Abstract
In many cultivated crop species there is limited genetic variation available for the development of new higher yielding varieties adapted to climate change and sustainable farming practises. The distant relatives of crop species provide a vast and largely untapped reservoir of genetic variation for a wide range of agronomically important traits that can be exploited by breeders for crop improvement. In this paper, in what we believe to be the largest introgression programme undertaken in the monocots, we describe the transfer of the entire genome of Festuca pratensis into Lolium perenne in overlapping chromosome segments. The L. perenne/F. pratensis introgressions were identified and characterised via 131 simple sequence repeats and 1612 SNPs anchored to the rice genome. Comparative analyses were undertaken to determine the syntenic relationship between L. perenne/F. pratensis and rice, wheat, barley, sorghum and Brachypodium distachyon. Analyses comparing recombination frequency and gene distribution indicated that a large proportion of the genes within the genome are located in the proximal regions of chromosomes which undergo low/very low frequencies of recombination. Thus, it is proposed that past breeding efforts to produce improved varieties have centred on the subset of genes located in the distal regions of chromosomes where recombination is highest. The use of alien introgression for crop improvement is important for meeting the challenges of global food supply and the monocots such as the forage grasses and cereals, together with recent technological advances in molecular biology, can help meet these challenges.
Keywords: Introgression mapping, comparative mapping, recombination, interspecific diversity, monocots, SNP markers
Introduction
Global population levels are set to increase from 6 to 9 billion by 2050 with the result that we need to increase food production by 70% just to maintain our present nutrition levels, which already includes one billion severely malnourished and a further 100 million at near starvation level (Gustafson et al., 2010). It takes 7–10 years to breed a new forage grass or wheat variety, that is, the crosses that will lead to the development of varieties in 2020 have already been made. Furthermore, although we need to increase yield in many countries, food production has levelled out either due to climate change, for example, increased temperature, etc, or, in crops such as wheat, a lack of sufficient genetic variation, thus limiting the ability of breeders to develop higher yielding plant varieties. The identification and exploitation of genetic variation for the development of superior high yielding varieties adapted to the changing environment is therefore of critical importance to meet the challenges of global food security.
The transfer of genetic variation from the near and distant relatives of our key crop species, so called alien introgression, provides a vast and virtually untapped reservoir of genetic variation for agronomically important traits that can be utilised by plant breeders for the development of superior high yielding adapted crops. In the monocots, alien introgression has already had a significant impact on agricultural production. Examples of previous introgressions from alien species in the forage grasses includes genes that confer resistance to disease and tolerance to abiotic stress, etc. (for example, Roderick et al., 2003; Humphreys et al., 2005; Armstead et al., 2006b). In addition, hybrids between the world's major temperate forage grass Lolium perenne (high yielding, highly digestible) and Festuca species (low digestibility, but highly tolerant to abiotic and biotic stresses) such as Festuca pratensis are being directly exploited commercially. In wheat, examples of key introgressions include those from Aegilops umbellulata (which saved US wheat production from catastrophic failure in 1960; Sears, 1956, 1972); resistance to a range of diseases, tolerance to acid soils, increased yield advantage and stability from rye (in the late 90 s a 1B/1R translocation was present in the majority of world wheat varieties and a number of the present global top varieties, for example, ‘Rialto', still carry it; Ammar et al., 2004); a gene from Ae. ventricosa conferring resistance to eyespot has been exploited in breeding programmes (Garcia-Olmedo et al., 1977); many of the top wheat varieties in Europe, for example, ‘Robigus', are derived from unknown introgressions from Triticum dicoccoides; 30% of all wheat varieties produced at International Maize and Wheat Improvement Center are derived from crosses between normal wheat and ‘synthetic' wheat (synthetic wheat is derived from crosses between Ae. squarrosa, DD genome, and tetraploid wheat, AABB genomes followed by chromosome doubling via colchicine; Dreisigacker et al., 2008).
