Abstract
The energy-dependent uptake of trace nutrients by Gram-negative bacteria involves the coupling of an outer membrane transport protein to the transperiplasmic protein TonB. In the present study, a soluble construct of Escherichia coli TonB (residues 33–239) was used to determine the affinity of TonB to the outer membrane transporters BtuB, FecA and FhuA. Using fluorescence anisotropy, TonB(33–239) was found to bind with high-affinity (tens of nM) to both BtuB and FhuA; however, no high-affinity binding was observed to FecA. In BtuB, the high affinity binding of TonB(33–239) was eliminated by mutations in the Ton box, which yield transport-defective protein, or by the addition of a Colicin E3 fragment, which stabilizes the Ton box in a folded state. These results indicate that transport requires a high-affinity transporter-TonB interaction that is mediated by the Ton box. Characterization of TonB(33–239) using double electron-electron resonance (DEER) demonstrates that a significant population of TonB(33–239) exists as a dimer; moreover, interspin distances are in approximate agreement with interlocked dimers observed previously by crystallography for shorter TonB fragments. When bound to the outer membrane transporter, DEER shows that the TonB(33–239) dimer is converted to a monomeric form, suggesting that a dimer-monomer conversion takes place at the outer membrane during the TonB-dependent transport cycle.
Gram-negative bacteria use outer membrane transporters for the high-affinity scavenging and active uptake of a range of nutrients including forms of iron, vitamin B12, nickel and carbohydrate.1, 2 These transporters require the inner membrane protein TonB, which is in a complex with two other inner membrane proteins, ExbD and ExbB. TonB possesses a rigid polyproline motif that may span the length of the periplasmic space,3 and a globular C-terminal domain that interacts with the transporter.4, 5 An inner membrane proton motive force is required for transport, which may be used by ExbB and ExbD to drive conformational changes in TonB.6–10 In addition to transporting substrates, TonB-dependent transporters are also utilized as receptors for phage and colicins.11
TonB-dependent transporters have homologous structures that are based upon a 22-stranded β–barrel.12–14 The interior of the barrel is occluded by an N-terminal domain (sometimes referred to as a plug or core domain) of approximately 130 to 150 residues consisting of several highly-conserved regions. One highly conserved motif is the Ton box (see Figure 1), which is believed to be the energy coupling motif for transport.15 In the Escherichia coli vitamin B12 transporter, BtuB, substrate binding unfolds the Ton box into the periplasmic space.16–19 This substrate-induced unfolding transition is thought to regulate the transporter-TonB interaction, and crystal structures of TonB fragments in complex with BtuB4 or the Escherichia coli ferrichrome transporter FhuA5 indicate that the Ton box must unfold for the transporter to engage TonB though a strand-exchange mechanism. However, despite a high level of sequence conservation, single amino acid substitutions in the Ton box do not strongly affect transport, and only proline or glycine mutations at positions 8, 9 or 10 in the BtuB Ton box are known to abrogate or greatly reduce transport.20 Two transport defective mutations, L8P and V10P, have been shown to unfold the BtuB Ton box21 and dramatically affect the pattern of disulfide cross-linking with TonB in vivo.22 Since an unfolded Ton box is thought to be important for association with TonB, these results suggest that defective transport might arise from either a weakened or strengthened interaction between TonB and the Ton box.
Figure 1.
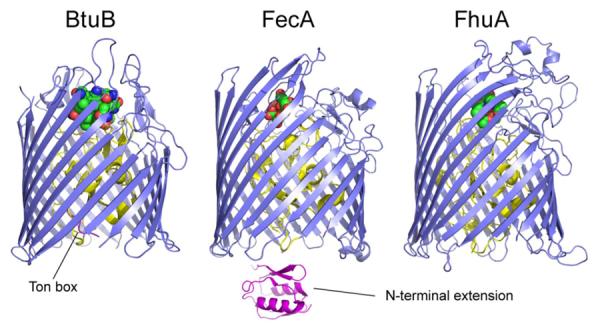
Three TonB-dependent transporters from Escherichia coli: the vitamin B12 transporter, BtuB (PDB ID: 1NQH); the ferric citrate transporter, FecA (PDB ID: 1KMP); and the ferrichrome transporter, FhuA (PDB ID: 1FCP).12–14 The transporters are based upon a 22-standed β-barrel (blue), where the N-terminal region forms a fold within the interior of the barrel (yellow). The position of the Ton box is indicated in BtuB. FecA possesses an N-terminal signaling domain (sometimes referred to as an N terminal extension). The N-terminal extension of FecA (magenta) was obtained by NMR spectroscopy of the isolated domain (PDB ID: 1ZZV).60
TonB-transporter crystal structures have yielded important insight into the nature of inter-membrane coupling, and indicate that several interactions are likely to stabilize the protein-protein complex. However, there have been relatively few quantitative binding studies that characterize TonB-transporter interactions in solution, and the results are not consistent. Reported binding affinities vary with the method used and span three orders of magnitude (10−9 M to 10−6 M). The affinity is not always modulated by substrate, and different approaches yield different binding stoichiometries.23–27 One source of variability may the choice of detergent, which has been shown to affect the energetics of the Ton box equilibrium in BtuB.28 Moreover, the breadth of these investigations has been narrow, in each case involving a single TonB-dependent transporter. Due to difficulties in working with full-length TonB, each of these studies utilized a different soluble TonB fragment, each of which has a different structure and tendency to dimerize.27, 29–32 Although in vivo data suggests that TonB cycles through several conformations,10, 33 at least one of which is a dimer,34 the role of the dimer in the transport cycle is not known.
In the present study we use fluorescence anisotropy to measure the binding affinity of a soluble TonB fragment lacking only the N-terminal 32 residues (the transmembrane domain). The binding of this fragment (TonBΔTMD) is measured to three Escherichia coli TonB-dependent transporters reconstituted into POPC/CHAPS mixed micelles: BtuB, FhuA, and the ferric citrate transporter FecA (Figure 1). This mixed micelle system was chosen because it maintains the native configuration of the Ton box and the substrate-induced unfolding observed previously in BtuB and FecA. We examine the effect of substrate, transport defective Ton box mutations and colicin E3 on the affinity of this interaction. The results provide strong evidence that high-affinity transporter-TonB binding is mediated by the Ton box, and that a loss of high-affinity binding observed for transport-defective Ton box mutants accounts for the resulting transport-defective phenotype. We demonstrate using site-directed spin labeling (SDSL) coupled with double electron-electron resonance (DEER) that a significant fraction of TonBΔTMD is isolated as a dimer. Moreover, the dimer of TonBΔTMD is converted to monomer upon binding to transporter. The results indicate how dimerization might participate in the transport cycle and mediate signaling between the inner and outer bacterial membrane.
EXPERIMENTAL PROCEDURES
Cloning and Mutagenesis
The DNA fragment corresponding to E.coli TonB residues 33–239 (TonBΔTMD) was PCR-amplified with primers designed to incorporate 5' NcoI and 3' XhoI sites for cloning the amplified product into pHIS-parallel1,35 which is a pET-22b derivative containing an N-terminal His6 tag followed by a TEV restriction site. The PCR product was digested, ligated into a pHIS-parallel1 vector that had been digested with the same enzymes, and sequenced to verify proper insertion. All mutations were introduced into TonB and BtuB plasmid DNA using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA), and subsequently verified by nucleotide sequencing. The Colicin E3R construct was prepared as described previously.36
Protein Expression and Purification
The plasmid encoding TonBΔTMD was transformed into T7 Express lysY/Iq competent cells (New England Biolabs, Ipswich, MA), and 1 L of 2xYT media containing 100mg/L ampicillin was inoculated with 10 ml overnight preculture grown to stationary phase. Cells were cultured at 37°C, induced with 0.5 mM isopropyl β-D-thiogalactopyranoside (IPTG) at OD600=0.7 and grown at 20°C for 5–6 hours post-induction. Cells were collected by centrifugation and resuspended in 25 mM Tris, pH 7.5, containing 1mM 4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF; Thermo Fisher Scientific, Pittsburgh, PA), 0.5 mM dithiothreitol (DTT; Avantor Performance Materials, Inc., Phillipsburg, NJ), 20 U/ml aprotinin (Calbiochem, Darmstadt, Germany), and 100 μM leupeptin (Roche Diagnostics, Indianapolis, IN). When possible, each of the following steps was carried out on ice or at 4°C, unless otherwise noted.
