Abstract
A major function of Rho-family GTPases is to regulate the organization of the actin cytoskeleton; filopodia, lamellipodia, and stress fiber are regarded as typical phenotypes of the activated Cdc42, Rac, and Rho, respectively. Using probes based on fluorescent resonance energy transfer, we report on the spatiotemporal regulation of Rac1 and Cdc42 at lamellipodia and membrane ruffles. In epidermal growth factor (EGF)-stimulated Cos1 and A431 cells, both Rac1 and Cdc42 were activated diffusely at the plasma membrane, followed by lamellipodial protrusion and membrane ruffling. Although Rac1 activity subsided rapidly, Cdc42 activity was sustained at lamellipodia. A critical role of Cdc42 in these EGF-induced morphological changes was demonstrated as follows. First, phorbol 12-myristate 13-acetate, which activated Rac1 but not Cdc42, could not induce full-grown lamellipodia in Cos1 cells. Second, a GTPase-activating protein for Cdc42, KIAA1204/CdGAP, inhibited lamellipodial protrusion and membrane ruffling without interfering with Rac1 activation. Third, expression of the Cdc42-binding domain of N-WASP inhibited the EGF-induced morphological changes. Therefore, Rac1 and Cdc42 seem to synergistically induce lamellipodia and membrane ruffles in EGF-stimulated Cos1 cells and A431 cells.
INTRODUCTION
Rho-family GTPases belong to the Ras superfamily of monomeric 20- to 30-kDa GTPases and regulate a variety of cellular functions (Hall, 1998; Ridley, 2001). Among them, a major function of Rho-family GTPases is to regulate the organization of the actin cytoskeleton; filopodia, lamellipodia, and stress fiber are regarded as typical phenotypes of the activated Cdc42, Rac, and Rho, respectively (Hall, 1998; Ridley and Hall, 1992; Ridley et al., 1992; Kozma et al., 1995). Rho-family GTPases also influence adhesion, motility, and proliferation of cells (Van Aelst and Symons, 2002). For example, in fibroblast, Rac1 plays a pivotal role for protrusion formation and forward motion, whereas Cdc42 regulates directionality of movement, including localization of lamellipodial activity to the leading edge (Nobes and Hall, 1999).
The Rho-family GTPases establish a complex network of stimulatory and/or inhibitory interactions among themselves (Bishop and Hall, 2000; Matozaki et al., 2000; Scita et al., 2000). In Swiss 3T3 fibroblasts, Cdc42, Rac, and Rho have been shown to act in a hierarchical cascade wherein Cdc42 activates Rac, which in turn activates Rho (reviewed in Hall, 1998). However, this prototypical cascade of Rho-family GT-Pase activation may not always be observed in other cell types. For example, Rac and Cdc42 antagonize Rho in N1E-115 neuroblastoma cells and NIH3T3 cells (Leeuwen et al., 1997; Sander et al., 1999; Caloca et al., 2003).
Rho-family GTPases are regulated by three factors; i.e., guanine nucleotide exchange factor (GEF), GTPase-activating protein (GAP), and guanine nucleotide dissociation inhibitor (GDI) (Takai et al., 2001). GEF activates Rho-family GTPases by promoting the exchange of GDP with GTP, resulting in the binding of the GTPases to their effectors. GAP activates the intrinsic GTP hydrolytic activity of the Rho-family GTPases, thereby leading to their conversion to the inactive GDP-bound states. A number of GEFs and GAPs have been shown to transduce signals from many growth factors to the Rho-family GTPases (Takai et al., 2001). In addition to the increasing number of GEFs and GAPs, the redundant specificity of GEFs and GAPs renders these signaling networks difficult to understand; many GEFs and GAPs have been shown to take multiple Rho-family GTPases as substrates, at least in vitro (Takai et al., 2001). The third group of regulators, GDIs, bind to the prenylated carboxy terminus of Rho-family GTPases and maintain their inactive state in the cytoplasm. Rho-family GTPases must be dissociated from GDI for translocation to the membrane and for activation by GEF (Olofsson, 1999).
To unravel the function of Rho-family GTPases in vivo, we need to visualize the spatiotemporal regulation of low-molecular-weight GTPases. For this purpose, our group and that of Hahn has developed in vivo probes based on the principle of fluorescent resonance energy transfer (FRET) (Kraynov et al., 2000; Itoh et al., 2002). Using these probes, Kraynov et al. (2000) have demonstrated high Rac activity at the membrane ruffles of serum-stimulated Swiss 3T3 cells. Itoh et al. (2002) have shown that, in migrating HT1080 cells, Rac1 activity is increased gradually toward the leading edge, whereas Cdc42 activity is highlighted at the tip of the leading edge. Here, we have videoimaged the activity of Rac1 and Cdc42 in cells stimulated by epidermal growth factor (EGF) and found that both Rac1 and Cdc42 are activated at the nascent lamellipodia. By using CdGAP, a GAP that preferentially acts on Cdc42, we show that not only Rac1 but also Cdc42 is indispensable for the EGF-induced lamellipodial protrusion.
MATERIALS AND METHODS
FRET Probes
The FRET probes used in this study have been described previously (Itoh et al., 2002). Briefly, Raichu-Rac1 and Cdc42 consist of truncated Rac1 or Cdc42 and the Cdc42/Rac1-interactive binding (CRIB) domain, sandwiched between a pair of green fluorescent protein (GFP) mutants, yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP). In these probes, the intramolecular binding of GTP-Rac1 or Cdc42 to the CRIB domain is expected to bring CFP into proximity to YFP, resulting in an increase in FRET from CFP to YFP. Thus, these probes monitor the local balance between the activities of GEFs and GAPs. Raichu-CRIB-X is another type of FRET probe consisting of the CRIB domain sandwiched between YFP and CFP. The binding of the endogenous GTP-Rac1 or Cdc42 to CRIB in the probe displaces YFP and CFP, thereby decreasing the FRET efficiency. Therefore, Raichu-CRIB-X reflects the local amount of GTP-Rac1 and/or GTP-Cdc42. In this study, the prototype probes Raichu-Rac1/1011x and Raichu-Cdc42/1054x were fused to the carboxy-terminal region of Ki-Ras4B. We first replaced the Ki-Ras4B–derived region with those of the authentic proteins: Raichu-Rac1 and Raichu-Cdc42 were fused to the carboxy-terminal region of Rac1 (aa 172–192) and Cdc42 (aa 171–191), respectively.
