pathogens with increased virulence and antibiotic resistance.60,80,81,82 Vaccination has possibly increased virulence of Plasmodium, Bordetella pertussis and infectious bursal disease virus.81,83,84 Recently, a vaccine escape variant of Streptococcus pneumoniae was identified that rapidly spread throughout the USA.85 In particular vaccines reducing pathogen growth, transmission or toxicity could select for increased virulence, in contrast to infection-blocking vaccines.80 A prime example of antimicrobial resistance affecting animal and human health forms the emergence of extended-spectrum beta-lactamases in Entero-bacteriaceae, conferring drug resistance to various classes of antibiotics.60
In addition, advancements in biomedical technology can in rare cases lead to the emergence of a pathogen through an unexpected route of transmission. One example is the recent multistate outbreak of fungal meningitis in the USA associated with contaminated steroid injection.86 Unexpected pathogen transmission through solid organ transplants and blood product transfusions has also been reported,87 but this concerned isolated cases rather than outbreaks.
Disease outbreaks emerging in a novel geographic area
The third major disease emergence category concerns the emergence of a disease complex in a novel area. Emergence may entail an expanding range, so-called ‘geographic expanders' or ‘geographic jumps', pathogens that access novel areas and host resources through saltation dispersal (‘virgin-soil' outbreaks). The first applies in particular to pathogens confronted with changing landscapes, as may be due to climate or land-use changes. Geographic jumps when successful may lead to rapid and widespread dissemination and major epidemics. The introduction of the mosquito-borne West Nile virus in the USA in 1999 forms an example.88 An example of a food-borne geographic jump forms the introduction of African swine fever in the Caucasus region and southern Russian Federation where pathogen introduction could be traced back to food scraps on board of a ship from Southern Africa, fed to pigs in Georgia, early 2007.89 Other recent examples include the emergence of Chikungunya virus in Italy, and Bluetongue virus in Northern Europe.90,91
Dynamics of emergence in a novel geographic area
Geographic range expanders encroach the landscape directly beyond the actual distribution limit. A gradual expansion of the geographic range may entail adjustment to an only slightly different landscape, at least during the initial phase. Adaptation during range expansion is mainly via ecological fitting and may translate into gradual genetic evolution.31,92 Eventually, all invasions come to a halt, with consolidation in the form of a novel geographic limit.93
Unlike range expansion, a long distance jump is usually governed by chance, as only few pathogens, vectors or infected hosts find access to the novel landscape.94 Hence, it is difficult to predict when and from where to where long distance jumps will occur. The initial establishment forms a major bottleneck presenting numerous compatibility issues concerning host and landscape. In addition, when the numbers of infected individuals are still low, extinction may easily occur by chance, even if the R0 of the pathogen in the new landscape is greater than 1. Once initial establishment is successful, an abundance of susceptible hosts may become within reach, facilitating rapid spread. Geographic jumps may be accompanied by a transient increase in virulence during the spread phase when the number of naive hosts within reach builds up rapidly (‘i' in Figure 1). The eventual net result of an invasion always entails a range expansion and may or may not be accompanied by more profound ecological dynamics and allopatric speciation of the pathogen.
Apart from geographic jumps and local expansions, many mixed forms might occur, with new colony establishment and growth leading to coalescence and an advancing pest or disease frontline, taking the form of stratified dispersal.95
Pathogens prone to emerge in a novel geographic area
Disease emergence involving new landscapes may concern the full range of pathogens, from viruses to bacteria, fungi, protozoa, helminths, ecto-parasites and other pest agents, and also arthropods. In principle, any type of pathogen may show up in a novel geographic area should the opportunity presents itself. Still, it remains that pathogens with flexibility are more dispersive and therefore, more prominent in this disease emergency category.
Drivers of emergence in a novel geographic area
The drivers of disease emergence in a novel landscape facilitate access by existing pathogens to host resources elsewhere distributed. More complex scenarios involve introduction of infected host populations or infected vectors becoming newly established upon arrival.