Cells were lysed using a French press, and the cleared supernatant was mixed at 4°C for 30 min. with 10 ml of Ni2+-NTA agarose resin that had been equilibrated in 25 mM Tris, pH 7.5, 300 mM NaCl, 20 mM imidazole. TonBΔTMD was eluted with 25 mM Tris, pH 7.5, 300 mM NaCl, 250 mM imidazole, and fractions containing protein were collected. In some cases, 1 mM DTT was added and allowed to react at room temperature for 30 min. to reduce disulfide bonds. The His6 tag was cut overnight at room temperature with 500U proTEV protease (Promega, Madison, WI) while TonBΔTMD was dialyzed against 25 mM Tris pH 7.5, 200 mM NaCl. TonBΔTMD was then buffer exchanged into 25 mM Tris pH 7.5, 100 mM NaCl and loaded onto an equilibrated HiTrap SP HP cation exchange column (GE Healthcare, Piscataway, NJ). TonBΔTMD was eluted with a gradient of 25 mM Tris pH 7.5, 1 M NaCl. Fractions containing pure TonBΔTMD were identified using SDS-PAGE, and as reported previously,37 TonBΔTMD migrated to a higher apparent molecular weight (~33 kDa) on SDS-PAGE gels, presumably due to its rigid polyproline motif. The protein was buffer exchanged into 25 mM Tris pH 7.5, 128 mM NaCl and concentrated using an Amicon 3,000 MWCO membrane (Millipore, Billerica, MA).
Expression, purification and reconstitution of BtuB,38 and FhuA and FecA,16 were performed as described previously. Reconstituted proteoliposomes were resuspended in 10 mM Hepes pH 6.5, 128 mM NaCl. After reconstitution, the 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) concentrations were determined using a standard phosphate assay, and 3-((3-Cholamidopropyl)dimethylammonio)-1-propanesulfonate (CHAPS; Anatrace, Santa Clara, CA) was added in excess to a 20:1 molar ratio for reconstitution into mixed micelles. Although some detergents are known to unfold the BtuB Ton box,28 this mixed micelle system does not affect the equilibrium of the Ton box and should also allow easy access of TonB to the transporters for binding studies.
Expression and purification of a 76-residue receptor binding fragment of colicin E3 (ColE3R) was carried out as described previously,36 except that purified ColE3R was buffer exchanged into 10 mM Hepes pH 6.5, 128 mM NaCl. All protein concentrations were measured in triplicate using the amido black assay,39 and the average concentrations from two separate assays were used for determination of binding affinites. The concentrations determined from amido black correlated well with those measured for the soluble proteins TonBΔTMD and ColE3R by absorbance spectroscopy at 280 nm.
Fluorescence Anisotropy and Data Analysis
For fluorescent labeling, purified TonBΔTMD L194C was concentrated to ~100 μM and reacted with an 8-fold molar excess of tris(2-carboxyethyl)phosphine (TCEP, Thermo Fisher Scientific, Pittsburgh, PA). Following incubation for 30 min. at room temperature, a 10-fold molar excess of BODIPY FL N-(2-Aminoethyl)Maleimide (Life Technologies, Grand Island, NY) at 5 mM dissolved in dimethyl sulfoxide and 25mM Tris pH 7.5 was added and allowed to react at room temperature for 2 hours. During labeling the dimethyl sulfoxide concentration was less than 5% (v/v). Excess fluorophore was removed by extensive dialysis in 25 mM Tris pH 7.5, 128 mM NaCl at 4°C. The labeling efficiency was estimated to be ~75% using absorbance spectroscopy and the extinction coefficient of BODIPY FL (~79,000 M−1cm−1). Although several different TonB structures alone and in complex with transporters have been reported, in all of these structures residue 194 is solvent-exposed and not positioned at a protein-protein interface.
For titrations in a total volume of 150 μl, approximately 1 μl TonBΔTMD was combined with a large excess of 10 mM Hepes pH 6.5, 128 mM NaCl, for a final TonBΔTMD concentration of 23 nM. For each titration, approximately 10–15 samples were prepared where various concentrations of BtuB, FhuA, or FecA were added in place of Hepes buffer. For substrate-bound titrations, the substrate was also added in place of Hepes buffer. For such BtuB titrations, CaCl2, which increases the affinity of BtuB for vitamin B12 by a factor of 1000,40 was added to 400 μM. Vitamin B12 was added to 2 μM in order minimize inner filter effects until the BtuB concentrations during the titration required larger amounts of substrate for saturation; in these cases a 1.1-fold molar excess was used. To examine the effect of ColE3R on the TonB:BtuB affinity, ColE3R was added to 750 μM. For substrate-bound FhuA titrations, 150 μM Fe3+-ferrichrome was added, and for substrate-bound FecA titrations, 100 μM ferric citrate was added. In all cases, the samples were allowed to equilibrate at room temperature for at least 30 minutes before transfer into 100 μl quartz sample cells for anisotropy measurements. At high vitamin B12 concentrations there were minor increases in the measured anisotropies (<0.02) attributed to scattering and the small Stokes shift of Bodipy FL. Care was taken to verify that neither viscosity nor scattering effects influenced the measured affinities, and that the mixed micelle suspension did not alter TonBΔTMD structure. The EPR spectra obtained from sites on TonBΔTMD (see below) provide a signature for local structure and dynamics, and they were not modified in the mixed micelle suspension.
Fluorescence anisotropy data were collected on a Fluorolog Modular Spectrofluorometer (Horiba Scientific, Edison, NJ). The excitation and emission wavelengths were 501 nm and 515 nm, respectively, with 3 nm monochromator slit widths and an integration time of 0.1 s or 1 s, depending on the counts per second. Each data point was measured in triplicate, and the average anisotropy was recorded. In each of our experiments the anisotropy approached a similar maximum value. Data were plotted and analyzed using OriginPro (Northampton, MA). The concentrations of free ligand were corrected for ligand-depletion, and in the worst cases this was less than a 15% correction. At concentrations of transporter higher than 10 μM, non-specific binding to TonB appears to take place and only the low concentration, high affinity portions of the binding curves were analyzed. To make an estimate of the dissociation constant for the TonB-transporter complex (KD), the anisotropy, y, was fit to Eq. 1.
| [1] |
Here, Bmax is a scaling factor, x is the concentration of transporter, a is the anisotropy in the absence of transporter, and n is an exponential term to account for any cooperativity. The binding was not highly cooperative and in all cases the data were fit with values of n between 1 and 1.5. The standard errors reported are those generated by the fitting.
Electron Paramagnetic Resonance and Data Analysis
For spin-labeling, TonBΔTMD was concentrated to ~150 μM before ion exchange chromatography and reacted with a ten-fold molar excess of S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate (MTSL; Toronto Research Chemicals Inc., North York, Ontario) at room temperature for 4 hours. Excess label was eliminated during ion exchange chromatography. As mentioned above, in some cases TonBΔTMD was reacted with 1 mM DTT at room temperature for 30 min. to reduce any disulfide bonds prior to spin-labeling. Labeling efficiencies of TonB appeared to be in excess of 80% as judged by double integration of a non-normalized EPR spectrum compared to a standard spin sample. Continuous-wave EPR spectroscopy was performed on a Bruker EMX spectrometer fitted with an ER4123D dielectric resonator (Bruker Biospin, Billerica, MA). All X-band spectra were taken using 2 mW incident microwave power, 1 G field modulation, and a sweep width of 100 G. Samples were adjusted to approximately 50–100 μM, and volumes of 5 μL were loaded into capillaries (0.60 mm i.d. × 0.84 mm o.d.; Vitrocom, Mountain Lakes, NJ). Spectra were baseline corrected and normalized using LabVIEW software provided by Christian Altenbach (UCLA).
