Abstract
Nonviral genetic therapeutic intervention strategies for neurological disorders hold great promise, but a lack of vector efficacy, coupled with vector toxicity, continue to hinder progress. Here we report the application of a newly developed class of polymer, distinctly different from conventional branched polymers, as a transfection agent for the delivery of glial cell line derived neurotrophic factor (GDNF) encoding gene. This new 2-(dimethylamino)ethyl methacrylate (DMAEMA) based cyclized knot polymer was studied for neuronal cell transfection applications, in comparison to branched polyethyleneimine (PEI). While showing a similar transfection profile over multiple cell types, the cyclized knot polymer showed far lower toxicity. In addition, transfection of Neu7 astrocytes with the GDNF encoding gene was able to cause neurite outgrowth when cocultured with dorsal root ganglia (DRGs). The cyclized knot polymer assessed here (PD-E 8%PEG), synthesized via a simple one-pot reaction, was shown to have great potential for neuronal gene therapy applications.
Keywords: Deactivation enhanced atom transfer radical polymerization (DE-ATRP), glial cell line-derived neurotrophic factor (GDNF), DMAEMA, transfection, astrocytes, dorsal root ganglia
Growth factors such as glial derived neurotrophic factor (GDNF)1 potentially provide a means of disease modifying therapeutic intervention for neurological diseases such as Parkinson’s disease (PD). While GDNF has been extensively proven to protect dopaminergic neurons in animal models of PD,2,3 the short protein half-life4 means that the timing of administration is critical.5 Translation to clinical trials for patients with PD has required direct cannulation of patients and a continual infusion (to overcome the short protein half-life) of GDNF.6−8 Varying efficiency (perhaps attributed to cannulae type or dosing9), effects being reversed post therapy,10 device related problems such as infection,6 excoriation,10 and migration8 all provide rationale for gene therapies based upon a one-off intervention strategy.
Nonviral gene vectors, while largely are not as efficacious as the viral counterpart, offer a means of transfecting neuronal cells11−13 without the perceived safety concerns often associated with the use of viral vectors.14,15 Trojan horse liposomes with surface coatings of poly(ethylene glycol) (PEG) and compacted DNA nanoparticles based on PEG substituted lysine peptides have been used to successfully produce GDNF transgene expression in the mammalian central nervous system (CNS).16,17 PEG modified poly(amido amine) (PAMAM) dendrimers have also been used to deliver the GDNF encoding gene,18 whereby conjugating targeting peptides (lactoferrin and transferrin) allowed brain uptake of 0.1% and 0.14% (respectively) post systemic delivery.19 Two different structures of the polymer vector, polyethyleneimine (PEI), have been used to deliver marker genes to the brain by direct injection. The earlier of these studies used branched PEIs, with the most successful results being the ones which used PEI of molecular weight 25 kDa.20 Later, a linear 22 kDa version of PEI was also used in the mouse brain and was found to transfect both neurons and glia cells.21
All of the aforementioned polymer vectors have differing structures, namely, linear,11,17,21 branched,20 or the symmetrical three-dimensional highly branched dendrimer.18 The dependence of gene vector efficacy upon polymer structure has previously been reported for cationic 2-(dimethylamino)ethyl methacrylate (DMAEMA) based polymers, whereby an increase in transfection capability was observed when branching was introduced into the structure.22,23 In addition, we have recently shown that an entirely new structure of polymer, termed a “cyclized knot”, can be synthesized, whereby the growing chains link upon themselves (intramolecular cross-links) instead of linking with neighboring chains (traditional branching, intermolecular cross-links).24 To form these soluble knotted structures, a high molar ratio of branching monomer (ethylene glycol dimethacrylate (EGDMA)) and a highly controlled polymerization reaction are required to avoid gelation, previously deemed both theoretically25 and experimentally impossible.26,27 Deactivation enhanced atom transfer radical polymerization (DE-ATRP)28 suppresses intermolecular reactions and allows a high molar ratio of EGDMA to be used.29 A series of DMAEMA based cyclized knot transfection agents were designed and synthesized by this reaction mechanism and analyzed over a range of cell types. The one containing 8% PEG with a molecular weight of 30 kDa, termed PD-E 8%PEG, showed a generally higher transfection efficiency profile with lower cytotoxicity than that of the SuperFect PAMAM dendrimer control.30
While initial studies with the cyclized knot polymer showed high transgene expression with luciferase and green fluorescent protein plasmids, it was desired to optimize this system for the delivery of plasmid GDNF (pGDNF) for neuronal applications. It is therefore hypothesized that this new class of polymer, with a high transfection capability, could successfully transfect a range of CNS specific cell types and deliver the GDNF encoding gene to produce a functional outcome, measured by dorsal root ganglion (DRG) neurite outgrowth. Furthermore, the existence of PEG in the polymer structure will render the polymer less cytotoxic than PEI, a polymeric transfection agent that has been studied for applications in the mammalian brain. Lastly, with a long-term view to a single therapeutic intervention strategy for neurological diseases such as PD, we aimed to study whether greater GDNF functionality could be achieved via a GDNF transfection strategy, or a recombinant protein administration strategy. To do this, a DRG neurite length assay was performed to measure the bioactivity of the transgene protein in comparison to administration of recombinant protein.
