Abstract
DNA alkylation damage can be repaired by nucleotide excision repair (NER), base excision repair (BER) or by direct removal of alkyl groups from modified bases by O 6-alkylguanine DNA alkyltransferase (AGT; E.C. 2.1.1.63). DNA mismatch repair (MMR) is also likely involved in this repair. We have investigated alkylation-induced mutagenesis in a series of NER- or AGT-deficient Escherichia coli strains, alone or in combination with defects in the MutS, MutL or MutH components of MMR. All strains used contained the Fʹprolac from strain CC102 (FʹCC102) episome capable of detecting specifically lac GC to AT reverse mutations resulting from O 6-alkylguanine. The results showed the repair of O 6-methylguanine to be performed by AGT ≫ MMR > NER in order of importance, whereas the repair of O 6-ethylguanine followed the order NER > AGT > MMR. Studies with double mutants showed that in the absence of AGT or NER repair pathways, the lack of MutS protein generally increased mutant frequencies for both methylating and ethylating agents, suggesting a repair or mutation avoidance role for this protein. However, lack of MutL or MutH protein did not increase alkylation-induced mutagenesis under these conditions and, in fact, reduced mutagenesis by the N-alkyl-N-nitrosoureas MNU and ENU. The combined results suggest that little or no alkylation damage is actually corrected by the mutHLS MMR system; instead, an as yet unspecified interaction of MutS protein with alkylated DNA may promote the involvement of a repair system other than MMR to avoid a mutagenic outcome. Furthermore, both mutagenic and antimutagenic effects of MMR were detected, revealing a dual function of the MMR system in alkylation-exposed cells.
Introduction
Alkylation is a common class of DNA damage, which can be induced by various environmental chemicals and possibly endogenous substances. From bacteria to humans, many types of DNA repair systems operate to repair the damage, such as nucleotide excision repair (NER), base excision repair (BER) and a system to remove alkyl groups directly from modified bases as in the case with O 6-alkylguanine DNA alkyltransferase (AGT; E.C. 2.1.1.63). Mismatch repair systems (MMRS) may also be involved in this repair, although their precise role is still unclear. MMRS play an important role in removing mismatched base pairs and small insertion/deletion loops during DNA replication and in maintaining high fidelity of genome DNA (1–4). MMRS also target alkylated base lesions. O 6-alkylguanine-containing base pairs caused by treatments with alkylating agents are recognised by MutS, a protein recognising mismatch lesions, and become subjects of MMRS (5). We previously demonstrated that deficiency of MutS increased the mutation frequency induced by methylating agents more than that of ethylating agents, a finding that correlated well with the observed higher MutS binding to methylated guanine base pairs than to corresponding ethylated base pairs (6). Recently, Lupari et al. (7) reported that translesion synthesis (TLS) polymerase-related rescue of cytotoxicity induced by methylating agents depended on MMR.
Among the DNA repair systems, AGT is thought to play a major role to protect organisms from alkylation damage (8,9). Escherichia coli AGT consists of an inducible enzyme, Ada, and a constitutive enzyme, Ogt. Both enzymes efficiently remove alkylated bases by transferring the alkyl group from DNA to themselves in a suicidal process resulting in enzyme inactivation (10). NER is also well known to repair a broad range of DNA lesions including alkylated bases (11). However, it is unclear which repair system is primarily involved in the repair of each alkylated damage created by various methylating and ethylating agents. In this study, we examine the mutant frequencies induced by alkylating agents in various repair backgrounds to estimate their relevant contributions. Rye et al. (12) reported that MMR proteins collaborate with AGT in the repair of O 6-methylguanine. On the other hand, lack of AGT is known to enhance methylation-induced cell damage such as apoptosis and chromosomal instability through futile cycles of MMR or through direct signalling by MMR proteins (8,13–16). As repair pathways likely compete, one informative way to examine the role of MMR is to study its effects on alkylation mutagenesis in the absence of AGT or NER. For this reason, we constructed MMR-deficient strains additionally deficient in AGT or NER activity and measured the mutant frequencies for a series of alkylating agents. The results suggested that little or no alkylation-containing base pairs are corrected by the MMRS itself. Nevertheless, alkylated base pairs are subject to interaction with MutS protein, as we observed that mutS deficiency increased the mutagenic activity of methyl methanesulphonate (MMS), ethyl methanesulphonate (EMS), N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) and N-ethyl-N′-nitro-N-nitroso-guanidine (ENNG). However, these increases were not seen in the case of mutL or mutH deficiencies. We, therefore, suggest that MutS interaction with alkylated base pairs leads indirectly to repair or mutation avoidance mediated by a repair system other than MMR.
Materials and methods
Chemicals
N-methyl-N-nitrosourea (MNU) (CAS 684-93-5) and ENNG (CAS 63885-23-4) were purchased from Nakarai Tesque (Kyoto, Japan) and EMS (CAS 62-50-0) from Wako Pure Chemicals (Osaka, Japan). N-ethyl-N-nitrosourea (ENU) (CAS 759-73-9), MMS (CAS 66-27-3) and MNNG (CAS 70-25-7) were purchased from Sigma–Aldrich Chemicals (St Louis, MO, USA).