For pulsed EPR measurements, 25 μL of ~125–175 μM TonBΔTMD diluted in 10 mM Hepes pH 6.5, 128 mM NaCl, 25% glycerol was loaded into quartz capillaries (1.5 mm i.d. × 1.8 mm o.d. Vitrocom, Mountain Lakes, NJ). For measurements with transporter, ~100–150 μM BtuB, FhuA, or FecA was added in place of Hepes buffer, and for samples with substrate, ~350–550 μM ferric citrate, Fe3+-ferrichrome, or vitamin B12 with 1.5 mM CaCl2 were also added in place of Hepes buffer. Samples for double electron-electron resonance (DEER) were flash-frozen in isopropanol cooled with dry ice, and the data were recorded at 80 K on an X-band Bruker Elexsys E580 spectrometer fitted with an ER4118X-MS3 split-ring resonator or a Q-band Bruker E580 spectrometer fitted with an EN5107D2 dielectric resonator. Data were acquired using the four pulse DEER sequence41 with a 16 ns π/2 and two 32 ns π observe pulses separated by a π pump pulse that was optimized at 28 ns at X-band and 32 ns at Q band. The dipolar evolution times were typically 0.7–2.0 μs, depending on the measured phase memory time of each sample. For X-band measurements, the pump frequency was set to the center maximum of the nitroxide spectrum, and the observe frequency was set to the low-field maximum (and vice versa for Q-band experiments). The phase-corrected dipolar evolution data were processed assuming a three-dimensional background and Fourier transformed, and the distance distributions were obtained with Tikhonov regularization using DeerAnalysis2011 software.42
RESULTS
EPR spectroscopy indicates that there are structural changes in TonBΔTMD upon association with BtuB
Single cysteine mutations were generated at five sites in TonBΔTMD, as shown in Fig. 2a, to incorporate the spin labeled side chain R1 (Fig. 2b). EPR spectroscopy was then used to monitor the interaction between TonBΔTMD and the outer-membrane transporters at these sites. To rule out slower protein rotational diffusion or chaotropic effects as an explanation for changes in EPR lineshape upon transporter binding, the solution viscosity was increased by the addition of Ficoll or mixed micelles. This did not significantly broaden or change the TonBΔTMD lineshapes, indicating that the observed changes may be attributed to protein protein-contact or changes in TonBΔTMD structure upon association with transporter.
Figure 2.
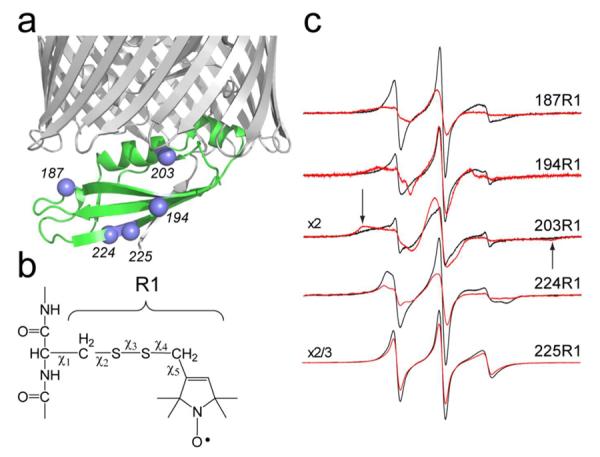
a) High-resolution model of a C-terminal fragment of TonB bound to BtuB (PDB ID 2GSK), showing the Cα-carbons (blue spheres) to which b) the spin-label side-chain R1 was attached. c) EPR spectra from the labeled sites in TonBΔTMD in the presence (red traces) and absence (black traces) of BtuB.
As seen in Fig. 2c, the EPR spectra from spin-labeled TonB broaden when bound to BtuB. For sites 187R1 and 203R1, this is likely due to direct contact of the R1 side chain with BtuB. This immobilization is particularly apparent in the bound spectrum for 203R1, where broad hyperfine extrema (arrows in Fig. 2c) are resolved upon binding to BtuB.43 In the crystal structure of the BtuB-TonB complex, the label at position 187 should contact BtuB at a region near the loop joining β-strands 18 and 19 in the BtuB barrel, and the label at site 203 should interact with residues in turn 7 (such as L425). The spectrum from 203R1 is very broad in the absence of BtuB, and this may in part be due to dipolar interactions resulting from a TonB dimer (see below). The spectra from sites 194 and 224 also change dramatically upon BtuB binding, but source of these changes is likely different. The label at site 194 faces the periplasm in the TonB:BtuB complex, and it does not interact directly with BtuB. In this case, the more immobile EPR lineshape upon association with BtuB must result from changes in backbone dynamics of TonBΔTMD and/or a change in the local structure so that the interactions made by 194R1 are altered. In the TonB-BtuB crystal structure, 194R1 lies on a β-strand with an open edge and it should interact with K177 on the adjacent strand. The label at site 224 also does not interact directly with BtuB and the change in lineshape upon transporter association reflects an increased order or reduced rate in R1 motion, perhaps due to a change in backbone motion or an interaction of this label with the N-terminal end of the TonB fragment. Thus, while the EPR spectra shown in Fig. 2 are consistent with the TonB-BtuB complex obtained by crystallography, they also indicate that changes take place in the structure and/or dynamics of TonBΔTMD upon association with transporter. As we discuss below, this may reflect the conversion of TonBδTMD from a dimer to a monomeric form upon binding transporter.
BtuB and FhuA exhibit high affinity binding to TonBΔTMD
To determine the affinity of TonBΔTMD for the outer membrane transporters, we measured the fluorescence anisotropy of TonBΔTMD L194C labeled with BODIPY FL upon association with transporter. Shown in Figure 3 are fluorescence anisotropy data for TonBΔTMD upon titration with either the apo or substrate-bound forms of BtuB, FhuA, and FecA. For both BtuB and FhuA, high affinity binding to TonBΔTMD is detected with apparent affinities of 61 and 64 nM, respectively. In these cases, approximately the same limiting anisotropy value was reached at high saturating concentrations of transporter. At concentrations of transporter above 10 μM, there is an additional increase in anisotropy that may result from non-specific association. The addition of substrate resulted in a modest two to three fold increase in affinity; however, this affinity is close to the concentration of labeled TonBΔTMD required to make the anisotropy measurement and may not be accurately determined. A summary of the binding data, including the errors to the fit, is shown in Table 1.
Figure 3.
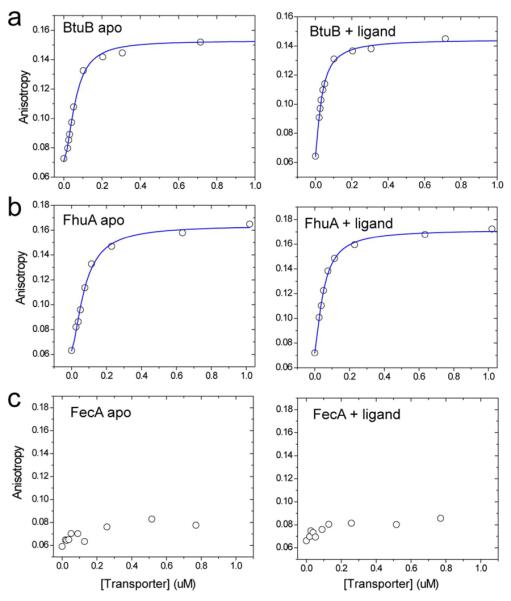
Fluorescence anisotropy data for the association of TonBΔTMD 194-Bodipy FL to the wild-type transporters a) BtuB b) FhuA and c) FecA. Anistropy measurements were performed both in the absence (left panel) and presence (right panel) of ligand. The solid (blue) traces are fits to the data using Eq. 1, and yeilds the parameters listed in Table 1. Further increases in fluorescence anisotropy are observed when each of these transporters is added to concentrations exceeding 10 μM. But the anisotropy fails to saturate at 100 μM transporter and is likely the result of non-specific association of transporter to the TonBΔTMD preparation.
