Abstract
Oxidative stress contributes to the pathogenesis of many diseases, including heart failure, but the role and regulation of oxidative DNA damage in many cases have not been studied. Here, we set out to examine how oxidative DNA damage is regulated in cardiomyocytes. Compared to normal healthy controls, human hearts in end-stage cardiomyopathy (EsCM) showed a high degree of DNA damage by histological evidence of damage markers, including 8-oxoG and γH2AX (8-oxoG: 4.7±0.88 vs. 99.9±0.11%; γH2AX: 2.1±0.33 vs. 85.0±13.8%; P<0.01) This raised the possibility that defective DNA repair may be partly responsible. Indeed, nutrient deprivation led to impaired base-excision repair (BER) in cardiomyocytes in vitro, accompanied by loss of the BER enzyme OGG1, while BER activity was rescued by recombinant OGG1 (control vs. nutrient deprived vs. nutrient deprived+OGG1; 100±2.96 vs. 68.2±7.53 vs. 94.0±0.72%; ANOVA, P<0.01). Hearts from humans with EsCM and two murine models of myocardial stress also showed a loss of OGG1 protein. OGG1 loss was inhibited by the autophagy inhibitor bafilomycin and in autophagy-deficient Atg5−/− mouse embryonic fibroblasts. However, pharmacological activation of autophagy, itself, did not induce OGG1 loss, suggesting that autophagy is necessary but not sufficient for OGG1 turnover, and OGG1 loss requires concurrent nutrient deprivation. Finally, we found that the role of autophagy in nutrient starvation is complex, since it balanced the positive effects of ROS inhibition against the negative effect of OGG1 loss. Therefore, we have identified a central role for OGG1 in regulating DNA repair in cardiomyopathy. The manipulation of OGG1 may be used in future studies to examine the direct contribution of oxidative DNA damage to the progression of heart failure.
Keywords: 8-oxoG, cardiomyopathy, oxidative genomic damage, heart failure
Heart failure is a significant public health burden with multiple causes, including genetic mutations, hypertension, coronary artery disease, metabolic disease, and chemotherapy. However, despite different inciting causes, all forms of heart failure show a collection of unifying “common pathway” changes that includes oxidative stress, adenosine-5′-triphosphate (ATP) depletion, fibrosis, inflammation, and cell death (1–4). Oxidative stress, particularly, is believed to play a causative role in the progression of pathology (5, 6). Unpaired electrons in reactive oxygen species (ROS) such as superoxide (O2·−), hydrogen peroxide (H2O2), the hydroxyl radical (OH·), nitric oxide (NO·−), and peroxynitrite (ONOO−) can damage cellular macromolecules, including lipids, proteins, and DNA (7). ROS-induced DNA damage results in a variety of lesions, including single-strand breaks (SSBs) and double-strand breaks (DSBs) and oxidized DNA nucleotides, the most abundant of which is 8-oxo-7,8-dihydroguanine (8-oxoG). Crucially, SSB/DSB generation leads to activation of the DNA damage response (DDR) followed by repair, an early feature of which is activation of the ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3-related (ATR) kinases (8).
The repair of DNA nucleotide damage, such as 8-oxoG is primarily through base-excision repair (BER), involving the repair enzyme, 8-oxoguanine DNA glycosylase (OGG1). After removal of the damaged 8-oxoG base moiety by OGG1, the remaining AP site is processed by apurinic/apyrimidinic endonuclease 1 (APE1). The single-nucleotide gap is then processed, filled, and religated by DNA polymerase, X-ray repair cross-complementing protein 1 (XRCC1), and DNA ligase (7). Nonhomologous end joining (NHEJ) is the primary repair pathway for DSB repair in mammalian cells (8).
Many steps in DNA repair are ATP dependent (9–11), and ATP depletion itself is a characteristic feature of heart failure (3). Mechanisms that contribute to reduced ATP in heart failure are complex and include myocardial ischemia, reduction in nutrient availability, and changes in mitochondrial coupling and function (3). However, it is currently unknown whether and how cardiomyocytes integrate these conditions with DNA damage and repair. In addition, few studies have examined DNA damage, DDR activation, and genomic stability in heart failure and cardiomyocytes in detail. Here, we report robust evidence of DNA damage and activation of the DDR in failing human hearts. In vitro, cardiomyocyte nutrient depletion regulated DNA repair and genomic stability that was mediated by decreased base-excision repair and reduced OGG1 protein abundance. Our work supports a novel relationship between DNA repair and nutrient availability that may explain genomic DNA damage accumulation in the heart during disease progression.
MATERIALS AND METHODS
Ethical statement and human tissues
Human myocardial tissue was collected under a protocol approved by the Papworth Hospital Tissue Bank Review Board and Cambridgeshire Research Ethics Committee (Cambridge, UK). Written, informed consent was obtained for human left ventricular (LV) myocardial tissue from male patients undergoing cardiac transplant for ischemic and idiopathic end-stage heart failure (male Caucasians, aged 42–58, n=10). In our preliminary assessment, levels of oxidative DNA damage (8-oxoG) and the repair enzyme (OGG1) did not differ between ischemic and idiopathic cardiomyopathic hearts. Therefore, we used all ischemic and idiopathic cardiomyopathic samples collectively as representative of end-stage cardiomyopathy (EsCM). Normal human LV tissues were obtained from UK Tissue Bank (de Montfort University, Leicester, UK), from healthy road traffic accident victims who did not have any prior history of cardiovascular disease (control; male Caucasians, aged 41–58; n=9). All human LV tissues were from whole ventricles, and samples were taken from full myocardial thickness. At the time of transplantation or donor harvest, whole hearts were removed after preservation and transported in cold cardioplegic solution, as described previously (12).