Bacterial strains and plasmids
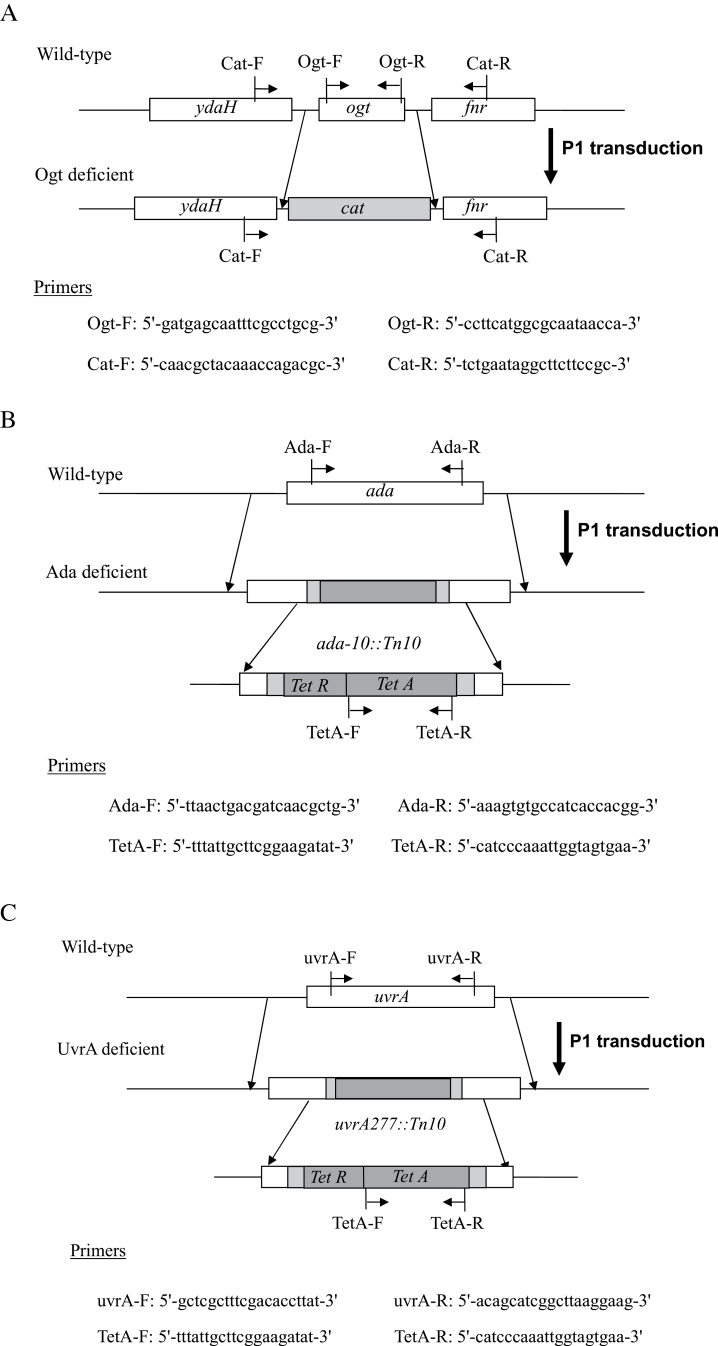
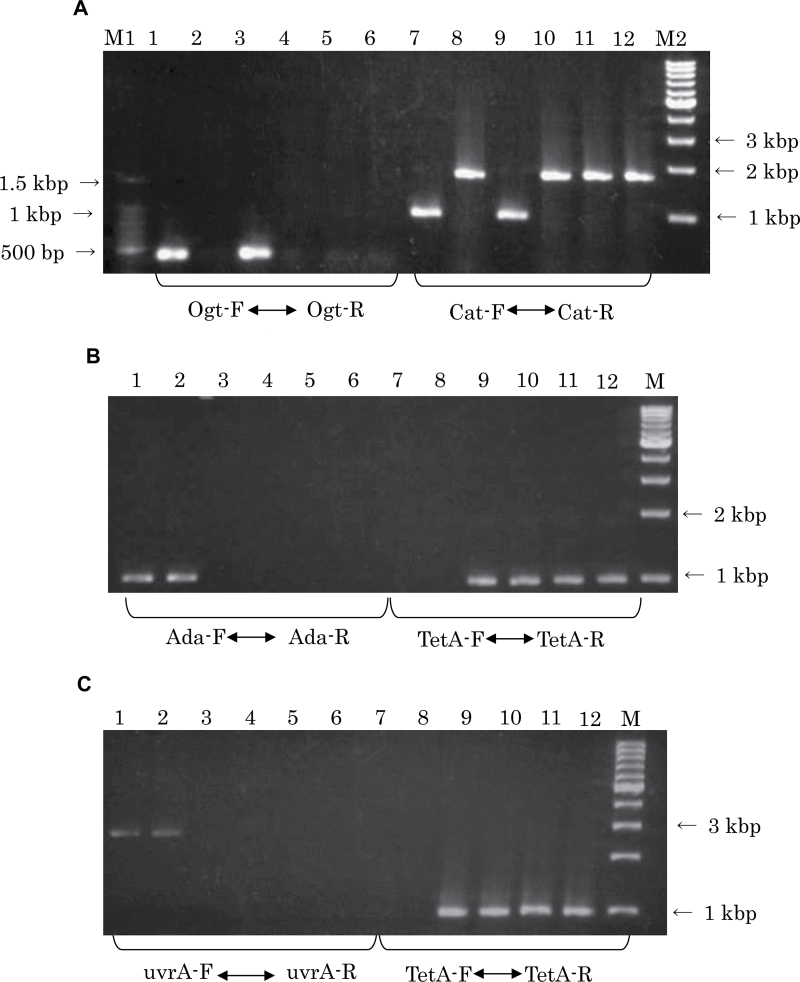
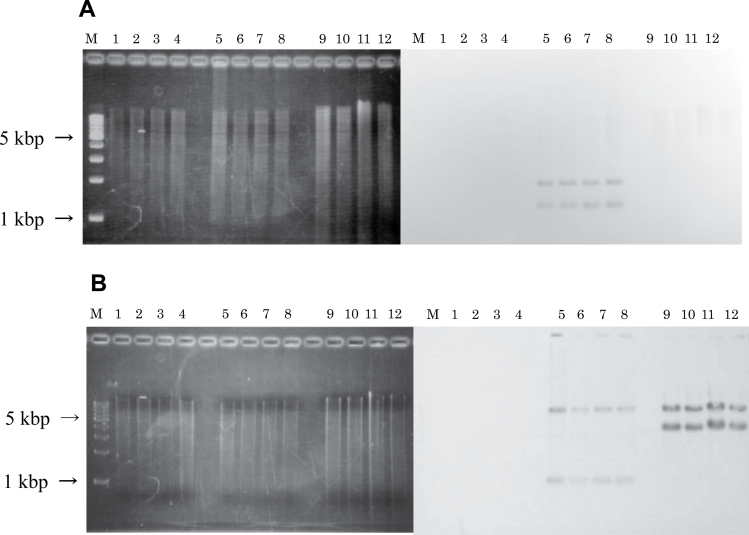
The bacterial strains used in this study are shown in Table I. All strains used in the mutagenesis experiments are derived from strain KA796 (ara, thi and Δpro-lac) (17) and also containing the Fʹprolac from strain CC102 (FʹCC102) (25) that permits scoring of lac GC to AT transitions, as described (17,25). The wild-type, NR10832; the MMR-deficient derivatives, NR12896 (mutS201::Tn5), NR11102 (mutL211::Tn5), NR12897 (mutH471::Tn5); and the NER-deficient derivative, NR12999 (uvrA277::Tn10) have been described by Negishi et al. (17). AGT-deficient strains GW5352 (ada-10::Tn10) (18) and KT233 (ada::kan and ogt::cat) (19) were gifts from Drs S. Cohen (Massachusetts Institute of Technology) and M. Sekiguchi (Kyushu University), respectively. Mutants doubly deficient in MMR and either AGT or NER were constructed in this study. P1 transductions, performed as described previously (20), were used to introduce the ada and ogt disruptions from GW5352 and KT233, respectively, and the uvrA277::Tn10 marker from NR12999, using selection for antibiotic resistance (tetracycline or chloramphenicol resistance for Tn10 or cat gene insertions, respectively) (see also Figure 1). Disruption of the genes in the final strains was confirmed by observed loss of PCR products amplified from the ada, ogt and uvrA genes, using as specific primers: 5′-gatgagcaatttcgcctgcg-3′ (forward) and 5′-ccttcatggcgcaataacca-3′ (reverse) for ogt; 5′-ttaactgacgatcaacgctg-3′ (forward) and 5′-aaagtgtgccatcaccacgg-3′ (reverse) for ada; and 5′-gctcgctttcgacaccttat-3′ (forward) and 5′-acagcatcggcttaaggaag-3′ (reverse) for uvrA (see Figure 2). To confirm that the transduced genes were inserted in the correct locations in the E. coli genome, we performed Southern blot analysis as previously described (20). Results are shown in Figure 3. Probes used were a 553-bp PCR-amplified region within the cat gene (forward primer 5′-atcccaatggcatcgtaaag-3′ and reverse primer 5′-atcacagacggcatgatgaa-3′) and a 1000-bp PCR-amplified region within the tetA gene (forward primer 5′-ttattgcttcggaagatat-3′ and reverse primer 5′-catcccaaattggtagtgaa-3′). PCR products were purified using the GenEluteTM Clean-Up Kit (Sigma–Aldrich Chemicals, St Louis, MO, USA) and labelled with digoxigenin using a PCR DIG Probe Synthesis Kit (Roche Diagnostics, Indianapolis, IN, USA) in accordance with the manufacturer’s protocols. Genomic DNA extracted from each E. coli strain was digested with two restriction enzymes, EcoRI and PstI (Takara Bio Inc., Otsu, Japan), at 37°C overnight. Digested samples were electrophoresed on a 0.8% agarose gel and transferred to a nitrocellulose membrane (Hybond-N+; GE Healthcare Life Sciences, Tokyo, Japan). The membrane was baked at 80°C for 2h and then subjected to pre-hybridisation and hybridisation in DIG Easy Hyb (Roche Diagnostics, Indianapolis, IN, USA) according to the manufacturer’s protocol. Two bands hybridising with cat gene-specific probes were observed only in strains in which the cat gene should be inserted (KT11141, KT21181 and KT31131), whereas the bands were not observed in the parental strains and uvrA-disrupted strains. For the Tn10-carrying strains, 7-kb and 1-kb bands were detected in ada-deficient strains and 7-kb and 4.5-kb bands in the uvrA-deficient strains. (As the sequence of the region annealed by the probes contained an EcoRI site, two bands hybridised to the probe.) Taken together, the results show that the newly constructed E. coli strains are, as expected, deficient in AGT or NER function, in addition to their MMR deficiency. The double-mutant strains were as resistant to the toxicity of MNNG as the single MMR-deficient strains (data not shown). The spontaneous mutant frequencies of E. coli strains used in this study are shown in Table I. MMR-deficient strains show mutant frequencies 20–100 times higher than the proficient strains.
Table I.