Table 1.
High affinity dissociation equilibrium constants (KD) estimated from fluorescence anisotropy data.†
| Transporter | KD | error |
|---|---|---|
| BtuB apo | 61 nM | ± 5 nM |
| BtuB +Ca2+B12 | 26 nM | ± 2 nM |
| FhuA apo | 64 nM | ± 4 nM |
| FhuA +FeChr | 38 nM | ± 3 nM |
| BtuB + ColE3R | 1.5μM | ± 0.2μM |
Errors given are standard errors based upon the fits to the data using Eqn. 1. See Experimental Procedures. In each case, the data were fit with n values that ranged from 1 to 1.5. Non-specific binding was detected at higher transporter concentrations (ca. > 10μM), and data taken at these higher concentrations was not used in the analysis.
Unlike the binding of TonBΔTMD to BtuB or FhuA, the anisotropy data for the TonBΔTMD/FecA interaction did not yield a high-affinity interaction in this experimental system. The source of this difference is not clear, but FecA possesses an additional globular transcriptional signaling motif N-terminal to the Ton box (residues 1–79).44, 45 This domain may interact with the Ton box of the transporter, as indicated by a crystal structure of FpvA46 (a related TonB-dependent transporter) and this interaction may interfere with high-affinity TonB binding or alter the energetics of the TonB-FecA interaction (see Discussion).
Transport-defective mutations in the BtuB Ton box eliminate high-affinity binding to TonBΔTMD
Substitution of proline or glycine at several positions along the Ton box is known to impair transport 22. Two of these mutations, L8P and V10P, alter the pattern of crosslinking between BtuB and TonB making it less specific,47 and they alter the Ton box configuration by promoting its unfolding.21 Here, we tested the ability of BtuB with either the L8P or V10P mutation to bind TonBΔTMD using fluorescence anisotropy.
Shown in Fig. 4a is the anisotropy data for wild-type BtuB, and Figs. 4c and 4d plot the anisotropy data for the interaction of TonBΔTMD with the mutant BtuB transporters, L8P and V10P, respectively. In each case, no high-affinity (nM) binding is detected. These data suggest that the high-affinity binding of TonB is associated with the Ton box, and that defective transport results from a failure of BtuB to bind TonB with high affinity.
Figure 4.
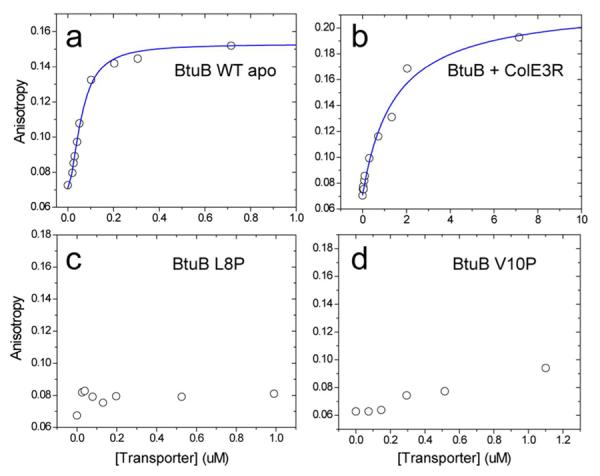
Fluorescence anisotropy data for the association of TonBΔTMD 194-Bodipy FL to a) wild-type BtuB, and b) BtuB in the presence of the receptor binding fragment of colicin E3. This colicin receptor fragment is known to trap the Ton box in a folded state (see text). Note the different concentration axis for this sample. Shown in c) and d) are fluorescence anisotropy data for the association of TonBΔTMD 194-Bodipy FL to the transport-defective mutants L8P and V10P, respectively.
Colicin E3R binding to the extracellular surface reduces the affinity of TonBΔTMD to BtuB
The colicin E3 receptor-binding domain (ColE3R) is a 76-residue fragment that binds to BtuB competitively with substrate. It has been shown by both chemical cross-linking36 and by EPR spectroscopy48 that ColE3R alters the configuration of the Ton box of BtuB and stabilizes the folded form of the Ton box. This ligand has the opposite effect of substrate, which promotes the unfolded form of the Ton box.
Shown in Figure 4b are fluorescence anisotropy data for the binding of TonBΔTMD to BtuB in the presence of bound ColE3R. Compared to the apo form of BtuB (Fig. 3a), the high affinity binding of TonBΔTMD has been reduced and the data are fit to a single dissociation constant of 1.5 μM (Table 1). The difference in affinity represents a shift in binding free energy for the association of TonBΔTMD to BtuB of about 2 kcal/mole, and colicin E3R was previously observed by EPR spectroscopy to shift the folded-to-unfolded Ton box equilibrium by an energy difference of similar magnitude.48 Both this colicin result and the effect of the Ton box proline mutations presented above indicate that high-affinity binding of BtuB to TonB is mediated by the Ton box.
TonBΔTMD can exist as a dimer in solution, which is converted to monomer upon transporter association
The TonB fragment used here, TonBΔTMD (residues 33–239), includes the C-terminal globular domain, a flexible linker (103–149) and the polyproline region. Structures of shorter fragments that possess just the globular domain are observed to form intertwined dimers,30, 32 whereas structures of longer fragments that include the flexible linker (103–149) appear to be monomeric.29, 31 To determine whether a dimer was present in our preparation, several singly spin-labeled TonBΔTMD mutants (Fig. 2a) were examined using double electron-electron resonance (DEER).
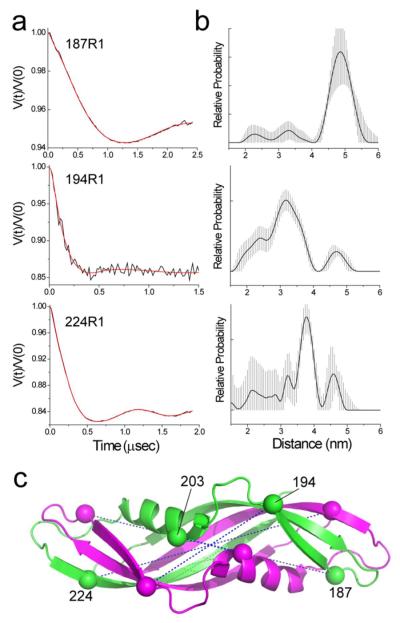
As seen in Fig. 5a, significant dipolar coupling between spins is observed for single spin-labeled sites at positions 187, 194 and 224, indicating that TonBΔTMD is oligomerized. The time-dependent change in intensity in these spin echo signals (the modulation depth) indicates that spin pairs are dipolar coupled in a defined structure. These signals were converted into distances and distance distributions, which are shown in Fig. 5b. We also observed significant dipolar coupling at site 203; however, distances extracted from this spin label may be questionable because the dipolar coupling becomes significant relative to the excitation bandwidth in the DEER experiment when distance separations are significantly less than 20 Angstroms.49
Figure 5.