The cyclized knot polymer was synthesized by DE-ATRP to a final molecular weight (Mw) of 32.4 kDa and a polydispersity index of 1.54,30 so as to be of similar molecular weight as the 25 kDa branched PEI control. As outlined in Scheme 1, PD-E 8%PEG contains EGDMA, 45% of which is used in intramolecular cyclization reactions, with the remaining containing free vinyl groups. These functional groups leave the potential open for a range of post synthesis modifications by simple chemistries.
Scheme 1.
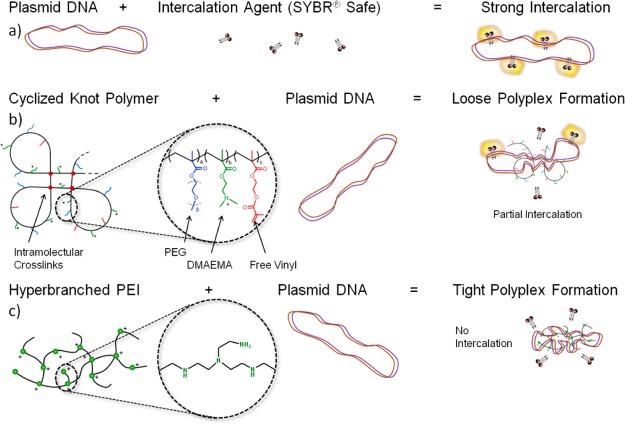
(a) Representation of the action of fluorescent intercalating agent SYBR Safe on naked plasmid DNA, where a strong intercalation leads to high fluorescence. (b) Representation of the monomers used in the synthesis and resulting cyclized knot structure, where PEG = poly(ethylene glycol) methyl ether methacrylate, DMAEMA = 2-(dimethylamino)ethyl methacrylate, and the branching agent used is ethylene glycol dimethacrylate (EGDMA). When this cyclized knot polymer interacts with DNA, a certain degree of intercalation still takes place, indicating loose polyplex formation. (c) The 25 kDa hyperbranched PEI, with every third atom protonizable, results in an almost complete loss of intercalation, which indicates strong condensation of DNA.
Polyplexes were formed with pGDNF at a variety of polymer to plasmid ratios (w/w). Characterization of the size and charge of the polyplexes formed using PD-E 8%PEG shows that at polymer/plasmid ratios between 2:1 and 5:1 a size of between 90 and 130 nm was formed (as determined by dynamic light scattering (DLS) with zeta potentials between 35 and 48 mV (Figure 1a; see Supporting Information (SI) Figure 1 for transmission electron microscopy (TEM) image). Figure 1b shows images taken from an agarose gel post electrophoresis, showing no mobility through the gel of the PD-E 8%PEG polyplexes or the PEI control at a 2:1 ratio (w/w). It should be noted here that the weight to weight ratio is used for greater accuracy; however, a conversion to the commonly used nitrogen/phosphate (N/P) ratio is as follows for PD-E 8%PEG: 2:1 w/w (0.96:1 N/P), 3:1 w/w (1.4:1 N/P), 4:1 w/w (1.9:1 N/P), and 5:1 w/w (2.4:1 N/P). Due to the inclusion of neutral EGDMA and PEG, PD-E 8%PEG has much lower charge density than PEI, which at a 2:1 w/w ratio equates to ∼15:1 N/P ratio).
Figure 1.
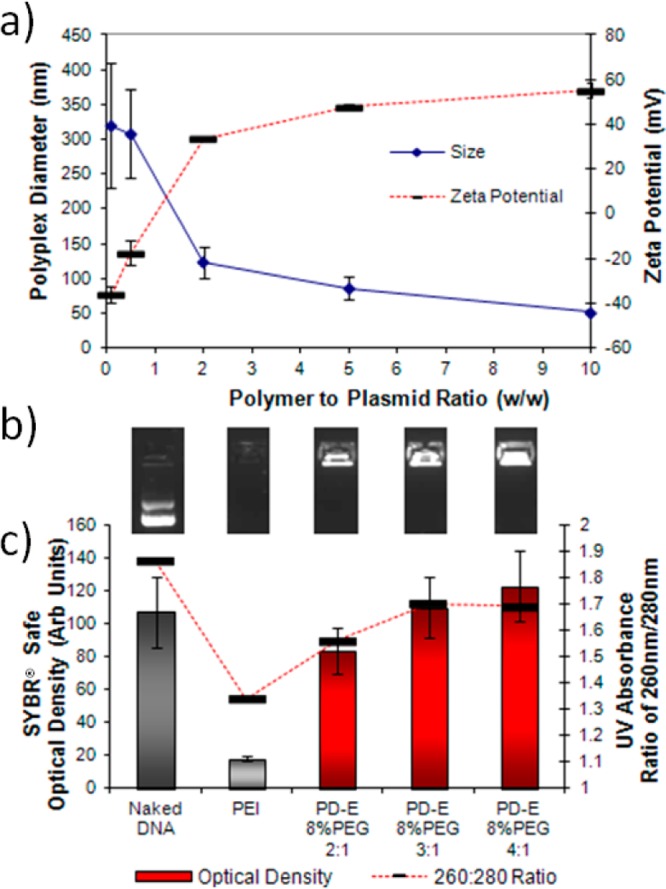
(a) Size and zeta potential analysis of the polyplexes formed with PD-E 8%PEG at various polymer/plasmid ratios (w/w) showing the condensation of plasmid DNA at 2:1 ratio with diameters between 90 and 130 nm. (b) Gel electrophoresis imaging showing no movement through the gel of polyplexes. (c) Optical density measurements (bars) of the above SYBR Safe fluorescence and 260/280 nm ratio (line), both indicating that there is a difference in polyplex formation between PEI and the cyclized knot polymer PD-E 8%PEG.