Plasmids
The coding regions of wild-type Rac1 and Cdc42 were subcloned into pCXN2-mRFP, which was an expression vector derived from pCXN2 and contained cDNA of the monomeric red fluorescent protein (RFP) before the cloning site (Campbell et al., 2002). pIRM21 was an expression vector derived from pCAGGS and contained a FLAG tag at the 5′ side of the cloning site, followed by an internal ribosomal entry site and the cDNA of dsFP593, a red fluorescent protein (Itoh et al., 2002). In pIRM21-KIAA1204/CdGAP, the coding region of KIAA1204/CdGAP was inserted into pIRM21 (Itoh et al., 2002). In pIRM21-N-WaspCRIB, the CRIB domain (aa 124–274) of N-Wasp was inserted into pIRM21 (Ono et al., 2000). pCAGGS-RFPN-PLC22 was a pCAGGS-derived expression vector encoding the two SH2 domains of bovine phospholipase C (PLC)-γ (aa 538–774) with a DsRed-tag (BD Biosciences Clontech, Palo Alto, CA).
Time-Lapse Imaging
Cos1 cells and A431 cells were purchased from the Japanese Cancer Research Resources Bank (Tokyo, Japan). Cells were maintained in DMEM supplemented with 10% fetal bovine serum. Cells were plated on a collagen-coated 35-mm-diameter glass-base dish (Asahi Techno Glass, Tokyo, Japan) and transfected with plasmids with Polyfect (QIAGEN, Valencia, CA). Cells were serum starved for 6 h and stimulated with 10 ng ml-1 EGF, 100 nM phorbol 12-myristate 13-acetate (PMA), or 100 nM 4-α-PMA. Cells were imaged on an Axiovert microscope (Carl Zeiss, Jena, Germany) equipped with a cooled MicroMax charge-coupled device camera (Roper Scientific, Trenton, NJ) controlled by MetaMorph software (Universal Imaging, West Chester, PA) as described previously (Mochizuki et al., 2001). For dual-emission ratio imaging, we used an 86436 excitation filter (Chroma Technology, Brattleboro, VT), a 455DRLP dichroic mirror (Omega Optical, Brattleboro, VT), and two emission filters, 86470 for CFP and 86535 for YFP, alternated by a filter changer. Cells were illuminated with a 75-W xenon lamp through a 10% ND filter (Omega Optical) and a 60× oil immersion objective lens. The exposure time was 0.5 s when binning was set to 4 × 4. In most experiments, cells were imaged every 30 s for 1 h. After background subtraction, the ratio image of YFP/CFP was created with the MetaMorph software and used to represent FRET efficiency. Kymographs were generated along 5-pixel-wide box regions oriented in the direction of individual protrusions using MetaMorph software as described previously (Bear et al., 2002). Each cell imaging was repeated at least five times to confirm the reproducibility.
Phalloidin Staining
A431 cells were serum starved for 8 h an stimulated with 10 ng ml-1 EGF. Before and 5 min after stimulation, cells were fixed with 3.7% formaldehyde in the medium, permeablized with 0.1% Triton-X 100 in phosphate-buffered saline, and stained with Alexa488-conjugated phalloidin (Molecular Probes, Eugene, OR).
Confocal Microscopy and Two-Photon Excitation Fluorescence Microscopy (TPEM)
Cells were observed with an Olympus Fluoview FV500 multiphoton excitation microscope (Olympus Optical, Tokyo, Japan) equipped with external photomultiplier tubes, an argon laser, a He:Ne laser, and a MAITAI Ti: sapphire laser (Spectra Physics, Mountain View, CA). The MAITAI laser was capable of generating >100-fs pulse at a repetition rate of 80 MHz. The output wavelength was tunable from 780 to 920 nm. The output laser beam, with a power of >0.7 W, was horizontally polarized. The excitation wavelength for the TPEM was 790 nm as described previously (Fan et al., 1999). For the FRET imaging, we used an IR-cut filter, RDM650 (Olympus Optical), a dichroic mirror, DM505 (Olympus Optical), and two emission filters, 480AF30 for CFP and 535AF26 for YFP (Omega Optical). For the conventional confocal imaging with the argon laser or He:Ne laser, we used two dichroic mirrors, DM 458/515 and DM488/543, and two emission filters, BA535-565 for YFP and BA560IF for mRFP (Olympus Optical).
Pull-Down Analysis of Rac1 and Cdc42
Quantification of GTP-bound Rac1 and Cdc42 was performed as described previously with slight modifications (Sander et al., 1999). Briefly, cells were lysed in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1% NP-40, 0.5% deoxycholic acid, 0.1% SDS, 1 mM Na3VO4), clarified by centrifugation, and incubated with bacterially produced GST-PAK-CRIB fusion protein. The proteins bound to the fusion protein were precipitated by use of glutathione-Sepharose beads and separated by SDS-PAGE, followed by immunoblotting with specific antibodies for Rac1 and Cdc42 (BD Transduction Laboratories, Lexington, KY). Bound antibodies were detected by an ECL chemiluminescence system (Amersham Biosciences, Piscataway, NJ) and analyzed with an LAS-1000 image analyzer (Fuji-Film, Tokyo, Japan).