Animal tissues
Mouse experiments were approved by the local Animal Ethics Committee (University of Cambridge) and subject to UK Home Office licensing. Transverse aortic constriction was performed on 3- to 4-month-old FVB/N mice, as described previously (13, 14). LV dilatation was confirmed by echocardiography (15). In vivo starvation was performed by removing access to feed for 48 h in CB57BL/6 mice with ad libitum water (16).
Cell culture and primary myocyte isolation
HL1 mouse cardiomyocytes were cultured in Claycomb medium (Sigma, St. Louis, MO, USA), as originally described (17). Neonatal rat ventricular myocytes were isolated from 1- to 3-d-old Sprague–Dawley rats, as described previously (18) in accordance with UK Home Office regulations. All treatment for cells was started 48 h after culture in normal maintenance medium containing full serum and glucose. Atg5+/+ and Atg5−/− immortalized mouse embryonic fibroblasts (MEFs) were obtained from Dr. Tamotsu Yoshimori (Osaka University, Osaka, Japan) and cultured in DMEM containing 10% FCS.
Immunohistochemistry
Immunohistochemistry was performed as described previously (19) using antibodies to γH2AX, p-ATM, NBS-1 (all 1:500; Cell Signaling Technology, Beverly, MA, USA), and 8-oxoG (1:5000; Japan Institute of Aging). Quantification of positively stained nuclei was assessed by counting ≥100 nuclei in >4 separate fields/section. For immunocytochemistry, rat neonatal ventricular myocytes were cultured on gelatin-coated coverslips, and immunostaining was performed using an antibody to 8-oxoG (1:100; Millipore, Bedford, MA, USA).
Western blot analysis
Human and mouse heart proteins were extracted using RIPA buffer (50 mM Tris, pH 7.5; 150 mM NaCl; 0.5% sodium deoxycholate; 0.1% SDS; 1% Nonidet P-40; and 1× Roche Complete Mini Protease Inhibitor Cocktail; Roche, Basel, Switzerland). Tissues were kept ice-cold and homogenized mechanically, followed by 3 rounds of sonication. Cell protein lysates were prepared using single lysis buffer (50 mM Tris, pH 8.0; 150 mM NaCl; 0.02% sodium azide; 1% Nonidet P-40; and 1× Roche Complete Mini Protease Inhibitor Cocktail). Briefly, cells were washed 3 times with ice-cold PBS and collected by scraping in single lysis buffer. Lysates were passed 5 times through a 21-gauge needle and then centrifuged at 10,000 g for 10 min at 4°C. Protein concentration was determined by the bicinchoninic acid method, according to manufacturer’s instructions (Pierce, Rockford, IL, USA). Proteins were boiled for 10 min in Laemmli buffer (4% SDS, 20% glycerol, 10% β-mercaptoethanol, 0.005% bromophenol blue, and 0.125 M Tris-HCl) resolved by SDS-PAGE and transferred to a polyvinylidene fluoride membrane. Primary antibodies were used at 1:1000 dilutions in 5% milk protein in PBS-Tween after membranes were preblocked for 1 h at room temperature with 5% milk protein in PBS-Tween. Primary antibodies used were OGG1 (Novus Biologicals, Littleton, CO, USA), sarcomeric actin (Sigma), Rho GDI (Santa Cruz Biotechnology, Santa Cruz, CA, USA), p-ATM, ATM/ATR substrate, and γH2AX (Cell Signaling Technologies).
Ex vivo 8-oxoG BER assay
8-OxoG excision was assessed using an extension of traditional duplex cleavage assays based on an 8-oxoG-containing molecular beacon adapted from Mirbahai et al. (20): 5′FAM-ACT8AAGCGCCGCACGCCATGTCGACGCGCTTCAGTGC-dabcyl-3′ (Alta Bioscience, Birmingham UK). The molecular beacon was heated to 95°C for 5 min and allowed to cool slowly to room temperature. Cell (30 μg) or heart tissue lysates (200 μg) were incubated with 400 fM of beacon in buffer (50 mM NaCl, 10 mM Tris-HCl, 10 mM MgCl2, and 1 mM dithiothreitol, pH 7.9) for 60 min at 37°C. Fluorescence was measured by excitation at 485 nm and emission at 528 nm. Data were normalized to background fluorescence per sample.
Myocytes (90% confluence) or MEFs (70% confluence) were treated with or without serum- and glucose-free (SGF) medium or 100 nM bafilomycin for the indicated times, followed by incubation for 60 min in PBS containing 10 μM DCFDA (Invitrogen, Carlsbad, CA, USA). Fluorescence was measured by excitation/emission at 485 nm/530 nm at time 0 and after 60 min. Cell viability was measured after incubation with 0.2 mg/ml MTT (Sigma) for 1 h at 37°C. Absorbance was read at 570 nm.
Statistical analysis
Statistical tests were performed using the Mann-Whitney U test, Student’s t test or 1-way ANOVA.