By use of the intercalating dye SYBR Safe, the degree of intercalation was used as an indicator of the change in DNA conformation during polyplex formation. As expected, high intercalation between naked DNA and SYBR Safe resulted in a high fluorescence intensity. However, as Scheme 1 visually represents, this fluorescence intensity is reduced upon changing the conformation of the DNA by being condensed into a polyplex.23 Interestingly, a low SYBR Safe fluorescence intensity was observed with PEI polyplexes (quantified in Figure 1c), but remains unchanged for PD-E 8%PEG polyplexes at a 5:1 ratio. Previous studies using PicoGreen, another intercalating agent, to observe complexed DNA also showed that the fluorescence intensity varies according to the transfection agent used, with PEI and SuperFect and poly-l-lysine causing a large reduction in DNA intercalation.31 As outlined in Scheme 1b and c, this reduction is likely due to a large change in DNA conformation upon complexing with PEI, which is less so when the cyclized knot polymer is used. In addition, when UV/vis spectroscopy was used to analyze the polyplexes, the 260/280 nm ratio was altered for the DNA contained within PEI polyplexes, but less so for cyclized knot polyplexes (Figure 1c and SI Figure 2). Although the 260/280 ratio is typically used to measure DNA purity, it has been shown that for lipoplexes reaching a size in the range of the wavelength measured (230–300 nm) Raleigh scattering can effect the UV spectra.32 However, the polyplexes formed here are of the same size magnitude as those of PEI (see the SI), indicating that perhaps a loose polyplex formation was occurring with cyclized knot polymers, as also indicated in the case of some other branched polymers.33 Despite this loose formation, the polyplexes still protect the DNA from enzymatic degradation (see SI Figure 3).
The transfection profile of PD-E 8%PEG across multiple cell lines was analyzed in comparison to naked G-Luciferase (G-Luc) DNA and PEI (Figure 2). The human dopaminergic neuroblastoma (SHSY-5Y), rat neuroblastoma (B104), the neuro-inhibitory astrocyte cell line (Neu7), primary astrocytes extracted from newborn rat pups and pheochromocytoma cells (PC12) were used as model systems for assessing the transfection capability of the cyclized knot polymer. The cytotoxicity of the polyplexes were also analyzed using these cell types. PEI gave statistically higher luciferase transgene expression in two of the cell types (Figure 2 – left panel). However, the right-hand panel of Figure 2 shows that the PD-E 8%PEG, at a polymer/plasmid ratio of 4:1, exhibits a statistically lower effect on cell viability that PEI at a ratio of 2:1. As previously shown, this cyclized knot polymer has a superior transfection/cytotoxicity profile when compared to the SuperFect polyamidoamine (PAMAM) dendrimer, and the toxicity profile is dependent upon the degree of PEGylation.30 Here we use the 8%PEGylated polymer, and show that this level allows over 80% cell viability to be obtained in all cell types tested. The high transfection capability of this cyclized knot polymer is likely due to the combination of having a dense three-dimensional structure (like that of the dendrimer), the low polydispersity index (reducing the number of less efficient variants), the inclusion of PEG (reduced toxicity to allow more viable cells to be transfected), and that the polyplexes formed have a loose association with plasmid DNA.
Figure 2.
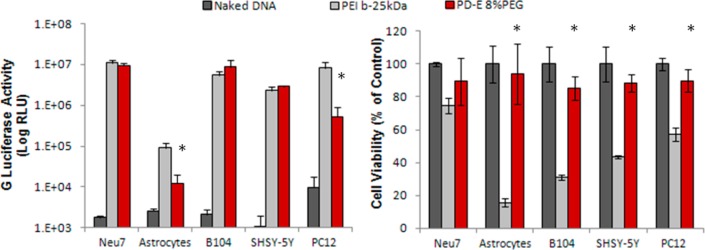
Transfection capability (left-hand panel) and cell viability analysis (right-hand panel) of PD-E 8%PEG in comparison to naked DNA (negative control) and PEI (positive control) over a range of cell types. PD-E 8%PEG exhibits a comparable transfection profile to PEI, but with markedly less cytotoxicity (* marks statistically significant difference from PEI group (P < 0.5), n = 4, error bars represent ± standard deviation).