E. coli strains used in this study and their spontaneous mutant frequencies
| Straina | Relevant genotype | Spontaneous mutant frequencyb (×107) | References |
|---|---|---|---|
| NR10832 | KA796, wild-type | 0.3±0.1 | [17] |
| NR12896 | KA796, mutS201::Tn5 | 34.3±3.3 | [17] |
| NR11102 | KA796, mutL211::Tn5 | 27.5±2.4 | [17] |
| NR12897 | KA796, mutH471::Tn5 | 28.9±8.1 | [17] |
| NR12999 | KA796, uvrA277::Tn10 | 0.4±0.1 | [17] |
| GW5352 | AB1157, ada-10::Tn10 | — | [18] |
| KT233 | AB1157, ada::kan, ogt::cat | — | [19] |
| KT01121 | NR10832, ada-10::Tn10, ogt::cat | 1.1±0.6 | This study |
| KT11141 | NR12896, mutS201::Tn5, ada-10::Tn10, ogt::cat | 41.7±7.4 | This study |
| KT21181 | NR11102, mutL211::Tn5, ada-10::Tn10, ogt::cat | 21.1±0.5 | This study |
| KT31131 | NR12897, mutH471::Tn5, ada-10::Tn10, ogt::cat | 32.6±5.8 | This study |
| KT10021U | NR12896, mutS201::Tn5, uvrA277::Tn10 | 43.0±9.4 | This study |
| KT20021U | NR11102, mutL211::Tn5, uvrA277::Tn10 | 13.1±2.1 | This study |
| KT30013U | NR12897, mutH471::Tn5, uvrA277::Tn10 | 34.7±3.7 | This study |
Fig. 1.
Ogt, Ada and UvrA knockouts of E. coli used in this study. Also shown are the primer sequences and their locations as used for their confirmation after P1 transduction into the relevant tester strains. See Materials and methods for details. (A) Disruption of the ogt gene by replacement of the chloramphenicol resistance gene (cat), (B) disruption of ada gene introduced by tetracycline resistant gene (tetR and tetA) and (C) disruption of the uvrA gene as in (B).
Fig. 2.
Analysis of gene disruptions genes in newly created strains by PCR. For primers used, see Figure 1. (A) ogt and cat genes: lanes 1 and 7, wild-type (NR10382); lanes 2 and 8, ogt (KT233); lanes 3 and 9, mutS (NR12896); lanes 4 and 10, mutS ada ogt (KT11141); lanes 5 and 11, mutL ada ogt (KT21181); lanes 6 and 12, mutH ada ogt (KT31131), M1, 100-bp marker; M2, 1-kb marker. (B) ada and tetA genes: lanes 1 and 7, wild-type (NR10382); lanes 2 and 8, ogt (KT233); lanes 3 and 9, mutS (NR12896); lanes 4 and 10, mutS ada ogt (KT11141); lanes 5 and 11, mutL ada ogt (KT21181); lanes 6 and 12, mutH ada ogt (KT31131); M, 1-kb marker. (C) uvrA and tetA genes: lanes 1 and 7, wild-type (NR10382); lanes 2 and 8, ogt (KT233); lanes 3 and 9, mutS (NR12896); lanes 4 and 10, mutS ada ogt (KT11141); lanes 5 and 11; mutL ada ogt (KT21181); lanes 6 and 12: mutH ada ogt (KT31131); M, 1-kb marker.
Fig. 3.
Southern blot analysis to confirm correct chromosomal location of gene disruptions following P1 transduction. Chromosomal DNA was digested with EcoRI and PstI and probed with gene-specific PCR products. See Materials and methods for details of procedures and primers used. (A) Confirmation of correct location of the cat gene. On the left, polyacrylamide gel stained with CBB (Coomassie Brilliant Blue); on the right, the corresponding membrane blotted with cat gene-derived PCR product. (B) Confirmation of tetA gene. On the left, polyacrylamide gel stained with CBB; on the right, the corresponding membrane blotted with a tetA-gene derived PCR product.
The plasmids containing each wild-type MMR gene were introduced into corresponding double mutants by electroporation (Electroporator EC-100, Thermo Quest, Milan, Italy). Plasmid pMQ315 contains the mutS + gene (21), plasmid pMQ350 contains the mutL + gene and pRH71-17 (22) contains the mutH + gene (23). All three plasmids are derivatives of plasmid pBR322 (24). Cells transformed with plasmid were selected on Luria–Bertani agar containing ampicillin. The transformation of plasmid pMQ315 (mutS +) into mutS-deficient E. coli (KT11141 or KT10021U) decreased the spontaneous mutant frequency, whereas the frequency was not influenced by the transfection of pBR322, although the extent of spontaneous mutation did not decrease to the level of parental AGT-deficient strain (KT01121). When plasmids pMQ350 (mutL +) and pRH71-17 (mutH +) were introduced in their corresponding deficient strains, spontaneous mutant frequencies also decreased (data not shown).