RESULTS
End-stage failing human hearts show increased levels of DNA damage and DDR activation
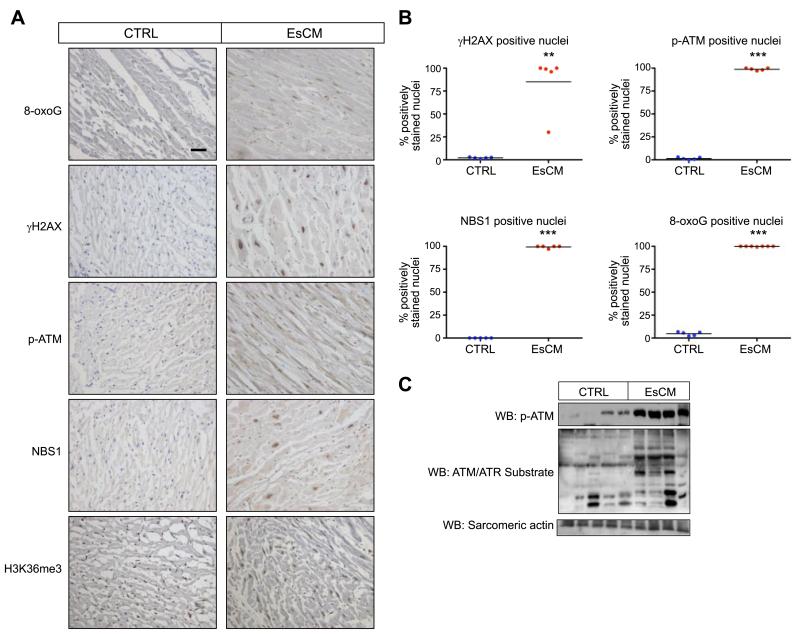
In keeping with the hypothesis that DNA damage is part of the generic and unifying set of features found in heart failure regardless of original inciting causes, evidence of DNA damage and DDR activation was found in human EsCM LV tissue both from patients with ischemic cardiomyopathy and those with idiopathic dilated cardiomyopathy, compared to healthy control age- and sex-matched victims of road traffic accidents (n=5). Using immunohistochemistry, we detected significantly higher counts of cells positive for DNA damage markers 8-oxoG and γH2AX and DDR activation marker phosphorylated ATM, as well as the repair protein NBS1 (8-oxoG: 4.7±0.88 vs. 99.9±0.11%; γH2AX: 2.1±0.33 vs. 85.0±13.8%; p-ATM: 1.4±0.58 vs. 98.8±0.58%; NBS1: 0 vs. 99.4±0.60%; Mann-Whitney U test, P<0.01; Fig. 1A, B). The ubiquitous trimethylated histone 3 lysine 36 (H3K36me3, control) showed no difference in nuclear staining. To verify the finding of marked DNA damage further, we performed Western blot analysis in a separate panel of LV tissue lysates (5 control and 4 EsCM) and found evidence of ATM activation by up-regulated ATM phosphorylation and ATM/ATR substrate phosphorylation, again reflecting increased DDR (Fig. 1C).
Figure 1.
DNA damage and activation of the DNA damage response (DDR) in human end-stage cardiomyopathy. A) Representative images from immunohistochemical detection of γH2AX, p-ATM, NBS1, and 8-oxoG in healthy control (CTRL) and EsCM human LV cardiac tissue (brown-stained nuclei). H3K36me3 antibody was used as a positive control. B) Quantification was performed by counting positively stained nuclei from ≥100 nuclei in each cardiac section. **P < 0.01, ***P < 0.001; Mann-Whitney U test. C) Western blot analysis of human cardiac lysates showed DDR and ATM activation by ATM phosphorylation (p-ATM) and significant levels of ATM/ATR substrate phorsphorylation in EsCM but not CTRL. Equal protein loading was demonstrated by blotting for sarcomeric actin.
Nutrient deprivation predisposes to impaired rates of DNA repair
On the basis of the high degree of DNA damage found in our myocardial tissue analysis, we hypothesized that defective DNA repair should be at least partly responsible. Nutrient depletion is another unifying characteristic found in all forms of heart failure (3). Therefore, we tested the link between reduced nutrient availability and DNA damage repair in cardiomyocytes. The in vitro model of serum and glucose deprivation (SGF conditions) has previously been used to mimic the stimulus of nutrient depletion in heart failure (21, 22).
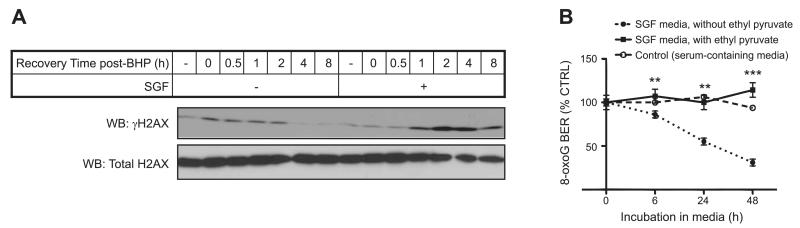
DSB repair efficacy in cells can be monitored by γH2AX abundance with Western blot analysis. Therefore, we examined the accumulation of γH2AX abundance in cardiomyocytes in response to the damage stimulus of pulsed tert-butyl hydrogen peroxide (BHP) exposure (Fig. 2A). Cardiomyocytes were precultured overnight either in serum-containing or SGF medium before being exposed to BHP. Notably, SGF itself did not lead to γH2AX accumulation, reflecting that at this time course, SGF itself was not responsible for generating DSB detectable by this assay. In contrast, following exposure to the pulse of BHP, cells that were in control serum-containing conditions showed γH2AX accumulation that returned to baseline by 4 h recovery; whereas those cultured in SGF showed delayed γH2AX induction, increased γH2AX accumulation and elevated γH2AX even at 8 h recovery, reflecting delayed repair of DSB. We proceeded to test 8-oxoG DNA damage repair under the same SGF conditions. The initial step of 8-oxoG DNA repair requires BER of the damaged DNA base moiety. Hence, we developed an ex vivo assay, in which 8-oxoG excision from an 8-oxoG-containing molecular beacon was quantified. Lysates from myocytes cultured in SGF had significantly less capacity to excise 8-oxoG over the indicated time course, but this was rescued by SGF conditions that included ethyl pyruvate, which is a cell-permeable Kreb’s cycle substrate and hence a supplemental source of ATP (Student’s t test, P<0.01 for 6 and 24 h, P<0.001 for 48 h; Fig. 2B). The latter suggested that loss of BER in the context of nutrient deprivation was ATP-dependent.
