Abstract
The study of melanocyte biology in the zebrafish presents a highly tractable system for understanding fundamental principles of developmental biology. Melanocytes are visible in the transparent embryo and in the mature fish following metamorphosis, a physical transformation from the larval to adult form. While early developing larval melanocytes are direct derivatives of the neural crest, the remainder of melanocytes develop from unpigmented precursors, or melanocyte stem cells (MSCs). The Tol2 transposable element has facilitated the construction of stable transgenic lines that label melanocytes. In another application, integration of Tol2 constructs makes possible clonal analysis of melanocyte and MSC lineages. Drugs that block melanin synthesis, ablate melanocytes, and block establishment of MSC populations allow the interrogation of this model system for mechanisms of adult stem cell development and regulation.
Keywords: Melanocyte, Stem cell, Clonal analysis, Lineages, Zebrafish, Tol2
1. Introduction
The distinctive pigment pattern of the adult zebrafish quickly led to the identification of spontaneous mutants (sparse, rose, and leopard) affecting melanocyte development, allowing initial exploration of the fundamental genes responsible for pigment stripe morphology (1– 3). Genetic screens have identified additional alleles that inform our understanding of embryonic melanocyte development (4, 5) and genes involved in adult metamorphosis (6).
The most fundamental technique for evaluating mutants or testing the effect of drugs on melanocyte development and homeostasis is the counting of melanocytes themselves. Melanocytes are first visible by 30 h post-fertilization (hpf) and quickly expand in number over the next 2 days (Fig. 1). Approximately 420 melanocytes are visible by 3 days post-fertilization (dpf), representing ~90% of all larval melanocytes that will develop prior to metamorphosis, which occurs around 14 dpf (7, 8). Wild-type melanocytes frequently abut one another, making accurate counts difficult to obtain. Epinephrine or norepinephrine treatment contracts the melanocytes after 4 dpf (4). Alternatively, the use of the mutant line mlphaj120 (9), which has constitutively contracted melanocytes, is particularly useful for phenocopying epinephrine treatment in large-scale applications, such as drug screens (10).
Fig. 1.
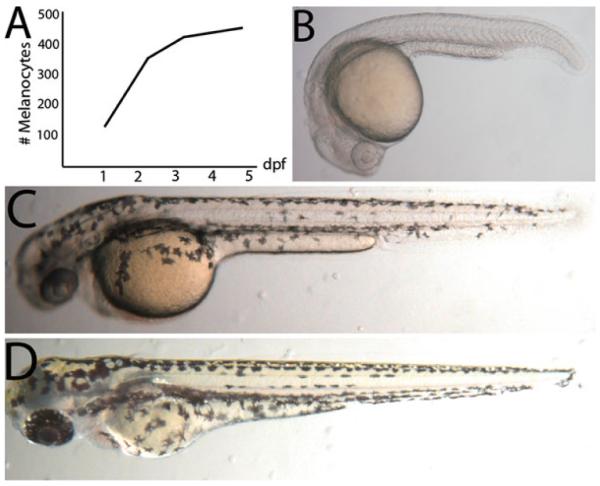
Melanocyte development in the zebrafish. (a) Timeline of melanocyte development in the larvae. (b) Zebrafish larvae at 1 dpf just prior to pigmented melanocytes becoming visible. (c) At 2 dpf, melanocytes are visible and migrating ventrally. (d) At 3 dpf, the ontogenetic, neural crest derived melanocytes have nearly reached the stereotypic dorsal, ventral, yolk, and lateral (right and left) stripes.
One property of some mutants, including sparse, is that melanocytes initially differentiate and melanize before ultimately undergoing apoptosis (2). Since melanocytes have a distinct cellular marker (melanin) and often take days to be extruded from the skin, it can be difficult to tell if the melanocytes are living or dead. The transgenic line Tg(fTyrp1 > eGFP)j900 expresses a GFP reporter driven by the Takifugu rubripes tyrosinase-related protein 1 (Tyrp1) promoter and is specifically expressed in melanocytes (11). Use of this transgenic line allows detection of cell death by looking for extinguishment of the GFP marker and can be combined with the mlphaj120 mutation to facilitate scoring of living melanocytes.
Zebrafish have a noted capacity for regenerating many tissue types, and properties of melanocyte regeneration were initially explored in fin amputation studies (12, 13). Following amputation of the caudal fin, the blastema regenerates all tissues including bone, artery, skin, and pigment cells, including melanocytes. Drugs that block melanin synthesis, like phenylthiocarbamide (PTU) (14), can be used to test hypotheses about melanocyte precursors in the regenerate. This work has shown that regenerated melanocytes in the fin arise from unpigmented precursors, or melanocyte stem cells (MSCs), rather than existing differentiated melanocytes (13).