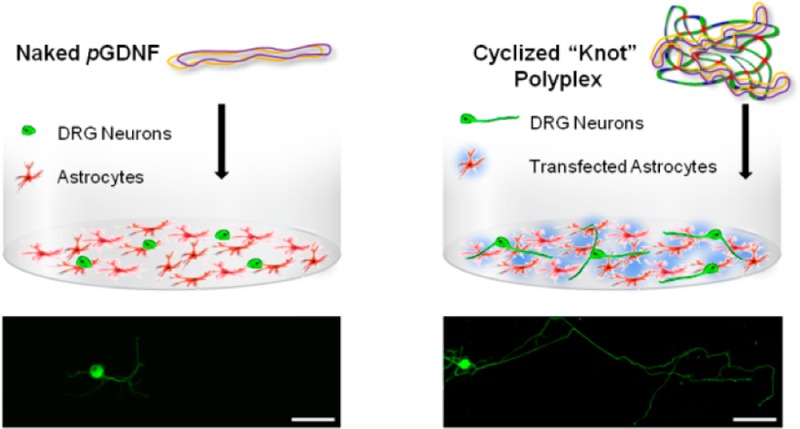
GDNF is a secreted protein, meaning that, for applications in neurological disorders such as Parkinson’s disease, specific targeting to dopaminergic neurons (those lost in Parkinson’s disease34) is not necessary. If cells of the striatum (where dopaminergic neurons terminate) are targeted, then the effects of GDNF transgene expression should be observed regardless of which cell type is transfected. With this in mind, we sought to create a coculture model of neurons seeded upon a bed of astrocytes. Neu7 immortalized astrocytes (Fawcett laboratory)35 showed the highest luciferase transgene level and so were chosen to be the substrate cell line. Dorsal root ganglia (DRGs) have been previously been shown to be affected by GDNF in terms of neurite extension,36 and were used to analyze if the resulting transgene activity post transfection was biologically active.
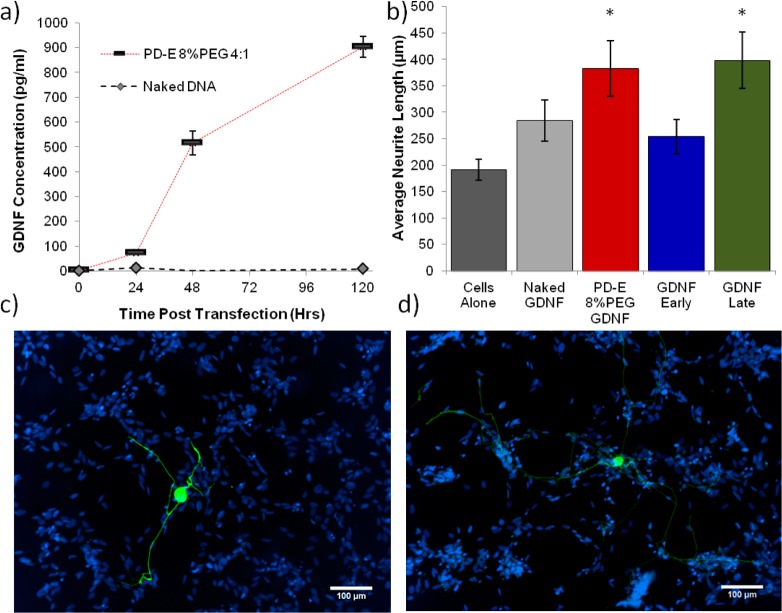
Transfection of Neu7 cells with the plasmid encoding for human GDNF (pGDNF) was then performed using PD-E 8%PEG and compared that of naked pGDNF DNA. The cumulative release of transgene GDNF was shown to increase during the studied period of 4 days (Figure 3a and SI Figure 4) as measured by ELISA. With increasing GDNF levels from day 1 to day 2, it was decided that, for the coculture model, DRGs would be added 30 h post transfection and analyzed at 60 h post transfection. Naked DNA, PD-E 8%PEG polyplexes, or recombinant GDNF (GDNF Early) were administered to Neu7 cells 30 h prior to the addition of the DRGs. A cell-alone group was also included that received no treatment. Upon addition of the DRGs, a further group received recombinant GDNF (GDNF Late). Figure 3b shows that transfecting a bed of astrocytes prior to the addition of DRGs results in a significant increase in the maximum neurite length of DRGs. As expected, the GDNF early group showed no increase in maximum neurite length as the media was changed 12 h post addition. Interestingly, the administration of 10 ng of recombinant GDNF at the time of seeding the DRGs (GDNF Late) resulted in neurite lengths comparable to the transfection group, highlighting the effect of a one-off transfection strategy.
Figure 3.
