Abstract
The small guanosine triphosphatase Rac1 is activated by E-cadherin-mediated cell-cell adhesion and is required for the accumulation of actin filaments, E-cadherin, and β-catenin at sites of cell-cell contact. However, the modes of activation and action of Rac1 remain to be clarified. We here found that suppression of IQGAP1, an actin-binding protein and an effector of Rac1, by small interfering RNA apparently reduced the accumulation of actin filaments, E-cadherin, and β-catenin at sites of cell-cell contact in Madin-Darby canine kidney II epithelial cells under the conditions in which knockdown of Rac1 reduced them. Knockdown of Rac1 did not affect the localization of these junctional components in cells expressing a constitutively active IQGAP1 mutant defective in Rac1/Cdc42 binding. Knockdown of either Rac1 or IQGAP1 accelerated the 12-O-tetradecanoylphorbol-13-acetate-induced cell-cell dissociation. The basal Rac1 activity, which was maintained by E-cadherin-mediated cell-cell adhesion, was inhibited in the IQGAP1-knocked down cells, whereas the Rac1 activity was increased in the cells overexpressing IQGAP1. Together, these results indicate that Rac1 enhances the accumulation of actin filaments, E-cadherin, and β-catenin by acting on IQGAP1 and suggest that there exists a positive feedback loop comprised of “E-cadherin-mediated cell-cell adhesion→Rac1 activation→actin-meshwork formation by IQGAP1→increasing E-cadherin-mediated cell-cell adhesion.”
INTRODUCTION
Dynamic rearrangements of cell-cell adhesion are essential for many physiological processes, including tissue rearrangement during development (such as the compaction of the eight-cell embryo), wound healing, synaptogenesis, and tumor metastasis (Takeichi, 1995; Adams and Nelson, 1998; Gumbiner, 2000). The cadherin family is a major group of cell-cell adhesion molecules that mediate intercellular adhesion by Ca2+-dependent homophilic interactions (Takeichi, 1995; Adams and Nelson, 1998; Gumbiner, 2000). Cadherins bind β-catenin, which in turn is linked to the actin cytoskeleton via α-catenin (Tsukita et al., 1992; Kemler, 1993). This linkage between cadherins and the actin-based cytoskeleton contributes to the development of a strong adhesive state. However, the mechanism underlying dynamic rearrangement of cell-cell adhesion remains to be clarified.
Rho family small guanosine triphosphatases (GTPases), including Rac1, Cdc42, and RhoA, are involved in the regulation of cadherin-mediated cell-cell adhesion (Fukata and Kaibuchi, 2001; Braga, 2002). It has been reported that microinjection of dominant negative Rac1 (Rac1N17) (which inhibits the activation of endogenous Rac1 by titrating out its guanine nucleotide exchange factors) and C3 botulinum toxin (which inactivates RhoA) into keratinocytes decreases the accumulation of actin filaments and cadherin at sites of cell-cell contact (Braga et al., 1997). It has also been shown that overexpressing constitutively active Rac1 (Rac1V12), a mutant that is defective in GTPase activity and is thought to exist constitutively in GTP-bound form in cells, in Madin-Darby canine kidney II (MDCKII) epithelial cells increases the accumulation of actin filaments, E-cadherin, and β-catenin at sites of cell-cell contact, whereas overexpressing Rac1N17 reduces their accumulation (Takaishi et al., 1997). Ectopic expression of Tiam1, a GDP/GTP exchange factor for Rac, in MDCKII cells increases the accumulation of actin filaments and E-cadherin at sites of cell-cell contact (Hordijk et al., 1997). Thus, Rac1 seems to be required for the accumulation of actin filaments at cell-cell contact sites and for establishment of cadherin-mediated cell-cell adhesion. On the other hand, we and others showed in an earlier study that E-cadherin-mediated cell-cell adhesion induces Rac1 activation in MDCKII cells and that basal levels of active Rac1 are still maintained even when cells become confluent (Nakagawa et al., 2001; Noren et al., 2001; Betson et al., 2002). E-cadherin-mediated cell-cell adhesion also induces Cdc42 activation in mouse L fibroblasts stably expressing E-cadherin (Kim et al., 2000). These observations suggest that bidirectional signaling pathways must exist, that is, a positive feedback loop between E-cadherin and Rac1. However, the modes of activation and action of Rac1 are not fully understood.
Because IQGAP1, an effector of Rac1 (Hart et al., 1996; Kuroda et al., 1996) and actin-binding protein (Bashour et al., 1997; Fukata et al., 1997), localizes at sites of cell-cell contact (Kuroda et al., 1998), IQGAP1 is a likely candidate for being an effector through which Rac1 can regulate E-cadherin activity. We previously found that overexpression of IQGAP1 reduces E-cadherin-mediated cell-cell adhesion by interacting with β-catenin, causing α-catenin to dissociate from the cadherin/catenin complex (Kuroda et al., 1998). Activated Rac1 and Cdc42 interact with IQGAP1 and thereby inhibit the interaction of IQGAP1 with β-catenin, leading to strong adhesive activity (Fukata et al., 1999). However, the loss of function of IQGAP1 remains unclear and the physiological meaning of the interaction of IQGAP1 with GTP-bound form of Rac1 and actin filaments at sites of cell-cell contact is unknown.
In the present study, we examined whether the suppression of IQGAP1 by RNA interference (RNAi) affects the accumulation of actin filaments, E-cadherin, and β-catenin at sites of cell-cell contact in MDCKII cells. We found that the reduction of IQGAP1 decreased these junctional components as in the case of Rac1-knocked down cells and that knockdown of IQGAP1 decreased the amount of GTP-bound form of Rac1.
MATERIALS AND METHODS
Materials and Chemicals
MDCKII cells and anti-E-cadherin rat monoclonal antibody (ECCD-2) were kindly provided by Dr. Akira Nagafuchi (Kumamoto University, Kumamoto, Japan) and Dr. Shoichiro Tsukita (Kyoto University, Kyoto, Japan). Rhodamine-phalloidin (P-1951) was purchased from Sigma-Aldrich (St. Louis, MO). Anti-ZO-1 rat monoclonal antibody was purchased from Chemicon International (Temecula, CA). Anti-E-cadherin and anti-β-catenin mouse monoclonal antibodies were obtained from BD Transduction Laboratories (Lexington, KY). Anti-Rac1 rabbit polyclonal antibody (C-11) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-IQGAP1 rabbit polyclonal antibody was generated against glutathione S-transferase (GST)-IQGAP1 (216-683 aa). We confirmed that antibodies against IQGAP1 and Rac1 antibodies specifically recognized IQGAP1 and Rac1, respectively, by using recombinant IQGAP1 and Rac1 proteins, and the cell lysates expressing IQGAP1 and Rac1. The immunoreactive bands corresponding to IQGAP1 and Rac1 were diminished by knockdown of IQGAP1 and Rac1, respectively, indicating that these antibodies specifically recognize the target proteins as described below. 12-O-Tetradecanoylphorbol-13-acetate (TPA) was purchased from Wako Pure Chemicals (Osaka, Japan). All materials used in the nucleic acid study were purchased from Takara Shuzo Co. (Kyoto, Japan). Other materials and chemicals were obtained from commercial sources.
Plasmid Constructs
EGFP-IQGAP1(T1050AX2) and EGFP-IQGAP1(G75Q) were produced by subcloning IQGAP1(T1050AX2) and IQGAP1(G75Q) fragments (Fukata et al., 2002) into pEGFP-C2. A fragment harboring a Cdc42/Rac1-interactive binding region (CRIB) of αPAK (70-106 aa) was inserted into pGEX-4T-1 as described previously (Nakagawa et al., 2001). The expression and purification of GST fusion proteins were performed as described previously (Fukata et al., 2002).