Since melanocytes are dispensable in a laboratory setting, they may be ablated via physical (15) or chemical means (11, 16). Chemicals such as 4-hydroxyanisole (4-HA) that molecularly mimic endogenous ligands specifically metabolized in melanocytes produce cytotoxic by-products that ablate melanocytes (16). Following cell death and washout of the drug, MSCs are able to regenerate the ablated melanocyte population and can be studied for their regenerative potential. Consequently, genetic screens using melanocyte ablating drugs have identified novel genes involved in melanocyte regeneration (17).
An ErbB3 mutant that has deficiencies in melanocyte development at metamorphosis has led to the use of ErbB inhibitors such as AG1478 to effect MSC populations (18). Combining AG1478 and 4-HA treatments has made possible the study of MSC establishment during development and regulation of MSCs in the larvae (19). Use of these drugs with PTU has shown that melanocytes found in the early embryo are direct derivatives of the neural crest, developing between 1 and 3 dpf. Following this developmental window, a small proportion (~10%) of embryonic melanocytes are added in a regulative fashion from MSCs (20).
The development of the Tol2 transposable element as a tool for generating transgenic lines in the zebrafish has revolutionized zebrafish genetics (21). This tool can similarly be used to generate clones for studying developmental questions of lineage. Injection of small amounts of transposon harboring a melanocytespecific reporter construct permits the study of individual precursor cells that give rise to both ontogenetic melanocytes and MSCs. In addition, the use of ubiquitous promoters, such as EF1 α (22), allows the study of precursor cells that may give rise to multiple pigment cell types including melanocytes, xanthophores, and iridiphores in the zebrafish (23). An alternative approach for studying clones in the zebrafish was pioneered using gamma-irradiation (24). In a similar fashion, the albino mutant has unpigmented melanocytes and can be utilized to study the lineages of melanocytes and MSCs. Disruption or ablation of the albino+ allele by applying X-irradiation to heterozygous individuals (albb4/+) expressing the fTyrp1 > eGFP transgene results in clonally related albino melanocytes that can be clearly visualized on a pigmented melanocyte background.
2. Materials
2.1. Counting Melanocytes
Egg Water: 0.06 ppt scientific grade marine salt (Coralife) in carbon- filtered water. Add 1.2 g of salt to 20 L carbon- filtered water. Store at room temperature (RT).
Tricaine methanesulfonate (3-amino benzoic acid ethyl ester or 3-aminobenzoate) (Sigma Aldrich). For stock solution dissolve 400 mg tricaine powder in 97.9 mL DD water and 2.1 mL 1 M Tris (pH 9). Adjust pH to ~7. Store at RT.
Epinephrine (Sigma Aldrich) or norepinephrine (Sigma Aldrich). Working solution: 5 mg/mL in H2O. Can be stored at RT for up to 1 month.
TC thumb operated tally counter.
Mutant zebrafish line mlphaj120, deficient in melanophilin a (9). Available from lab of Steve Johnson (Washington University in St. Louis) and Zebrafish International Research Consortium (ZIRC).
Transgenic zebrafish line Tg(fTyrp1 > eGFP)j900, expressing eGFP under the control of the Takifugu rubripes tyrosinase-related protein 1 promoter (25). Available from lab of Steve Johnson (Washington University in St. Louis) and ZIRC.
2.2. Reverse Labeling of Melanocytes Using PTU
Phenylthiocarbamide (Sigma-Aldrich). Stock solution: 200 mM in ethanol (store at RT). Working solution: Dilute to 200 μ M in carbon- filtered water (adults) or egg water (embryos) prior to use.
Tricaine methanesulfonate (see Subheading 2.1).
Razor blade.
2.3. Birthdating Melanocytes Using fTyrp1 > eGFP Expression and PTU
Tg(fTyrp1 > eGFP)j900 (see Subheading 2.1 ).
PTU (see Subheading 2.2 ).
Stereomicroscope with fluorescence filters for GFP detection.
2.4. Drugs for Ablating Melanocytes and Melanocyte Stem Cells
Dimethyl sulfoxide (DMSO) (Sigma-Aldrich).
4-hydroxyanisole (4-HA) (Sigma-Aldrich). Stock solution: 10 mg/mL in DMSO. Store at −20°C in 50 μ L aliquots. Do not refreeze after thawing for use.
AG1478 (4-(3-Chloroanilino)-6,7-dimethoxyquinazoline, Calbiochem), an ErbB kinase inhibitor. Stock solution: 20 mM in DMSO. Store at −20°C in 20 μ L aliquots. Do not refreeze after thawing for use.