(a) Analysis of the cumulative GDNF concentration in the supernatant media of Neu7 cells at indicated time points post transfection with PD-E 8%PEG. (b) Results of the coculture experiment whereby the maximum neurite length of DRGs seeded upon a bed of Neu7 was measured. Neu7 cells alone or those treated with naked pGDNF showed the smallest neurite outgrowth. The transfection group (PD-E 8%PEG) showed an average maximum length of ∼3 μm. The group that received recombinant GDNF at the time of DRG seeding also showed long neurite extensions (* marks a statistically significant difference from naked DNA group (P < 0.5), n = 4, minimum of 20 images obtained, error bars represent ± standard error of the mean). (c,d) Typical micrographs from the groups naked DNA (c) and PD-E 8%PEG GDNF (d), where blue (DAPI) is the nuclear stain of the bed of astrocytes and green (βIII tubulin immunostain) shows the DRG cell body and neurite extension.
In conclusion, the 2-(dimethylamino)ethyl methacrylate based cyclized knot polymer shows a comparable neuronal cell transfection profile in comparison to PEI, but with reduced toxicity. The use of DE-ATRP allows the incorporation of branching monomers and PEG moieties in situ, the latter of which has been shown to reduce the toxicity of the polymer. Study of the polyplex formation reveals that there is a difference in the intercalating ability of intercalating agents between polyplexes formed with the cyclized knot polymer and a commonly used transfection agent, PEI. This study also shows that a cyclized knot polymer can be used to overexpress GDNF in an astrocyte cell line which has a functional effect on coseeded DRGs. The versatile nature of DE-ATRP and the encouraging initial results show this chemistry to be a platform technology from which to build on, through the incorporation of degradable monomers to further increase efficiency.
Methods
Formation and Characterization of Polyplexes
The cyclized knot polymer (PD-E 8%PEG) was synthesized as previously reported,30 with a brief outline being described in the Supporting Information. Two plasmids were used for these studies. The plasmid encoding GDNF (pGDNF) (see SI for details of plasmids) was used for polyplex formation analysis and subsequent functional assays. A reporter gene encoding the Gaussia Princeps Luciferase (pG-Luc) was used for the transfection and cytotoxicity assessment over a range of cell types. Polyplexes were formed by adding varying weight ratios of plasmid and polymer made up as solutions in RNase free water (Sigma), and incubating at room temperature for 30 min prior to use. Using 25 μg of DNA per polyplex sample, size and zeta potential measurements were carried out using a Zetasizer Nano-2590 instrument (Malvern Instruments) as described previously.37 Using 1 μg of DNA per sample, 15 μL polyplex solutions were made up for gel electrophoresis (5 μL), UV spectroscopy (3× μL), and transmission electron microscopy sample preparation (3 μL) (for further details, see the SI). Negative and positive controls of a plasmid solution alone (Naked DNA) and polyplexes formed with the branched PEI polymer (at a 2:1 w/w ratio (∼15:1 N/P ratio)) were used, respectively. Optical density measurements were obtained from the gel electrophoresis images using ImageJ software to determine the level of intercalation occurring with the complexed DNA.
Transfection Analysis
All cells were cultured using standard sterile cell culture techniques at 37 °C, 5% CO2 and in a humidified incubator. The media used for the individual cell lines is described in the SI. Twenty-four hours prior to experimentation cells were seeded into 96-well plates at a density of 10 000 cells per well. Polyplexes were made up fresh prior to use as described above, using the plasmid encoding a cell secreted form of G-luciferase. Polyplexes containing 1 μg of DNA were added to the existing fetal bovine serum supplemented media of each well, so all experiments were carried out in the presence of serum. A weight of 1 μg of naked DNA was added to each well as a negative control for both the transfection and the cytotoxicity studies. Twenty-four hours post addition of the samples, the cell supernatant was removed for transfection analysis. A volume of 50 μL of this supernatant was transferred to each well of opaque black 96-well plates, and the BioLux Gaussia Luciferase Assay Kit (New England BioLabs) was used according to the manufactures protocol. Luminescence values were read immediately using a Varioskan Flash plate reader (Thermo Scientific) equipped with SkanIt software, and the average of the four readings was plotted.
Cell Viability Assessment
The effect of the polyplex solutions on the cells metabolic activity was measured immediately post transfection, that is, once the media was removed for transfection analysis. The cells were washed twice with Hank’s balanced salt solution (HBSS) (Sigma), and 100 μL of a 10% solution of the alamarBlue reagent (made up in HBSS) was added to each well and incubated for a further 3 h. The absorbance values at 595 and 550 nm were then read using the Varioskan Flash plate reader according to the manufacturers’ protocol and converted to a percentage of cell viability by normalizing to the Naked DNA treatment.
GDNF Transfection and Neurite Outgrowth Assay
Neu7 cells were cultured and seeded 24 h prior to assessment at 10 000 cells per well of a 24 well plate containing 2 mL of cell media. Naked DNA controls or cyclized knot polyplexes were prepared as outlined above, but with 1 μg of pGDNF, and added to the cell medium (followed by a gentle tap to improve distribution). A volume of 50 μL was then immediately removed from each well and stored at −20 °C for later enzyme-linked immunosorbent assay (ELISA) analysis. Then at each time point a further 50 μL was removed and stored at −20 °C. Once all the samples were collected, a human GDNF duo set ELISA kit (R&D Systems) was used per protocol to determine the cumulative amount of GDNF production.