Mutation assay
The lac allele of F’CC102 reverts to lac + exclusively by G·C to A·T transition (25). We have previously used this lac reversion system to detect mutagenicity of alkylating agents in wild-type and MMR-deficient strains (6,25). For mutagenesis, 0.1ml of overnight cultures of each strain were incubated with 0.5ml of 0.1M sodium phosphate buffer (pH 7.4) and 0.1ml of mutagen solution dissolved in dimethyl sulphoxide or water for 1h at 37°C. Next, 0.1ml of the treated cultures were spread onto three minimal lactose plates to determine the number of revertants, and adequately diluted cultures were also spread onto three minimal glucose plates to determine the total viable cell numbers. The doses of mutagen used in these assays were largely non-toxic. In most cases, survival was >80%, and in only a few cases survival decreased to 60–70% at the highest dose used. Mutant frequencies were calculated by dividing the number of lac + revertants by the number of total viable cells. Experiments were repeated two or three times. Typical results are shown in Figures 4–7. Statistical analysis was performed using the Student’s t test.
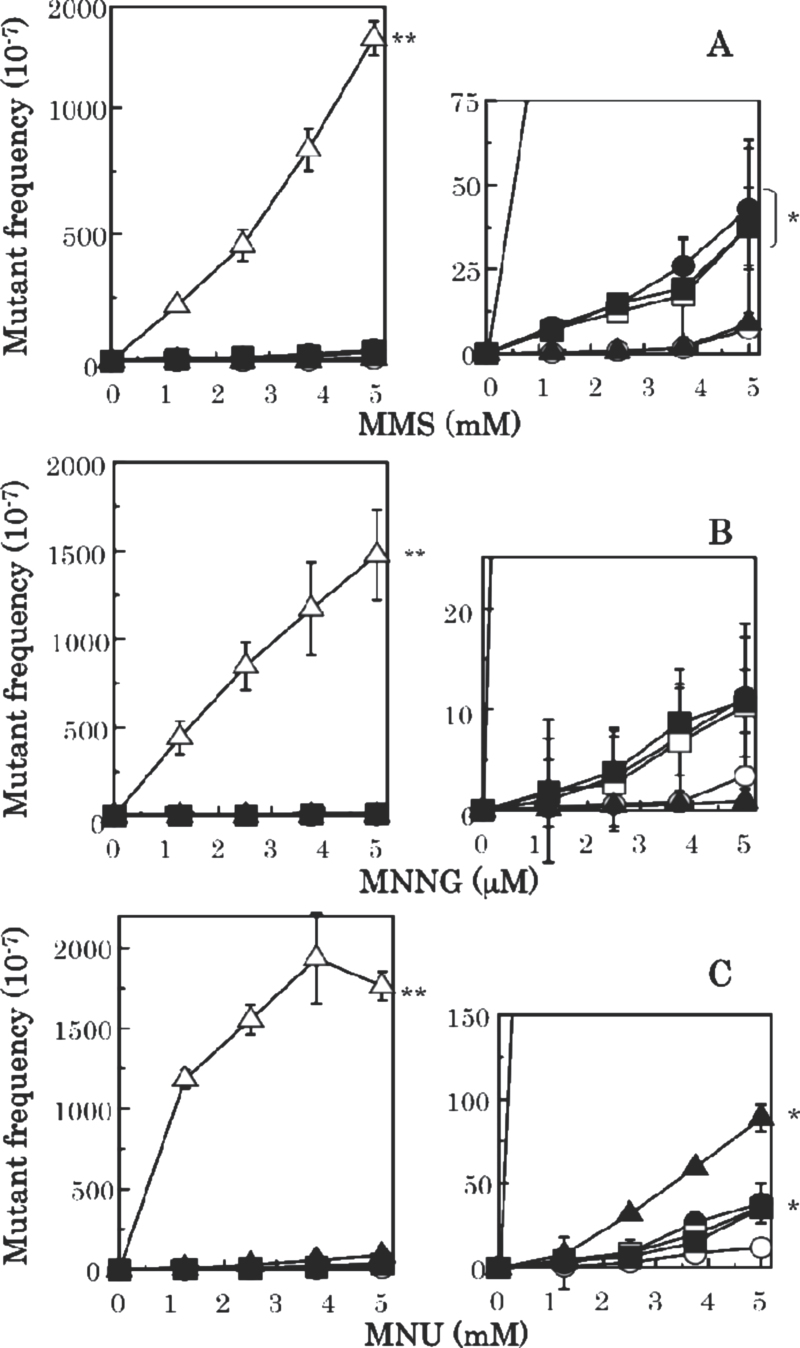
Fig. 4.
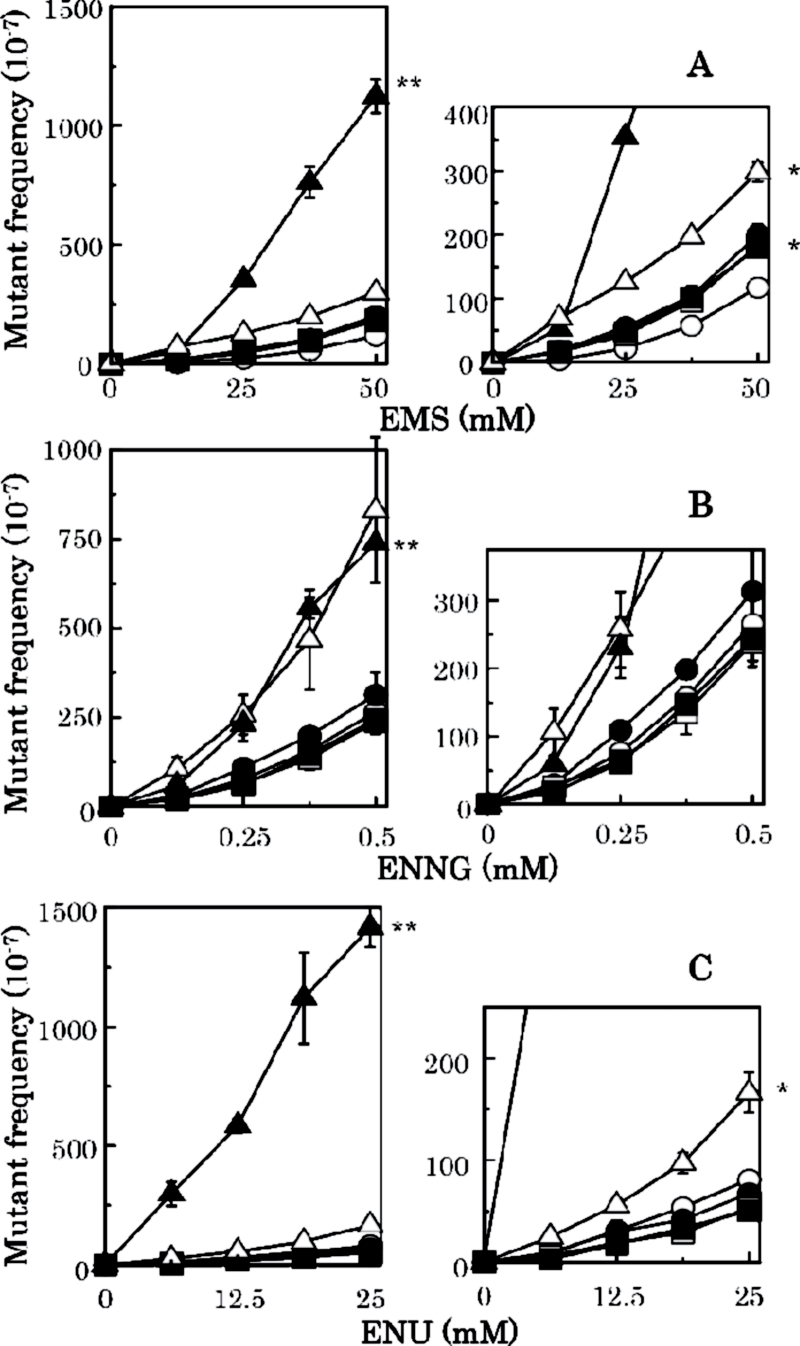
Mutant frequencies induced by methylating agents: (A) MMS, (B) MNNG and (C) MNU. In the small-sized graphs described at right side of each figure from (A) to (C), the mutant frequencies are shown with a small scale. Strains are NR10832 (wild-type), open circle; KT01121 (ada ogt), open triangle; NR12896 (mutS), closed circle; NR11102 (mutL), open square; NR12897 (mutH), closed square; and NR12999 (uvrA), closed triangle. Mutant frequencies are corrected by subtracting spontaneous mutant frequencies. Statistical analysis was performed using the Student’s t test. **P > 0.01 and *P > 0.05 compared with the mutant frequency for the wild-type.