RESULTS
EGF Activates Rac1 and Cdc42 at the Plasma Membrane
To study the role of Rac1 and Cdc42 in the growth factor-induced morphological change, we visualized their activities in Cos1 cells, where EGF induces prominent membrane ruffling via Rac (Michiels et al., 1997; Hardt et al., 1998; Miki et al., 2000). First, intracellular localizations of the probes Raichu-Rac1 and Raichu-Cdc42 were compared with those of the authentic Rac1 and Cdc42 tagged with monomeric red fluorescent protein (mRFP) (Campbell et al., 2002). The distribution of mRFP-tagged Rac1 and Cdc42 was in general agreement with that of the previous report (Michaelson et al., 2001); Rac1 was localized predominantly at the plasma membrane and to a lesser extent at the intracellular membrane compartment. In contrast, Cdc42 was localized largely at the Golgi and intracellular vesicles and to a lesser extent at the plasma membrane (Figure 1A). The localizations of Raichu-Rac1 and Raichu-Cdc42, which were fused to the carboxy-terminal regions of Rac1 and Cdc42, respectively, were indistinguishable from those of the mRFP-tagged Rac1 and Cdc42 (Figure 1A), supporting the previous observations that the carboxy-terminal region primarily determines the intracellular distribution of Rac1 and Cdc42 and that overexpressed Rac1 and Cdc42 localized mostly on the plasma membrane and on the endomembrane, respectively (Michaelson et al., 2001).
Figure 1.
Activation of Rac1 and Cdc42 upon EGF stimulation. (A) Localization of mRFP-Rac1, mRFP-Cdc42, Raichu-Rac1, and Raichu-Cdc42. Cells were imaged for mRFP or YFP with a confocal microscope. Stacked horizontal images and vertical sections along the white lines are presented with scale bars (10 μm). (B) Cells expressing Raichu probes were excited at a wavelength of 440 nm, and fluorescent images of YFP and CFP were obtained through band-pass filters every 30 s for 30 min. Ratio images of YFP/CFP were generated to demonstrate FRET efficiency in intensity-modulated display mode. Cells were stimulated with 10 ng/ml EGF and representative images before and after EGF stimulation are shown (video files are available as supplementary information). Ratio ranges are shown on the right. Each cell imaging was repeated at least five times.
For the detection of FRET, Cos1 cells expressing Raichu probes were imaged for YFP and CFP at an excitation wavelength of 440 nm (Figure 1B). The emission ratio, YFP/CFP, reflects the FRET efficiency from CFP to YFP and parallels the GTP/GDP ratio on the probe (Itoh et al., 2002). In the following figures, representative images from time-lapse data are presented in intensity-modulated display mode, which associates color hue with YFP/CFP and the intensity of each hue with the mean value of YFP and CFP. On EGF stimulation, Rac1 and Cdc42 were rapidly activated at the peripheral region (Figure 1B and supplementary video). We did not observe any change in YFP/CFP ratio in the cells expressing Raichu-Rac1-Y40C, an effector loop mutant used as a negative control.
To discriminate signals at the plasma membrane from those at the endomembrane, we obtained tangential FRET images by TPEM (Fan et al., 1999). The EGF-induced activation of Rac1 and Cdc42 was found to occur mostly at the plasma membrane, but not at the endomembrane (Figure 2A). Furthermore, we noticed that Rac1 and Cdc42 were activated not only at the peripheral plasma membrane but also at the central plasma membrane over and below the nuclear and perinuclear regions. This observation suggests that the seeming lack of EGF-induced Rac1 and Cdc42 activities at the perinuclear region in Figure 1B might be caused by low FRET-signals from the intracellular membrane compartments. If we know that most of the changes in Rac1 and Cdc42 activities occur at the plasma membrane, we can overcome this problem and improve the sensitivity and spatial resolution by placing probes only at the plasma membrane. Thus, we examined whether we could observe the EGF-induced activation of Rac1 and Cdc42 with the Raichu probes fused to the carboxy-terminal region of Ki-Ras4B, which we designated Raichu-Rac1/1011X and Raichu-Cdc42/1054X. As shown in Figure 2B, Raichu-Rac1/1011X and Raichu-Cdc42/1054X were localized preferentially at the plasma membrane and readily detected EGF-induced activation of Rac1 and Cdc42 at the plasma membrane in TPEM, as did the Raichu-Rac1 and Raichu-Cdc42 used in Figure 2A. Furthermore, we confirmed that these probes improved both the sensitivity and spatial resolution of FRET images, as will be shown in the following figures. Although TPEM allowed us to obtain tangential FRET images, it could not be used for the time-lapse experiments due to its long scanning time. Considering these restrictions, we concluded that the use of Raichu-Rac1/1011X and Raichu-Cdc42/1054X with conventional epifluorescent microscopes is most suitable for the imaging of EGF-induced activation of Rac1 and Cdc42 at the plasma membrane. Notably, however, the difference in the membrane targeting signals may lead to different conclusions. To overcome this problem, we used both types of probes, i.e., those fused to the carboxyl terminus of Ki-Ras4B and those fused to the carboxyl termini of authentic GTPases, for most of the following.
Figure 2.
Plasma membrane activation of Rac1 and Cdc42 detected by two-photon excitation fluorescence microscopy. (A) Images of YFP and CFP of Cos1 cells expressing Raichu probes fused to the authentic carboxy termini were obtained by TPEM before and 3 min after EGF stimulation. Stacked XY sections and XZ sections of YFP and YFP/CFP (FRET) images in pseudocolor mode are shown. (B) Cos1 cells expressing Raichu-Rac1/1011X and Raichu-Cdc42/1054X were imaged as in A. Ratio ranges are shown at the bottom of each panel.