Cell Culture
MDCKII cells were maintained at 37°C in a humidified atmosphere of 5% CO2 and 95% air in DMEM containing 10% calf serum. For RNAi experiment, MDCKII cells were seeded at a density of 1 × 105 cells/13-mm coverglass in 24-well dishes. At 24 h after seeding, the cells were transfected with plasmids or small interfering RNA (siRNA). At 48 h after transfection, the cells were fixed and stained with the indicated antibodies (see below). For immunoblot analysis, the cells were scraped at 48 h after transfection. The transfection efficiency of plasmid into MDCK cells is usually 30-50% under our condition. The transfection efficiency of siRNA is close to 90% judging from fluorescein isothiocyanate-labeled silamin and fluorescein isothiocyanate-labeled siIQ-GAP1 RNAs.
To generate MDCKII cells stably expressing EGFP-IQGAP1(T1050AX2), MDCKII cells (5 × 105 cells/10-cm dish) were transfected with 2 μg of pEGFP-C2-IQGAP1(T1050AX2), by using LipofectAMINE 2000 reagent (Invitrogen, Carlsbad, CA), and cultured them in the presence of 0.6 mg/ml G418 to select stable transformants. Colonies of G418-resistant cells were isolated as described previously (Fukata et al., 2001). Three stable clones were isolated.
Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
To determine the target sequences of Rac1 and IQGAP1 in MDCKII cells, RT-PCR was performed on mRNA isolated from MDCKII cells by using ReverTra Ace-α-(Toyobo, Osaka, Japan). Primers were used that were specific for human Rac1 and IQGAP1.
RNA Interference
RNA oligomers containing 21 nucleotides were synthesized in sense and antisense directions corresponding to human IQGAP1 at nucleotides 363-381 (5′-UGCCAUGGAUGAGAUUGGA-3′) and human Rac1 at nucleotides 552-570 (5′-GAGGAAGAGAAAAUGCCUG-3′) with dTdT overhangs at each 3′ terminus (JBioS, Saitama, Japan). The selected sequences were submitted to a BLAST search against the human genome to ensure that only IQGAP1 or Rac1 gene was targeted. As an unspecific siRNA control, (5′-CAGUCGCGUUUGCGACUGG-3′) with dTdT overhangs at 3′ terminus was used. Annealing was performed as described previously (Elbashir et al., 2001). Transfection was performed using LipofectAMINE 2000 reagent. At 48 h after transfection, cells were fixed for immunofluorescence analysis or scraped for immunoblot analysis.
Immunofluorescence Analysis
The cells were fixed with 3.0% formaldehyde in phosphate-buffered saline (PBS) for 10 min and then treated with PBS containing 0.2% Triton X-100 and 2 mg/ml bovine serum albumin for 10 min. The fixed cells were stained with the indicated antibodies as described previously (Fukata et al., 2002). The fluorescence was examined using a confocal laser scanning microscope (LSM 510; Carl Zeiss, Oberkochene, Germany) built around an Axiovert 100M (Carl Zeiss), and series of images from 1-μm YZ sections were collected. Images were stacked using the LSM 510 software and processed by Adobe Photoshop (Adobe Systems, San Jose, CA). The intensity of the fluorescence was calculated by the LSM 510 software.
Immunoblot Analysis
The cells were washed with PBS and then suspended in 200 μl of SDS sample solution [60 mM Tris-Cl, pH 6.8, 2% SDS, 10% glycerol, 0.025% bromphenol blue (wt/vol water), and 5% 2-mercaptoethanol]. Then, 50 μl of each sample from a 13-mm coverslip was resolved by SDS-PAGE and subjected to immunoblot analysis by using the indicated antibodies.
Subcellular Fractionation
MDCKII cells were seeded at a density of 3 × 105 cells in 10-cm dishes. At 24 h after seeding the cells, the indicated siRNA was transfected. At 48 h after transfection, transfected MDCKII cells were fractionated. The cells from a 10-cm dish were washed and homogenized with homogenizing buffer (20 mM HEPES, pH 7.4, 5 mM KCl, 1 mM MgCl2, 50 mM NaF, 30 mM sodium pyrophosphate, 20 μg/ml leupeptin, 50 μg/ml PMSF, and 1 mM dithiothreitol) in a Dounce homogenizer. The homogenates were centrifuged at 1000 × g for 5 min. The resulting pellets were used as nuclear fraction. The supernatant fluids were further centrifuged at 100,000 × g for 30 min. The resulting supernatants were used as the cytoplasmic fraction. The resulting pellets were used as the membrane fraction. Then, 50 μl of each fraction was subjected to immunoblot analysis. The subcellular fractionation was confirmed by marker proteins (nuclear fraction, CREB; cytoplasmic fraction, Rho GDI; membrane fraction, E-cadherin).
Cosedimentation Assay
F-actin was purified from an acetone powder prepared from rabbit skeletal muscle as described previously (Pardee and Spudich, 1982). F-actin was mixed with either GST-IQGAP1-CHD (1-216 aa) or GST-IQGAP1-CHD-G75Q (1-216 aa), and incubated at room temperature for 1 h in buffer A [20 mM Tris-HCl, pH 7.4, 0.5 mM dithiothreitol, 2 mM MgCl2, 100 mM NaCl, 1 mM EDTA, 10% (wt/vol) sucrose, 0.5 mM ATP, 10 μg/ml leupeptin, and 10 μM PMSF]. After the incubation, 50 μl of each reaction mixture was layered onto a 100-μl sucrose barrier [20% (wt/vol) sucrose in buffer B] and was centrifuged at 200,000 × g for 1 h at room temperature. The supernatants and pellets were separated and subjected to SDS-PAGE, followed by silver staining.
Time-Lapse Imaging and Image Analysis
MDCK cells were seeded at a density of 1 × 104 cells/3.5-cm-diameter glass-bottom dishes. At 24 h after seeding, the cells were transfected with the indicated siRNAs. At 48 h after transfection, the cells were stimulated with TPA (200 nM) and observed with an inverted microscope (IX71-Image; Olympus, Tokyo, Japan) built around Olympus IBMU. Olympus IX71-IMAGE was equipped with a cooled charge-coupled device camera (CoolSNAP HQ; Roper Scientific, Chiba, Japan) controlled by MetaMorph software (Universal Imaging, Downingtown, PA) at 5-min intervals by using a 40× Uapo40× oil iris 3/340 (numerical aperture 1.35).
Detection of GTP-bound Rac1 by Use of GST-CRIB
MDCKII cells (3 × 105 cells/10-cm dish) were seeded in subconfluent. At 24 h after seeding, the cells were transfected with the indicated siRNAs. At 48 h after transfection, the cells were washed twice with ice-cold HEPES-buffered saline (20 mM HEPES, pH 7.4, 137 mM NaCl, and 3 mM KCl), and lysed with 800 μl of lysis buffer (50 mM Tris-HCl at pH 7.4, 10 mM MgCl2, 1% NP-40, 150 mM NaCl, 10 μg/ml leupeptin, 10 μg/ml aprotinin, and 10 μM PMSF). The lysates were then centrifuged at 20,000 × g for 2 min at 4°C, and the supernatant was incubated with purified GST or GST-CRIB immobilized beads at 4°C for 1 h. The beads were washed three times with an excess of lysis buffer and eluted with 45 μl of SDS sample solution. Then, 40 μl of each eluate was subjected to SDS-PAGE, followed by immunoblotting with anti-Rac1 antibody.