Fig. 7.
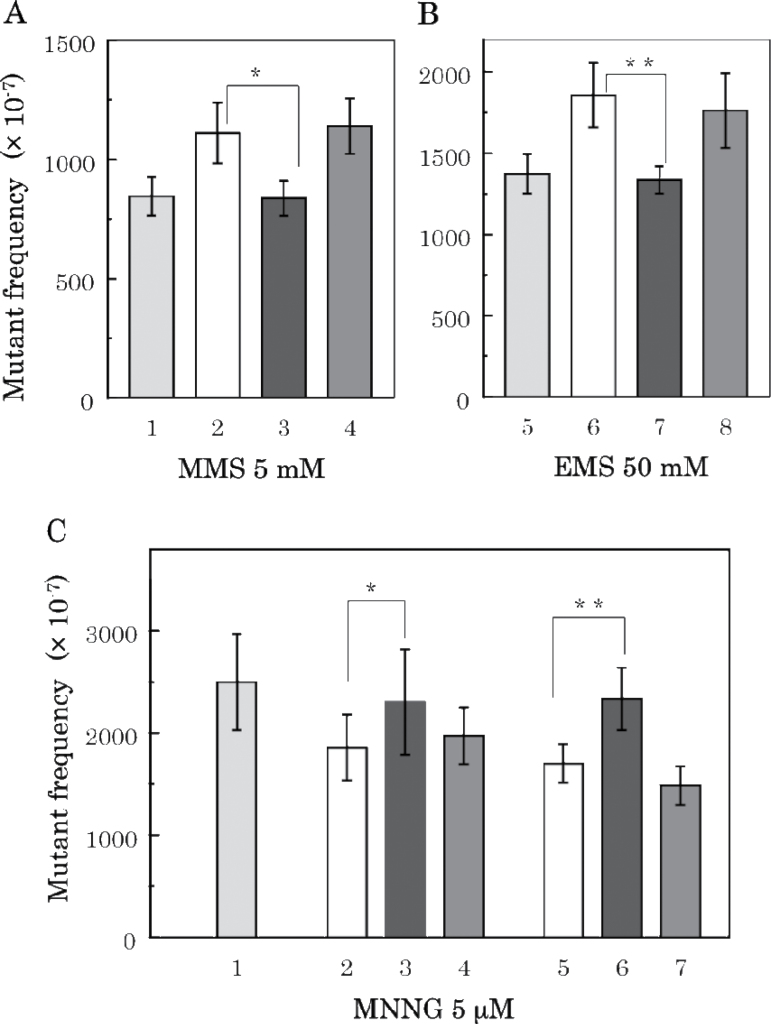
Complementation of MMR deficiencies by plasmid-carried wild-type mutS, mutL and mutH genes. (A) MMS mutagenesis. Strains used are 1 (grey bar) KT01121 (ada ogt); 2 (white bar) KT11141 (mutS ada ogt); 3 (black bar) KT11141 (mutS ada ogt) with pMQ315 (mutS +); and 4 (dark grey bar) KT11141 (mutS ada ogt) with pBR322. (B) EMS mutagenesis. Strains are 5 (grey bar) NR12999 (uvrA); 6 (white bar) KT10021U (mutS uvrA); 7 (black bar) KT10021U (mutS uvrA) with pMQ315 (mutS +); 8 (dark grey bar) KT10021U (mutS uvrA) with pBR322. (C) MNNG mutagenesis. Strains are 1 (grey bar) KT01121 (ada ogt); 2 (white bar) KT21181 (mutL ada ogt); 3 (black bar) KT21181 (mutL ada ogt) with pMQ350 (mutL+); 4 (dark grey bar) KT21181 (mutL ada ogt) with pBR322; 5 (white bar) KT31131 (mutH ada ogt); 6 (black bar) KT31131 (mutH ada ogt) with pRH71-17 (mutH+); 7 (dark grey bar) KT31131 (mutH ada ogt) with pBR322. Statistical analysis was performed using the Student’s t test. **P > 0.01 and *P > 0.05 compared with the mutant frequency for each parental strain with MMR deficiency.
Results
Mutagenic activities of alkylating agents in MMR-deficient strains
To estimate how much the various repair systems contribute to the repair of alkylated bases, we measured mutant frequencies induced by methylating agents (MMS, MNU and MNNG) and ethylating agents (EMS, ENU and ENNG) in E. coli strains deficient in AGT, NER or MMR. As shown in Figure 4 and Table II, mutation frequencies induced by each methylating agent increased >100 times in an AGT-deficient strain compared with the wild-type exposed to the same agent. The frequencies were also elevated modestly (3- to 5-fold) in the single MMR-deficient strains (mutS, mutL and mutH). The NER deficiency enhanced the mutagenesis induced by MNU (7.5-fold) but not by MMS or MNNG (Table II and Figure 4).
Table II.