Land-use changes and changes in climate and weather modulating host and vector habitats or environmental pathogen survival may drive a local range expansion of pathogens.59,96,97,98 An example is the expansion of the tick-borne pathogen B. burgdorferi sensu strictu, the cause of Lyme disease, in the USA, probably driven by a combination of changes in climate, agriculture and land use.99
Geographic jumps typically result from international trade and travel, including transport of live animals, food items, plants and accompanying insects, thus enabling disease agents to hitchhike and establish in novel places.59,97,100 For instance, a major epizootic of Rift Valley Fever virus in the Arabian Peninsula in 2000–2001 was attributed to shipments involving live animals and mosquitoes from mainland Africa.101 Similarly, in 2003, the bacterium Ralstonia (Pseudomonas) solanacearum race 3 biovar 2 jumped from Kenya to greenhouses in the USA via imported geranium plants.77 The globalization of fisheries production and supply explains how white spot disease in shrimps became first introduced in Mozambique from Asia in 2011. Geographic jumps may also result from the active migration of wild species including a range of mammals and also birds, fish and arthropods, in the process introducing entire microbial reservoirs into novel geographic areas.46
Concluding remarks and perspectives
Pathogens will continue to find ways to exploit novel host resources, be it humans, domestic animals, plants, marine life or natural ecosystems.102 The framework presented here comprehensively categorizes the great diversity of EID agents and events on the basis of the changes in the interplay between pathogens, hosts and environment. EID events are grouped into three main, distinct and only slightly intersecting categories that each features typical disease ecological dynamics, sets of drivers of emergence and pathogen trait profiles. The framework is not restricted to infectious disease emergence in human and livestock as it would also apply to pest and disease challenges emerging in plant production, fisheries and bee keeping. The framework thus has potential to assist in disentangling and structuring a formidable amount of information on infectious diseases and may be used to help identify the circumstances in food and agriculture, natural resource management and other forms of human behavior that enhance pathogen emergence. This, in turn, may lead to adjustments in food and agriculture, land use, physical planning, trade practices and so on, based on more thorough understanding of the interplay and feedback between human action, pathogen–host–environment dynamics, and pathogen evolution.
Certain extremely flexible pathogens fit several emergence categories. For example, influenza A viruses collectively have shown capable of species jumps, virulence jumps, and inter-continental scale invasions. H5N1 HPAI emerged as a virulence jumper in avian hosts in 1996, paving the way for a presumably migratory waterfowl vectored panzootic of the H5N1 subclade 2.2 viruses in 2006, and at the same time, led to continual spill-over and even rare events of human-to-human transmission, posing the threat of a species jump. In addition, the build-up of influenza A virus genetic diversity in the human–swine–avian host reservoir continues to increase.
As shown in Figure 1, the divide between an existing and emerging disease complex is not always clear. Flare-ups of existing diseases may interfere with, overlap with or assist new disease emergence given the fluctuations in disease prevalence and incidence. Figure 1 also elaborates on the grey areas between the three emergence categories. The disease emergence narratives presented here are mostly of recent origin, stretching over just a few decades, and matching the turbulence created by a global set of contemporary drivers bringing the rapid changes in the availability, use and management of the earth's terres-trial resources. Naturally, in such a time frame, disease emergence and pathogen evolution are based on variations of already existing forms of microbial life. A different temporal resolution would have changed the picture. All currently existing pathogens at some stage in their evolutionary history have switched host species, spread geographically and underwent character changes. Hence, when fitting EID events in the current framework, the geospatial and temporal scales of the event should be made explicit and accounted for. Using the framework here presented, most EID pathogens and disease emergence dynamics are captured with relative ease. In some cases, precise information on changes in pathogen genetics, host specificity, transmission modes, incidence and pattern or invasion dynamics relative to the pre-emergence situation is missing. Here, the framework may assist in gap analysis to reveal critical information that is missing and/or research yet to be performed.
While the existing, well-established disease complexes are causing most of the actual human suffering in the form of chronic disease burdens, the respective pathogens are relatively inflexible (e.g., specialist infectious disease pathogens or macroparasites) and more responsive to health protection measures.18 Yet, in poor countries, there is usually a lack of investment to address even these old diseases. In fact, in these countries, the fight against old and new diseases would present a twin objective. In contrast, in the wealthier countries, many of the old diseases have been eliminated, are being effectively suppressed or never occurred. Naturally, this does not apply to EID events and pandemic threats remain a major global concern. Disrupting the transmission of EID events may turn notoriously difficult and a more preventative approach is therefore called for, with creation of robustness and resilience at the human/food and agriculture/ecosystems interfaces, collectively involving health professionals, in particularly at the community level, farmers, hunters, tourists, consumers and the public at large.103 It is in this sense that the here presented framework for emerging infectious disease analysis may be usefully applied.