RESULTS
Knockdown of Rac1 and IQGAP1 by RNAi
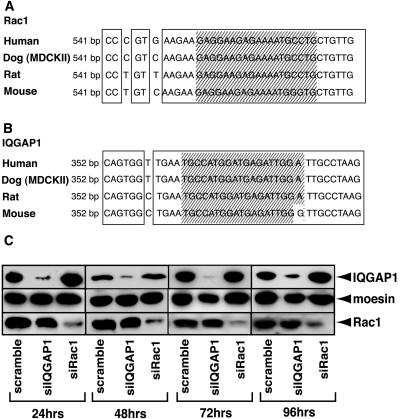
Several lines of evidence using dominant negative mutant and constitutively active mutant suggest that Rac1 is required for cadherin-mediated cell-cell adhesion. We also found that Rac1 and its effector IQGAP1, an actin-binding protein, are involved in cadherin-mediated cell-cell adhesion (zharvKuroda et al., 1998; Fukata et al., 1999). However, direct evidence has yet to be presented that Rac1 and IQGAP1 are required for cadherin-mediated cell-cell adhesion. We, therefore, examined whether Rac1 and IQGAP1 affect the cadherin-mediated cell-cell adhesion in MDCKII cells by using RNAi. We first determined the sequences of Rac1 and IQGAP1 in MDCKII cells by RT-PCR, chose the target sequences common between human and canine sequences for RNAi (Figure 1, A and B) and evaluated the effects of RNAi in MDCKII cells by immunoblot analysis in various periods of time (24, 48, 72, and 96 h) (Figure 1C). Subconfluent MDCKII cells were transfected with siRNA. At that time, the cells still remained subconfluent and adhered to each other steadily (i.e., not the initial stage of cell-cell contact). Immunoblot analysis revealed that Rac1 was specifically knocked down by Rac1-specific siRNA (Figure 1C), whereas the IQGAP1-specific and scramble siRNA did not affect the expression level of Rac1. Under these conditions, the expression level of moesin as a control protein did not change among them. As in the case of Rac1-knockdown, IQGAP1-suppression by siRNA was examined by immunoblot analysis. We could confirm that IQGAP1 was specifically knocked down (Figure 1C). Because we could observe the knockdown of Rac1 and IQGAP1 from 24 h to 96 h, we performed all experiments by using siRNA at 48 h after transfection.
Figure 1.
Knockdown of Rac1 and IQGAP1. (A) Sequence alignment of Rac1 in various species. Shaded box indicates the target sequence for RNAi. (B) Sequence alignment of IQGAP1 in various species. Shaded box indicates the target sequence for RNAi. (C) Knockdown of Rac1 and IQGAP1. Total cell extracts from scramble-, Rac1-, or IQGAP1-siRNA-transfected MDCKII cells were subjected to immunoblot analysis by using anti-IQGAP1, -moesin, and -Rac1 antibodies, which revealed that Rac1 and IQGAP1 were specifically knocked down at 24, 48, 72, and 96 h after transfection.
Rac1 Suppression by siRNA Reduces Actin Accumulation at Sites of Cell-Cell Contact
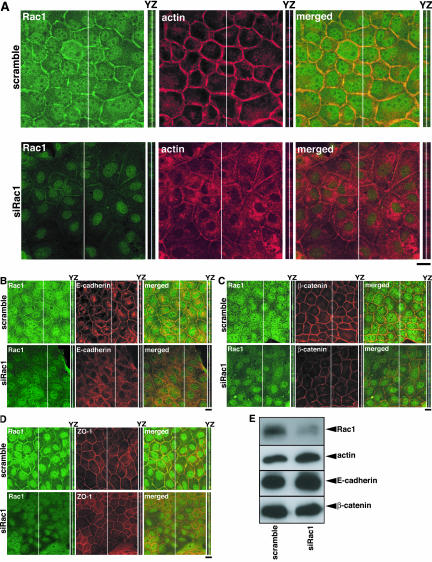
We first examined the effect of Rac1-knockdown on the accumulation of actin filaments at sites of cell-cell contact in MDCKII epithelial cells. Subconfluent MDCKII cells were transfected with scramble- or Rac1-siRNA. At 48 h after transfection, the intensity of actin filaments at sites of cell-cell contact was examined by immunofluorescence analysis. Under these conditions, the cells still remained subconfluent and adhered to each other steadily but were not fully polarized. Immunofluorescence analysis showed that Rac1 localized at sites of cell-cell contact, cytoplasm, and nuclei in scramble-siRNA-transfected cells, whereas Rac1 was reduced at sites of cell-cell contact, cytoplasm, and nuclei in Rac1-siRNA-transfected cells (Figure 2A). This indicates that anti-Rac1 antibody specifically recognizes endogenous Rac1 localization in MDCKII cells. Suppression of Rac1 reduced the accumulation of actin filaments at sites of cell-cell contact, whereas actin accumulation at sites of cell-cell contact was observed in scramble-siRNA-transfected cells (Figure 2A). We also examined the effect of Rac1 abrogation on the localization of E-cadherin, β-catenin, and ZO-1. ZO-1 is known to localize at the apical junction and was used as an apical membrane marker. The intensities of E-cadherin and β-catenin at sites of cell-cell contact were apparently reduced (Figure 2, B and C). These changes in actin filaments, E-cadherin, and β-catenin at sites of cell-cell contact were also observed in the YZ section (Figure 2, A-C). In comparison with the intensities of actin, E-cadherin, and β-catenin, the intensity of ZO-1 was slightly reduced (Figure 2D). When the cells were stained by 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI), a marker of plasma membranes, the intensity of DiI did not change in Rac1-knocked down cells (our unpublished data). The intensities of actin filaments, E-cadherin, and β-catenin at sites of cell-cell contact decreased in Rac1-siRNA-transfected cells to 46, 53, and 49%, respectively (Supplemental Figure 1). These results were consistent with previous observations using the dominant negative mutant of Rac1 (Braga et al., 1997; Takaishi et al., 1997). We also confirmed that the expression levels of actin, E-cadherin, and β-catenin did not alter upon knockdown of Rac1 by immunoblot analysis (Figure 2E). Under these condition, the expression level of moesin as a control protein did not change as described in Figure 1C (our unpublished data).
Figure 2.
Rac1 suppression by siRNA reduces the accumulation of actin filaments, E-cadherin, and β-catenin at sites of cell-cell contact. The transfected cells with siRNA for Rac1 were fixed at 48 h after transfection and stainedbyanti-Rac1antibody(green)andrhodamine-phalloidin (red) (A), E-cadherin (B), β-catenin (C), or ZO-1 (D) and analyzed with a confocal microscope. A stack of images of XY sections is shown. The image of YZ section was taken at the level indicated by a white line in XY section and is shown on the right of the corresponding image. Bars, 10 μm. (E) Expression levels of Rac1, actin, E-cadherin, and β-catenin in control (scramble-siRNA-transfected) and Rac1-suppressed cells were examined by immunoblot analysis.
IQGAP1 Abrogation by siRNA Reduces Actin Accumulation at Sites of Cell-Cell Contact
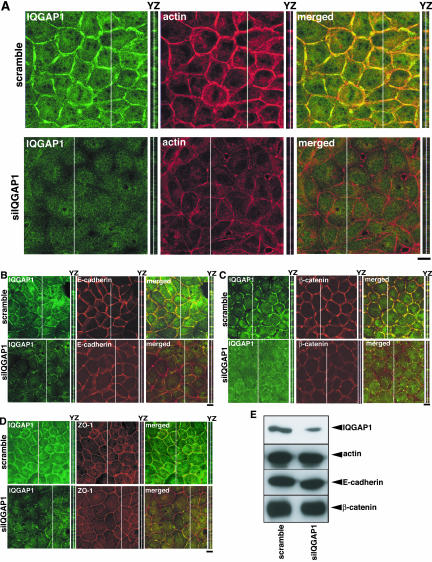
IQGAP1 is an effector of Rac1 (Hart et al., 1996; Kuroda et al., 1996) and accumulates at sites of cell-cell contact (Kuroda et al., 1998). IQGAP1 has actin-cross-linking activity and its activity is enhanced by activated Rac1(Bashour et al., 1997; Fukata et al., 1997). Thus, it is conceivable that IQGAP1 cross-links actin filaments at sites of cell-cell contact downstream of Rac1, contributing to the establishment of actin meshwork at sites of cell-cell contact. We knocked down IQGAP1 by RNAi in MDCKII cells. Immunofluorescence analysis showed that IQGAP1 was localized at sites of cell-cell contact and cytoplasm in scramble-siRNA-transfected cells, whereas IQGAP1 was reduced at sites of cell-cell contact and cytoplasm in IQGAP1-siRNA-transfected cells (Figure 3A). This indicates that anti-IQGAP1 antibody specifically recognizes endogenous IQGAP1 localization in MDCKII cells. As in the case of Rac1-knockdown, the intensity of actin filaments at sites of cell-cell contact was markedly reduced in IQGAP1-siRNA-transfected cells (Figure 3A). We also examined the effect of IQGAP1 abrogation on the localization of E-cadherin, β-catenin, and ZO-1. The intensities of E-cadherin and β-catenin at sites of cell-cell contact were markedly reduced (Figure 3, B and C). These changes in actin filaments, E-cadherin, and β-catenin at sites of cell-cell contact were also observed in YZ section (Figure 3, A-C). The intensity of ZO-1 was slightly reduced (Figure 3D). When the cells were stained by DiI, the intensity of DiI did not change in IQGAP1-knocked down cells (our unpublished data). The intensities of actin filaments, E-cadherin, and β-catenin at sites of cell-cell contact decreased in IQGAP1-siRNA-transfected cells to 47, 46, and 45%, respectively (Supplemental Figure 2). The expression levels of actin, E-cadherin, and β-catenin did not alter upon suppression of IQGAP1 (Figure 3E). Under these condition, the expression level of moesin as a control protein did not change as described in Figure 1C (our unpublished data). These results were essentially similar to those seen in Rac1-knocked down cells.