EGF-induced Activation Is Transient for Rac1 but Sustained for Cdc42 at Lamellipodia
Before imaging analysis, we confirmed the EGF-induced activation of Rac1 and Cdc42 by Bos' pull-down assay by using the GST-fused CRIB domain of PAK. As shown in Figure 3, A and B, the amounts of GTP-Rac1 and GTP-Cdc42 started increasing within 1 min of EGF stimulation, peaked at 3 min, and decreased thereafter. The time courses of Rac1 and Cdc42 activation were very similar, but the level of activation was usually modest in this assay. Then, we obtained time-lapse images of EGF-induced activation of Rac1 and Cdc42 in Cos1 cells and A431cells (Figure 3, C and D, and supplementary videos). Rac1 was activated rapidly and diffusely within 1 min. The Rac1 activity peaked at 3 min, and returned to the basal level in 10–15 min. Lamellipodial protrusion was also initiated within 1 min of EGF inoculation and reached its zenith within 5 min. After this period, protrusion and retraction of lamellipodia were observed repeatedly and randomly until ∼20 min after EGF stimulation. Notably, when membrane ruffling was most prominent at 5 min, Rac1 activity had already dropped near to the basal level in most areas, except at the nascent lamellipodia (Figure 3C, yellow arrows). Retraction of lamellipodia did not necessarily follow the decrease in Rac1 activity (Figure 3C, white arrowheads). Cdc42 also showed a peculiar pattern of activation (Figure 3C). EGF activated Cdc42 within 1 min, with the level of activity being higher at the peripheral plasma membrane and lower at the central plasma membrane. In contrast to Rac1, the activity of Cdc42 remained high at the lamellipodia until its retraction. We obtained essentially the same result in the EGF-stimulated A431 cells (Figure 3D and supplementary video). In these cells, actin polymerization at the EGF-induced lamellipodia and membrane ruffles were confirmed by the staining with phalloidin (Figure 3E).
Figure 3.
Time-lapse images of Rac1 and Cdc42 activity in the EGF-stimulated Cos1 and A431 cells. (A) Serum-starved Cos1 cells were stimulated with EGF for the indicated periods and analyzed by Bos' pull-down method by using GST-PAK-CRIB. Proteins bound to GST-PAK-CRIB, and total lysates were subjected to SDS-PAGE and probed with anti-Rac1 or anti-Cdc42 antibody. (B) The intensity of each band in A was quantified and plotted against time with SD. The highest intensity in each experiment is set arbitrarily to 1.0 (n = 4). (C) Cos1 cells expressing Raichu-Rac1/1011x and Raichu-Cdc42/1054x were stimulated with EGF (n = 10). FRET images were obtained every 30 s for 30 min, and data at the indicated time-points are shown (video files are available as supplementary information). Yellow arrows and white arrowheads indicate nascent and retracting lamellipodia, respectively. (D) A431 cells expressing Raichu-Rac1/1011x or Raichu-Cdc42/1054x were stimulated with EGF. FRET images were obtained every 30 s for 30 min, and data at the indicated time points are shown. Each cell imaging was repeated at least five times. (E) A431 cells were stained with Alexa488-conjugated phalloidin before and 5 min after EGF stimulation. The right panel shows the higher magnification of the boxed region. Bars indicate 10 μm.
In the experiments shown here, we used probes fused to the carboxy terminus of Ki-Ras4B. We confirmed our observation by kymograph analysis with probes fused to the authentic carboxy termini (Figure 4). Again, the activation of Rac1 at lamellipodia was transient, whereas Cdc42 activation tended to be sustained at lamellipodia and membrane ruffles. The different time course in the activation of Rac1 and Cdc42 may implicate Rac1 in the induction of lamellipodia but might exclude it from the maintenance of lamellipodia and membrane ruffling. Cdc42, by contrast, might be required for both the induction and the maintenance of lamellipodia and membrane ruffles upon EGF stimulation.
Figure 4.
Kymographic analysis of Rac1 and Cdc42 activity. FRET images of Cos1 cells expressing Raichu-Rac1 and Raichu-Cdc42 (shown in Figure 1B) were used for kymographic analysis. Presented data show activities of Rac1 and Cdc42 at the lamellipodial protrusion in the boxed regions of the right panels. Ascending and descending contours of the edge indicate protrusion and withdrawal events, respectively. Ratio ranges are shown on the right.
Raichu-Rac1 and Raichu-Cdc42 monitor the balance between GEF and GAP activities at the plasma membrane, but they are insensitive to RhoGDI. To compensate for this defect, we next used Raichu-CRIB-X. In this probe consisting of the CRIB domain of PAK, YFP, and CFP, FRET efficiency inversely correlated with the local concentration of the endogenous Rac-GTP and Cdc42-GTP, enabling us to take the RhoGDI activity into consideration (Itoh et al., 2002). The YFP/CFP ratio of Raichu-CRIB-X decreased diffusely within 1 min after EGF stimulation, reached its nadir at 3 min, and returned to the basal level at 10 min (Figure 5). This observation was consistent with the findings obtained with Raichu-Rac1 and Raichu-Cdc42 and validated the use of these probes for the monitoring of Rac1 and Cdc42 activities after EGF stimulation. Because overexpression of Raichu-CRIB-X inhibited lamellipodial protrusion, probably by sequestering the active Rac1 and Cdc42, we had to minimize the expression level of Raichu-CRIB-X for the imaging. This limitation rendered this probe inapplicable to most of the following experiments.
Figure 5.
Time-lapse images of Rac1 and Cdc42 activity taken using Raichu-CRIB-X. Cos1 cells expressing Raichu-CRIB-X were stimulated with EGF. FRET images were obtained every 30 s for 1 h, and data at the indicated time points are shown as in Figure 1 (n = 5). Note that, in the Raichu-CRIB-X-expressing cells, the FRET efficiency inversely correlated with the local concentration of the endogenous Rac-GTP and Cdc42-GTP. Bars, 10 μm.