For functional analysis, Neu7 cells were seeded at 10 000 cells per well in 8-well glass chamber slides that were precoated with poly-l-lysine. Although higher than the above study, this low seeding density was chosen so as to allow for proliferation over the 4 days due to the tendency of Neu7 astrocytes to lift off from the culture substrate when nearing/reaching confluency. For this study, the smaller 8-well glass chamber slides were used with a higher cell density, the same polyplex quantity, and smaller volume of media, thereby increasing the transgene expression concentration to ∼1 ng/mL (see SI Figure 5). Twenty-four hours later, the treatment groups of either; 1 μg of Naked DNA (negative control), cyclized knot polyplexes (1 μg of pGDNF), or 10 ng/mL of human recombinant GDNF (GDNF Early) were added to the culture medium. After 12 h, the media was replaced with fresh media, and at 30 h post treatment dorsal root ganglia (extracted from adult rats according to the protocol in ref (38)) were added to each well at a concentration of 1000 cells per well. A further treatment group was added here, that received 10 ng of human recombinant GDNF at the time of DRG addition (GDNF Late). A further 30 h post addition of DRGs, cells were washed, fixed, and immunostained using the neuronal specific βIII tubulin monoclonal antibody (Tuj1; Abcam) as described previously.39 After the final washes, the walls of the chambers were removed and the slides were mounted using VECTASHEILD (Vector Laboratories, U.K.) mounting medium containing the nuclear counter stain DAPI. A minimum of 20 images were taken using an upright fluorescence microscope (Olympus BX51, Mason Technologies), and NeuronJ (within ImageJ software) was used to trace and measure the longest DRG neurite.
Statistical Analysis
To compare the transfection capability/cytotoxicity of PD-E 8%PEG with PEI, Student t tests were performed for each cell line. One-way ANOVA was used to compare the neurite lengths. In both cases, P < 0.05 was considered as a statistically significantly difference between groups.
Acknowledgments
We thank Professor J. Fawcett for the kind gift of the Neu7 cell line.
Glossary
Abbreviations
- GDNF
glial derived neurotrophic factor
- DRGs
dorsal root ganglia
- PEI
polyethylenimine
Supporting Information Available
Description of materials used, polymer synthesis details, polyplex characterization methods, cell culture techniques, plasmid preparation and SI Figures 1–5. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
B.N. and M.A.-R. contributed equally. B.N., M.A.-R., and M.N. conceived the idea and carried out cell experiments with A.V.P. W.W. designed the reaction for the preparation of knot polymer. Y.Z. and W.W. were instrumental in the cyclized knot polymer/polyplex characterization. E.C. assisted with plasmid preparation and polyplex characterization. E.D., W.W., and A.V.P. contributed ideas throughout the project and to manuscript preparation.
Science Foundation of Ireland, Strategic Research Cluster (SRC) (Grant No. 07/SRC/B1163) for financial support of this research.
The authors declare no competing financial interest.
Supplementary Material
References
- Lin L.; Doherty D.; Lile J.; Bektesh S.; Collins F. (1993) GDNF: a glial cell line-derived neurotrophic factor for midbrain dopaminergic neurons. Science 260, 1130–1132. [DOI] [PubMed] [Google Scholar]
- Sauer H.; Rosenblad C.; Björklund A. (1995) Glial cell line-derived neurotrophic factor but not transforming growth factor beta 3 prevents delayed degeneration of nigral dopaminergic neurons following striatal 6-hydroxydopamine lesion. Proc. Natl. Acad. Sci.U.S.A. 92, 8935–8939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, Opacka J.; Blunt. (1998) Long-term protection of the rat nigrostriatal dopaminergic system by glial cell line-derived neurotrophic factor against 6-hydroxydopamine in vivo. Eur. J. Neurosci. 10, 57–63. [DOI] [PubMed] [Google Scholar]
- Kearns C. M.; Gash D. M. (1995) GDNF protects nigral dopamine neurons against 6-hydroxydopamine in vivo. Brain Res. 672, 104–111. [DOI] [PubMed] [Google Scholar]