TonBΔTMD dimerizes in solution. a) Dipolar evolution data obtained by DEER for TonBΔTMD in solution for spin labels at sites 187, 194 and 224. The data for 187 and 224 were recorded at Q-band. The amplitude in the modulation varied between samples, and either reflects different levels of dimerization in the TonBΔTMD preparation or differences in spin-labeling efficiency. The red traces represent the best fit to the data and yield the distance distributions shown in b). The error bars, which demark the shaded region in the distance distribution, indicate the range of solutions that can be obtained given the maximum error in the background subtraction. c) Structure of the intertwined dimer (PDB ID: 1IHR). The two high-resolution structures (PDB ID: 1IHR or 1QXX) provide reasonable matches to the pulse EPR data (Table 2).
Table 2 summarizes the distances measured using pulse EPR and compares them with the expected Cα-Cα distances based upon two intertwined dimer structures (PDB ID: 1IHR and 1QXX). The EPR-derived distances are consistent with either intertwined dimer structure,30, 32 but do not fit a third high-resolution structure reported for a TonB dimer31 or models of TonB dimers created from published monomeric structures. In most cases, the differences between the inter-spin and the Cα-Cα distances may be accounted for by the likely rotameric states of the R1 side-chain. However, the label at site 224 yields a distance that is 8 Å shorter than the predicted Cα-Cα distance, and this difference cannot be accounted for by the likely R1 rotamers at this site. This suggests that the solution structure for the TonBΔTMD oligomer is similar but not identical to the crystal structures for the intertwined dimer. As seen in Fig. 5b and Table 2, the distance distributions measured from these sites are broader than those typically seen for the R1 label when attached to well-folded proteins.50 Although a portion of this distribution is likely a result of more than one label rotamer, these broad distributions suggest that the TonBΔTMD dimer is conformationally heterogenous. Some conformational heterogeneity would not be unexpected for a protein domain that participates in a reversible protein-protein interaction.51
Table 2.
Distances for TonBΔTMD obtained from single spin labeled sites.†
| Mutant | Distance; distribution Å | Cα-Cα (1IHR) Å | Cα-Cα(1QXX) Å |
|---|---|---|---|
| R187R1 | 47.8; σ(r)=10.8 | 46.0 | 47.2 |
| L194R1 | 32.1; σ(r)=7.5 | 25.3 | 24.9 |
| E203R1 | ~ 17 Ň | 11.5 | 11.2 |
| I224R1 | 36.2; σ(r)=8.8 | 44.1 | 43.7 |
The EPR spectra shown in Fig. 2 indicate that there are conformational changes in TonBΔTMD upon transporter association, and pulse EPR was used to examine the state of the TonB dimer when associated with the outer membrane transporters. Examples of the DEER data obtained are shown in Figure 6. The modulation depths observed for 203R1 and 194R1 (Fig. 6a, b) are typical of those seen in our preparations and reflect the number of interacting spin pairs that are excited in the pulse EPR experiment. When bound to transporter these modulation depths are dramatically reduced, and Fig. 6c compares the changes in modulation depth for three sites when bound to BtuB. Since the relaxation parameters of the labels and the excitation parameters used in the experiment are unchanged, the data indicate that binding of TonBΔTMD to the transporter reduces the number of interacting spin pairs, and converts the dimer into monomer. This change in modulation depth is correlated with a high-affinity TonB-transporter interaction. As seen in Fig, 6d, the reduction in modulation depth (conversion to monomer) was not as significant for transport-defective mutants, such as BtuB L8P, or when bound to FecA.
Figure 6.
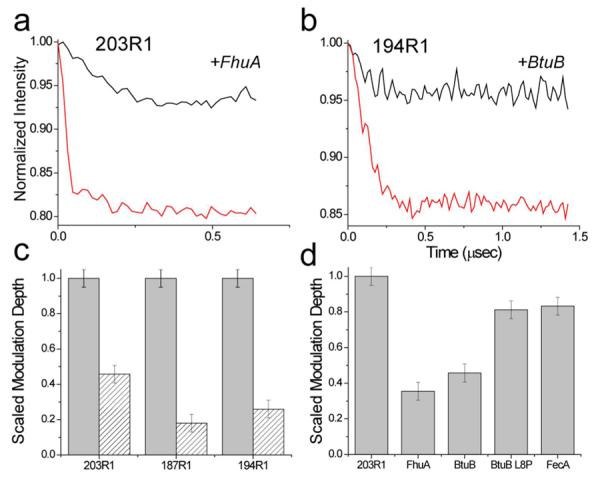
The binding of TonBΔTMD to transporter dissociates the TonB dimer. Dipolar evolution data for two spin-labeled TonBΔTMD mutants: a) 203R1 and b) 194R1, when bound to the transporters FhuA or BtuB in the presence of substrate. For these examples, the data were recorded at X-band, and the pulse excitation parameters were not varied, allowing a comparison of TonBΔTMD oligomerization. Binding of TonBΔTMD to the transporter dramatically reduces the modulation depth in these DEER signals, indicating a loss of dipolar-coupled spins and a dissociation of the TonB oligomer (see Text). In c) the changes in the scaled modulation depth are plotted are plotted for three TonBΔTMD spin labels, 203R1, 197R1 and 194R1 in the absence (grey bars) and presence (hatched bars) of BtuB. In d) the modulation depth of 203R1 when bound to FhuA, BtuB, BtuB L8P, and FecA is compared. The conversion to monomer is less efficient for transporters or mutants that do not exhibit high-affinity TonB binding. Under the conditions of this experiment all added transporter (which was stoichiometrically limiting) was bound to TonB.
To provide a more quantitative assessment of this phenomenon, the modulation depths in these DEER signals may be used to make a rough estimate of the fraction of dimerized protein.52 Based upon the excitation parameters reported previously52 and assuming 100% labeling efficiency, we estimate from the X-band modulation depths of 187R1 and 194R1 that roughly 50% of TonBΔTMD is dimerized, and that binding to transporter (which is stoichiometrically limiting) reduces the amount of dimerized TonB to about 10%. Measurements on samples labeled less than 100% will lead to an underestimate of the fraction of dimer.
DISCUSSION
In the present study, a combination of fluorescence anisotropy and EPR spectroscopy was used to characterize the interaction between TonB and the transporters BtuB, FhuA and FecA. For BtuB and FhuA, fluorescence anisotropy (Fig. 3) indicates that the TonB construct TonBΔTMD binds with high-affinity to the transporter and that this affinity increases in the presence of substrate (Table 1). These increases were less than expected based upon the energetics of the Ton box unfolding transition in BtuB,53 or based upon the substrate-enhanced binding observed in-vivo.47, 54 However, high affinity binding with a KD on the order of a few nM might be difficult to detect in our assay due to the minimum level of labeled TonB required to make the measurement. In addition, TonB will not have unrestricted 3-dimensional diffusion in the intact system as it does in our reconstituted model system. As a result, the exposure and unfolding of the Ton box observed previously for BtuB may be much more important when TonB is undergoing limited or dimensionally restricted diffusion relative to the periplasmic surface of the transporter.
The measurements made here provide a strong indication that the high-affinity (tens of nM) binding of TonB is mediated by the transporter Ton box. When the transport-defective Ton box mutants L8P and V10P of BtuB were examined,20 no high-affinity binding was observed to either mutant (Fig. 4). This suggests that the failure of these mutants to transport is associated with a lack of high-affinity binding to TonB. Binding of the colicin E3 receptor fragment to wild type BtuB also dramatically reduced TonB affinity, which is consistent with previous reports that colicin E3 stabilizes the Ton box in a folded or buried state.36, 48 Taken together, these data provide strong evidence that an unfolding of the Ton box in BtuB triggers a high affinity interaction with TonB, and that a reversible high-affinity interaction between the Ton box and TonB mediates transport.