PMA Activates Rac1, but Not Cdc42, in Cos1 Cells
Next, we searched for an upstream regulator of Rac1 and Cdc42 by using various SH2 peptides that are known to bind to the EGF receptor. Among them, we found that PLC-22, which consists only of the two SH2 domains of PLC-γ, inhibited EGF-induced lamellipodial protrusion and membrane ruffling, as well as activation of Rac1 and Cdc42 (Figure 6, A and B). This observation was further confirmed by treating the cells with a PLC inhibitor, U73122 (our unpublished data). Thus, in the Cos1 cells used here, PLC-γ seemed to regulate EGF-induced activation of Rac1 and Cdc42 and the resulting lamellipodial protrusion and membrane ruffling.
Figure 6.
Requirement of PLC-γ for the EGF-induced activation of Rac1 and Cdc42 and membrane ruffling. (A) Cos1 cells expressing the Raichu probes Raichu-Rac1/1011x and Raichu-Cdc42/1054x, and a dominant negative mutant of PLC-γ, PLC-22, were stimulated with EGF at time point zero. Cells transfected with pIRM or pIRM-PLC-22 were identified by the IRES-driven expression of a red fluorescent protein, dsFP593 (shown in red). Bars, 10 μm. (B) The relative emission ratio (YFP/CFP) of the whole cell area is shown with SD (n = 5). (C) FRET images of Cos1 cells expressing the Raichu probes Raichu-Rac1/1011x and Raichu-Cdc42/1054x and stimulated with PMA at time point zero. Bars, 10 μm. Each cell imaging was repeated at least five times. (D) At least thirty Cos1 cells were videoimaged with a 20× objective lens in each experiment and the induction of membrane ruffling was scored at 5 and 20 min after stimulation. Averaged data from five independent experiments are shown with SD (n = 5).
PLC-γ exerts its effect via two products, inositol triphosphate and diacylglycerol, which in turn increase the cytoplasmic calcium levels and stimulate many diacylglycerol-responsive enzymes, respectively. Thus, we examined the effect of PMA, an analogue of diacylglycerol. We found that PMA activated Rac1 transiently and diffusely (Figure 6C), but it did not induce membrane ruffling (Figure 6D). An inactive analog of diacylglycerol, 4-α-PMA, did not activate Rac1. Probably due to the short duration of activation, we could not detect PMA-induced Rac1 activation by the pull-down analysis (our unpublished data). Notably, PMA did not activate Cdc42, but rather suppressed Cdc42 activity (Figure 6C). This observation suggested that the coactivation of Cdc42 and Rac1 was required for the EGF-induced lamellipodial protrusion and membrane ruffling. Alternatively, the duration of Rac1 activation after PMA stimulation might not have been long enough for the induction of lamellipodial protrusion.
Requirement of Cdc42 Activation
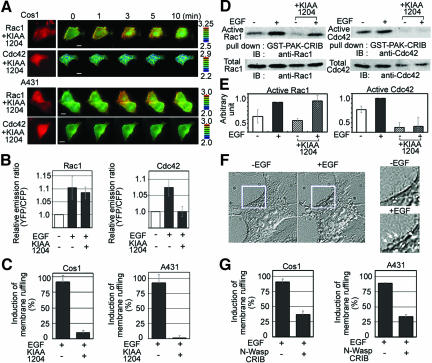
To address this question more directly, we used a GAP for Cdc42, KIAA1204/CdGAP, a human homolog of mouse CdGAP (Lamarche-Vane and Hall, 1998; Itoh et al., 2002). Although the GAP domain of CdGAP has been shown to stimulate GTPase activity of Rac1 and Cdc42 in vitro, we have found that full-length KIAA1204/CdGAP acts more potently on Cdc42 than on Rac1 in culture cells (Itoh et al., 2002). Expression of KIAA1204/CdGAP completely suppressed the EGF-induced Cdc42 activation without suppressing Rac1 activation as examined by FRET imaging (Figure 7, A and B). Moreover, CdGAP almost completely suppressed the EGF-induced membrane ruffling both in Cos1 cells and in A431 cells (Figure 7C). The specificity of the effect of CdGAP was also confirmed by the pull-down analysis (Figure 7, D and E). Interestingly, cells expressing KIAA1204/CdGAP often showed bubble-like membrane protrusions at the periphery and also at the dorsal surface of the EGF-stimulated cells, suggesting that Cdc42 activity was required for the efficient initiation of Rac-dependent morphological changes (Figure 7F). Finally, to confirm the requirement of Cdc42 activity, we examined the effect of the expression of the CRIB domain of N-WASP, which has been shown to block the Cdc42 signaling pathway (Ono et al., 2000). As expected, expression of the N-WASP CRIB inhibited the EGF-induced membrane ruffling both in Cos1 and in A431 cells (Figure 7G). These observations demonstrated that Cdc42 was not required for EGF-induced Rac1 activation but that Cdc42 played an essential role in the EGF-induced lamellipodial protrusion and membrane ruffling.
Figure 7.
Effect of Cdc42 inhibition on EGF-induced activation of Rac1. (A) Cos1 cells and A431 cells expressing Raichu probes Raichu-Rac1/1011x and Raichu-Cdc42/1054x and a Cdc42-specific GAP, CdGAP/KIAA1204, were stimulated with EGF at time point zero. CdGAP/KIAA1204-expressing cells were identified by the expression of dsFP593 (shown in red). Bars, 10 μm. (B) The relative emission ratio (YFP/CFP) of the whole cell area is shown with SD (n = 5). (C) At least thirty Cos1 cells and A431 were videoimaged with a 20× objective lens in each experiment and the induction of membrane ruffling was scored at 5 min after stimulation. Averaged data from five independent experiments are shown with SD. (D) Cos1 cells expressing KIAA1204/CdGAP were serum starved, stimulated with EGF for 3 min, and analyzed by Bos' pull-down method by using GST-PAK-CRIB. Proteins bound to GST-PAK-CRIB and total lysates were subjected to SDS-PAGE and immunoblotting by using anti-Rac1 or anti-Cdc42 antibody. (E) The intensity of each band was quantified by an image analyzer. The level of GTP-Rac1 or GTP-Cdc42 is corrected by the amount of the total GT-Pases and is shown with SD (n = 3). (F) Differential interference contrast images of the Cos1 cells expressing KIAA1204/CdGAP before and 5 min after EGF stimulation. (G) Cos1 cells and A431 cells expressing Raichu probes Raichu-Rac1/1011x and Raichu-Cdc42/1054x and the CRIB domain of N-WASP were stimulated with EGF. N-WASP-CRIB–expressing cells were identified by the expression of dsFP593. At least 30 Cos1 cells and A431 were videoimaged with a 20× objective lens in each experiment, and the induction of membrane ruffling was scored at 5 min after stimulation.