Acknowledgments
We thank Nienke Hartemink (Utrecht University, Faculty of Veterinary Medicine, The Netherlands) and Giuliano Cecchi (Food and Agriculture Organization of the United Nations, Italy) for discussions and comments on the manuscript.
References
- Woolhouse ME, Dye C. Population biology of emerging and re-emerging pathogens—preface. Philos Trans R Soc Lond B Biol Sci. 2001;356:981–982. [Google Scholar]
- Jones KE, Patel NG, Levy MA, et al. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor LH, Latham SM, Woolhouse ME. Risk factors for human disease emergence. Philos Trans R Soc Lond B Biol Sci. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse MEJ, Gowtage-Sequeria S. Host range and emerging and reemerging pathogens. Emerg Infect Dis. 2005;11:1842–1847. doi: 10.3201/eid1112.050997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaveland S, Laurenson MK, Taylor LH. Diseases of humans and their domestic mammals: pathogen characteristics, host range and the risk of emergence. Philos Trans R Soc Lond B Biol Sci. 2001;356:991–999. doi: 10.1098/rstb.2001.0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvell CD, Kim K, Burkholder JM, et al. Emerging marine diseases—climate links and anthropogenic factors. Science. 1999;285:1505–1510. doi: 10.1126/science.285.5433.1505. [DOI] [PubMed] [Google Scholar]
- Daszak P, Cunningham AA, Hyatt AD. Emerging infectious diseases of wildlife—threats to biodiversity and human health. Science. 2000;287:443–449. doi: 10.1126/science.287.5452.443. [DOI] [PubMed] [Google Scholar]
- Woolhouse M, Gaunt E. Ecological origins of novel human pathogens. Crit Rev Microbiol. 2007;33:231–242. doi: 10.1080/10408410701647560. [DOI] [PubMed] [Google Scholar]
- Lloyd-Smith JO, George D, Pepin KM, et al. Epidemic dynamics at the human–animal interface. Science. 2009;326:1362–1367. doi: 10.1126/science.1177345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe ND, Dunavan CP, Diamond J. Origins of major human infectious diseases. Nature. 2007;447:279–283. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederberg J, Shope RE, Oaks SC. Washington, DC; National Academy Press; 1992. Emerging infections: microbial threats to health in the United States. [PubMed] [Google Scholar]
- Smolinski MS, Hamburg MA, Lederberg J. Washington, DC; National Academies Press; 2003. Microbial threats to health: emergence, detection, and response. [PubMed] [Google Scholar]
- Morse SS. Factors in the emergence of infectious diseases. Emerg Infect Dis. 1995;1:7–15. doi: 10.3201/eid0101.950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daszak P, Cunningham AA, Hyatt AD. Anthropogenic environmental change and the emergence of infectious diseases in wildlife. Acta Trop. 2001;78:103–116. doi: 10.1016/s0001-706x(00)00179-0. [DOI] [PubMed] [Google Scholar]
- Dobson A. Population dynamics of pathogens with multiple host species. Am Nat. 2004;164:S64–S78. doi: 10.1086/424681. [DOI] [PubMed] [Google Scholar]
- Peeler EJ, Feist SW. Human intervention in freshwater ecosystems drives disease emergence. Freshw Biol. 2011;56:705–716. [Google Scholar]
- Perry BD, Grace D, Sones K. Current drivers and future directions of global livestock disease dynamics. Proc Natl Acad Sci USA. 2011;108:1–8. doi: 10.1073/pnas.1012953108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RA, McMichael AJ. Social and environmental risk factors in the emergence of infectious diseases. Nat Med. 2004;10:S70–S76. doi: 10.1038/nm1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keusch G, Pappaioanou M, Gonzales MC, Scott KA, Tsai P. Washington, DC; National Academies Press; 2009. Sustaining global surveillance and response to emerging zoonotic diseases. [PubMed] [Google Scholar]
- Barrett LG, Thrall PH, Burdon JJ, Linde CC. Life history determines genetic structure and evolutionary potential of host–parasite interactions. Trends Ecol Evol. 2008;23:678–685. doi: 10.1016/j.tree.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright RK, Sokolow SH, Gorman ME, Daszak P, Foley JE. Causal inference in disease ecology: investigating ecological drivers of disease emergence. Front Ecol Environ. 2008;6:420–429. [Google Scholar]