2.5. Clonal Analysis of Melanocyte Lineages Using Tol2 Transposon Labeling
Plasmid DNA containing fTyrp1 > eGFP reporter flanked by Tol2 transposon elements or EF1 α > GFP reporter flanked by Tol2 transposon elements.
Ambion mMessage mMachine SP6 kit (Ambion, Inc.).
Plasmid containing transposase open reading frame.
Phenol red (Sigma Aldrich); 1% solution in sterile milliQ water.
Sterile milliQ water.
Sutter P-87 Micropipette puller (Sutter Instrument Co.).
Glass thin walled capillary with filament;1.0 mm O.D, 0.75 mm I.D., 4 in. length (World Precision Instruments, Inc.).
Glass plate or slide covered with paraffin film.
Razor blade.
Dissecting scope (40× magnification) with reticle.
Sterile 30 G1 precision glide needle (Becton Dickinson).
Glass syringe, 25 μ l capacity.
MPPI-3 Pressure Injector with micropipette holder kit (Applied Scientific Instrumentation, Inc.).
Tank with compressed N2.
Micro-manipulator (World Precision Instruments, Inc.).
Mineral oil (Sigma Aldrich) in Petri dish.
Grooved silicon pad for holding fertilized eggs.
Stereomicroscope with fluorescence filters for GFP detection.
2.6. Lineage Analysis Using X-Ray Induced Clones
Albino mutant zebrafish line albb4/b4 (ZIRC).
Tg(fTyrp1 > eGFP)j900 (see Subheading 2.1).
X-ray machine (Faxitron Cabinet X-Ray System—Model 43855D) (see Note 1).
Stereomicroscope with fluorescence filters for GFP detection.
3. Methods
3.1. Counting Melanocytes
To anesthetize fish, use 1 mL stock tricaine solution per 25 mL egg water.
Once embryos are anesthetized, transfer five fish to a clean Petri dish and remove excess egg water. Add 100 μl of 5 mg/mL solution of epinephrine or norepinephrine to the embryos and wait 5–10 min for melanocytes to contract (Fig. 2a, b).
Once melanocytes are con firmed to be contracted, transfer embryos to fresh egg water containing tricaine so fish stay immobilized (see Note 2). Count melanocytes under a dissecting microscope with a TC thumb counter.
To facilitate counting, focus on the five embryonic melanocyte stripes (one dorsal, two lateral, one ventral, one yolk) one at a time.
Adult zebra fish can be anesthetized in a one-half strength tricaine solution (0. 5 mL per 25 mL) and epinephrine treated (5 mg/mL) concurrently. By the time fish are anesthetized melanocytes are typically contracted. If not fully anesthetized by time of melanocyte contraction, add a small additional amount of tricaine.
Transfer fish from tricaine/epinephrine solution to a Petri dish with a slotted spoon and quickly proceed to count melanocytes on a dissecting microscope. If fish begins to wake from anesthetic, return to water until immobile and proceed counting.
The mlphaj120 line (Fig. 2c) is particularly useful for counting melanocytes without the need for epinephrine treatment. In combination with fTyrp1 > eGFP expression (Fig 2d) melanocytes can be both counted and determined if alive when evaluating mutant phenotypes or effects of drugs on melanocyte development.
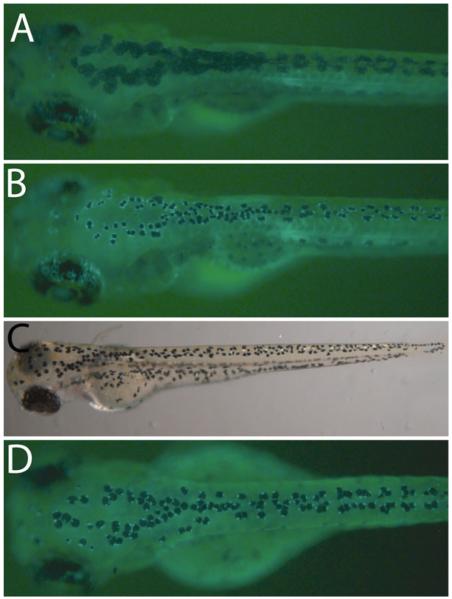
Fig. 2.
Techniques for visualizing individual melanocytes. (a) Wild-type zebrafish at 5 dpf expressing fTyrp1 > eGFP transgene. (b) 10 min epinephrine treatment (5 mg/mL) of fish shown in (a), which facilitates counting cells and clearly shows cells expressing GFP. (c) The mlphaj120 mutant line has constitutively contracted melanocytes that facilitate counting cells. (d) The fTyrp1 > eGFP transgene on a mlphaj120 background allows unambiguous identification of GFP + melanocytes.
3.2. Reverse Labeling of Melanocytes Using PTU
Anesthetize adult fish in tricaine.