Figure 2.
Nutrient deprivation regulates DNA repair. A) DNA DSB damage and repair was tracked by Western blot for γH2AX following the recovery from a stimulus with 100 μM tert-butyl hydroperoxide (BHP). Lysates were taken from HL1 myocytes cultured either in normal or SGF conditions before and during the recovery from the stimulus with BHP. B) An ex vivo assay of BER activity was set up using a fluorescent 8-oxoG-containing molecular beacon. BER activity was determined using lysates from HL1 myocytes cultured in SGF over the time course indicated. Decreased BER activity in SGF-treated cells was inhibited when these were cultured with ethyl pyruvate, a cell-permeable supplemental source of ATP. Rescue of BER activity reached similar levels as found in cells in control serum-containing medium. **P < 0.01, *** P < 0.001; Student’s t test.
Impaired BER is due to a decrease in OGG1 protein
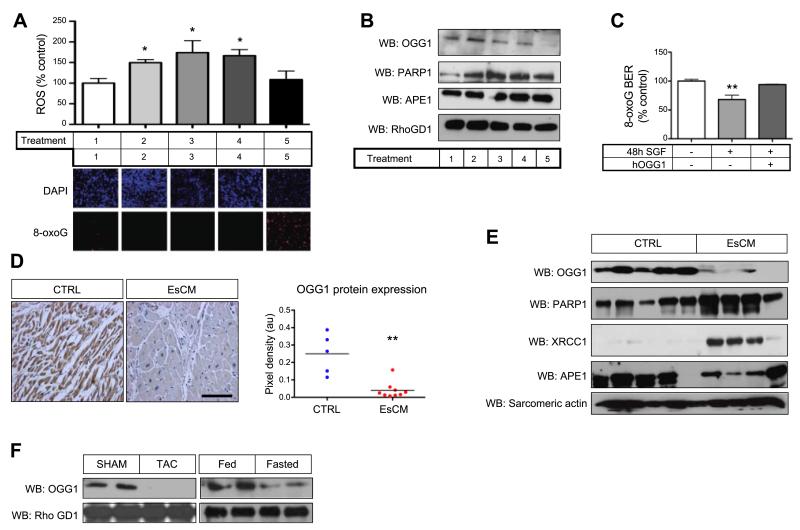
To assess the importance of nutrient availability relative to ROS production in more detail, we examined well-established models of hypertrophic and inflammatory stimuli known to increase ROS abundance both in cardiomyocytes and in vivo (23). Hypertrophic agonists phenylephrine (PE) and angiotensin II (AngII) and proinflammatory lipopolysaccharide (LPS) increased ROS abundance, but it had no effect on 8-oxoG accumulation. In contrast, 8-oxoG accumulation was pronounced only in SGF-treated cardiomyocytes (Fig. 3A). Because we suspected that defective DNA repair might have a significant role in DNA damage accumulation, we examined lysates from similarly treated myocytes for abundance of the key 8-oxoG base-excision repair enzyme OGG1, and other repair enzymes [poly (ADP-ribose) polymerase 1 (PARP1) and APE1] by Western blot analysis. While PE and LPS led only to a minimal decrease in OGG1, SGF treatment, in contrast, led to significantly reduced OGG1 protein but not PARP1 and APE1 (Fig. 3B). Moreover SGF-induced reduction of BER activity was rescued by the addition of recombinant OGG1 (100±2.96 vs. 68.2±7.53 vs. 94.0±0.72%; ANOVA, P<0.01; Fig. 3C). Next, we examined the expression of OGG1 in human EsCM hearts. OGG1 protein, by immunohistochemistry (0.25±0.05 vs. 0.04±0.02 pixels, P<0.01; Fig. 3D) and Western blot analysis (Fig. 3E), was significantly reduced in EsCM compared to control hearts. But OGG1 mRNA levels were unchanged (data not shown). The lack of similarly significant reduction in protein levels of other repair enzymes (PARP1, XRCC1, and APE1) often found in complex with OGG1, strengthened our conclusion for the importance of OGG1 in the context of our study. To demonstrate whether OGG1 protein abundance was similarly regulated in different forms of myocardial stress, Western blot analysis was also performed using murine cardiac lysates from 2 different in vivo models. Lysates from murine hearts subjected to transverse aortic constriction (8 wk) and in vivo starvation (48 h) both displayed reduced OGG1 protein content (Fig. 3F). For these in vivo models, myocardial ischemia has been demonstrated in the former (24), and in the latter, nutrient deprivation leads to the activation of cardiac autophagy (25–27). Hence, taken together, nutrient depletion promotes the loss of OGG1 protein both in vitro and in vivo. Therefore, we proceeded to examine the regulation of OGG1 specifically at the level of protein turnover.
Figure 3.