Alien introgression, in its simplest form, involves the sexual hybridisation of different species to form an inter-specific F1 hybrid. Alien introgression occurs in the F1 hybrid (or its derivatives) when related, that is, homoeologous, chromosomes from the two parental species (that is, chromosomes that carry orthologous genes in essentially the same order) recombine at meiosis resulting in the generation of inter-specific recombinant chromosomes. These recombinant chromosomes are then transmitted to the next generation through the gametes. The repeated backcrossing of the F1 hybrid, or its derived amphiploid, to one of the parental genotypes results in the generation of lines which carry the majority of the genome of one species but also carry one or more chromosome segments from the other parental species.
Although the value of genetic variation from alien species has been clearly demonstrated, only a fraction of their full potential in breeding has so far been exploited in breeding programmes. In the past, this has been as a direct result of the lack of robust high throughput screening procedures that enable the rapid identification and characterisation of introgressed chromosome segments. However, recent advances in conventional and next generation sequencing platforms and technology in combination with the sequencing of the crop and model plant genomes, for example, rice and Brachypodium (Febrer et al., 2010), are now enabling the development of strategies to fully exploit the potential of alien species for crop improvement, for example, (Griffiths et al., 2006).
The forage grasses are one example where modern technology is being exploited to introgress interspecific genetic variation into an important crop species, that is, the L. perenne/F. pratensis introgression system (King et al., 1998, 1999, 2002a, 2002b; Moore et al., 2005; Armstead et al., 2006a, 2006b; King et al., 2007; Armstead et al., 2007a; Harper et al., 2011). The Lolium/Festuca introgression system is based on a series of seven monosomic substitution lines. In each of these seven lines one of the chromosomes of L. perenne (2n=2x=14; DNA content=2034 Mbp; Bennett and Smith, 1976), has been replaced by its equivalent (homoeologous) chromosome from F. pratensis (2n=2x=14; DNA content=2181 Mbp; Bennett et al., 1982). The monosomic substitution line carrying F. pratensis chromosome 3 was selected and backcrossed to the normal L. perenne parental genotype. Recombination occurred between F. pratensis chromosome 3 and its L. perenne group 3 homoeologous partner during meiosis in the monosomic substitution. The resulting progeny from the cross therefore carried a range of different size F. pratensis chromosome 3 segments. The F. pratensis chromosome segments in each of the progeny were then identified and characterised using genetic markers and genomic in situ hybridisation. Thus, as a result a series of lines carrying overlapping F. pratensis chromosome segments were developed for F. pratensis chromosome 3, that is, an introgression series (King et al., 2007). Further characterisation of the L. perenne/F. pratensis introgressions was achieved via the bin mapping of sequences derived from functionally annotated gene models on circa every 5th–10th BAC/PAC clone (BP) from rice chromosome 1 minimum tiling path (MTP), that is, thus providing a means of establishing the syntenic relationship between L. perenne/F. pratensis chromosome 3 and the sequenced rice chromosome 1 pseudomolecule (PM).
In this paper, we describe the exploitation of genomic strategies to transfer the entire genome of F. pratensis into L. perenne in overlapping segments. In addition, we describe the relationship between recombination frequency throughout the genome relative to gene content and describe the relationship between key monocot relatives and L. perenne and F. pratensis. The availability of these genetic stocks and their relationship to other species has major implications for food production and security in the monocots.
Materials and methods
Plant material
Seven L. perenne/F. pratensis monosomic substitution lines were developed with L. perenne as the recipient and F. pratensis as the donor (as previously described in King et al., 2002a, 2002b; King et al., 2007; Harper et al., 2011). Thus, each monosomic substitution was diploid carrying 13 L. perenne chromosomes and 1 F. pratensis chromosome (each substitution line containing a F. pratensis chromosome from a different linkage group). Each substitution line was backcrossed (as both the male and female parent) to the diploid L. perenne parent to produce BC2 mapping populations (King et al., 2002a, 2002b; King et al., 2007). During meiosis in the monosomic substitution lines recombination occurs between the F. pratensis chromosome and its L. perenne homoeologue. As a result of this homoeologous recombination, a proportion of individuals making up the seven BC2 mapping populations were expected to carry L. perenne/F. pratensis recombinant chromosomes, (the size and position of the F. pratensis chromosome segment in each BC2 individual being determined by the site of homoeologous recombination at meiosis in the gametes of the monosomic substitution).