Once fish are unresponsive, blot the fish several times on a paper towel to remove excess water, and place on a Petri dish.
Using a sharp razor blade, firmly press down on the caudal fin approximately one-half the distance between the distal tip of the fin and where the fin emerges from the body. Detach excised fin tissue and discard.
Place fish back into water to recover from anesthetic.
Following recovery, transfer fish to 200 μM solution of PTU to block melanin synthesis in newly developing melanocytes (Fig. 3b). PTU solution should be replaced every third day to ensure that melanin synthesis remains blocked.
To reveal pigmentation in regenerated melanocytes, remove fish from PTU and allow several days for melanin synthesis to resume and pigmentation to develop (Fig. 3c).
Fig. 3.
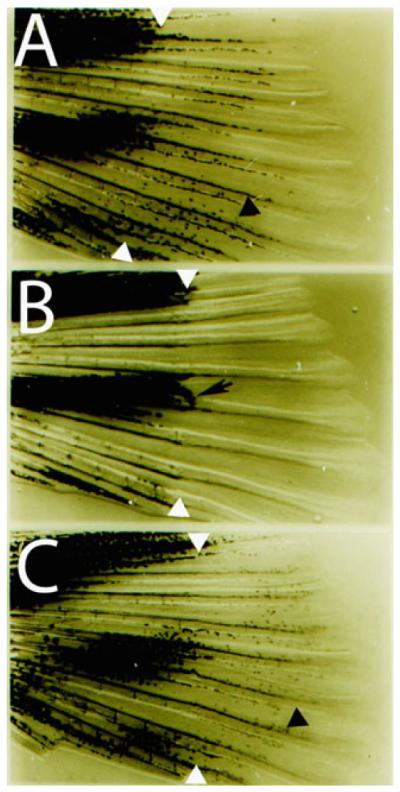
Reverse labeling of melanocytes using PTU. (a) Fin regeneration in an untreated caudal fin. White arrowheads indicate amputation plane and black arrowhead indicates regenerated pigmented melanocyte. (b) Fin regeneration in a PTU treated caudal fin. Note the lack of pigmented melanocytes in regenerate, except for a few melanocytes immediately distal to the amputation plane that preceded the amputation (black arrow). (c) Washout of PTU following fin regeneration (similar to b) reveals the presence of melanocytes (black arrowhead) that develop from unpigmented precursors.
3.3. Birthdating Melanocytes Using Tg(fTyrp1 > eGFP)j900 and PTU
Breed Tg(fTyrp1 > eGFP)j900 adults and rear embryos (26).
By 2 dpf, hundreds of ontogenetic melanocytes are visible and embryos can be sorted for those that inherited the transgenic GFP marker (see Note 3).
By 3 dpf, the majority (~90%) of ontogenetic embryonic melanocytes have developed. At this point, transfer embryos to a solution of egg water containing 200 μM PTU to block melanin synthesis in remaining melanocytes yet to differentiate.
Change egg water with PTU every 2 days.
At 7 dpf, screen embryos for GFP+, unmelanized pigment cells (Fig. 4).
Fig. 4.
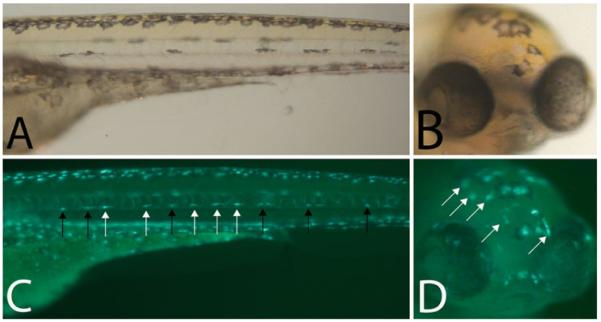
Birthdating melanocytes using Tg(fTyrp1 > eGFP)j900 in conjunction with PTU (a) Trunk and (b) head of 5 dpf zebrafish larvae treated with PTU at 2.5 dpf, blocking melanin synthesis in MSC derived embryonic melanocytes in the lateral stripe. (c, d) Epifluorescence of (a, b) reveals additional melanocytes that are not visible in bright field. Black arrows are previously differentiated melanized, GFP + melanocytes. White arrows are more recently differentiated unmelanized, GFP + melanocytes.
3.4. Drugs for Ablating Melanocytes and Melanocyte Stem Cells
Embryos can be produced either by in vitro fertilization or natural breeding (26).
Dilute stock solution of AG1478 to 3 μM in egg water prior to application. AG1478 treatment is optimal between 8 and 48 hpf (Fig. 5a). While melanocytes that are derived directly from the neural crest develop normally, later developing melanocytes that require a MSC intermediate fail to develop (Fig. 5e).