- Kearns C. M.; Cass W. A.; Smoot K.; Kryscio R.; Gash D. M. (1997) GDNF protection against 6-OHDA: time dependence and requirement for protein synthesis. J. Neurosci. 17, 7111–7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S. S.; Patel N. K.; Hotton G. R.; O’Sullivan K.; McCarter R.; Bunnage M.; Brooks D. J.; Svendsen C. N.; Heywood P. (2003) Direct brain infusion of glial cell line-derived neurotrophic factor in Parkinson disease. Nat. Med. 9, 589–595. [DOI] [PubMed] [Google Scholar]
- Slevin J. T.; Gerhardt G. A.; Smith C. D.; Gash D. M.; Kryscio R.; Young B. (2005) Improvement of bilateral motor functions in patients with Parkinson disease through the unilateral intraputaminal infusion of glial cell line—derived neurotrophic factor. J. Neurosurg. 102, 216–222. [DOI] [PubMed] [Google Scholar]
- Lang A. E.; Gill S.; Patel N. K.; Lozano A.; Nutt J. G.; Penn R.; Brooks D. J.; Hotton G.; Moro E.; Heywood P.; Brodsky M. A.; Burchiel K.; Kelly P.; Dalvi A.; Scott B.; Stacy M.; Turner D.; Wooten V. G. F.; Elias W. J.; Laws E. R.; Dhawan V.; Stoessl A. J.; Matcham J.; Coffey R. J.; Traub M. (2006) Randomized controlled trial of intraputamenal glial cell line–derived neurotrophic factor infusion in Parkinson disease. Ann. Neurol. 59, 459–466. [DOI] [PubMed] [Google Scholar]
- Sherer T. B.; Fiske B. K.; Svendsen C. N.; Lang A. E.; Langston J. W. (2006) Crossroads in GDNF therapy for Parkinson’s disease. Mov. Disord. 21, 136–141. [DOI] [PubMed] [Google Scholar]
- Slevin J. T.; Gash D. M.; Smith C. D.; Gerhardt G. A.; Kryscio R.; Chebrolu H.; Walton A.; Wagner R.; Young A. B. (2006) Unilateral intraputaminal glial cell line–derived neurotrophic factor in patients with Parkinson disease: response to 1 year each of treatment and withdrawal. Neurosurg. Focus 20, 1–7. [DOI] [PubMed] [Google Scholar]
- Tzeng S. Y.; Guerrero-Cázares H.; Martinez E. E.; Sunshine J. C.; Quiñones-Hinojosa A.; Green J. J. (2011) Non-viral gene delivery nanoparticles based on Poly(β-amino esters) for treatment of glioblastoma. Biomaterials 32, 5402–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Mahony A. M.; Godinho B. M. D. C.; Ogier J.; Devocelle M.; Darcy R.; Cryan J. F.; O’Driscoll C. M. (2012) Click-modified cyclodextrins as nonviral vectors for neuronal siRNA delivery. ACS Chem. Neurosci. 3, 744–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H.; Schellinger J. G.; Chu D. S. H.; Pun S. H. (2012) Neuron-targeted copolymers with sheddable shielding blocks synthesized using a reducible, RAFT-ATRP double-head agent. J. Am. Chem. Soc. 134, 16554–16557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. E.; Ehrhardt A.; Kay M. A. (2003) Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 4, 346–358. [DOI] [PubMed] [Google Scholar]
- Newland B.; Moloney T. C.; Fontana G.; Browne S.; Abu-Rub M. T.; Dowd E.; Pandit A. S. (2013) The neurotoxicity of gene vectors and its amelioration by packaging with collagen hollow spheres. Biomaterials 34, 2130–2141. [DOI] [PubMed] [Google Scholar]
- Xia C.-F.; Boado R. J.; Zhang Y.; Chu C.; Pardridge W. M. (2008) Intravenous glial-derived neurotrophic factor gene therapy of experimental Parkinson’s disease with Trojan horse liposomes and a tyrosine hydroxylase promoter. J. Gene Med. 10, 306–315. [DOI] [PubMed] [Google Scholar]
- Yurek D. M.; Fletcher A. M.; Smith G. M.; Seroogy K. B.; Ziady A. G.; Molter J.; Kowalczyk T. H.; Padegimas L.; Cooper M. J. (2009) Long-term transgene expression in the central nervous system using DNA nanoparticles. Mol. Ther. 17, 641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R.; Han L.; Li J.; Ren F.; Ke W.; Jiang C.; Pei Y. (2009) Neuroprotection in a 6-hydroxydopamine-lesioned Parkinson model using lactoferrin-modified nanoparticles. J. Gene Med. 11, 754–763. [DOI] [PubMed] [Google Scholar]
- Huang R.; Ke W.; Liu Y.; Jiang C.; Pei Y. (2008) The use of lactoferrin as a ligand for targeting the polyamidoamine-based gene delivery system to the brain. Biomaterials 29, 238–246. [DOI] [PubMed] [Google Scholar]
- Abdallah B.; Hassan A.; Benoist C.; Goula D.; Behr J. P.; Demeneix B. A. (1996) A powerful nonviral vector for in vivo gene transfer into the adult mammalian brain: polyethylenimine. Hum. Gene Ther. 1996(7), 1947–1954. [DOI] [PubMed] [Google Scholar]
- Goula D.; Remy J. S.; Erbacher P.; Wasowicz M.; Levi G.; Abdallah B.; Demeneix B. A. (1988) Size, diffusibility and transfection performance of linear PEI/DNA complexes in the mouse central nervous system. Gene Ther. 5, 712–717. [DOI] [PubMed] [Google Scholar]