Reduced OGG1 protein abundance regulates DNA damage accumulation. A) Rat neonatal ventricular myocytes were cultured in 5% myocyte medium (control; lane 1) or in control medium plus 1 μM AngII (lane 2), 100 μM PE (lane3), 2 μg/ml LPS (lane 4), or SGF medium (lane 5) for 72 h. ROS production was assessed using DCFDA probe, and 8-oxoG was assessed by immunofluorescence using DAPI as a nuclear counterstain. *P < 0.05; ANOVA. B) Lysates from myocytes treated with the same conditions (lanes 1–5) were assessed at 48 h by Western blot for OGG1, PARP1, and APE1 protein abundance. RhoGDI was used to show equal protein loading. C) As in Fig. 2B, 8-oxoG BER ex vivo excision was assayed using lysates from myocytes treated for 48 h in control or SGF conditions, with and without 10 U of recombinant human OGG1 supplemented in the ex vivo reaction. **P < 0.01; ANOVA. D, E) OGG1 protein abundance in human control (CTRL) and EsCM cardiac tissue was assessed by immunohistochemistry (D) and Western blot analysis (E). Other repair enzymes (PARP1, XRCC1, and APE1) were also assessed by Western blot to highlight the significance of OGG1 in this context. Sarcomeric actin was used to show equal protein loading. **P < 0.01; Student’s t test. F) OGG1 protein abundance was assessed by Western blot using cardiac lysates from mice that had undergone transverse aortic constriction (pressure-overload cardiomyopathy) and 48 h in vivo starvation, compared to sham operation and normal fed conditions, respectively.
Autophagy is necessary, but not sufficient, for the loss of OGG1
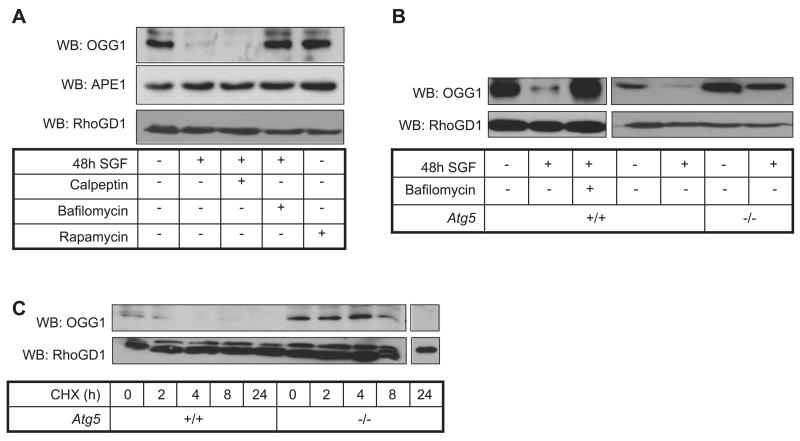
OGG1 protein abundance was examined in SGF-treated myocytes with and without inhibitors of calpain and autophagy. Cleavage of OGG1 protein by calpain has previously been described (28), and autophagy participates in protein turnover during nutrient starvation (29). Rapamycin was used as a pharmacological agonist of autophagy. OGG1 abundance was unaffected by the calpain inhibitor calpeptin under SGF treatment but was rescued by bafilomycin A1, a specific inhibitor of autophagosome-lysosome fusion (Fig. 4A). In contrast, rapamycin did not induce OGG1 down-regulation (Fig. 4A, lane 5), suggesting that autophagy is necessary for OGG1 turnover in SGF treatment but not sufficient by itself to induce changes in OGG1 expression. The latter may reflect the requirement for concurrent nutrient depletion. Levels of another repair enzyme, APE1, were unchanged by SGF or autophagy (Fig. 4A).
Figure 4.
Autophagy regulates OGG1 protein abundance under conditions of nutrient deprivation. A) Western blot analysis of OGG1 and APE1 protein abundance in myocytes cultured in SGF conditions for 48 h, with or without 50 μM calpeptin or 100 nM bafilomycin, or control medium with or without 100 nM rapamycin. B) Culture in SGF conditions for 48 h induced OGG1 protein loss in Atg5+/+ MEFs, which was inhibited by 100 nM bafilomycin, and SGF-induced OGG1 loss was not observed in autophagy-deficient Atg5−/− MEFs. RhoGDI was used to show equal protein loading. C) OGG1 protein half-life was prolonged in Atg5−/− MEFs compared to Atg5+/+ MEFs, as determined by treatment with the translation inhibitor cycloheximide (100 μg/ml).
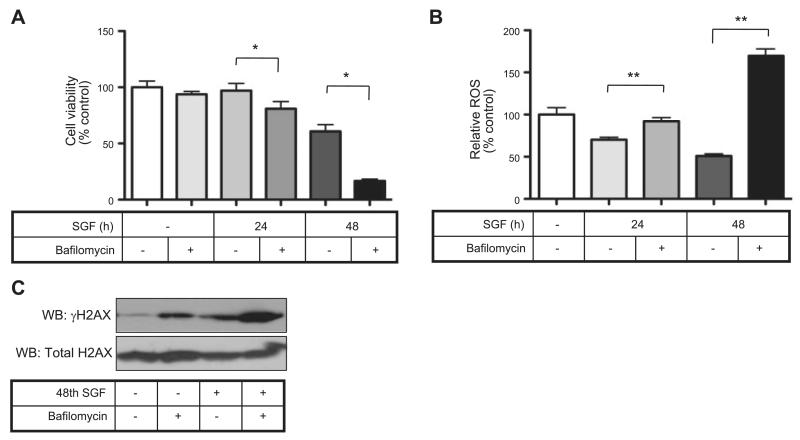
To confirm that autophagy influences OGG1 turnover in general, we examined the effect of SGF treatment on MEFs that were null for the autophagy gene 5 (Atg5). SGF treatment led to reduced OGG1 abundance in wild-type MEFs, which was reversible again with bafilomycin, but this was not seen in Atg5−/− MEFs (Fig. 4B). In addition, the protein half-life of OGG1 was prolonged in Atg5−/− MEFs compared to wild-type MEFs (Fig. 4C). Our data suggest that autophagy may in these conditions contribute to reduced OGG1 expression. However, others have demonstrated a protective role for autophagy under conditions of cell stress (26). Furthermore autophagy promotes ATP regeneration and would, therefore, be expected to maintain genomic stability and cell viability (26). We, therefore, further investigated the role of autophagy in SGF conditions. Autophagy inhibition with bafilomycin resulted in the loss of cell viability (Fig. 5A) and a relative increase in ROS production (Fig. 5B) in SGF-treated myocytes. Bafilomycin treatment also increased DSB formation, as monitored by γH2AX accumulation, particularly under SGF conditions (Fig. 5C). Hence, although autophagy appears to promote loss of OGG1 protein under conditions of nutrient starvation and impaired BER is OGG1 dependent, there is a critical balance because inhibition of autophagy itself also drives a sustained state of nutrient depletion, ROS up-regulation, and cell death (Fig. 6).