Dilute stock solution of 4-HA to 4 μg/mL in egg water prior to application. To block or kill ontogenetic melanocytes prior to their development, apply 4-HA at 1 dpf for 2 days (Fig. 5a, c). To kill previously differentiated melanocytes, apply 4-HA for 2 days (Fig. 5a) until melanocytes appear small and punctate and signs of detritus are evident.
To regenerate melanocytes from MSCs, washout 4-HA. Between 1 and 2 days post-washout, new melanocytes should be visible in all canonical embryonic zebra fish stripes, except for the yolk stripe which fails to regenerate (Fig. 5d) (16).
AG1478 and 4-HA treatments can be combined in the larvae for testing models of MSC regulation (Fig. 5f) (19).
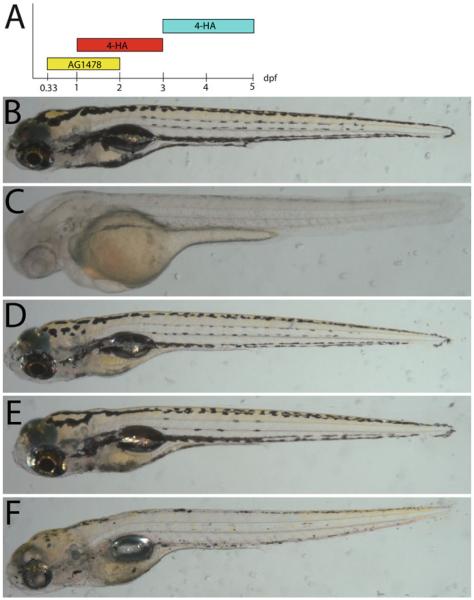
Fig. 5.
Drugs that ablate melanocytes and MSCs (a) Timeline of drug applications. Early 4-HA treatment ablates melanocytes prior to their melanization. Late 4-HA treatment ablates differentiated melanocytes. AG1478 treatment from 8 to 48 hpf blocks establishment of MSCs. (b) 8 dpf untreated zebrafish. (c) 2 dpf zebrafish treated with 4-HA beginning at 1 dpf. (d) 8 dpf zebrafish treated with 4-HA between 1 and 3 dpf fully regenerates. (e) 8 dpf zebrafish treated with AG1478 has normal development of neural-crest derived ontogenetic melanocytes but fails to fill in the lateral stripes as seen in (b) and (d). (f) 8 dpf zebrafish treated with AG1478 and 4-HA largely fails to regenerate melanocytes. The majority of pigment visible is detritus that has not yet cleared from the embryo.
3.5. Clonal Analysis of Melanocyte Lineages Using Tol2 Transposon Labeling
Prepare capped transposase mRNA using Ambion mMessege mMachine SP6 kit. Dilute to 75 ng/ μL and store at −70°C in 5 μL aliquots.
Prepare injection needles by pulling 4 in. 1 mm thin capillary needles under the following conditions: [H = 329, P = 150, V = 100, T = 200, Psi = 150], resulting in two usable injection needles per pull (see Note 4). Place pulled needle on a glass slide covered with paraf fin and view under a dissecting microscope. At full magnification (40×), align sharp tip of pulled needle with reticle markings and cut at a 45° angle with a razor blade so that the sharp, cut end is approximately one reticle tick wide.
Prepare injection solution consisting of: 1 μL fTyrp1 > eGFP: Tol2 plasmid (15 ng/ μL), 1 μL transposase mRNA (75 ng/ μL), 1 μL phenol red, and 12 μL sterile milliQ water (see Note 5).
Load 5–7 μL of injection solution into a glass syringe with a sterile 30 gauge needle. Insert 30 gauge needle into the blunt side of prepared injection needle and transfer injection solution. Solution should wick to the sharp, pulled point of the injection needle.
Insert injection needle into micropipette holder and tighten firmly without breaking the needle.
Open tank of compressed N2 and set pressure to 30 psi. Turn on pressure injector unit.
Adjust micro-manipulator to ~45 ° angle, and place tip of needle into a Petri dish one-half full of mineral oil, just below the surface. Center the tip of the injection needle immediately next to reticle and pulse one time to release a single injection of labeled solution. Adjust pressure on pressure injector unit until desired volume, approximately five reticle ticks wide at 40× magni fication, is consistently released on multiple pulses. Phenol red makes the injection solution clearly visible as it is injected into the mineral oil. Leave the injection needle submerged in mineral oil to avoid drying out and clogging the tip of needle when waiting for embryos to be bred.
Produce one clutch of zebra fish embryos via in vitro fertilization to ensure consistent, uniform timing of development. 15–20 min following application of sperm to eggs, cytoplasmic flow of the yolk into the one-cell embryo should be visible.