Unlike BtuB and FhuA, FecA did not show evidence for a high-affinity binding mode. This could be due to the conformation of the Ton box in FecA, which may interact with the N-terminal extension or transcriptional domain in this transporter. FecA belongs to a family of TonB-dependent transporters that regulate their own transcription, and as shown in Fig. 1, they contain an N-terminal extension not seen in other TonB-dependent transporters. The lack of high-affinity TonB binding to FecA observed here is consistent with an NMR chemical shift analysis indicating that the FecA N-terminal signaling domain interferes with TonB binding to the FecA Ton box.55 In FpvA, a related transporter from Pseudomonas aeruginosa, the affinity of the corresponding TonB fragment was found to be in the low micromolar range,23 similar to that seen here for FecA (see Fig. 3). Interference by the N-terminal extension is consistent with a structure obtained for FpvA where the N-terminus interacts with the Ton box,46 and it has been proposed that TonB interacts with the Ton box of FpvA through a β-strand exchange mechanism. Substrate is found to enhance the binding of TonB to FecA in-vivo,56 and the failure here to observe a substrate-induced affinity increase suggests that model systems have not completely captured important features of the in-vivo system.
As indicated above (see Experimental Procedures), the TonBΔTMD preparation was pure by SDS gel electrophoresis, but EPR data indicate that a significant fraction of TonBΔTMD is present as a dimer. This finding is consistent with previous sedimentation equilibrium data on TonBΔTMD (containing an N-terminal His-6 tag), which indicated that the protein preparation contains a level of TonB dimer that may be 10 to 20%.37 In our preparations, the fraction of dimer makes up approximately 50% of the protein as indicated by the modulation depths in the DEER signals (Figs. 5 and 6). While there is evidence that TonB dimers function during the transport cycle, the fraction of dimerized TonB within the native inner membrane environment may be highly dynamic and different from that observed here in an in vitro model system.
TonB appears to cycle between at least two conformations,7, 10, 33 at least one of which is a dimer;8, 34 as a result, the monomeric and intertwined dimeric structures reported for TonB may both represent physiologically relevant states. At the present time, the role of TonB dimerization in the transport cycle and the stage at which dimerization takes place remain poorly characterized. TonB dimers are detected in inner membrane fractions from sucrose density gradient separations, and transport is impaired when TonB is covalently cross-linked to form dimer.8 This indicates that dimerization might occur prior to an interaction with transporter. On the other hand, it has been proposed that TonB binds transporter as a monomer, but recruits a second TonB to associate as a dimer that is primed for substrate transport.25, 26 The data obtained here (Fig. 6) argue against this model and indicate that dimerization occurs at a stage separable from the substrate translocation step, since the loss of modulation depth in the DEER data shows that dimer is converted to monomer upon TonB association with the transporter. It should be noted that the physiological relevance of the intertwined dimeric crystal structure has been questioned based upon in vivo disulfide cross linking;57 however, cross-linking between residues that appear distant in the crystal structure may result from protein dynamics or trapped conformational intermediates, and the broad distance distributions seen in the DEER data obtained here suggest that TonBΔTMD has some disorder that is either static or dynamic.
Dimerization might play one of several roles in the TonB-dependent transport cycle. For example, the dimer may function to position TonB for interaction with the transporter by interacting with the peptidoglycan layer. Peptidoglycan-binding domains from Escherichia coli share sequence homology with the C-terminal domain of TonB and are structurally homologous with the intertwined dimer of TonB; moreover, the C-terminal domain of TonB has been shown to bind peptidoglycan.58 TonB dimerization might also act as a mechanism to provide signaling between the inner and outer membranes. Conversion of the dimer to a monomer upon transporter binding might be the signal that initiates the energy transduction step that is driven by the TonB/ExbB/ExbD complex.
A model for how a dimer-monomer conversion might function in TonB-dependent transport is shown in Fig. 7. Once TonB is assembled into a functional complex with ExbD and ExbB, the proton motive force is used to assemble TonB into an intertwined dimeric structure that is kinetically trapped. As an intertwined dimer, TonB associates with the peptidoglycan layer and diffuses laterally as proposed previously in a “membrane surveillance” model.58 As it diffuses, TonB will encounter the periplasmic surface of the transporter, which also associates with the peptidoglycan layer or with peptidoglycan-binding proteins such as OmpF and OmpA.58 When the extracellular surface of the transporter binds substrate, the periplasmic exposure of the Ton box is increased and dimerized TonB engages the transporter. The unfolded Ton box facilitates dissociation of the TonB dimer and the formation of a high-affinity 1:1 TonB-transporter complex, which may resemble complexes seen by crystallography. This dimer-monomer conversion functions as a signal to the inner membrane complex to dissociate the TonB-transporter complex and reform the TonB dimer. This step, which requires energy from the TonB/ExbB/ExbD system, facilitates the release of substrate from its binding site and substrate movement through the transporter, perhaps as a result of a transient or partial unfolding of the protein core.59
Figure 7.
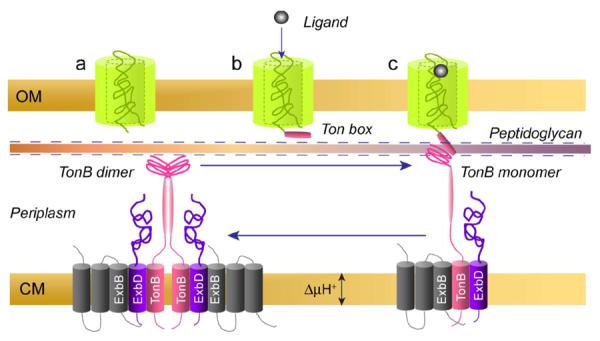
A model for TonB-dependent transport. Dimerization of TonB is driven by the proton-motive force acting through ExbB and ExbD (the exact stoichiometry of this complex is unknown). TonB is kinetically trapped in this dimerized form, which associates with and diffuses along the peptidoglycan layer as proposed previously.58 Transporter also directly interacts with the peptidoglycan or associates with peptidoglycan binding proteins such as OmpF or OmpA. a) Transporter in its apo form rarely interacts with TonB, but upon binding ligand b) the exposure of the Ton box to the periplasmic space is increased and the transporter Ton box interacts with TonB to form c) a monomeric transport-ready complex. A transient unfolding of the N-terminal luminal domain facilitates the transport of ligand, which may be driven by the TonB binding and dissociation step.
In summary, the soluble TonB fragment, TonBΔTMD, is found to bind with 10−8 M affinity to either BtuB or FhuA. The binding of colicin E3R to BtuB or the presence of a transport-defective mutation in BtuB eliminates high-affinity binding, indicating that this interaction is mediated by the Ton box. The FecA N-terminal domain also appears to abrogate high-affinity binding, which is consistent with previously reported binding measurements in FpvA. A large fraction of TonBΔTMD in solution is present as a dimer, which resembles two intertwined dimer structures observed previously by crystallography for a shorter TonB fragment. The binding of this fragment to transporter results in the conversion of dimer into monomer in a process that is apparently mediated by the Ton box. As a result, the transport cycle may involve conformational changes in TonB that are mediated both by the inner membrane complex and by the outer membrane transporters.
Acknowledgements
We thank Prof. Robert Nakamoto for helpful discussions and for assistance with the fluorescence anisotropy instrumentation and measurements.
1) Abbreviations used
- CHAPS
3-((3-Cholamidopropyl)dimethylammonio)-1-propanesulfonate
- EPR
electron paramagnetic resonance
- MTSL
methanethiosulfonate spin label
- POPC
palmitoyloleoylphosphatidylcholine
- R1
spin labeled side chain produced by derivatization of a cysteine with the MTSL
- SDSL
site-directed spin labeling
- TCEP
tris(2-carboxyethyl)phosphine
Footnotes
This work was supported by a grant from the National Institutes of Health, NIGMS, GM 035215.