- Schrag SJ, Wiener P. Emerging infectious disease: what are the relative roles of ecology and evolution. Trends Ecol Evol. 1995;10:319–324. doi: 10.1016/s0169-5347(00)89118-1. [DOI] [PubMed] [Google Scholar]
- Conraths FJ, Schwabenbauer K, Vallat B, et al. Animal health in the 21st century—a global challenge. Prev Vet Med. 2011;102:93–97. doi: 10.1016/j.prevetmed.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Burke DS.Evolvability of emerging virusesIn: Nelson AM, Horsburgh CR (eds.)Pathology of emerging infections 2. Washington, DC; American Society for Microbiology; 19981–12. [Google Scholar]
- Alexander HK, Day T. Risk factors for the evolutionary emergence of pathogens. J R Soc Interface. 2010;7:1455–1474. doi: 10.1098/rsif.2010.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaveland S, Haydon DT, Taylor L. Overviews of pathogen emergence: which pathogens emerge, when and why. Curr Top Microbiol Immunol. 2007;315:85–111. doi: 10.1007/978-3-540-70962-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slingenbergh JI, Gilbert M, de Balogh KI, Wint W. Ecological sources of zoonotic diseases. Rev Sci Tech. 2004;23:467–484. doi: 10.20506/rst.23.2.1492. [DOI] [PubMed] [Google Scholar]
- Woolhouse MEJ, Haydon DT, Antia R. Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol Evol. 2005;20:238–244. doi: 10.1016/j.tree.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann O, Heesterbeek JA. Chichester; John Wiley & Sons Ltd; 2000. Mathematical epidemiology of infectious diseases: model building, analysis, and interpretation. [Google Scholar]
- Antia R, Regoes RR, Koella JC, Bergstrom CT. The role of evolution in the emergence of infectious diseases. Nature. 2003;426:658–661. doi: 10.1038/nature02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agosta SJ, Janz N, Brooks DR. How specialists can be generalists: resolving the ‘parasite paradox' and implications for emerging infectious disease. Zoologia (Curitiba) 2010;27:151–162. [Google Scholar]
- Pulliam JR. Viral host jumps: moving toward a predictive framework. Ecohealth. 2008;5:80–91. doi: 10.1007/s10393-007-0149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes EC, Drummond AJ. The evolutionary genetics of viral emergence. Curr Top Microbiol Immunol. 2007;315:51–66. doi: 10.1007/978-3-540-70962-6_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepin KM, Lass S, Pulliam JR, Read AF, Lloyd-Smith JO. Identifying genetic markers of adaptation for surveillance of viral host jumps. Nat Rev Microbiol. 2010;8:802–813. doi: 10.1038/nrmicro2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, Tamuri AU, Hay AJ, Goldstein RA. Charting the host adaptation of influenza viruses. Mol Biol Evol. 2011;28:1755–1767. doi: 10.1093/molbev/msq317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulliam JR, Dushoff JG, Levin SA, Dobson AP. Epidemic enhancement in partially immune populations. PLoS ONE. 2007;2:e165. doi: 10.1371/journal.pone.0000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulliam JRC, Epstein JH, Dushoff J, et al. Agricultural intensification, priming for persistence and the emergence of Nipah virus: a lethal bat-borne zoonosis. J R Soc Interface. 2012;9:89–101. doi: 10.1098/rsif.2011.0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulliam JRC, Dushoff J. Ability to replicate in the cytoplasm predicts zoonotic transmission of livestock viruses. J Infect Dis. 2009;199:565–568. doi: 10.1086/596510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse ME, Taylor LH, Haydon DT. Population biology of multihost pathogens. Science. 2001;292:1109–1112. doi: 10.1126/science.1059026. [DOI] [PubMed] [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GJ, Vijaykrishna D, Bahl J, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- Crawford PC, Dubovi EJ, Castleman WL, et al. Transmission of equine influenza virus to dogs. Science. 2005;310:482–485. doi: 10.1126/science.1117950. [DOI] [PubMed] [Google Scholar]