Figure 3.
IQGAP1 suppression by siRNA reduces the accumulation of actin filaments, E-cadherin, and β-catenin at sites of cell-cell contact. (A) The transfected cells with siRNA for IQGAP1 were fixed at 48 h after transfection and stained by anti-IQGAP1 antibody (green) and rhodamine-phalloidin (red) (A), E-cadherin (B), β-catenin (C), or ZO-1 (D) and analyzed with a confocal microscope. A stack of images of XY sections is shown. The image of YZ section was taken at the level indicated by a white line in XY section, and is shown on the right of the corresponding image. Bars, 10 μm. (E) Expression levels of IQGAP1, actin, E-cadherin, and β-catenin in control (scramble-siRNA-transfected) and IQGAP1-suppressed cells were examined by immunoblot analysis.
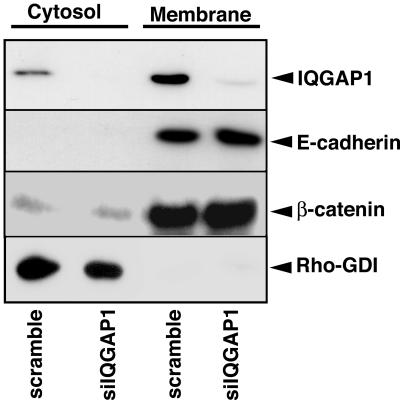
Next, we biochemically investigated the subcellular distribution of E-cadherin and β-catenin in IQGAP1-suppressed cells. Under our conditions, E-cadherin was detected only in the membrane fraction in control (scramble-siRNA-treated) cells, whereas IQGAP1 and β-catenin were detected in both cytosol and membrane fractions (Figure 4). The levels of IQGAP1 and β-catenin in the cytosol fraction were much lower than those in the membrane fraction. When IQGAP1 was knocked down, the amounts of IQGAP1 in both cytosol and membrane fractions were reduced, whereas the amounts of E-cadherin and β-catenin in the membrane and cytosol fractions did not change (Figure 4). Where did E-cadherin and β-catenin go in the IQGAP1-knocked down cells? E-cadherin/β-catenin complex is actively internalized and recycled back to the plasma membrane (Le et al., 1999). The pool of E-cadherin undergoing endocytosis and recycling is markedly increased without stable cell-cell contacts, i.e., in preconfluent cells and after cell contacts are disrupted by depletion of extracellular Ca2+. Thus, it can be speculated that E-cadherin and β-catenin were translocated from cell-cell contact sites to endocytic vesicles that could not be visualized by immunofluorescence under the conditions (Le et al., 1999). Together, these results indicate that IQGAP1 is involved in the accumulation of actin filaments, E-cadherin, and β-catenin at sites of cell-cell contact in MDCKII cells. Because IQGAP1 is an effector of Rac1 and cross-links actin filaments, these results also strongly suggest that Rac1 increases the accumulation of actin filaments and thereby the accumulation of E-cadherin and β-catenin at sites of cell-cell contact acting on IQGAP1.
Figure 4.
Subcellular distribution of IQGAP1, E-cadherin, and β-catenin in IQGAP1-knocked down MDCKII cells. MDCKII cells were transfected with scramble- or IQGAP1-siRNA. At 48 h after transfection, MDCKII cells were separated into nuclear (nucleus; our unpublished data), cytoplasmic (cytosol), and membrane (membrane) fractions, and the fractions were subjected to immunoblot analysis by using anti-IQGAP1, E-cadherin, β-catenin, and Rho GDI antibodies. These results are representative of three independent experiments.
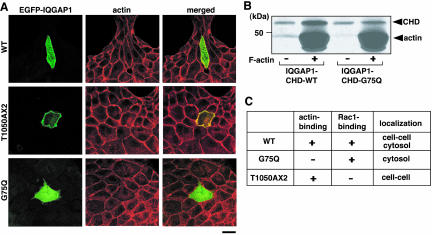
To strengthen this rationale, we used an IQGAP1 mutant (named T1050AX2) that is defective in Rac1/Cdc42 binding by mutation at the ras GAP-related domain (Fukata et al., 2002). It has recently been shown that residues 1054-1077 of IQGAP1 are essential for the Cdc42-binding (Mataraza et al., 2003b). This observation is consistent with the fact that IQGAP1(T1050AX2) does not bind Rac/Cdc42 because the mutated portion is very close to residues 1054-1077 of IQGAP1. We previously showed that IQGAP1(T1050AX2) induces multiple leading edges and colocalizes with actin filaments at the leading edge in Vero fibroblasts (Fukata et al., 2002). We also found that IQGAP1(T1050AX2) bypasses the stimulatory effect of activated Rac1 on the CLIP-170/IQGAP1 interaction (Fukata et al., 2002). Based on these results and the fact that IQGAP1(T1050AX2) does not interact with Rac1, EGFP-IQGAP1(T1050AX2) is considered to function as a constitutively active mutant.
When EGFP-IQGAP1(T1050AX2) was transiently introduced into MDCKII cells, EGFP-IQGAP1(T1050AX2) specifically localized at sites of cell-cell contact (Figure 5A). Under the same conditions, EGFP-IQGAP1(WT) localized at sites of cell-cell contact and in cytosol (Figure 5A). When cells expressing IQGAP1(T1050AX2) were treated with cytochalasin D, both IQGAP1(T1050AX2) and actin filaments delocalized from sites of cell-cell contacts as in the case of IQGAP1(WT) (our unpublished data). Thus, it is likely that IQGAP1(T1050AX2) interacts with actin filaments and its localization depends on actin filaments. Because IQGAP1 is thought to interact with F-actin through the calponin homology domain (Bashour et al., 1997; Fukata et al., 1999), we produced IQGAP1(G75Q) that was mutated at the calponin homology domain (with substitution of Gln for Gly-75) and showed less binding activity to F-actin (Figure 5B). EGFP-IQGAP1(G75Q) did not localize at sites of cell-cell contact (Figure 5A). We also confirmed that the double mutant EGFP-IQGAP1(G75Q/T1050AX2) did not localize at sites of cell-cell contact (our unpublished data). These observations indicate that the interaction of IQGAP1 with actin filaments is necessary for the localization of IQGAP1 at sites of cell-cell contact in MDCKII cells. The characters of IQGAP1 and its mutants are summarized in Figure 5C.
Figure 5.