DISCUSSION
Rac plays a pivotal role in the growth-factor-induced membrane ruffling (Ridley et al., 1992). One report has proposed that not only Rac but also Cdc42 is required for the membrane ruffling in leukocytes (Cox et al., 1997). However, because the Cdc42-N17 mutant used in this previous study may inhibit not only GEFs for Cdc42 but also GEFs for Rac, the inhibitory effect by Cdc42-N17 does not necessarily confirm the involvement of Cdc42. In the present study, the requirement of Cdc42 for the EGF-induced lamellipodial protrusion and membrane ruffling was suggested by the observations that EGF-induced Rac1 activation could not induce lamellipodia in the presence of a GAP for Cdc42 and that PMA-stimulated Rac1 activation did not trigger lamellipodial protrusion in Cos1 cells. Furthermore, the sustained activation of Cdc42 at lamellipodia and membrane ruffles strongly supports its involvement in these morphological changes. Our results seem to be consistent with the previous report that a constitutively active Cdc42 mutant cooperates with Rac to induce lamellipodia in Swiss3T3 cells (Nobes and Hall, 1995). However, in disagreement with the previous model in which Cdc42 activates Rac to transform filopodial protrusion to lamellipodia (Nobes and Hall, 1995), we suggest that Rac1 is activated independently of Cdc42 but that it requires Cdc42 activity for the lamellipodial protrusion and membrane ruffling, at least in EGF-stimulated Cos1 and A431 cells. Without reagents that specifically block Rac activation, we cannot rule out the possibility that Cdc42 activation is sufficient for the EGF-induced morphological changes. However, the idea that Rac is required for the EGF-induced morphological changes is supported by many previous studies (Michiels et al., 1997; Hardt et al., 1998; Miki et al., 2000) and also by our observation that Rac1 is activated at nascent lamellipodia.
Of note, our proposal does not necessarily disagree with the previous observations that expression of constitutively active Rac1 is sufficient for the induction of lamellipodia and membrane ruffling (Ridley et al., 1992; Nobes and Hall, 1999) and that Cdc42 is not required for such Rac-dependent morphological changes (Kozma et al., 1996; Allen et al., 1997; Kozma et al., 1997; Gauthier-Rouviere et al., 1998). It has been shown that Cdc42 determines the localization of lamellipodial activity during cell migration (Nobes and Hall, 1999); therefore, Cdc42 seems to play a critical role in the initiation of Rac-dependent lamellipodial protrusion. Without such Cdc42 activation, EGF may not induce lamellipodia and membrane ruffling, because the duration of Rac activation is relatively short (<15 min). In contrast, sustained Rac1 activation in other experimental conditions may somehow overcome such requirement for Cdc42 activation in the induction of lamellipodia. Interestingly, we have found that Cdc42 activity is locally elevated at the active membrane ruffles in the cells expressing constitutively active Rac1 (our unpublished result). Although the mechanism underlying this Cdc42 activation remains unknown, sustained Rac1 activation may locally stimulate Cdc42 to initiate lamellipodial protrusion.
Many signaling molecules have been proposed to be responsible for the growth factor-induced activation of Rac and also for the accompanying lamellipodia formation and membrane ruffling (Bar-Sagi and Hall, 2000; Scita et al., 2000; van Leeuwen et al., 2003). Among them, PLC-γ is known to transduce signals of EGF receptor to the membrane ruffling and cell motility (Chen et al., 1996; Wells et al., 1999). In agreement with this, we found that a dominant negative form of PLC-γ consisting only of the regulatory domain or a PLC inhibitor U73122 blocked Rac and Cdc42 activation in EGF-stimulated Cos1 cells. Of the two signaling molecules released by PLC-γ, diacylglycerol was found to activate Rac1. This observation partly agrees with the previous report that PMA-induced membrane ruffling is dependent on Rac in Swiss 3T3 cells (Ridley et al., 1992). However, in Cos1 cells, PMA-induced Rac1 activation could not fully elicit lamellipodia and membrane ruffling. The discrepancy between Swiss 3T3 cells and Cos1 cells may be due to the requirement of additional Cdc42 activation or the difference in duration of PMA-induced Rac1 activation.
The mechanism underlying the difference in the spatial pattern of Rac1 and Cdc42 activities is under investigation. We have recently shown that GAPs play a pivotal role in the spatial regulation of Ras activity (Ohba et al., 2003); therefore, a similar mechanism may also apply to Rac and Cdc42. Another issue that should be further studied is the role of Cdc42 in the membrane ruffles. Persistent Cdc42 activity at lamellipodia may be required for the polymerization and depolymerization of actin filaments, which causes membrane ruffling by the mechanism proposed by Bear et al. (2002).