- Song DS, An DJ, Moon HJ, et al. Interspecies transmission of the canine influenza H3N2 virus to domestic cats in South Korea, 2010. J Gen Virol. 2011;92:2350–2355. doi: 10.1099/vir.0.033522-0. [DOI] [PubMed] [Google Scholar]
- Herfst S, Schrauwen EJ, Linster M, et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe ND, Daszak P, Kilpatrick AM, Burke DS.Bushmeat hunting, deforestation, and prediction of zoonotic emergence Emerg Infect Dis2005; 111822–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altizer S, Bartel R, Han BA. Animal migration and infectious disease risk. Science. 2011;331:296–302. doi: 10.1126/science.1194694. [DOI] [PubMed] [Google Scholar]
- Chua KB, Chua BH, Wang CW. Anthropogenic deforestation, El Nino and the emergence of Nipah virus in Malaysia. Malays J Pathol. 2002;24:15–21. [PubMed] [Google Scholar]
- Shortridge KF, Zhou NN, Guan Y, et al. Characterization of avian H5N1 influenza viruses from poultry in Hong Kong. Virology. 1998;252:331–342. doi: 10.1006/viro.1998.9488. [DOI] [PubMed] [Google Scholar]
- Martin V, Sims L, Lubroth J, Pfeiffer D, Slingenbergh J, Domenech J. Epidemiology and ecology of highly pathogenic avian influenza with particular emphasis on South East Asia. Dev Biol (Basel) 2006;124:23–36. [PubMed] [Google Scholar]
- Rohde H, Qin J, Cui Y, et al. Open-source genomic analysis of Shiga-toxin-producing E. coli O104:H4. N Engl J Med. 2011;365:718–724. doi: 10.1056/NEJMoa1107643. [DOI] [PubMed] [Google Scholar]
- Anderson RM, May RM. Coevolution of hosts and parasites. Parasitology. 1982;85:411–426. doi: 10.1017/s0031182000055360. [DOI] [PubMed] [Google Scholar]
- Ewald PW. Host–parasite relations, vectors, and the evolution of disease severity. Annu Rev Ecol Syst. 1983;14:465–485. [Google Scholar]
- Day T. Virulence evolution and the timing of disease life-history events. Trends Ecol Evol. 2003;18:113–118. [Google Scholar]
- Alizon S, Hurford A, Mideo N, van Baalen M. Virulence evolution and the trade-off hypothesis: history, current state of affairs and the future. J Evol Biol. 2009;22:245–259. doi: 10.1111/j.1420-9101.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2005;439:344–348. doi: 10.1038/nature04388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock DM, Zuccotti G. The evolution of influenza resistance and treatment. JAMA. 2009;301:1066–1069. doi: 10.1001/jama.2009.324. [DOI] [PubMed] [Google Scholar]
- An TQ, Tian ZJ, Leng CL, Peng JM, Tong GZ. Highly pathogenic porcine reproductive and respiratory syndrome virus, Asia. Emerg Infect Dis. 2011;17:1782–1784. doi: 10.3201/eid1709.110411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Liu Y, Calvert L, et al. Evidence that DNA-A of a geminivirus associated with severe cassava mosaic disease in Uganda has arisen by interspecific recombination. J Gen Virol. 1997;78:2101–2111. doi: 10.1099/0022-1317-78-8-2101. [DOI] [PubMed] [Google Scholar]
- Greger M. The human/animal interface: emergence and resurgence of zoonotic infectious diseases. Crit Rev Microbiol. 2007;33:243–299. doi: 10.1080/10408410701647594. [DOI] [PubMed] [Google Scholar]
- Gootz TD. The global problem of antibiotic resistance. Crit Rev Immunol. 2010;30:79–93. doi: 10.1615/critrevimmunol.v30.i1.60. [DOI] [PubMed] [Google Scholar]
- Lim JY, Yoon J, Hovde CJ. A brief overview of Escherichia coli O157:H7 and its plasmid O157. J Microbiol Biotechnol. 2010;20:5–14. [PMC free article] [PubMed] [Google Scholar]
- Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Entero-bacteriaceae. . Emerg Infect Dis. 2011;17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Costa VM, King CE, Kalan L, et al. Antibiotic resistance is ancient. Nature. 2011;477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- Friesen TL, Stukenbrock EH, Liu Z, et al. Emergence of a new disease as a result of interspecific virulence gene transfer. Nat Genet. 2006;38:953–956. doi: 10.1038/ng1839. [DOI] [PubMed] [Google Scholar]