Transfer fertilized embryos to a grooved silicone pad lying in a Petri dish with a Pasteur pipet and remove excess egg water to reduce excessive rolling and movement of embryos while attempting to inject.
Carefully pierce the chorion of the one cell embryo with the injection needle and continue to puncture into the yolk, aiming for a location just underneath the visible single cell. Apply a single pulse to inject plasmid-transposase solution. Carefully remove the needle from the embryo (see Note 6).
Continue to inject remainder of eggs from the clutch until done (see Note 7).
Transfer injected embryos back into a Petri dish with egg water and place at 28.5° (standard temperature).
By 6–8 hpf, clean up embryos and remove all damaged, dead, or dying embryos. Allow embryos to grow at ~50 per Petri dish in egg water under standard conditions.
At 3 dpf, screen through injected embryos with a stereomicro-scope and set aside all fish with GFP + melanocytes (Fig. 6a–c).
Procedure for EF1 α > GFP:Tol2 plasmid is identical. Clones generated with this transposon are not limited to melanocytes since the promoter is ubiquitously expressed (Fig. 6d–f).
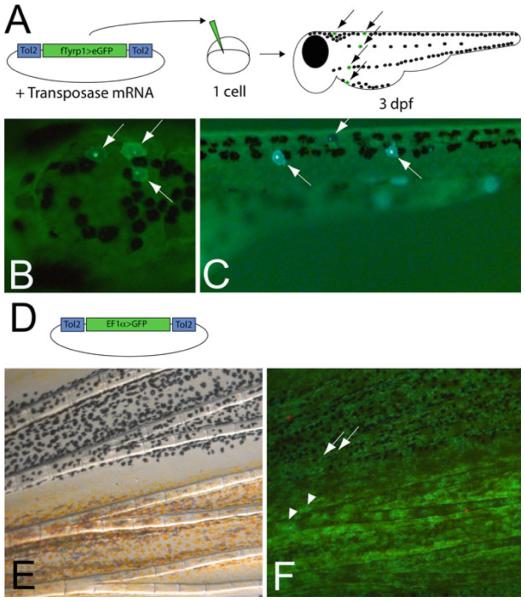
Fig. 6.
Clonal analysis of melanocyte lineages using Tol2 transposon labeling (a) Protocol for generating clones with the fTyrp1 > eGFP expressing melanocytes. Plasmid and transposase mRNA are coinjected into 1-cell embryo and screened at 3 dpf for GFP + melanocytes. (b) Epifluorescence of 3 GFP + melanocytes in the head and (c) trunk. (d) EF1 α > GFP transposon lineage construct used for generating clones that can be visualized in either melanocytes or other cells. (e) Bright field image of adult caudal fin. (f) Epifluorsecence image of fin. This experiment reveals that melanocytes (arrows) and xanthophores (arrowheads) develop from the same precursor.
3.6. Lineage Analysis Using X-Ray Induced Clones
Breed albino homozygotes (albb4/b4) with Tg(fTyrp1 > eGFP)j900 (see Note 8).
Sort embryos using a dissecting microscope and Pasteur pipet, removing unfertilized eggs and con firming approximately equivalent developmental staging of fertilized embryos.
Prepare X-ray cabinet by warming up X-ray tube incrementally.
When working with embryos, place up to 500 embryos in the center of a Petri dish and place in the X-ray cabinet directly underneath X-ray source (Fig. 7a). When working with adults, anesthetize fish in tricaine, quickly blot dry on paper towels, and transfer to Petri dish with a slotted spoon just prior to application of X-rays.
For low rates of clone production, embryos are placed ~30 cm from the X-ray source and exposed to 2 min of 120 kVp, equivalent to ~787 Rads (see Note 9).
At 2 dpf, once melanocytes have begun clearly expressing the fTyrp1 > eGFP marker, remove all fish lacking GFP expression (see Note 3).
At 3 dpf, sort through GFP-positive embryos for GFP+, albino melanocytes lacking pigment (Fig. 7b) (see Note 10).
Fig. 7.