Figure 5.
Autophagy is adaptive and essential under conditions of nutrient deprivation. A) Autophagy inhibition with bafilomycin decreased cell viability under SGF conditions. *P < 0.05. B) Autophagy inhibition with bafilomycin up-regulates ROS production under SGF conditions. **P < 0.01. C) Western blot for γH2AX showed that genotoxic stress under SGF conditions was augmented by the presence of autophagy inhibition with bafilomycin.
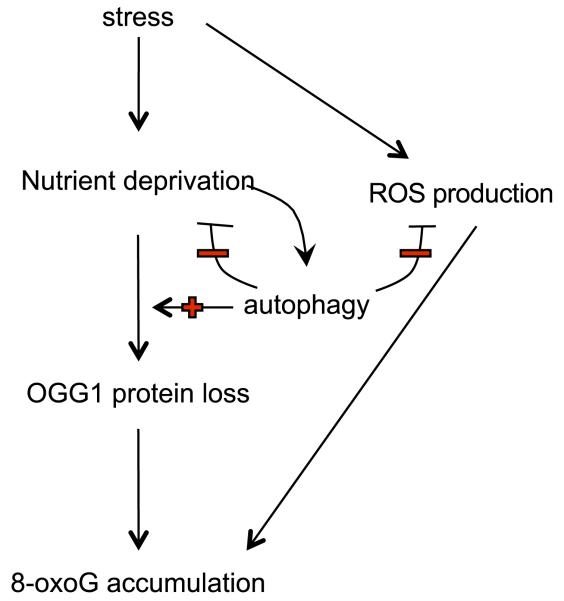
Figure 6.
Model of 8-oxoG accumulation in cardiac stress. 8-oxoG accumulation is induced by nutrient deprivation through OGG1 protein loss that is dependent on autophagy, but autophagy also promotes nutrient regeneration and inhibits ROS production.
DISCUSSION
Oxidative stress plays a key role in the progression of heart failure (5), but the regulation of DNA damage in cardiomyocytes is unclear. Although intuitively assumed, whether and how oxidative DNA damage contributes to disease progression is also unclear. Indeed, the significance of ROS-related genomic damage in human heart failure has till now not been examined in detail. Although there are many different causes of heart failure, oxidative stress is a unifying common pathway feature that can be found in all forms of heart failure, regardless of the initial inciting cause (5). Therefore, we have made a careful histological investigation of both ischemic and idiopathic EsCM human hearts and found that DNA damage accumulation and activation of the DDR occurred in >90% of cells in EsCM, with only low background levels in healthy age- and sex-matched control hearts. We have further found that nutrient availability regulates both base-excision repair and DSB repair. In the case of the former, BER was limited by the loss of the BER enzyme OGG1 both in vitro and in vivo.
Cardiomyocyte ROS production from hypertrophic and inflammatory agonists is well characterized (30), but our data suggest that 8-oxoG accumulation is regulated through reduced repair as opposed to ROS-inducing agonists. OGG1 protein loss and OGG1-dependent impairment of 8-oxoG excision suggest that OGG1 protein is the rate-limiting factor in 8-oxoG accumulation in myocytes. Moreover, 8-oxoG excision repair was effectively restored by exogenous OGG1. Our ex vivo results suggest that the remaining components of the base excision repair pathway are intact.
Autophagy is an adaptive cellular response to multiple stressors, including DNA damage, oxidative stress, nutrient limitation, and perturbations in protein quality control (29). Cardiomyocyte autophagy is also thought to be an adaptive response to nutrient deprivation, although the role of autophagy in vivo in murine models of heart failure progression is more complex (31). In starvation in vivo, autophagy preserves cardiac function; however, other data demonstrate the possibility of autophagic cell death as a contributor to cardiac remodeling and dilation (32). Our results suggest that cardiomyocyte adaptation to nutrient deprivation leads to autophagy-driven OGG1 protein loss. Autophagy induction alone has no effect on OGG1 expression in the absence of nutrient starvation. We also found that autophagy regulates OGG1 half-life in Atg5−/− MEFs. But paradoxically, autophagy is essential for maintaining cell viability and the inhibition of ROS in nutrient-deprived cardiomyocytes. Indeed inhibition of autophagy can have negative effects in multiple models of cardiac function in vivo (33, 34). In our context, although inhibition of autophagy during nutrient deprivation inhibits the loss of OGG1, it also up-regulates ROS abundance and reduces cell viability. The balance of these opposing effects of autophagy, therefore, appears to be finely tuned, and whether there are specific means to maintain OGG1 levels without affecting the beneficial effects of autophagy will require further investigation.