Alkylation-induced mutability of repair-deficient E. coli strainsa
| Genotype | MMS (5mM) | MNNG (0.005mM) | MNU (5mM) | EMS (50mM) | ENNG (0.5mM) | ENU (25mM) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ratio vs. wild-type (MF × 107) | Survivalb (%) | 1 (7.4±1.0) | 91 | 1 (3.4±0.9) | 86 | 1 (11.9±3.8) | 81 | 1 (117.5±4.7) | 83 | 1 (226.2±55.3) | 64 | 1 (81.6±6.8) | 86 |
| ada, ogt | 173.3 | 68 | 433.9 | 87 | 148.2 | 95 | 2.5 | 86 | 3.1 | 72 | 2.1 | 79 | |
| uvrA | 1.2 | 88 | 0.3 | 101 | 7.5 | 48 | 9.6 | 96 | 2.8 | 55 | 17.5 | 78 | |
| mutS | 5.8 | 87 | 3.3 | 97 | 3.2 | 77 | 1.7 | 81 | 1.2 | 63 | 0.8 | 97 | |
| mutL | 5.1 | 81 | 3.0 | 91 | 3.0 | 84 | 1.6 | 84 | 0.9 | 62 | 0.7 | 85 | |
| mutH | 5.1 | 91 | 3.2 | 98 | 2.9 | 85 | 1.5 | 86 | 0.9 | 76 | 0.6 | 79 | |
MF: mutant frequency.
aShown are mutant frequencies obtained at the highest dose of each mutagen tested relative to the frequency for the wild-type strain at the same dose (i.e. wild-type = 1).
bViabilities are shown by the percentages of colony forming units in the presence of mutagen compared with that in the absence of mutagen.
For the ethylating agents, AGT is much less effective, whereas NER becomes more important (Figure 5 and Table II): EMS- and ENU-induced mutant frequencies were elevated in the uvrA strain by 9.6- and 17.5-fold, respectively. The frequency of ENNG-induced mutagenesis was increased to a similar extent (about 3-fold) in AGT- and NER-deficient E. coli (Table II). Mutagenesis by the ethylating agents was little affected by MMR deficiency. Taken together, methylated guanine residues appear repaired mainly by the AGT system, whereas ethylated guanines are subject to effective repair by the NER system. The contribution of MMR to the repair of alkylated lesions is modest, but appears to be significant at least in the repair of methylated lesions, as summarised in Table II.
Fig. 5.
Mutant frequencies induced by ethylating agents: (A) EMS, (B) ENNG and (C) ENU. Strains are NR10832 (wild-type), open circle; KT01121 (ada ogt), open triangle; NR12896 (mutS), closed circle; NR11102 (mutL), open square; NR12897 (mutH), closed square; and NR12999 (uvrA), closed triangle. Mutant frequencies are corrected by subtracting spontaneous mutant frequencies. Statistical analysis was performed using the Student’s t test. **P > 0.01 and *P > 0.05 compared with the mutant frequency for the wild-type.
Mutagenic activities of alkylating agents in mutants doubly deficient in MMR and AGT/NER
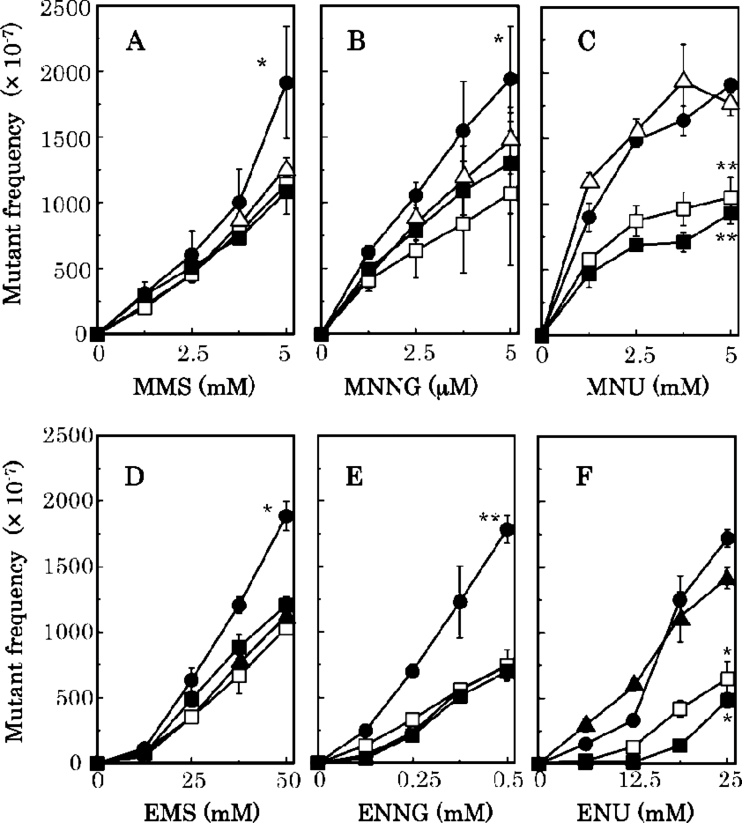
The precise involvement of MMR in mutagenesis induced by alkylating agents is difficult to estimate because it appears significantly smaller than AGT in case of methylation damage or than NER for ethylation damage (Table II). To gain more insight into this question, we thought it helpful to measure the effects of MMR deficiency in the absence of either main repair pathway. To do so, we constructed AGT- or NER-deficient mutants that were additionally defective in either MutH, MutL or MutS activity. The results presented in Figure 6 show that the mutant frequency induced by MMS and MNNG increased in the AGT-deficient mutS strain, but not in the corresponding mutL or mutH strains (Figure 6A and B and Table III). In fact, for the case of MNU, the addition of the mutL or mutH defect decreased the mutant frequency (Figure 6C and Table III).
Fig. 6.
Mutant frequencies induced by alkylating agents. Top: MMS (A), MNNG (B) and MNU (C) were tested using KT01121 (ada ogt), open triangle; KT11141 (ada ogt and mutS), closed circle; KT21181 (ada ogt mutL), open square; and KT31131 (ada ogt and mutH), closed square. Bottom: EMS (D), ENNG (E) and ENU (F) were tested using NR12999 (uvrA), closed triangle; KT10021U (uvrA mutS), closed circle; KT20021U (uvrA mutL), open square; and KT30013U (uvrA mutH), closed square. Statistical analysis was performed using the Student’s t test. **P > 0.01 and *P > 0.05 compared with the mutant frequency for the wild-type.
Table III.
Alkylation-induced mutability of E. coli strains containing multiple repair deficienciesa
| Genotype | MMS (5mM) | MNNG (0.005mM) | MNU (5mM) | EMS (50mM) | ENNG (0.5mM) | ENU (25mM) |
|---|---|---|---|---|---|---|
| Wild-type (mutant frequency ×107) | 1 (7.4±1.0) | 1 (3.4±0.9) | 1 (11.9±3.8) | 1 (117.5±4.7) | 1 (226.2±55.3) | 1 (81.6±6.8) |
| ada, ogt | 173.3 | 433.9 | 148.2 | 2.5 | 3.1 | 2.1 |
| ada, ogt, mutS | 258.8 | 571.6 | 160.3 | 5.3 | 4.9 | 1.8 |
| ada, ogt, mutL | 154.1 | 315.0 | 88.4 | 2.5 | 1.8 | 1.3 |
| ada, ogt, mutH | 146.5 | 382.7 | 78.1 | 2.9 | 2.1 | 1.4 |
| uvrA | 1.2 | 0.3 | 7.5 | 9.6 | 2.8 | 17.5 |
| uvrA, mutS | 5.5 | 4.0 | 8.5 | 16.0 | 7.9 | 21.1 |
| uvrA, mutL | 4.5 | 1.7 | 1.8 | 8.8 | 3.3 | 8.0 |
| uvrA, mutH | 5.1 | 2.3 | 0.5 | 10.3 | 3.1 | 5.9 |
aShown are mutant frequencies obtained at the highest dose of each mutagen tested relative to the frequency for the wild-type strain at the same dose (i.e. wild-type = 1).