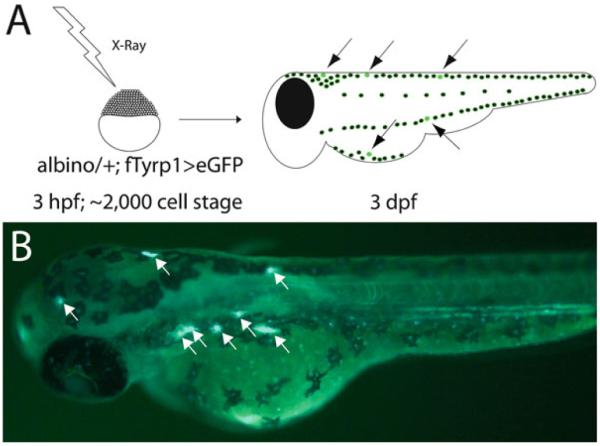
Lineage analysis using X-ray induced clones on albino heterozygous background. (a) Protocol for inducing albino melanocytes with X-rays. X-rays can be applied with temporal precision at various time points in development and screened after 3 dpf following completion of melanocyte differentiation. (b) Epifluorescence image of 3 dpf zebrafish following X-irradition to generate albino clones. fTyrp1 > eGFP reveals albino melanocytes (white arrows).
Footnotes
Any source of ionizing radiation, such as gamma rays from a cesium source, will suf fice to generate double strand breaks for the purpose of generating hemizygous clones.
Larval fish will die or visibly begin to degrade if left in epinephrine or norepinephrine for periods longer than 10 min. Adult fish can typically withstand up to 30 min in epinephrine without negative effects.
In embryos with wild-type melanocytes, sorting after 2 dpf makes identifying GFP expression more dif ficult as melanin levels increase. Treatment with epinephrine (see Subheading 3.1) to contract melanocytes can facilitate sorting in older embryos.
H = heat; P = pull; V = velocity; T = time, Psi = Pressure.
The amount or concentration of plasmid and transposase can be varied to increase or decrease the number of clones generated per injected clutch. The concentrations indicated result in approximately 5–10% of injected fish containing melanocyte clones.
If properly injected, the phenol red in the injection solution will cause a small area of yolk to be distinctly dyed red. If dye quickly wicks away and does not stain the yolk, the needle was not sufficiently inserted and the transposon will likely fail to integrate.
We find that a single investigator can perform the in vitro fertilization and transposon injection using 3–4 clutches and labeling 500–1,000 embryos per morning.
If application of X-rays at specific early developmental time points is essential, in vitro fertilization is the optimal method for breeding (26). Alternatively, if natural breeding is used, barriers separating males and females should be removed in the morning to avoid rare, accidental late night breeding.
Amount of X-rays applied can be varied depending on the number of precursors to be induced per fish. Empirical testing of X-ray dosage in relation to the number of clones generated should be done.
While melanocytes in the dorsal and lateral stripes are particularly easy to identify, care should be taken when examining the ventral strip near the yolk, which is more difficult to screen.
References
- 1.Johnson SL, Africa D, Walker C, Weston JA. Genetic control of adult pigment stripe development in Zebrafish. Dev Biol. 1995;167:27–33. doi: 10.1006/dbio.1995.1004. [DOI] [PubMed] [Google Scholar]
- 2.Parichy DM, Rawls JF, Pratt SJ, Whitfield TT, Johnson SL. Zebrafish sparse corresponds to an orthologue of c-kit and is required for the morphogenesis of a subpopulation of melanocytes, but is not essential for hematopoiesis or primordial germ development. Development. 1999;126:3425–36. doi: 10.1242/dev.126.15.3425. [DOI] [PubMed] [Google Scholar]
- 3.Parichy DM, Mellgren EM, Rawls JF, Lopes SS, Kelsh RN, Johnson SL. Mutational analysis of endothelin receptor b1 (rose) during neural crest and pigment pattern development in the zebrafish Danio rerio. Dev Biol. 2000;227:294–306. doi: 10.1006/dbio.2000.9899. [DOI] [PubMed] [Google Scholar]
- 4.Rawls JF, Johnson SL. Temporal and molecular separation of the kit receptor tyrosine kinase's roles in melanocyte migration and survival. Dev Biol. 2003;262:152–61. doi: 10.1016/s0012-1606(03)00386-5. [DOI] [PubMed] [Google Scholar]
- 5.Mellgren EM, Johnson SL. A requirement for kit in embryonic zebrafish melanocyte differentiation is revealed by melanoblast delay. Dev Gene Evol. 2004;214:493–502. doi: 10.1007/s00427-004-0428-y. [DOI] [PubMed] [Google Scholar]
- 6.Parichy DM, Ransom DG, Paw B, Zon LI, Johnson SL. An orthologue of the kit-related gene fms is required for development of neural crest-derived xanthophores and a sub- population of adult melanocytes in the zebrafish, Danio rerio. Development. 2000;127:3031–3044. doi: 10.1242/dev.127.14.3031. [DOI] [PubMed] [Google Scholar]