IQGAP1 mutant defective in Rac1/Cdc42 binding or in actin binding. (A) To visualize the localization of IQGAP1 mutants in a single cell, EGFP-IQGAP1(WT), EGFP-IQGAP1(T1050AX2), and EGFP-IQGAP1(G75Q) were transiently transfected into MDCKII cells at a low transfection efficiency, and the cells were stained by rhodamine-phalloidin (red). The representative images are shown. Bar, 10 μm. (B) GST-IQGAP1-CHD (1-216 aa) or GST-IQGAP1-CHD-G75Q (100 nM each) was mixed with F-actin (3 μM) and incubated for 1 h at room temperature. After the incubation, 50 μl of each reaction mixture was layered onto a 100-μl 20% (wt/vol) sucrose barrier and was centrifuged at 200,000 × g for 1 h at room temperature. Pellet samples were resolved by SDS-PAGE and visualized by silver staining. (C) Characters of IQGAP1 mutants.
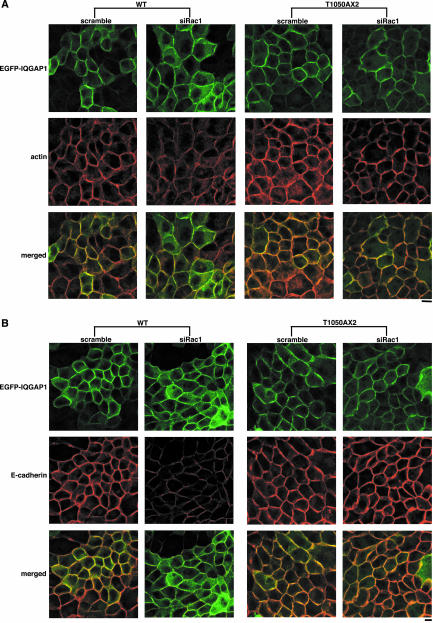
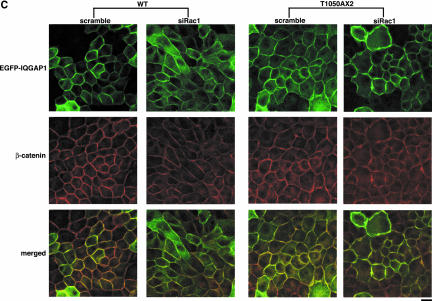
Next, we examined the effect of Rac1-knockdown on the accumulation of actin filaments at sites of cell-cell contact by using the cells stably expressing EGFP-IQGAP1(WT) or EGFP-IQGAP1(T1050AX2). Both EGFP-IQGAP1(WT) and EGFP-IQGAP1(T1050AX2)-expressing cells express the GFP-fusion protein to extents similar to those of endogenous IQGAP1 (our unpublished data). Transfection of Rac1-siRNA reduced the accumulation of actin filaments at sites of cell-cell contact in EGFP-IQGAP1(WT)-expressing cells, whereas transfection of Rac1-siRNA in EGFP-IQGAP1(T1050AX2)-expressing cells did not affect actin accumulation (Figure 6A). Under the conditions, we confirmed that Rac1 was knocked down by siRNA in both EGFP-IQGAP1(WT)- and EGFP-IQGAP1(T1050AX2)-expressing cells (our unpublished data). Similar results were obtained toward E-cadherin and β-catenin (Figure 6, B and C). These results indicate that EGFP-IQGAP1(T1050AX2), the constitutively active mutant, bypasses Rac1 and maintains the accumulation of actin filaments, E-cadherin, and β-catenin at sites of cell-cell contact, whereas overexpression of EGFP-IQGAP1(WT) cannot rescue the Rac1-knocked down phenotype.
Figure 6.
IQGAP1 mutant defective in Rac1/Cdc42 binding suppresses the effect of Rac1-knockdown on actin filaments, E-cadherin, and β-catenin. The cells expressing EGFP-IQGAP1(WT) or EGFP-IQGAP1(T1050AX2) were transfected with siRNA for Rac1, fixed at 48 h after transfection and stained by rhodamine-phalloidin (red) (A), E-cadherin (B), and β-catenin (C). Essentially similar results were obtained when two other cell lines expressing EGFP-IQGAP1(WT) or EGFP-IQGAP1(T1050AX2) were used. Bar, 10 μm.
IQGAP1 Suppression by siRNA Promotes TPA-induced Cell Scattering
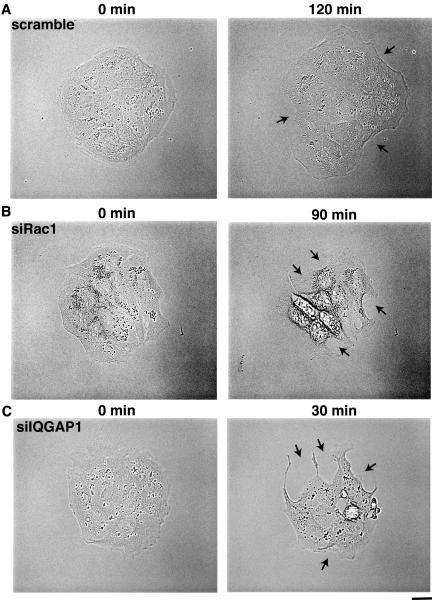
Cell scattering provides an example of dynamic rearrangement of E-cadherin mediated cell-cell adhesion (Adams and Nelson 1998: Gumbiner 2000). TPA or HGF (hepatocyte growth factor) induces centrifugal spreading of MDCK cells in colonies, cell-cell dissociation, and ultimately cell scattering. TPA-induced cell-cell dissociation and subsequent cell scattering are inhibited in MDCK cells expressing constitutively active Rac1 (Kodama et al., 1999; Fukata et al., 2001). We have previously shown that the level of GTP-bound form of Rac1 and the Rac1/IQGAP1 complex decrease after stimulation with TPA in MDCK cells (Fukata et al., 2001). These results indicate that active Rac1 is important for cell-cell adhesion in MDCK cells and that the inactivation of Rac1 is necessary for cell-cell dissociation during action of TPA. To further investigate the roles of IQGAP1 and Rac1 in cell-cell adhesion, we examined the cell-cell dissociation in IQ-GAP1- and Rac1-suppressed cells by time-lapse microscopic observation. At 120 min after the stimulation with TPA, some cells started detaching from one another in scramble-siRNA-transfected cells (Figure 7A; see Supplemental Movie S 1). The average time for cell-cell dissociation (50% cells in colonies show cell-cell dissociation) is ∼140 min. Similar result was observed in Rac1-siRNA-transfected cells (Figure 7B; see Supplemental Movie S 2), but the average time for cell-cell dissociation is ∼100 min, indicating that Rac1-suppressed cells dissociate more rapidly than the control cells do. Some cells in IQGAP1-siRNA-transfected cells started detaching at 30 min after the stimulation with TPA (Figure 7C; see Supplemental Movie S 3). The average time for cell-cell dissociation in IQGAP1-siRNA-transfected cells is ∼35 min. These results indicate that knockdown of either IQGAP1 or Rac1 accelerates cell-cell dissociation in TPA-stimulated MDCK Cells.
Figure 7.
TPA-induced cell scattering is enhanced by knockdown of IQGAP1. (A) MD-CKII cells transfected with siRNA for scramble were stimulated with TPA (200 nM) at 48 h after transfection. (B) MDCKII cells transfected with siRNA for Rac1 were stimulated with TPA (200 nM) at 48 h after transfection. (C) MDCKII cells transfected with siRNA for IQGAP1 were stimulated with TPA (200 nM) at 48 h after transfection. Representative time-lapse images are shown. Elapsed time is indicated on the top. Arrowheads indicate the representative cells dissociated by TPA stimulation. Bar, 10 μm.