Similarly, EMS and ENNG induced higher mutant frequencies in the NER-deficient mutS strain compared with either single defect (Figure 6D and E and Table III), whereas no such effect was seen for the corresponding mutL or mutH defects. The case of ENU appeared somewhat different, as no enhancing effect of mutS was observed in the NER background. However, in this case the mutL or mutH defect decreased mutability (Figure 6F and Table III). Thus, an interesting parallel is uncovered for the case of alkylation by MNU and ENU (Figure 6C and F); no increase was observed for the mutS deficiency, but a decrease for the mutL and mutH deficiencies was observed (see also Table III).
Recovery of MMR function by introduction of plasmids containing MMR genes
To confirm that the deficiency of MMR function is responsible for the effects observed above, the relevant MMR genes were reintroduced into the various strains on pBR322-based plasmids. When mutS + gene was reintroduced in mutS-deficient KT11141 (AGT-deficient) or KT10021U (NER deficient), MMS- and EMS-induced mutation decreased to the level of parent AGT- or NER-deficient strains KT01121 or NR12999, as shown in Figure 7A and B. Introduction of the mutL + or mutH + genes to the corresponding mutL or mutH-deficient strains KT21181 (AGT deficient) or KT31131 (NER deficient) increased the MNNG-induced mutant frequencies to the level of the MMR-proficient strain (Figure 7C). In all cases, empty vector pBR322 showed no effects (Figure 7A–C).
Discussion
Many alkylating agents induce mutation via formation of O 6-alkylguanine, resulting in GC to AT transitions, and the repair pathway(s) of O 6-alkylguanine are critical for mutagenesis. Previously, we demonstrated that MutS protein, whose function involves the recognition of mismatched lesions in DNA, recognises O 6-MeG:T base pairs more efficiently than O 6-EtG:T pairs (6), and further that, consistent with this notion, MutS deficiency increased mutagenesis by methylating agents, while affecting mutagenesis by ethylating agents to a much smaller extent (6).
In this study, we used a set of E. coli strains, containing the Fʹprolac (FʹCC102) episome that can detect specifically the GC to AT transition induced by O 6-alkylguanine (25). We examined which repair pathways work mainly to repair O 6-alkylguanine in DNA, using AGT-, NER- and MMR-deficient strains. Mutant frequencies for methylating agents were highest (increased by several 100-fold) in the AGT-deficient strain, followed by the MMR- and NER-deficient strains, whereas for the ethylating agents frequencies were the highest in the NER-defective strain, followed by the AGT and MMR deficiencies. Thus, the repair of O 6-methylguanine appears to be performed by AGT ≫ MMR > NER in order of contribution, whereas the repair of O 6-ethylguanine may be performed in the order of NER > AGT > MMR.
It is well documented that AGT protects cells and organisms from genotoxicity and carcinogenicity due to the presence of O 6-methylguanine (9). However, the role of AGT in repair of O 6-ethylguanine is less clear. Our results indicate that the role of AGT in O 6-ethylguanine repair is small compared with that of O 6-methylguanine. In contrast, the NER system works inefficiently for O 6-methylguanine, whereas it is the most efficient pathway for O 6-ethylguanine (Table II). With regard to the role of MMR, a deficiency in this process affects mutagenesis by methylating agents more strongly than for ethylating agents (Table II), in agreement with our previous observations on MutS protein binding to O 6-MeG:T versus O 6-EtG:T base pairs (6). Feitsma et al. (26) reported that ENU-induced mutation in the zebrafish germ line was not affected by deficiency of Msh6, a MutS homologue. In our experiments with ENU, we likewise found that the lack of MMR proteins did not affect mutagenesis. In fact, a modest reduction (up to 2-fold) may be seen (Table II). On the other hand, lack of MMR proteins promoted mutagenesis by the methylating agents significantly (up to 5-fold) (Table II).
To further dissect the role of MMR, we investigated its effect in the absence of the major AGT or NER repair pathway. Interestingly, in newly constructed strains lacking these major repair pathways we observed that mutant frequencies with MMS, EMS, MNNG and ENNG were increased by a MutS deficiency, suggesting a clearly protective or antimutagenic role of MutS protein. On the other hand, for these cases no increases in mutagenesis were observed in mutL- or mutH-deficient strains (Figure 6 and Table III). In case of normally functioning MMR, MutS protein would recognise an alkylated base or mismatch at the replication fork and complete repair through recruitment of MutL and MutH proteins, all three being indispensable for the process. Therefore, our results suggest that MutS might play an antimutagenic role that is different from triggering MutHLS-dependent MMR. For example, it is possible that the recognition of the alkylated site by MutS could lead to initiation or promotion of another (error-free) repair pathway, such as excision repair or recombinational repair.
On the other hand, the case of MNU and ENU is clearly different. The MNU- and ENU-induced mutant frequencies in mutS-deficient and -proficient E. coli are similar, but frequencies decreased in the mutL- and mutH-deficient strains. This suggests that the full action of MMR might actually be mutagenic for the case of the two nitrosoureas at least when the major AGT or NER repair pathways are absent. Lehner and Jinks-Robertson (27) in a study of spontaneous mutagenesis in Saccharomyces cerevisiae reported that the MMR system was responsible for a subgroup of the observed mutations. Specifically, MMR was proposed to increase mutagenesis by diverting replication forks stalled at an (as yet hypothetical) spontaneous DNA lesion away from error-free resolution by DNA recombination into an error-prone, Pol zeta-dependent TLS pathway. The antirecombinational properties of the MMR system, through its break down of recombination intermediates, have been described (28). A similar mechanism may underlie the current observations in E. coli. In this study, replication forks are likely to be stalled at least some of the DNA alkylation sites induced be MNU and ENU. Strains lacking the major AGT or NER repair pathways will suffer from high levels of unrepaired alkylation damage, and the recombinational repair of stalled replication forks may be a major mode of damage and mutation avoidance. If so, the antirecombinogenic action of the MutHLS system will likewise promote TLS of the alkylated bases. Such TLS might involve the replicative polymerase Pol III holoenzyme or any one of the E. coli accessory DNA polymerases Pol II, Pol IV or Pol V. TLS by the latter two would be expected to be particularly mutagenic as these enzymes lack exonucleolytic proofreading. These two polymerases are also induced as part of the SOS response, which can be activated by high levels of alkylation damage. The validity of this model may be investigated by further genetic studies with recombination-defective strains, like recA, and/or strains lacking one or more of the TLS polymerases.