A major weakness of our FRET-based imaging techniques involves the GFP-based ratiometry, in which the fluorescence ratio of YFP to CFP is used to monitor the FRET efficiency. In this method, the signal-to-noise ratio of the image deteriorates in the presence of an excess amount of the probe. Therefore, when we observe events on the plasma membrane, reduction of the probes in the intracellular membrane compartments is essential to achieve a high signal-to-noise ratio. For this purpose, use of the carboxy terminus of Ki-Ras4B is convenient because it delivers the probes evenly on the plasma membrane. However, recent studies have shown that carboxy termini of different GTPases may drive proteins to distinct subdomains on the plasma membrane (reviewed in Hancock, 2003). Hence, it is critical to confirm the results by using probes fused to the authentic carboxy termini, as we show in Figure 4.
Another weakness that is specific to Raichu-Rac1 and Raichu-Cdc42 is their inability to bind to RhoGDI, which is known to sequester Rac1 and Cdc42 in the cytoplasm. Therefore, what we could observe with Raichu-Rac1 and Raichu-Cdc42 was the local activity balance between GEFs and GAPs for Rac1 and/or Cdc42, but we could not observe the quantity of GTP-bound Rac1 or Cdc42. To overcome this, we used Raichu-CRIB-X, which directly monitors the level of the endogenous GTP-Rac1 and GTP-Cdc42 (Itoh et al., 2002), and confirmed that Rac1 and/or Cdc42 were activated at the plasma membrane in EGF-stimulated Cos1 cells (Figure 5).
In conclusion, the present analysis strongly suggested that local and synergistic activation of Rac1 and Cdc42 was required for the EGF-induced lamellipodial protrusion and membrane ruffling in Cos1 and A431 cells. Apparently, the importance of spatial and temporal regulation of Rac1 and Cdc42 is not limited to EGF-induced morphological change. Extensive use of in vivo probes, including Raichu, will further help to clarify the complex signaling network of Rac1 and Cdc42.
Supplementary Material
Acknowledgments
We thank A. Miyawaki, J. Miyazaki, Y. Takai, M. Tanaka, R.Y. Tsien, and the KAZUSA DNA Institute for provision of plasmids, and N. Yoshida, N. Fujimoto, and Y. Matsuura for technical assistance. This work was supported in part by Special Coordination Funds for Promoting Science and Technology from the Ministry of Education, Science, Sports, and Culture of Japan, and by grants from YASUDA Medical Research Foundation and the Health Science Foundation, Japan.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–08–0609. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-08-0609.
Abbreviations used: CFP, cyan fluorescent protein; EGF, epidermal growth factor; FRET, fluorescent resonance energy transfer; GAP, GTPase-activating protein; GDI, guanine nucleotide dissociation inhibitor; GEF, guanine nucleotide exchange factor; GFP, green fluorescent protein; mRFP, monomeric red fluorescent protein; PMA, phorbol 12-myristate 13-acetate; TPEM, two-photon excitation fluorescence microscopy; YFP, yellow fluorescent protein.
Online version of this article contains supplementary videos for some figures. Online version is available at www.molbiolcell.org.
References
- Allen, W.E., Jones, G.E., Pollard, J.W., and Ridley, A.J. (1997). Rho, Rac and Cdc42 regulate actin organization and cell adhesion in macrophages. J. Cell Sci. 110, 707-720. [DOI] [PubMed] [Google Scholar]
- Bar-Sagi, D., and Hall, A. (2000). Ras and Rho GTPases: a family reunion. Cell 103, 227-238. [DOI] [PubMed] [Google Scholar]
- Bear, J.E., et al. (2002). Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell 109, 509-521. [DOI] [PubMed] [Google Scholar]
- Bishop, A.L., and Hall, A. (2000). Rho GTPases and their effector proteins. Biochem. J. 348, 241-255. [PMC free article] [PubMed] [Google Scholar]
- Caloca, M.J., Zugaza, J.L., Matallanas, D., Crespo, P., and Bustelo, X.R. (2003). Vav mediates Ras stimulation by direct activation of the GDP/GTP exchange factor Ras GRP1. EMBO J. 22, 3326-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, R.E., Tour, O., Palmer, A.E., Steinbach, P.A., Baird, G.S., Zacharias, D.A., and Tsien, R.Y. (2002). A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99, 7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, P., Murphy-Ullrich, J.E., and Wells, A. (1996). A role for gelsolin in actuating epidermal growth factor receptor-mediated cell motility. J. Cell Biol. 134, 689-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, D., Chang, P., Zhang, Q., Reddy, P.G., Bokoch, G.M., and Greenberg, S. (1997). Requirements for both Rac1 and Cdc42 in membrane ruffling and phagocytosis in leukocytes. J. Exp. Med. 186, 1487-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, G.Y., Fujisaki, H., Miyawaki, A., Tsay, R.K., Tsien, R.Y., and Ellisman, M.H. (1999). Video-rate scanning two-photon excitation fluorescence microscopy and ratio imaging with cameleons. Biophys. J. 76, 2412-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier-Rouviere, C., Vignal, E., Meriane, M., Roux, P., Montcourier, P., and Fort, P. (1998). RhoG GTPase controls a pathway that independently activates Rac1 and Cdc42Hs. Mol. Biol. Cell 9, 1379-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, A. (1998). Rho GTPases and the actin cytoskeleton. Science 279, 509-514. [DOI] [PubMed] [Google Scholar]
- Hancock, J.F. (2003). Ras proteins: different signals from different locations. Nat. Rev. Mol. Cell. Biol. 4, 373-385. [DOI] [PubMed] [Google Scholar]
- Hardt, W.D., Chen, L.M., Schuebel, K.E., Bustelo, X.R., and Galan, J.E. (1998). S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 93, 815-826. [DOI] [PubMed] [Google Scholar]