DNA damage is a critical regulator of cell death and senescence, and the latter processes are found in heart failure. Hence, there is a clear potential for DNA damage and the DDR to influence key cellular processes that contribute to heart failure pathology. Apart from the role of DNA damage in regulating cell death and senescence, the impact specifically of 8-oxoG accumulation in nondividing cells, such as cardiomyocytes is unknown. Typically, lesions such as 8-oxoG are believed to contribute to genomic instability and tumorigenesis in dividing cells based on replication-associated mispairing. 8-oxoG pairs opposite adenine during replication, promoting G:C to T:A transversion mutations (35). Similarly, 8-oxoG may mispair with adenine during transcription, leading to transcriptional mutagenesis (36). While OGG1-dependent BER is the major 8-oxoG excision system in mammalian cells, backup repair pathways, such as long-patch BER and nucleotide excision repair may also contribute to oxidative DNA damage repair (37). The sufficiency of these latter pathways in heart failure remains to be tested. Under constant conditions of oxidative stress in heart failure, we expect that modest reductions in a single pathway, such as deficiency in OGG1-mediated repair, may have important implications, particularly over time frames, during which cardiac decompensation develops in human disease.
It is already well established that OGG1 protein is protective against oxidative stress-induced injury and cell death in cellular models (38). Hence, the benefit of OGG1 may also be predicted in myocardial stress. Our study is important not only because it highlights a possible therapeutic target, OGG1, but also because manipulating OGG1 in future studies may allow us to understand the specific contribution of DNA damage in the disease development or progression of heart failure.
Acknowledgments
L.S. is supported by a Wellcome Trust Studentship in Metabolic and Cardiovascular Disease, M.R.B. is supported by a British Heart Foundation (BHF) professorship, and R.S.F. is supported by a BHF intermediate fellowship. This study is also supported by the UK National Institute for Health Research Cambridge Biomedical Research Centre.
Abbreviations
- 8-oxoG
8-oxo-7,8-dihydroguanine
- APE1
apurinic/apyrimidinic endonuclease 1
- Atg5
autophagy gene 5
- ATM
ataxia telangiectasia mutated
- ATP
adenosine-5′-triphosphate
- ATR
ataxia telangiectasia and Rad3-related
- BER
base-excision repair
- DDR
DNA damage response
- DSB
double-strand break
- EsCM
end-stage cardiomyopathy
- H3K36me3
trimethylated histone 3 lysine 36
- LPS
lipopoly-saccharide
- LV
left ventricular
- MEF
mouse embryonic fibroblast
- NHEJ
nonhomologous end joining
- OGG1
8-oxoguanine DNA glycosylase
- PARP1
poly (ADP-ribose) polymerase 1
- PE
phenylephrine
- ROS
reactive oxygen species
- SSB
single-strand break
- XRCC1
X-ray repair cross-complementing protein 1
REFERENCES
- 1.Bozkurt B, Mann DL, Deswal A. Biomarkers of inflammation in heart failure. Heart Fail. Rev. 2010;15:331–341. doi: 10.1007/s10741-009-9140-3. [DOI] [PubMed] [Google Scholar]
- 2.Whelan RS, Kaplinskiy V, Kitsis RN. Cell death in the pathogenesis of heart disease: mechanisms and significance. Annu. Rev. Physiol. 2010;72:19–44. doi: 10.1146/annurev.physiol.010908.163111. [DOI] [PubMed] [Google Scholar]
- 3.Neubauer S. The failing heart—an engine out of fuel. N. Engl. J. Med. 2007;356:1140–1151. doi: 10.1056/NEJMra063052. [DOI] [PubMed] [Google Scholar]
- 4.Giordano FJ. Oxygen, oxidative stress, hypoxia, and heart failure. J. Clin. Invest. 2005;115:500–508. doi: 10.1172/JCI200524408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hare JM. Oxidative stress and apoptosis in heart failure progression. Circ. Res. 2001;89:198–200. [PubMed] [Google Scholar]
- 6.Webster KA. Hypoxia: life on the edge. Antioxid. Redox Signal. 2007;9:1303–1307. doi: 10.1089/ars.2007.1730. [DOI] [PubMed] [Google Scholar]
- 7.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 8.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lammens K, Bemeleit DJ, Mockel C, Clausing E, Schele A, Hartung S, Schiller CB, Lucas M, Angermuller C, Soding J, Strasser K, Hopfner KP. The Mre11: Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell. 2011;145:54–66. doi: 10.1016/j.cell.2011.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osley MA, Tsukuda T, Nickoloff JA. ATP-dependent chromatin remodeling factors and DNA damage repair. Mutat. Res. 2007;618:65–80. doi: 10.1016/j.mrfmmm.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang JB, Goellner EM, Wang XH, Trivedi RN, St Croix CM, Jelezcova E, Svilar D, Brown AR, Sobol RW. Bioenergetic metabolites regulate base excision repair-dependent cell death in response to DNA damage. Mol. Cancer Res. 2010;8:67–79. doi: 10.1158/1541-7786.MCR-09-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Movassagh M, Choy MK, Goddard M, Bennett MR, Down TA, Foo RS. Differential DNA methylation correlates with differential expression of angiogenic factors in human heart failure. PLoS One. 2010;5:e8564. doi: 10.1371/journal.pone.0008564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tachibana H, Naga Prasad SV, Lefkowitz RJ, Koch WJ, Rockman HA. Level of beta-adrenergic receptor kinase 1 inhibition determines degree of cardiac dysfunction after chronic pressure overload-induced heart failure. Circulation. 2005;111:591–597. doi: 10.1161/01.CIR.0000142291.70954.DF. [DOI] [PubMed] [Google Scholar]