It is worth re-emphasising that, at least for the case of MNU and ENU exposure, the apparent effects of the MMR system are different in the repair-deficient backgrounds (AGT and NER) and the wild-type background. Although in the wild-type background the MMR effect is clearly antimutagenic (Figures 4 and 5), it is mutagenic in the repair-deficient backgrounds (Figures 6 and 7). In the simplest model, this reflects the dual aspects of MMR that are likely operating concurrently: (i) the correction of replication errors (a mutation-prevention function) and (ii) the breakdown of recombination intermediates (a mutagenic function). With increasing DNA damage, the balance between the two components might shift from an overall antimutagenic role to a mutagenic role. It will be worthwhile to further investigate this balance, including the possible role of the SOS system, as well as the separate, protective role of MutS protein outside of its role within the MutHLS system.
Finally, it is an important question to ask why the effect of MMR deficiencies varied so significantly among the different alkylating agents used. DNA can be alkylated at a variety of base oxygen and nitrogen atoms, and each alkylating agent will produce its own profile of DNA modifications. In addition, the agents may modify proteins as well as the NTP or dNTP nucleotide pools that serve as RNA and DNA precursors. It is likely that each of these factors may affect, differentially, the manner in which cells can recover from the exposure and resume DNA replication with either more or less mutations.
In summary, our data have revealed distinct pathways for repairing mutagenic lesions induced by methylating and ethylating agents, including differential mutagenic and antimutagenic effects of the MutHLS system, and have suggested the existence of a novel, additional role of MutS protein in maintaining genome stability.
Funding
This research was supported by in part by project ES065086 of the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences (NIEHS).
References
- 1. Marti T. M., Kunz C., Fleck O. (2002). DNA mismatch repair and mutation avoidance pathways. J. Cell. Physiol., 191, 28–41 [DOI] [PubMed] [Google Scholar]
- 2. Schofield M. J., Hsieh P. (2003). DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol., 57, 579–608 [DOI] [PubMed] [Google Scholar]
- 3. Kunkel T. A., Erie D. A. (2005). DNA mismatch repair. Annu. Rev. Biochem., 74, 681–710 [DOI] [PubMed] [Google Scholar]
- 4. Arana M. E., Kunkel T. A. (2010). Mutator phenotypes due to DNA replication infidelity. Semin. Cancer Biol., 20, 304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rasmussen L. J., Samson L. (1996). The Escherichia coli MutS DNA mismatch binding protein specifically binds O(6)-methylguanine DNA lesions. Carcinogenesis, 17, 2085–2088 [DOI] [PubMed] [Google Scholar]
- 6. Taira K., Nakamura S., Nakano K., et al. (2008). Binding of MutS protein to oligonucleotides containing a methylated- or an ethylated guanine residue, and correlation with mutation frequency. Mutation Res., 640, 107–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lupari E., Ventura I., Marcon F., Aquilina G., Dogliotti E., Fortini P. (2012). Pol kappa partially rescues MMR-dependent cytotoxicity of O6-methylguanine. DNA Repair (Amst.), 11, 579–586 [DOI] [PubMed] [Google Scholar]
- 8. Mitra S. (2007). MGMT: a personal perspective. DNA Repair (Amst.), 6, 1064–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kaina B., Christmann M., Naumann S., Roos W. P. (2007). MGMT: key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents. DNA Repair (Amst.), 6, 1079–1099 [DOI] [PubMed] [Google Scholar]
- 10. Drabløs F., Feyzi E., Aas P. A., et al. (2001). Alkylation damage in DNA and RNA-repair mechanisms and medical significance. DNA Repair (Amst.), 3, 59–66 [DOI] [PubMed] [Google Scholar]
- 11. Nouspikel T. (2009). Nucleotide excision repair: variations on versatility. Cell. Mol. Live Sci., 66, 994–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rye P. T., Delaney J. C., Netirojjanakul C., Sun D. X., Liu J. Z., Essigmann J. M. (2008). Mismatch repair proteins collaborate with methyltransferases in the repair of O(6)-methylguanine. DNA Repair (Amst.), 7, 170–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Galloway S. M., Greenwood S. K., Hill R. B., Bradt C. I., Bean C. L. (1995). A role for mismatch repair in production of chromosome aberrations by methylating agents in human cells. Mutat. Res., 346, 231–245 [DOI] [PubMed] [Google Scholar]
- 14. Mojas N., Lopes M., Jiricny J. (2007). Mismatch repair-dependent processing of methylation damage gives rise to persistent single-stranded gaps in newly replicated DNA. Genes Dev., 21, 3342–3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cejka P., Jiricny J. (2008). Interplay of DNA repair pathways controls methylation damage toxicity in Saccharomyces cerevisiae. Genetics, 179, 1835–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feitsma H., Akay A., Cuppen E. (2008). Alkylation damage causes MMR-dependent chromosomal instability in vertebrate embryos. Nucleic Acids Res., 36, 4047–4056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Negishi K., Loakes D., Schaaper R. M. (2002). Saturation of DNA mismatch repair and error catastrophe by a base analogue in Escherichia coli. Genetics, 161, 1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lemotte P. K., Walker G. C. (1985). Induction and autoregulation of ada, a positively acting element regulating the response of Escherichia coli K-12 to methylating agents. J. Bacteriol., 161, 888–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Takano K., Nakamura T., Sekiguchi M. (1991). Roles of two types of O6-methylguanine-DNA methyltransferases in DNA repair. Mutat. Res., 254, 37–44 [DOI] [PubMed] [Google Scholar]
- 20. Takahashi E., Okamoto K., Arimoto S., Yamanaka H., Negishi T. (2006). Involvement of the drug efflux protein TolC in mutagenicity induced by MNNG or Trp-P-2. Mutat. Res., 605, 42–50 [DOI] [PubMed] [Google Scholar]
- 21. Wu T. H., Marinus M. G. (1994). Dominant negative mutator mutations in the mutS gene of Escherichia coli. J. Bacteriol., 176, 5393–5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aronshtam A., Marinus M. G. (1996). Dominant negative mutator mutations in the mutL gene of Escherichia coli . Nucleic Acids Res., 24, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grafstrom R. H., Hoess R. H. (1987). Nucleotide sequence of the Escherichia coli mutH gene. Nucleic Acids Res., 15, 3073–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bolivar F., Rodriguez R. L., Greene P. J., Bethlach M. C., Heyneker H. L., Boyer H. W. (1977). Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene, 2, 95–113 [PubMed] [Google Scholar]
- 25. Cupples C. G., Miller J. H. (1989). A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc. Natl Acad. Sci. USA, 86, 5345–5349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Feitsma H., de Bruijn E., van de Belt J., Nijman I. J., Cuppen E. (2008). Mismatch repair deficiency does not enhance ENU mutagenesis in the zebrafish germ line. Mutagenesis, 23, 325–329 [DOI] [PubMed] [Google Scholar]
- 27. Lehner K., Jinks-Robertson S. (2009). The mismatch repair system promotes DNA polymerase zeta-dependent translesion synthesis in yeast. Proc. Natl Acad. Sci. USA, 106, 5749–5754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Surtees J. A., Argueso J. L., Alani E. (2004). Mismatch repair proteins: key regulators of genetic recombination. Cytogenet. Genome Res., 107, 146–159 [DOI] [PubMed] [Google Scholar]