- Itoh, R.E., Kurokawa, K., Ohba, Y., Yoshizaki, H., Mochizuki, N., and Matsuda, M. (2002). Activation of Rac and Cdc42 video-imaged by FRET-based singlemolecule probes in the membrane of living cells. Mol. Cell. Biol. 22, 6582-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozma, R., Ahmed, S., Best, A., and Lim, L. (1995). The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol. Cell. Biol. 15, 1942-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozma, R., Ahmed, S., Best, A., and Lim, L. (1996). The GTPase-activating protein n-chimaerin cooperates with Rac1 and Cdc42Hs to induce the formation of lamellipodia and filopodia. Mol. Cell. Biol. 16, 5069-5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozma, R., Sarner, S., Ahmed, S., and Lim, L. (1997). Rho family GTPases and neuronal growth cone remodelling: relationship between increased complexity induced by Cdc42Hs, Rac1, and acetylcholine and collapse induced by RhoA and lysophosphatidic acid. Mol. Cell. Biol. 17, 1201-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraynov, V.S., Chamberlain, C., Bokoch, G.M., Schwartz, M.A., Slabaugh, S., and Hahn, K.M. (2000). Localized rac activation dynamics visualized in living cells. Science 290, 333-337. [DOI] [PubMed] [Google Scholar]
- Lamarche-Vane, N., and Hall, A. (1998). CdGAP, a novel proline-rich GTPase-activating protein for Cdc42 and Rac. J. Biol. Chem. 273, 29172-29177. [DOI] [PubMed] [Google Scholar]
- Leeuwen, F.N., Kain, H.E., Kammen, R.A., Michiels, F., Kranenburg, O.W., and Collard, J.G. (1997). The guanine nucleotide exchange factor Tiam1 affects neuronal morphology; opposing roles for the small GTPases Rac and Rho. J. Cell Biol. 139, 797-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matozaki, T., Nakanishi, H., and Takai, Y. (2000). Small G-protein networks: their crosstalk and signal cascades. Cell Signal. 12, 515-524. [DOI] [PubMed] [Google Scholar]
- Michaelson, D., Silletti, J., Murphy, G., D'Eustachio, P., Rush, M., and Philips, M.R. (2001). Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J. Cell Biol. 152, 111-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels, F., Stam, J.C., Hordijk, P.L., van der Kammen, R.A., Ruuls-Van Stalle, L., Feltkamp, C.A., and Collard, J.G. (1997). Regulated membrane localization of Tiam1, mediated by the NH2-terminal pleckstrin homology domain, is required for Rac-dependent membrane ruffling and C-Jun NH2-terminal kinase activation. J. Cell Biol. 137, 387-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki, H., Yamaguchi, H., Suetsugu, S., and Takenawa, T. (2000). IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature 408, 732-735. [DOI] [PubMed] [Google Scholar]
- Mochizuki, N., Yamashita, S., Kurokawa, K., Ohba, Y., Nagai, T., Miyawaki, A., and Matsuda, M. (2001). Spacio-temporal images of growth factor-induced activation of Ras and Rap1. Nature 411, 1065-1068. [DOI] [PubMed] [Google Scholar]
- Nobes, C.D., and Hall, A. (1995). Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81, 53-62. [DOI] [PubMed] [Google Scholar]
- Nobes, C.D., and Hall, A. (1999). Rho GTPases control polarity, protrusion, and adhesion during cell movement. J. Cell Biol. 144, 1235-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba, Y., Kurokawa, K., and Matsuda, M. (2003). Mechanism of the spatio-temporal regulation of Ras and Rap1. EMBO J. 22, 859-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olofsson, B. (1999). Rho guanine dissociation inhibitors: pivotal molecules in cellular signalling. Cell Signal. 11, 545-554. [DOI] [PubMed] [Google Scholar]
- Ono, Y., Nakanishi, H., Nishimura, M., Kakizaki, M., Takahashi, K., Miyahara, M., Satoh-Horikawa, K., Mandai, K., and Takai, Y. (2000). Two actions of frabin: direct activation of Cdc42 and indirect activation of Rac. Oncogene 19, 3050-3058. [DOI] [PubMed] [Google Scholar]
- Ridley, A.J. (2001). Rho family proteins: coordinating cell responses. Trends Cell Biol. 11, 471-477. [DOI] [PubMed] [Google Scholar]
- Ridley, A.J., and Hall, A. (1992). The small GTP-binding protein Rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 70, 389-399. [DOI] [PubMed] [Google Scholar]
- Ridley, A.J., Paterson, H.F., Johnston, C.L., Diekmann, D., and Hall, A. (1992). The small GTP-binding protein Rac regulates growth factor-induced membrane ruffling. Cell 70, 401-410. [DOI] [PubMed] [Google Scholar]
- Sander, E.E., ten Klooster, J.P., van Delft, S., van der Kammen, R.A., and Collard, J.G. (1999). Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J. Cell Biol. 147, 1009-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scita, G., Tenca, P., Frittoli, E., Tocchetti, A., Innocenti, M., Giardina, G., and Di Fiore, P.P. (2000). Signaling from Ras to Rac and beyond: not just a matter of GEFs. EMBO J. 19, 2393-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai, Y., Sasaki, T., and Matozaki, T. (2001). Small GTP-binding proteins. Physiol. Rev. 81, 153-208. [DOI] [PubMed] [Google Scholar]
- Van Aelst, L., and Symons, M. (2002). Role of Rho family GTPases in epithelial morphogenesis. Genes Dev. 16, 1032-1054. [DOI] [PubMed] [Google Scholar]
- van Leeuwen, F.N., Olivo, C., Grivell, S., Giepmans, B.N., Collard, J.G., and Moolenaar, W.H. (2003). Rac activation by lysophosphatidic acid LPA1 receptors through the guanine nucleotide exchange factor Tiam1. J. Biol. Chem. 278, 400-406. [DOI] [PubMed] [Google Scholar]
- Wells, A., Ware, M.F., Allen, F.D., and Lauffenburger, D.A. (1999). Shaping up for shipping out: PLCgamma signaling of morphology changes in EGF-stimulated fibroblast migration. Cell Motil. Cytoskeleton 44, 227-233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.