- Phyo AP, Nkhoma S, Stepniewska K, et al. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet. 2012;379:1960–1966. doi: 10.1016/S0140-6736(12)60484-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulkkinen K, Suomalainen LR, Read AF, Ebert D, Rintamäki P, Valtonen ET. Intensive fish farming and the evolution of pathogen virulence: the case of columnaris disease in Finland. Proc Biol Sci. 2010;277:593–600. doi: 10.1098/rspb.2009.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald PW. Evolution of virulence. Infect Dis Clin North Am. 2004;18:1–15. doi: 10.1016/S0891-5520(03)00099-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald PW. Guarding against the most dangerous emerging pathogens. Emerg Infect Dis. 1996;2:245–257. doi: 10.3201/eid0204.960401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy G, Lynch OA, Cagney C. Tracking emerging zoonotic pathogens from farm to fork. Meat Sci. 2008;78:34–42. doi: 10.1016/j.meatsci.2007.05.023. [DOI] [PubMed] [Google Scholar]
- Horm SV, Gutiérrez RA, Sorn S, Buchy P. Environment: a potential source of animal and human infection with influenza A (H5N1) virus. Influenza Other Respi Viruses. 2012;6:442–448. doi: 10.1111/j.1750-2659.2012.00338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domanska-Blicharz K, Minta Z, Smietanka K, Marché S, van Den Berg T. H5N1 high pathogenicity avian influenza virus survival in different types of water. Avian Dis. 2010;54:734–737. doi: 10.1637/8786-040109-ResNote.1. [DOI] [PubMed] [Google Scholar]
- Saif YM. Infectious bursal disease and hemorrhagic enteritis. Poult Sci. 1998;77:1186–1189. doi: 10.1093/ps/77.8.1186. [DOI] [PubMed] [Google Scholar]
- Mennerat A, Nilsen F, Ebert D, Skorping A. Intensive farming: evolutionary implications for parasites and pathogens. Evol Biol. 2010;37:59–67. doi: 10.1007/s11692-010-9089-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M, Xiao X, Domenech J, Lubroth J, Martin V, Slingenbergh J. Anatidae migration in the western Palearctic and spread of highly pathogenic avian influenza H5NI virus. Emerg Infect Dis. 2006;12:1650–1656. doi: 10.3201/eid1211.060223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M, Xiao X, Pfeiffer DU, et al. Mapping H5N1 highly pathogenic avian influenza risk in Southeast Asia. Proc Natl Acad Sci USA. 2008;105:4769–4774. doi: 10.1073/pnas.0710581105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loo I, Huijsdens X, Tiemersma E, et al. Emergence of methicillin-resistant Staphylococcus aureus of animal origin in humans. Emerg Infect Dis. 2007;13:1834–1839. doi: 10.3201/eid1312.070384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange RN, Scott PR. Plant disease: a threat to global food security. Annu Rev Phytopathol. 2005;43:83–116. doi: 10.1146/annurev.phyto.43.113004.133839. [DOI] [PubMed] [Google Scholar]
- Wheeler RW, Davies RL, Dalsgaard I, et al. Yersinia ruckeri biotype 2 isolates from mainland Europe and the UK likely represent different clonal groups. Dis Aquat Organ. 2009;84:25–33. doi: 10.3354/dao02039. [DOI] [PubMed] [Google Scholar]
- Nylund A, Devold M, Plarre H, Isdal E, Aarseth M. Emergence and maintenance of infectious salmon anaemia virus (ISAV) in Europe: a new hypothesis. Dis Aquat Organ. 2003;56:11–24. doi: 10.3354/dao056011. [DOI] [PubMed] [Google Scholar]
- Gandon S, Mackinnon MJ, Nee S, Read AF. Imperfect vaccines and the evolution of pathogen virulence. Nature. 2001;414:751–756. doi: 10.1038/414751a. [DOI] [PubMed] [Google Scholar]
- Williams PD. Darwinian interventions: taming pathogens through evolutionary ecology. Trends Parasitol. 2010;26:83–92. doi: 10.1016/j.pt.2009.11.009. [DOI] [PubMed] [Google Scholar]
- Malim MH, Emerman M. HIV-1 sequence variation: drift, shift, and attenuation. Cell. 2001;104:469–472. doi: 10.1016/s0092-8674(01)00234-3. [DOI] [PubMed] [Google Scholar]