- 14.Foo RS, Chan LK, Kitsis RN, Bennett MR. Ubiquitination and degradation of the anti-apoptotic protein ARC by MDM2. J. Biol. Chem. 2007;282:5529–5535. doi: 10.1074/jbc.M609046200. [DOI] [PubMed] [Google Scholar]
- 15.Wencker D, Chandra M, Nguyen K, Miao W, Garantziotis S, Factor SM, Shirani J, Armstrong RC, Kitsis RN. A mechanistic role for cardiac myocyte apoptosis in heart failure. J. Clin. Invest. 2003;111:1497–1504. doi: 10.1172/JCI17664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Claycomb WC, Lanson NA, Jr., Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, Izzo NJ., Jr. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc. Natl. Acad. Sci. U. S. A. 1998;95:2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foo RS, Siow RC, Brown MJ, Bennett MR. Heme oxygenase-1 gene transfer inhibits angiotensin II-mediated rat cardiac myocyte apoptosis but not hypertrophy. J. Cell. Physiol. 2006;209:1–7. doi: 10.1002/jcp.20723. [DOI] [PubMed] [Google Scholar]
- 19.Choy MK, Movassagh M, Siggens L, Vujic A, Goddard M, Sanchez A, Perkins N, Figg N, Bennett M, Carroll J, Foo R. High-throughput sequencing identifies STAT3 as the DNA-associated factor for p53-NF-κB- complex-dependent gene expression in human heart failure. Genome Med. 2010;2:37. doi: 10.1186/gm158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mirbahai L, Kershaw RM, Green RM, Hayden RE, Meldrum RA, Hodges NJ. Use of a molecular beacon to track the activity of base excision repair protein OGG1 in live cells. DNA Repair (Amst.) 2010;9:144–152. doi: 10.1016/j.dnarep.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 21.Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ. Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 22.Bialik S, Cryns VL, Drincic A, Miyata S, Wollowick AL, Srinivasan A, Kitsis RN. The mitochondrial apoptotic pathway is activated by serum and glucose deprivation in cardiac myocytes. Circ. Res. 1999;85:403–414. doi: 10.1161/01.res.85.5.403. [DOI] [PubMed] [Google Scholar]
- 23.Molkentin JD, Dorn GW., 2nd Cytoplasmic signaling pathways that regulate cardiac hypertrophy. Annu Rev Physiol. 2001;63:391–426. doi: 10.1146/annurev.physiol.63.1.391. [DOI] [PubMed] [Google Scholar]
- 24.Sano M, Minamino T, Toko H, Miyauchi H, Orimo M, Qin Y, Akazawa H, Tateno K, Kayama Y, Harada M, Shimizu I, Asahara T, Hamada H, Tomita S, Molkentin JD, Zou Y, Komuro I. p53-induced inhibition of Hif-1 causes cardiac dysfunction during pressure overload. Nature. 2007;446:444–448. doi: 10.1038/nature05602. [DOI] [PubMed] [Google Scholar]
- 25.Zhu H, Tannous P, Johnstone JL, Kong Y, Shelton JM, Richardson JA, Le V, Levine B, Rothermel BA, Hill JA. Cardiac autophagy is a maladaptive response to hemodynamic stress. J. Clin. Invest. 2007;117:1782–1793. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takemura G, Kanamori H, Goto K, Maruyama R, Tsujimoto A, Fujiwara H, Seishima M, Minatoguchi S. Autophagy maintains cardiac function in the starved adult. Autophagy. 2009;5:1034–1036. doi: 10.4161/auto.5.7.9297. [DOI] [PubMed] [Google Scholar]
- 27.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 28.Hill JW, Hu JJ, Evans MK. OGG1 is degraded by calpain following oxidative stress and cisplatin exposure. DNA Repair (Amst.) 2008;7:648–654. doi: 10.1016/j.dnarep.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu. Rev. Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 30.Suliman HB, Welty-Wolf KE, Carraway M, Tatro L, Piantadosi CA. Lipopolysaccharide induces oxidative cardiac mitochondrial damage and biogenesis. Cardiovasc. Res. 2004;64:279–288. doi: 10.1016/j.cardiores.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 31.Gottlieb RA, Mentzer RM. Autophagy during cardiac stress: joys and frustrations of autophagy. Annu. Rev. Physiol. 2010;72:45–59. doi: 10.1146/annurev-physiol-021909-135757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knaapen MW, Davies MJ, De Bie M, Haven AJ, Martinet W, Kockx MM. Apoptotic versus autophagic cell death in heart failure. Cardiovasc. Res. 2001;51:304–312. doi: 10.1016/s0008-6363(01)00290-5. [DOI] [PubMed] [Google Scholar]
- 33.Kubota C, Torii S, Hou N, Saito N, Yoshimoto Y, Imai H, Takeuchi T. Constitutive reactive oxygen species generation from autophagosome/lysosome in neuronal oxidative toxicity. J. Biol. Chem. 2010;285:667–674. doi: 10.1074/jbc.M109.053058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Mizote I, Matsumura Y, Asahi M, Nishida K, Hori M, Mizushima N, Otsu K. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat. Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 35.Epe B. Role of endogenous oxidative DNA damage in carcinogenesis: what can we learn from repair-deficient mice? Biol. Chem. 2002;383:467–475. doi: 10.1515/BC.2002.049. [DOI] [PubMed] [Google Scholar]
- 36.Saxowsky TT, Meadows KL, Klungland A, Doetsch PW. 8-Oxoguanine-mediated transcriptional mutagenesis causes Ras activation in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 2008;105:18877–18882. doi: 10.1073/pnas.0806464105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oka S, Ohno M, Tsuchimoto D, Sakumi K, Furuichi M, Nakabeppu Y. Two distinct pathways of cell death triggered by oxidative damage to nuclear and mitochondrial DNAs. EMBO J. 2008;27:421–432. doi: 10.1038/sj.emboj.7601975. [DOI] [PMC free article] [PubMed] [Google Scholar]