REFERENCES
- 1.Schauer K, Rodionov DA, de Reuse H. New substrates for TonB-dependent transport: do we only see the `tip of the iceberg'? Trends Biochem Sci. 2008;33:330–338. doi: 10.1016/j.tibs.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 2.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol. 2010;64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohler SD, Weber A, Howard SP, Welte W, Drescher M. The proline-rich domain of TonB possesses an extended polyproline II-like conformation of sufficient length to span the periplasm of Gram-negative bacteria. Protein Sci. 2010;19:625–630. doi: 10.1002/pro.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shultis DD, Purdy MD, Banchs CN, Wiener MC. Outer membrane active transport: structure of the BtuB:TonB complex. Science. 2006;312:1396–1399. doi: 10.1126/science.1127694. [DOI] [PubMed] [Google Scholar]
- 5.Pawelek PD, Croteau N, Ng-Thow-Hing C, Khursigara CM, Moiseeva N, Allaire M, Coulton JW. Structure of TonB in complex with FhuA, E-coli outer membrane receptor. Science. 2006;312:1399–1402. doi: 10.1126/science.1128057. [DOI] [PubMed] [Google Scholar]
- 6.Jana B, Manning M, Postle K. Mutations in the ExbB Cytoplasmic Carboxy Terminus Prevent Energy-Dependent Interaction between the TonB and ExbD Periplasmic Domains. J Bacteriol. 2011;193:5649–5657. doi: 10.1128/JB.05674-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ollis AA, Manning M, Held KG, Postle K. Cytoplasmic membrane protonmotive force energizes periplasmic interactions between ExbD and TonB. Mol Microbiol. 2009;73:466–481. doi: 10.1111/j.1365-2958.2009.06785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh J, Postle K. Disulphide trapping of an in vivo energy-dependent conformation of Escherichia coli TonB protein. Mol Microbiol. 2005;55:276–288. doi: 10.1111/j.1365-2958.2004.04384.x. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh J, Postle K. Evidence for dynamic clustering of carboxy-terminal aromatic amino acids in TonB-dependent energy transduction. Mol Microbiol. 2004;51:203–213. doi: 10.1046/j.1365-2958.2003.03816.x. [DOI] [PubMed] [Google Scholar]
- 10.Larsen RA, Thomas MG, Postle K. Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol Microbiol. 1999;31:1809–1824. doi: 10.1046/j.1365-2958.1999.01317.x. [DOI] [PubMed] [Google Scholar]
- 11.Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, Cavard D. Colicin biology. Microbiol Mol Biol R. 2007;71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferguson AD, Chakraborty R, Smith BS, Esser L, van der Helm D, Deisenhofer J. Structural basis of gating by the outer membrane transporter FecA. Science. 2002;295:1715–1719. doi: 10.1126/science.1067313. [DOI] [PubMed] [Google Scholar]
- 13.Ferguson AD, Hofmann E, Coulton JW, Diederichs K, Welte W. Siderophore-mediated iron transport: Crystal structure of FhuA with bound lipopolysaccharide. Science. 1998;282:2215–2220. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- 14.Chimento DP, Mohanty AK, Kadner RJ, Wiener MC. Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nat Struct Biol. 2003;10:394–401. doi: 10.1038/nsb914. [DOI] [PubMed] [Google Scholar]
- 15.Chimento DP, Kadner RJ, Wiener MC. Comparative structural analysis of TonB-dependent outer membrane transporters: implications for the transport cycle. Proteins. 2005;59:240–251. doi: 10.1002/prot.20416. [DOI] [PubMed] [Google Scholar]
- 16.Kim M, Fanucci GE, Cafiso DS. Substrate-dependent transmembrane signaling in TonB-dependent transporters is not conserved. Proc Natl Acad Sci U S A. 2007;104:11975–11980. doi: 10.1073/pnas.0702172104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Q, Ellena JF, Kim M, Cafiso DS. Substrate-dependent unfolding of the energy coupling motif of a membrane transport protein determined by double electron-electron resonance. Biochemistry. 2006;45:10847–10854. doi: 10.1021/bi061051x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fanucci GE, Coggshall KA, Cadieux N, Kim M, Kadner RJ, Cafiso DS. Substrate-induced conformational changes of the periplasmic N-terminus of an outer-membrane transporter by site-directed spin labeling. Biochemistry. 2003;42:1391–1400. doi: 10.1021/bi027120z. [DOI] [PubMed] [Google Scholar]
- 19.Merianos HJ, Cadieux N, Lin CH, Kadner RJ, Cafiso DS. Substrate-induced exposure of an energy-coupling motif of a membrane transporter. Nat Struct Biol. 2000;7:205–209. doi: 10.1038/73309. [DOI] [PubMed] [Google Scholar]
- 20.Gudmundsdottir A, Bell PE, Lundrigan MD, Bradbeer C, Kadner RJ. Point Mutations in a Conserved Region (Tonb Box) of Escherichia-Coli Outer-Membrane Protein Btub Affect Vitamin-B12 Transport. J Bacteriol. 1989;171:6526–6533. doi: 10.1128/jb.171.12.6526-6533.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coggshall KA, Cadieux N, Piedmont C, Kadner RJ, Cafiso DS. Transport-defective mutations alter the conformation of the energy-coupling motif of an outer membrane transporter. Biochemistry. 2001;40:13964–13971. doi: 10.1021/bi015602p. [DOI] [PubMed] [Google Scholar]
- 22.Cadieux N, Bradbeer C, Kadner RJ. Sequence changes in the Ton box region of BtuB affect its transport activities and interaction with TonB protein. J Bacteriol. 2000;182:5954–5961. doi: 10.1128/jb.182.21.5954-5961.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adams H, Zeder-Lutz G, Schalk I, Pattus F, Celia H. Interaction of TonB with the outer membrane receptor FpvA of Pseudomonas aeruginosa. J Bacteriol. 2006;188:5752–5761. doi: 10.1128/JB.00435-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lefevre J, Delepelaire P, Delepierre M, Izadi-Pruneyre N. Modulation by substrates of the interaction between the HasR outer membrane receptor and its specific TonB-like protein, HasB. J Mol Biol. 2008;378:840–851. doi: 10.1016/j.jmb.2008.03.044. [DOI] [PubMed] [Google Scholar]
- 25.Khursigara CM, De Crescenzo G, Pawelek PD, Coulton JW. Enhanced binding of TonB to a ligand-loaded outer membrane receptor - Role of the oligomeric state of TonB in formation of a functional FhuA center dot TonB complex. J Biol Chem. 2004;279:7405–7412. doi: 10.1074/jbc.M311784200. [DOI] [PubMed] [Google Scholar]
- 26.Khursigara CM, De Crescenzo G, Pawelek PD, Coulton JW. Kinetic analyses reveal multiple steps in forming TonB-FhuA complexes from Escherichia coli. Biochemistry. 2005;44:3441–3453. doi: 10.1021/bi047882p. [DOI] [PubMed] [Google Scholar]
- 27.Khursigara CM, De Crescenzo G, Pawelek PD, Coulton JW. Deletion of the proline-rich region of TonB disrupts formation of a 2 : 1 complex with FhuA, an outer membrane receptor of Escherichia coli. Protein Sci. 2005;14:1266–1273. doi: 10.1110/ps.051342505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fanucci GE, Lee JY, Cafiso DS. Membrane mimetic environments alter the conformation of the outer membrane protein BtuB. J Am Chem Soc. 2003;125:13932–13933. doi: 10.1021/ja0376442. [DOI] [PubMed] [Google Scholar]
- 29.Peacock RS, Weljie AM, Howard SP, Price FD, Vogel HJ. The solution structure of the C-terminal domain of TonB and interaction studies with TonB box peptides. J Mol Biol. 2005;345:1185–1197. doi: 10.1016/j.jmb.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 30.Chang CS, Mooser A, Pluckthun A, Wlodawer A. Crystal structure of the dimeric C-terminal domain of TonB reveals a novel fold. J Biol Chem. 2001;276:27535–27540. doi: 10.1074/jbc.M102778200. [DOI] [PubMed] [Google Scholar]