- Schallon A.; Jérôme V.; Walther A.; Synatschke C. V.; Müller A. H. E.; Freitag R. (2010) Performance of three PDMAEMA-based polycation architectures as gene delivery agents in comparison to linear and branched PEI. React. Funct. Polym. 70, 1–10. [Google Scholar]
- Newland B.; Tai H.; Zheng Y.; Velasco D.; Di Luca A.; Howdle S. M.; Alexander C.; Wang W.; Pandit A. (2010) A highly effective gene delivery vector - hyperbranched poly(2-(dimethylamino)ethyl methacrylate) from in situ deactivation enhanced ATRP. Chem. Commun. 46, 4698–4700. [DOI] [PubMed] [Google Scholar]
- Zheng Y.; Cao H.; Newland B.; Dong Y.; Pandit A.; Wang W. (2011) 3D single cyclized polymer chain structure from controlled polymerization of multi-vinyl monomers: beyond Flory-Stockmayer theory. J. Am. Chem. Soc. 133, 13130–13137. [DOI] [PubMed] [Google Scholar]
- Flory P. J. (1941) Molecular size distribution in three dimensional polymers. I. Gelation1. J. Am. Chem. Soc. 63, 3083–3090. [Google Scholar]
- Wang L.; Li C.; Ryan A. J.; Armes S. P. (2006) Synthesis and peptide-induced degradation of biocompatible fibers based on highly branched poly(2-hydroxyethyl methacrylate). Adv. Mater. 18, 1566–1570. [Google Scholar]
- Bouhier M.-H.; Cormack P. A. G.; Graham S.; Sherrington D. C. (2007) Synthesis of densely branched poly(methyl methacrylate)s via ATR copolymerization of methyl methacrylate and ethylene glycol dimethacrylate. J. Polym. Sci., Part A: Polym. Chem. 45, 2375–2386. [Google Scholar]
- Wang W.; Zheng Y.; Roberts E.; Duxbury C. J.; Ding L.; Irvine D. J.; Howdle S. M. (2007) Controlling chain growth: a new strategy to hyperbranched materials. Macromolecules 40, 7184–7194. [Google Scholar]
- Saeed A. O.; Newland B.; Pandit A.; Wang W. (2012) The reverse of polymer degradation: in situ crosslinked gel formation through disulfide cleavage. Chem. Commun. 48, 585–587. [DOI] [PubMed] [Google Scholar]
- Newland B.; Zheng Y.; Jin Y.; Abu-Rub M.; Cao H.; Wang W.; Pandit A. (2012) Single cyclized molecule versus single branched molecule: a simple and efficient 3D “Knot” polymer structure for nonviral gene delivery. J. Am. Chem. Soc. 134, 4782–4789. [DOI] [PubMed] [Google Scholar]
- Holladay C.; Keeney M.; Newland B.; Mathew A.; Wang W.; Pandit A. (2010) A reliable method for detecting complexed DNA in vitro. Nanoscale 2, 2718–2723. [DOI] [PubMed] [Google Scholar]
- Braun C. S.; Kueltzo L. A.; Russell Middaugh C.; Findeis M. A. (2001) Ultraviolet absorption and circular dichroism spectroscopy of nonviral gene delivery complexes. Methods Mol. Med. 65, 253–284. [DOI] [PubMed] [Google Scholar]
- Tu C.; Li N.; Zhu L.; Zhou L.; Su Y.; Li P.; Zhu X. (2012) Cationic long-chain hyperbranched poly(ethylene glycol)s with low charge density for gene delivery. Polym. Chem. 4, 393–402. [Google Scholar]
- Bernheimer H.; Birkmayer W.; Hornykiewicz O.; Jellinger K.; Seitelberger F. (1973) Brain dopamine and the syndromes of Parkinson and Huntington Clinical, morphological and neurochemical correlations. J. Neurol. Sci. 20, 415–455. [DOI] [PubMed] [Google Scholar]
- Smith-Thomas L. C.; Fok-Seang J.; Stevens J.; Du J. S.; Muir E.; Faissner A.; Geller H. M.; Rogers J. H.; Fawcett J. W. (1994) An inhibitor of neurite outgrowth produced by astrocytes. J. Cell Sci. 107, 1687–1695. [DOI] [PubMed] [Google Scholar]
- Fine E. G.; Decosterd I.; Papaloïzos M.; Zurn A. D.; Aebischer P. (2002) GDNF and NGF released by synthetic guidance channels support sciatic nerve regeneration across a long gap. Eur. J. Neurosci. 15, 589–601. [DOI] [PubMed] [Google Scholar]
- Velasco D.; Réthoré G.; Newland B.; Parra J.; Elvira C.; Pandit A.; Rojo L.; San Román J. (2012) Low polydispersity (N-ethyl pyrrolidine methacrylamide-co-1-vinylimidazole) linear oligomers for gene therapy applications. Eur. J. Pharm. Biopharm. 82, 465–474. [DOI] [PubMed] [Google Scholar]
- Hall A. K. (2006) Rodent sensory neuron culture and analysis In Current Protocols in Neuroscience, John Wiley & Sons, Inc.: New York [DOI] [PubMed] [Google Scholar]
- Abu-Rub M. T.; Billiar K. L.; van Es M. H.; Knight A.; Rodriguez B. J.; Zeugolis D. I.; McMahon S.; Windebank A. J.; Pandit A. (2011) Nano-textured self-assembled aligned collagen hydrogels promote directional neurite guidance and overcome inhibition by myelin associated glycoprotein. Soft Matter 7, 2770–2781. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.