IQGAP1 Suppression by siRNA Decreases GTP-bound Form of Rac1
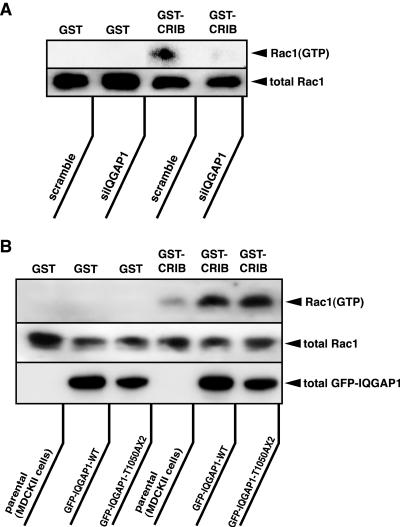
It has been recently proposed that a positive feedback loop comprised of phosphatidylinositol 3-OH kinase (PI3-kinase), phosphatidyl inositol-3,4,5-triphospahte (PIP3) accumulation, Rac1, and actin filaments at the leading edge exists in neutrophils during chemoattractant-induced cell polarization (Servant et al., 2000; Weiner et al., 2002; Srinivasan et al., 2003). We also proposed that epithelial cells have a positive feedback loop comprised of “cadherin-mediated adhesion→Rac1 activation” and “Rac1 activation→cadherin-mediated adhesion” (Fukata and Kaibuchi, 2001). Similarly, Rac1 and PI3-kinase are key molecules in this pathway. These previous observations led us to speculate that IQGAP1 is involved in this positive feedback loop in MDCKII cells. We directly addressed this possibility. We measured the Rac1 activity in the cells in which IQGAP1 was knocked down (Figure 8A). The levels of GTP-bound form of Rac1 were biochemically measured by affinity precipitation using the Rac1-binding region of its specific effector: the CRIB of PAK (Nakagawa et al., 2001). As reported previously (Nakagawa et al., 2001; Noren et al., 2001), Rac1 in its active GTP-bound form could be detected in MDCKII cells cultured in medium containing a normal Ca2+ level. When IQGAP1 was knocked down, the level of active Rac1 decreased. Total Rac1 in lysates did not change between IQGAP1-suppressed cells and control (scramble-siRNA-treated) cells. No Rac1 was detected in samples incubated with GST alone. Furthermore, we measured the Rac1 activity in the cells stably expressing EGFP-IQGAP1(WT) and -IQGAP1(T1050AX2) (Figure 8B). The level of active Rac1 increased in the cells expressing IQGAP1(WT) in comparison with that of the parental cells. The level of active Rac1 in the cells IQGAP1(T1050AX2) was slightly higher than that in the cells expressing IQGAP1(WT). Total Rac1 in lysates did not change among control (parental MDCK) cells and cells stably expressing EGFP-IQGAP1(WT) and -IQGAP1(T1050AX2).
Figure 8.
IQGAP1 is involved in basal Rac1 activity. (A) MDCKII cells were transfected with scramble- or IQGAP1-siRNA. At 48 h after transfection, the cells were lysed with lysis buffer, and the lysates were incubated with GST or GST-CRIB to measure the level of GTP-bound Rac1. (B) MDCKII cells expressing EGFP-IQGAP1(WT) or EGFP-IQGAP1(T1050AX2) were seeded and cultured for 24 h, then the cells were serum-starved. At 24 h after serum starvation, the cells were lysed with lysis buffer, and the lysates were incubated with GST or GST-CRIB to measure the level of GTP-bound Rac1. These results are representative of three independent experiments.
DISCUSSION
Modes of Action of Rac1 and IQGAP1 in Cadherin-mediated Cell-Cell Contact
We previously found that overexpression of IQGAP1 reduces E-cadherin-mediated cell-cell adhesion by interacting with β-catenin, causing α-catenin to dissociate from the cadherin/catenin complex (Kuroda et al., 1998). Activated Rac1 and Cdc42 interact with IQGAP1 and thereby inhibit the interaction of IQGAP1 with β-catenin, leading to strong adhesive activity (Fukata et al., 1999). However, several aspects of this process require further exploration: 1) the loss of function of IQGAP1 in epithelial cells; 2) the physiological meaning of interaction of IQGAP1 with GTP-bound form of Rac1; and 3) the physiological meaning of the interaction of IQGAP1 with actin filaments.
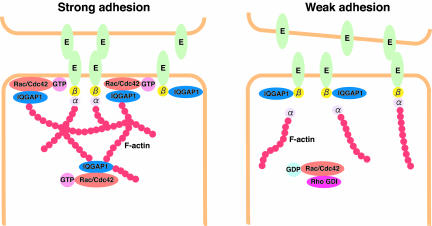
In the present study, we found that the abrogation of either Rac1 or IQGAP1 by RNAi reduced the accumulation of actin filaments, E-cadherin, and β-catenin at sites of cell-cell contact in MDCKII cells and that the effect of Rac1 suppression was reduced by the expression of constitutively active IQGAP1(T1050AX2), which does not bind Rac1. We also found that TPA-induced cell scattering in IQGAP1- or Rac1-suppressed cells occurred faster than that of scramble-siRNA-transfected cells. These results indicate that IQGAP1 as well as Rac1 are necessary for cell-cell adhesion. Based on these observations, we suggest that IQGAP1 has at least two different actions upon the nucleotide state of Rac1 (Figure 9). When the amount of GTP-bound form of Rac1 increases, Rac1 directly binds IQGAP1, thereby cross-linking actin filaments, which are linked to the cadherin/β-catenin complex via α-catenin. Under these conditions, IQGAP1 does not bind to β-catenin and cannot dissociate α-catenin from the cadherin/catenins complex, leading to strong adhesion. Because IQGAP1 has anti-GTPase activity (Hart et al., 1996), it is possible that IQGAP1 maintains the amount of GTP-bound form of Rac1 at sites of cell-cell contact, leading to stable adhesion. By contrast, when the amount of the GDP-bound form of Rac1 increases, IQGAP1 is freed from Rac1 and actin filaments, and interacts with β-catenin to dissociate α-catenin from the cadherin/catenin complex. This results in weak adhesion and cell-cell dissociation. Thus, IQGAP1 behaves both as a positive and a negative regulator downstream of Rac1. It may be noted that IQGAP1(T1050AX2) binds to β-catenin as IQGAP1(WT) does both in vivo and in vitro (our unpublished data). This raises the possibility that IQGAP1(T1050AX2) interacts with β-catenin and induces the dissociation of α-catenin from β-catenin to weaken the cell-cell adhesion. Nevertheless, the ectopic expression of IQGAP1(T1050AX2) seems to strengthen the cell-cell adhesion in MDCK cells. This apparent discrepancy may be explained by the notion that IQGAP1(T1050AX2) can connect actin filaments to the E-cadherin/β-catenin complex without α-catenin. Further studies are necessary to fully understand the mode of actions of IQGAP1(T1050AX2).
Figure 9.
Role of Rac1 and IQGAP1 in the regulation of E-cadherin-mediated cell-cell adhesion. E, E-cadherin; α, α-catenin; β, β-catenin.
Although we tried to knock down Cdc42 in MDCKII cells, we could not clearly detect endogenous Cdc42 by immunoblot and immunofluorescence analysis under the same conditions that we used to detect Rac1, probably due to the fact that the amount of Cdc42 expressed in MDCKII cells is smaller than that of Rac1. Previously we calculated the intracellular concentrations of Rac1 and Cdc42 in MDCKII cells to be ∼500 and 100 nM, respectively (Fukata et al., 2001). In MDCKII cells, Rac1 may play a more important role in the regulation of cadherin-mediated cell-cell adhesion than Cdc42.