- 31.Kodding J, Killig F, Polzer P, Howard SP, Diederichs K, Welte W. Crystal structure of a 92-residue C-terminal fragment of TonB from Escherichia coli reveals significant conformational changes compared to structures of smaller TonB fragments. J Biol Chem. 2005;280:3022–3028. doi: 10.1074/jbc.M411155200. [DOI] [PubMed] [Google Scholar]
- 32.Koedding J, Howard P, Kaufmann L, Polzer P, Lustig A, Welte W. Dimerization of TonB is not essential for its binding to the outer membrane siderophore receptor FhuA of Escherichia coli. J Biol Chem. 2004;279:9978–9986. doi: 10.1074/jbc.M311720200. [DOI] [PubMed] [Google Scholar]
- 33.Ollis AA, Postle K. ExbD Mutants Define Initial Stages in TonB Energization. J Mol Biol. 2011;415:237–247. doi: 10.1016/j.jmb.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sauter A, Howard SP, Braun V. In vivo evidence for TonB dimerization. J Bacteriol. 2003;185:5747–5754. doi: 10.1128/JB.185.19.5747-5754.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expres Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 36.Cadieux N, Phan PG, Cafiso DS, Kadner RJ. Differential substrate-induced signaling through the TonB-dependent transporter BtuB. Proc Natl Acad Sci U S A. 2003;100:10688–10693. doi: 10.1073/pnas.1932538100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moeck GS, Letellier L. Characterization of in vitro interactions between a truncated TonB protein from Escherichia coli and the outer membrane receptors FhuA and FepA. J Bacteriol. 2001;183:2755–2764. doi: 10.1128/JB.183.9.2755-2764.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fanucci GE, Cadieux N, Piedmont CA, Kadner RJ, Cafiso DS. Structure and dynamics of the beta-barrel of the membrane transporter BtuB by site-directed spin labeling. Biochemistry. 2002;41:11543–11551. doi: 10.1021/bi0259397. [DOI] [PubMed] [Google Scholar]
- 39.Kaplan RS, Pedersen PL. Determination of Microgram Quantities of Protein in the Presence of Milligram Levels of Lipid with Amido Black B-10. Anal Biochem. 1985;150:97–104. doi: 10.1016/0003-2697(85)90445-2. [DOI] [PubMed] [Google Scholar]
- 40.Cadieux N, Barekzi N, Bradbeer C. Observations on the calcium dependence and reversibility of cobalamin transport across the outer membrane Escherichia coli. J Biol Chem. 2007;282:34921–34928. doi: 10.1074/jbc.M707426200. [DOI] [PubMed] [Google Scholar]
- 41.Pannier M, Veit S, Godt A, Jeschke G, Spiess HW. Dead-time free measurement of dipole-dipole interactions between electron spins. J Magn Reson. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 42.Jeschke G, Chechik V, Ionita P, Godt A, Zimmermann H, Banham J, Timmel CR, Hilger D, Jung H. DeerAnalysis2006 - a comprehensive software package for analyzing pulsed ELDOR data. Appl Magn Reson. 2006;30:473–498. [Google Scholar]
- 43.McHaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 44.Enz S, Mahren S, Stroeher UH, Braun V. Surface signaling in ferric citrate transport gene induction: Interaction of the FecA, FecR, and FecI regulatory proteins. J Bacteriol. 2000;182:637–646. doi: 10.1128/jb.182.3.637-646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Welz D, Braun V. Ferric citrate transport of Escherichia coli: Functional regions of the FecR transmembrane regulatory protein. J Bacteriol. 1998;180:2387–2394. doi: 10.1128/jb.180.9.2387-2394.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brillet K, Journet L, Celia H, Paulus L, Stahl A, Pattus F, Cobessi D. A beta strand lock exchange for signal transduction in TonB-dependent transducers on the basis of a common structural motif. Structure. 2007;15:1383–1391. doi: 10.1016/j.str.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 47.Cadieux N, Kadner RJ. Site-directed disulfide bonding reveals an interaction site between energy-coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc Natl Acad Sci U S A. 1999;96:10673–10678. doi: 10.1073/pnas.96.19.10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fanucci GE, Cadieux N, Kadner RJ, Cafiso DS. Competing ligands stabilize alternate conformations of the energy coupling motif of a TonB-dependent outer membrane transporter. Proc Natl Acad Sci U S A. 2003;100:11382–11387. doi: 10.1073/pnas.1932486100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Polyhach Y, Godt A, Bauer C, Jeschke G. Spin pair geometry revealed by high-field DEER in the presence of conformational distributions. J Magn Reson. 2007;185:118–129. doi: 10.1016/j.jmr.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 50.Kim M, Xu Q, Murray D, Cafiso DS. Solutes alter the conformation of the ligand binding loops in outer membrane transporters. Biochemistry. 2008;47:670–679. doi: 10.1021/bi7016415. [DOI] [PubMed] [Google Scholar]
- 51.Mittag T, Kay LE, Forman-Kay JD. Protein dynamics and conformational disorder in molecular recognition. J Mol Recognit. 2010;23:105–116. doi: 10.1002/jmr.961. [DOI] [PubMed] [Google Scholar]
- 52.Hilger D, Jung H, Padan E, Wegener C, Vogel KP, Steinhoff HJ, Jeschke G. Assessing oligomerization of membrane proteins by four pulse DEER: pH-dependent dimerization of NhaA Na+/H+ antiporter of E. coli. Biophys J. 2005;89:1328–1338. doi: 10.1529/biophysj.105.062232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Freed DM, Horanyi PS, Wiener MC, Cafiso DS. Conformational Exchange in a Membrane Transport Protein Is Altered in Protein Crystals. Biophysical journal. 2010;99:1604–1610. doi: 10.1016/j.bpj.2010.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moeck GS, Coulton JW, Postle K. Cell envelope signaling in Escherichia coli. Ligand binding to the ferrichrome-iron receptor fhua promotes interaction with the energy-transducing protein TonB. J Biol Chem. 1997;272:28391–28397. doi: 10.1074/jbc.272.45.28391. [DOI] [PubMed] [Google Scholar]
- 55.Peacock RS, Andrushchenko VV, Demcoe AR, Gehmlich M, Lu LS, Herrero AG, Vogel HJ. Characterization of TonB interactions with the FepA cork domain and FecA N-terminal signaling domain. Biometals. 2006;19:127–142. doi: 10.1007/s10534-005-5420-0. [DOI] [PubMed] [Google Scholar]
- 56.Ogierman M, Braun V. Interactions between the outer membrane ferric citrate transporter FecA and TonB: studies of the FecA TonB box. J Bacteriol. 2003;185:1870–1885. doi: 10.1128/JB.185.6.1870-1885.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Postle K, Kastead KA, Gresock MG, Ghosh J, Swayne CD. The TonB dimeric crystal structures do not exist in vivo. mBio. 2010;1:1–7. doi: 10.1128/mBio.00307-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaserer WA, Jiang X, Xiao Q, Scott DC, Bauler M, Copeland D, Newton SM, Klebba PE. Insight from TonB hybrid proteins into the mechanism of iron transport through the outer membrane. J Bacteriol. 2008;190:4001–4016. doi: 10.1128/JB.00135-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flores Jimenez RH, Cafiso DS. The N-Terminal Domain of a TonB-Dependent Transporter Undergoes a Reversible Stepwise Denaturation. Biochemistry. 2012;51:3642–3650. doi: 10.1021/bi300118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ferguson AD, Labunskyy VM, Fomenko DE, Arac D, Chelliah Y, Amezcua CA, Rizo J, Gladyshev VN, Deisenhofer J. NMR structures of the selenoproteins Sep15 and SelM reveal redox activity of a new thioredoxin-like family. J Biol Chem. 2006;281:3536–3543. doi: 10.1074/jbc.M511386200. [DOI] [PubMed] [Google Scholar]