- 7.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 8.Parichy DM, Elizondo MR, Mills MG, Gordon TN, Engeszer RE. Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Dev Dyn. 2009;238:2975–3015. doi: 10.1002/dvdy.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheets L, Ransom DG, Mellgren EM, Johnson SL, Schnapp BJ. zebrafish melanophilin facilitates melanosome dispersion by regulating dynein. Curr Biol. 2007;17:1721–34. doi: 10.1016/j.cub.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hultman KA, Scott AW, Johnson SL. Small molecule modifier screen for kit-dependent functions in zebrafish embryonic melanocytes. zebrafish. 2008;5:279–287. doi: 10.1089/zeb.2008.0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Reilly-Pol T, Johnson SL. Neocuproine ablates melanocytes in adult zebrafish. zebrafish. 2008;5:257–264. doi: 10.1089/zeb.2008.0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rawls JF, Johnson SL. zebrafish kit mutation reveals primary and secondary regulation of melanocyte development during fin stripe regeneration. Development. 2000;127:3715–24. doi: 10.1242/dev.127.17.3715. [DOI] [PubMed] [Google Scholar]
- 13.Rawls JF, Johnson SL. Requirements for the kit receptor tyrosine kinase during regeneration of zebrafish fin melanocytes. Development. 2001;128:1943–9. doi: 10.1242/dev.128.11.1943. [DOI] [PubMed] [Google Scholar]
- 14.Milos N, Dingle A. Dynamics of pigment pattern formation in the zebrafish, Brachydanio rerio: I. Establishment and regulation of the lateral line melanophore stripe during the first eight days of development. J Exp Zool. 1978;205:205–16. [Google Scholar]
- 15.Yang CT, Sengelmann RD, Johnson SL. Larval melanocyte regeneration following laser ablation in zebrafish. J Invest Dermatol. 2004;123:924–29. doi: 10.1111/j.0022-202X.2004.23475.x. [DOI] [PubMed] [Google Scholar]
- 16.Yang CT, Johnson SL. Small molecule-induced ablation and subsequent regeneration of larval zebrafish melanocytes. Development. 2006;133:3563–73. doi: 10.1242/dev.02533. [DOI] [PubMed] [Google Scholar]
- 17.Yang CT, Hindes A, Hultman KA, Johnson SL. Mutations in gfpt1 and skiv2l2 cause distinct stage-specific defects in larval melanocyte regeneration in zebrafish. PLoS Genet. 2007;3:e88. doi: 10.1371/journal.pgen.0030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Budi EH, Patterson LB, Parichy DM. Embryonic requirements for ErbB signaling in neural crest development and adult pigment pattern formation. Development. 2008;135:2603–14. doi: 10.1242/dev.019299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hultman K, Budi E, Teasley D, Gottlieb A, Parichy D, Johnson SL. Defects in ErbB-dependent establishment of adult melanocyte stem cells reveals independent origins for embryonic and regeneration melanocytes. PLoS Genet. 2009;5:e1000544. doi: 10.1371/journal.pgen.1000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hultman KA, Johnson SL. Differential contribution of direct-developing and stem cell-derived melanocytes to the zebrafish larval pigment pattern. Dev Biol. 2010;337:425–3. doi: 10.1016/j.ydbio.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawakami K, Koga A, Hori H, Shima A. Excision of the tol2 transposable element of the medaka fish, Oryzias latipes, in zebrafish, Danio rerio. Gene. 1998;225:17–22. doi: 10.1016/s0378-1119(98)00537-x. [DOI] [PubMed] [Google Scholar]
- 22.Johnson AD, Krieg PA. A Xenopus laevis gene encoding EF-1 alpha S, the somatic form of elongation factor 1 alpha: sequence, structure, and identification of regulatory elements required for embryonic transcription. Dev Genet. 1995;17:280–90. doi: 10.1002/dvg.1020170313. [DOI] [PubMed] [Google Scholar]
- 23.Tu S, Johnson SL. Clonal analyses reveal roles of organ founding stem cells, melanocyte stem cells, and melanoblasts in establishment, growth, and regeneration of the adult zebrafish fin. Development. 2010;137:3931–3939. doi: 10.1242/dev.057075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Streisinger G, Coale F, Taggart C, Walker C, Grunwald DJ. Clonal origins of cells in the pigmented retina of the zebrafish eye. Dev Biol. 1989;131:60–69. doi: 10.1016/s0012-1606(89)80038-7. [DOI] [PubMed] [Google Scholar]
- 25.Zou J, Beermann F, Wang J, Kawakami K, Wei X. The Fugu TYRP11 promoter directs specific GFP expression in zebrafish: tools to study the RPE and the neural crest derived melanophores. Pigment Cell Res. 2006;19:615–627. doi: 10.1111/j.1600-0749.2006.00349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Westerfeld M. A guide for the laboratory use of zebrafish (Danio rerio) 4th edn. University of Oregon Press; Eugene, OR: 2000. The zebrafish book. [Google Scholar]