Positive Feedback Loop in Cadherin-mediated Cell-Cell Adhesion
We previously showed that E-cadherin-mediated cell-cell adhesion activates Rac1 through PI3-kinase (Nakagawa et al., 2001). Activated Rac1 increases the accumulation of actin filaments and E-cadherin at sites of cell-cell contact (Takaishi et al., 1997). It has also been reported that E-cadherin that is activated upon homophilic ligation directly signals to regulate E-cadherin-mediated cell-cell adhesion through Rac and PI3-kinase (Kovacs et al., 2002). These results suggest that a positive feedback loop exists between E-cadherin-mediated cell-cell adhesion and Rac1 activation. However, the molecular mechanism remains to be clarified. In the present study, we showed that knock-down of IQGAP1 decreased the amount of GTP-bound form of Rac1, which is thought to localize at sites of cell-cell contact (Nakagawa et al., 2001), and that overexpression of IQGAP1(WT) or IQGAP1(T1050AX2) increased the amount of GTP-bound form of Rac1. Judging from the facts that IQGAP1 cross-links actin filaments and that IQGAP1 is necessary for the accumulation of actin filaments at sites of cell-cell contact, IQGAP1 may be one of the components of the feedback loop (i.e., E-cadherin-mediated cell adhesion→PI3-kinase→Rac1 activation→actin-meshwork formation by IQGAP1→participation of IQGAP1 in E-cadherin-mediated cell adhesion). This hypothesis is consistent with previous theories about neutrophils. Bourne's group proposed that amplification of PIP3 at the leading edge requires Rac1 and PI3-kinase, and that actin filaments and these molecules establish a positive feedback loop (Servant et al., 2000; Weiner et al., 2002; Srinivasan et al., 2003).
Determination of IQGAP1 Localization at Sites of Cell-Cell Contact
Several factors play pivotal roles in determining the localization of IQGAP1 at sites of cell-cell contact. We previously showed that IQGAP1 accumulates at sites of cell-cell contact in EL cells, which are L cells stably expressing wild-type E-cadherin. The accumulation of IQGAP1 at sites of cell-cell contact could not be observed in nEαCL cells, which are L cells stably expressing an E-cadherin mutant in which the cytoplasmic domain is deleted and replaced by the C-terminal domain of α-catenin (Kuroda et al., 1998; Nagafuchi et al., 1994). Based on this result, IQGAP1 seems to be recruited at sites of cell-cell contact in EL cells through β-catenin. In the present study, we showed that the interaction of IQGAP1 with actin filaments is necessary for its localization at sites of cell-cell contact by using IQGAP1(G75Q) and IQGAP1(G75Q/T1050AX2), which do not bind actin and localizes in cytosol. It has recently reported that IQGAP1 associates with actin filaments and nectin in MDCKII cells (Katata et al., 2003). Together, it raises the possibility that IQGAP1 is not recruited at the cell-cell contact in nEαCL cells because of the lack of activation of Rac1, which causes the lack of actin meshwork at the cell-cell contact. Further studies are necessary to address this issue.
Other Physiological Processes in which IQGAP1 Functions
Several groups have reported that IQGAP1 accumulates at leading edges in directionally migrating cells (Hart et al., 1996; Kuroda et al., 1996; Mataraza et al., 2003b). Targeting and capturing of microtubule plus ends at special cortical regions are essential for directional cell migration. Recent studies revealed that CLIP-170 family (Perez et al., 1999; Fukata et al., 2002), EB1 family (Mimori-Kiyosue et al., 2000; Tirnauer and Bierer, 2000), and dynein/dynactin complex (termed +Tips) (Vaughan et al., 1999; Schuyler and Pellman, 2001) accumulate at the plus ends of microtubules and play an essential role in targeting of microtubules at cortical capture sites. We have recently found that IQGAP1 interacts with microtubules through CLIP-170 and plays a crucial role in cell polarization by capturing microtubules through CLIP-170 at the cortical region in Vero fibroblasts (Fukata et al., 2002). Ectopic expression of IQGAP1(T1050AX2), a constitutively active IQGAP1 mutant, induces multiple leading edges and showed multipolarized morphology (Fukata et al., 2002). It has recently reported that knock down of IQ-GAP1 by siRNA and transfection of dominant negative IQ-GAP1 mutant markedly reduced cell motility (Mataraza et al., 2003a). Thus, IQGAP1 is considered as a key molecule not only in cadherin-mediated cell-cell adhesion but also in directional cell migration.
In conclusion, we found that IQGAP1 is one of the key molecules in accumulating actin filaments at sites of cell-cell contact and in establishing E-cadherin-mediated cell-cell adhesion downstream of Rac1. The next step is to clarify the modes of action of other regulatory components in cadherin-mediated cell-cell adhesion, such as Cdc42, Rap1, and Rab family GTPases, by using RNAi.
Supplementary Material
Acknowledgments
We thank Drs. Akira Nagafuchi and Shoichiro Tsukita for providing MDCKII cells and antibody against E-cadherin (ECCD-2). We also thank T. Ishii for secretarial assistance. This work was supported by grants-in-aid for scientific research from the Ministry of Education, Science, and Culture of Japan, Japan Society for the Promotion of Science, the Novartis Foundation for the Promotion of Science, the Organization for Pharmaceutical Safety and Research (OPSR/Kiko), and Special Coordination Funds for Promoting Science and Technology.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-08-0582. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-08-0582.
Accession numbers. Human IQGAP1: L33075, Mouse IQGAP1: AF240630, Human Rac1: AF498964, Mouse Rac1: BC003828
Abbreviations used: CRIB, Cdc42/Rac1-interactive binding region; DiI, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate; MDCKII cells, Madin-Darby canine kidney II cells; PI3-kinase, phosphatidylinositol 3-OH kinase; RNAi, RNA interference; siRNA, small interfering RNA; TPA, 12-O-tetradecanoylphorbol-13-acetate; WT, wild type.
Online version of this article contains supplementary figures and video material. Online version available at www.molbiolcell.org.