- van Boven M, Mooi FR, Schellekens JF, de Melker HE, Kretzschmar M. Pathogen adaptation under imperfect vaccination: implications for pertussis. Proc Biol Sci. 2005;272:1617–1624. doi: 10.1098/rspb.2005.3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinnon MJ, Gandon S, Read AF. Virulence evolution in response to vaccination: the case of malaria. Vaccine. 2008;26:C42–C52. doi: 10.1016/j.vaccine.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueggemann AB, Pai R, Crook DW, Beall B. Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. e168; PLoS Pathog. 2007;3 doi: 10.1371/journal.ppat.0030168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous Multistate fungal meningitis outbreak investigation. Atlanta, GA; Centers for Disease Control and Prevention; 2012. Available at http://www.cdc.gov/hai/outbreaks/meningitis.html (accessed 29 November 2012). [Google Scholar]
- Kotton CN. Zoonoses in solid-organ and hematopoietic stem cell transplant recipients. Clin Infect Dis. 2007;44:857–866. doi: 10.1086/511859. [DOI] [PubMed] [Google Scholar]
- Murray KO, Mertens E, Desprès P. West Nile virus and its emergence in the United States of America. Vet Res. 2010;41:67. doi: 10.1051/vetres/2010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlands RJ, Michaud V, Heath L, et al. African swine fever virus isolate, Georgia, 2007. Emerg Infect Dis. 2008;14:1870–1874. doi: 10.3201/eid1412.080591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezza G, Nicoletti L, Angelini R, et al. Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet. 2007;370:1840–1846. doi: 10.1016/S0140-6736(07)61779-6. [DOI] [PubMed] [Google Scholar]
- ProMED-mail Bluetongue—Netherlands, Belgium, Germany (06): BTV-8.ProMED-mail: 28 Aug 2006 (20060828.2448). Available at http://www.promedmail.org (accessed 29 November 2012).
- Brooks DR, Hoberg EP. How will global climate change affect parasite–host assemblages. Trends Parasitol. 2007;23:571–574. doi: 10.1016/j.pt.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Blackburn TM, Pyšek P, Bacher S, et al. A proposed unified framework for biological invasions. Trends Ecol Evol. 2011;26:333–339. doi: 10.1016/j.tree.2011.03.023. [DOI] [PubMed] [Google Scholar]
- Wilson JR, Dormontt EE, Prentis PJ, Lowe AJ, Richardson DM. Something in the way you move: dispersal pathways affect invasion success. Trends Ecol Evol. 2009;24:136–144. doi: 10.1016/j.tree.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Hengeveld R. Dynamics of biological invasions. London; Chapman & Hall Ltd; 1989. [Google Scholar]
- Randolph SE, Rogers DJ. The arrival, establishment and spread of exotic diseases: patterns and predictions. Nat Rev Microbiol. 2010;8:361–371. doi: 10.1038/nrmicro2336. [DOI] [PubMed] [Google Scholar]
- Pfeffer M, Dobler G. Emergence of zoonotic arboviruses by animal trade and migration. Parasit Vectors. 2010;3:35. doi: 10.1186/1756-3305-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de La Rocque S, Rioux JA, Slingenbergh J. Climate change: effects on animal disease systems and implications for surveillance and control. Rev Sci Tech. 2008;27:339–354. [PubMed] [Google Scholar]
- Kurtenbach K, Hanincová K, Tsao JI, Margos G, Fish D, Ogden NH. Fundamental processes in the evolutionary ecology of Lyme borreliosis. . Nat Rev Microbiol. 2006;4:660–669. doi: 10.1038/nrmicro1475. [DOI] [PubMed] [Google Scholar]
- de La Rocque S, Balenghien T, Halos L, et al. A review of trends in the distribution of vector-borne diseases: is international trade contributing to their spread. Rev Sci Tech. 2011;30:119–130. doi: 10.20506/rst.30.1.2018. [DOI] [PubMed] [Google Scholar]
- Miller BR, Godsey MS, Crabtree MB, et al. Isolation and genetic characterization of Rift Valley fever virus from Aedes vexans arabiensis, Kingdom of Saudi Arabia. Emerg Infect Dis. 2002;8:1492–1494. doi: 10.3201/eid0812.020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse M. How to make predictions about future disease risks. Philos Trans R Soc Lond B Biol Sci. 2011;366:2045–2054. doi: 10.1098/rstb.2010.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos S, Slingenbergh J. Thoughts on human–animal–ecosystems interface. Transbound Emerg Dis. 2011;58:372–373. doi: 10.1111/j.1865-1682.2011.01214.x. [DOI] [PubMed] [Google Scholar]