References
- Adams, C.L., and Nelson, W.J. (1998). Cytomechanics of cadherin-mediated cell-cell adhesion. Curr. Opin. Cell Biol. 10, 572-577. [DOI] [PubMed] [Google Scholar]
- Bashour, A.M., Fullerton, A.T., Hart, M.J., and Bloom, G.S. (1997). IQGAP1, a Rac- and Cdc42-binding protein, directly binds and cross-links microfilaments. J. Cell Biol. 137, 1555-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betson, M., Lozano, E., Zhang, J., and Braga, V.M. (2002). Rac activation upon cell-cell contact formation is dependent on signaling from the epidermal growth factor receptor. J. Biol. Chem. 277, 36962-36969. [DOI] [PubMed] [Google Scholar]
- Braga, V. (2002). Cell-cell adhesion and signalling. Curr. Opin. Cell Biol. 5, 546-556. [DOI] [PubMed] [Google Scholar]
- Braga, V., Machesky, L.M., Hall, A., and Hotchin, N.A. (1997). The small GTPases rho and rac are required for the establishment of cadherin-dependent cell-cell contacts. J. Cell Biol. 137, 1421-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- Fukata, M., and Kaibuchi, K. (2001). Rho-family GTPases in cadherin-mediated cell-cell adhesion. Nat. Rev. Mol. Cell. Biol. 2, 887-897. [DOI] [PubMed] [Google Scholar]
- Fukata, M., Kuroda, S., Fujii, K., Nakamura, T., Shoji, I., Matsuura, Y., Okawa, K., Iwamatsu, A., Kikuchi, A., and Kaibuchi, K. (1997). Regulation of cross-linking of actin filament by IQGAP1, a target for Cdc42. J. Biol. Chem. 272, 29579-29583. [DOI] [PubMed] [Google Scholar]
- Fukata, M., Kuroda, S., Nakagawa, M., Kawajiri, A., Itoh, N., Shoji, I., Matsuura, Y., Yonehara, S., Kikuchi, A., and Kaibuchi, K. (1999). Cdc42 and Rac1 regulate the interaction of IQGAP1 with b-catenin. J. Biol. Chem. 274, 26044-26050. [DOI] [PubMed] [Google Scholar]
- Fukata, M., Nakagawa, M., Itoh, N., Kawajiri, A., Yamaga, M., Kuroda, S., and Kaibuchi, K. (2001). Involvement of IQGAP1, an effector of Rac1 and Cdc42 GTPases, in cell-cell dissociation during cell scattering. Mol. Cell. Biol. 21, 2165-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata, M., Watanabe, T., Noritake, J., Nakagawa, M., Yamaga, M., Kuroda, S., Matsuura, Y., Iwamatsu, A., Perez, F., and Kaibuchi, K. (2002). Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell 109, 873-885. [DOI] [PubMed] [Google Scholar]
- Gumbiner, B.M. (2000). Regulation of cadherin adhesive activity. J. Cell Biol. 148, 399-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart, M.J., Callow, M.G., Souza, B., and Polakis, P. (1996). IQGAP1, a calmodulin-binding protein with a rasGAP-related domain, is a potential effector for cdc42Hs. EMBO J. 15, 2997-3005. [PMC free article] [PubMed] [Google Scholar]
- Hordijk, P.L., ten Klooster, J.P., van der Kammen, R.A., Michiels, F., Oomen, L.C., and Collard, J.G. (1997). Inhibition of invasion of epithelial cells by Tiam1-Rac signaling. Science 278, 1464-1466. [DOI] [PubMed] [Google Scholar]
- Katata, T., Irei, K., Fukuhara, A., Kawakatsu, T., Yamada, A., Shimizu, K., and Takai, Y. (2003). Involvement of nectin in the localization of IQGAP1 at cell-cell adhesion sites through the actin cytoskeleton in Madin-Darby canine kidney cells. Oncogene 22, 2097-2109. [DOI] [PubMed] [Google Scholar]
- Kemler, R. (1993). From cadherin to catenins: cytoplasmic protein interactions and regulation of cell adhesion. Trends Genet. 9, 317-321. [DOI] [PubMed] [Google Scholar]
- Kim, S.H., Li, Z., and Sacks, D.B. (2000). E-cadherin-mediated cell-cell attachment activates Cdc42. J. Biol. Chem. 275, 36999-37005. [DOI] [PubMed] [Google Scholar]
- Kodama, A., Takaishi, K., Nakano, K., Nishioka, H., and Takai, Y. (1999). Involvement of Cdc42 small G protein in cell-cell adhesion, migration and morphology of MDCK cells. Oncogene. 18, 3996-4006. [DOI] [PubMed] [Google Scholar]
- Kovacs, E.M., Ali, R.G., McCormack, A.J., and Yap, A.S. (2002). E-cadherin homophilic ligation directly signals through Rac and phosphatidylinositol 3-kinase t regulate adhesive contacts. J. Biol. Chem. 277, 6708-6718. [DOI] [PubMed] [Google Scholar]
- Kuroda, S., Fukata, M., Kobayashi, K., Nakafuku, M., Nomura, N., Iwamatsu, A., and Kaibuchi, K. (1996). Identification of IQGAP as a putative target for the small GTPases, Cdc42 and Rac1. J. Biol. Chem. 271, 23363-23367. [DOI] [PubMed] [Google Scholar]
- Kuroda, S., et al. (1998). Role of IQGAP1, a target of the small GTPases Cdc42 and Rac1, in regulation of E-cadherin-mediated cell-cell adhesion. Science 281, 832-835. [DOI] [PubMed] [Google Scholar]
- Le, T.L., Yap, A.S., and Stow, J.L. (1999). Recycling of E-cadherin: a potential mechanism for regulating cadherin dynamics. J. Cell Biol. 146, 219-232. [PMC free article] [PubMed] [Google Scholar]
- Mataraza, J.M., Briggs, M.W., Li, Z., Entwistle, A., Ridley, A.J., and Sacks, D.B. (2003a). IQGAP1 promotes cell motility and invasion. J. Biol. Chem. 278, 41237-41245. [DOI] [PubMed] [Google Scholar]
- Mataraza, J.M., Briggs, M.W., Li, Z., Frank, R., and Sacks, D.B. (2003b). Identification and characterization of the Cdc42-binding site of IQGAP1. Biochem. Biophys. Res. Commun. 305, 315-321. [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue, Y., Shiina, N., and Tsukita, S. (2000). The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr. Biol. 10, 865-868. [DOI] [PubMed] [Google Scholar]
- Nagafuchi, A., Ishihara, S., and Tsukita, S. (1994). The roles of catenins in the cadherin-mediated cell adhesion: functional analysis of E-cadherin-alpha catenin fusion molecules. J. Cell Biol. 127, 235-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, M., Fukata, M., Yamaga, M., Itoh, N., and Kaibuchi, K. (2001). Recruitment and activation of Rac1 by the formation of E-cadherin-mediated cell-cell adhesion sites. J. Cell Sci. 114, 1829-1838. [DOI] [PubMed] [Google Scholar]
- Noren, N.K., Niessen, C.M., Gumbiner, B.M., and Burridge, K. (2001). Cadherin engagement regulates Rho family GTPases. J. Biol. Chem. 276, 33305-33308. [DOI] [PubMed] [Google Scholar]
- Pardee, J.D., and Spudich, J.A. (1982). Purification of muscle actin. Methods Enzymol. 85, 164-181. [DOI] [PubMed] [Google Scholar]
- Pece, S., Chiariello, M., Murga, C., and Gutkind, J.S. (1999). Activation of the protein kinase Akt/PKB by the formation of E-cadherin-mediated cell-cell junctions. Evidence for the association oh Phosphatidylinositol 3-kinase with the E-cadherin adhesion complex. J. Biol. Chem. 274, 19347-19351. [DOI] [PubMed] [Google Scholar]
- Perez, F., Diamantopoulos, G.S., Stalder, R., and Kreis, T.E. (1999). CLIP-170 highlights growing microtubule end in vivo. Cell 96, 517-527. [DOI] [PubMed] [Google Scholar]
- Servant, G., Weiner, O.D., Herzmark, P., Balla, T., Sedat, J.W., and Bourne, H.R. (2000). Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science. 287, 1037-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler, S.C., and Pellman, D. (2001). Microtubule “plus-end-tracking proteins”: the end is just the beginning. Cell 105, 421-424. [DOI] [PubMed] [Google Scholar]
- Srinivasan, S., Wang, F., Glavas, S., Ott, A., Hofmann, F., Aktories, K., Kalman, D., and Bourne, H.R. (2003). Rac and Cdc42 play distinct roles in regulating PI(3,4,5)P3 and polarity during neutrophil chemotaxis. J. Cell Biol. 160, 375-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaishi, K., Sasaki, T., Kotani, H., Nishioka, H., and Takai, Y. (1997). Regulation of cell-cell adhesion by rac and rho small G proteins in MDCK cells. J. Cell Biol. 139, 1047-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi, M. (1995). Morphogenetic roles of classic cadherins. Curr. Opin. Cell Biol. 7, 619-627. [DOI] [PubMed] [Google Scholar]
- Tirnauer, J.S., and Bierer, B.E. (2000). EB1 proteins regulate microtubule dynamics, cell polarity, and chromosome stability. J. Cell Biol. 149, 761-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita, S., Tsukita, S., Nagafuchi, A., and Yonemura, S. (1992). Molecular linkage between cadherins and actin filaments in cell-cell adherens junctions. Curr. Opin. Cell Biol. 4, 834-839. [DOI] [PubMed] [Google Scholar]
- Vaughan, K.T., Tynan, S.H., Faulkner, N.E., Echeverri, C.J., and Vallee, R.B. (1999). Colocalization of cytoplasmic dynein with dynactin and CLIP-170 at microtubule distal ends. J. Cell Sci. 112, 1437-1447. [DOI] [PubMed] [Google Scholar]
- Weiner, O.D., Neilsen, P.O., Prestwich, G.D., Kirschner, M.W., Cantley, L.C., and Bourne, H.R. (2002). A PtdInsP(3)- and Rho GTPase-mediated positive feedback loop regulates neutrophil polarity. Nat. Cell Biol. 4, 509-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.