Development of introgression maps
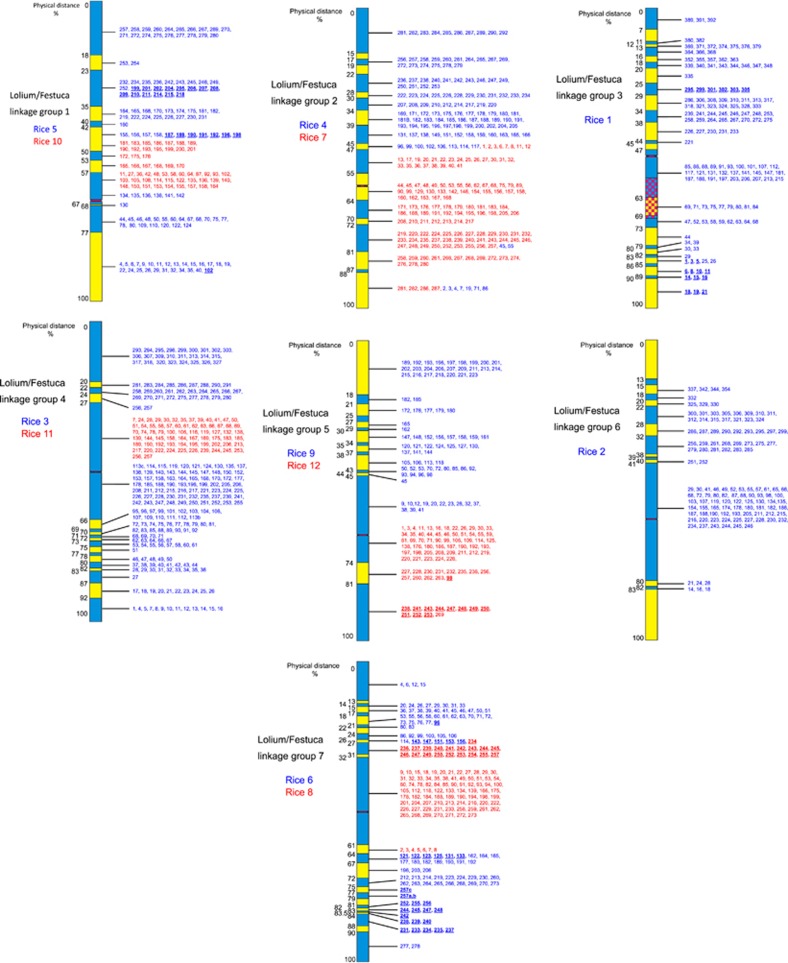
The BC2 populations were genetically mapped using simple sequence repeats generated by Vialactia Biosciences, Auckland, New Zealand (Gill et al., 2006), the Institute of Grassland and Environmental Research (King et al., 2008) and those available in the public domain. The genetic profiles of each of the substitution lines were used to select recombinant genotypes where recombination between L. perenne and the F. pratensis had occurred at different locations along the chromosomes. Mitotic root tip preparations of the selected genotypes were then analysed using genomic in situ hybridisation and the F. pratensis segments measured as in King et al. (2002a) except slides were analysed using a Leica DM5500B epifluorescence microscope (Leica Microsystems (UK) Ltd, Milton Keynes, UK), photographs were taken using a Leica DFC 350FX digital camera (Leica Microsystems (UK) Ltd) and images analysed and measured using the ImageJ package (http://rsbweb.nih.gov/ij/). The measured Festuca segments were used to produce introgression maps of all seven Festuca linkage groups (Figure 1) as described in King et al., (2002a).
Figure 1.
Introgression maps of L. perenne/F. pratensis linkage groups 1–7. Physical distances are shown on the left. Alternating blue and yellow colours are used to show the points of recombination between Lolium and Festuca delineating the physical bins. Centromeres are drawn in red. Bin allocation of rice BPs is shown on the right of each diagram (GenBank accession numbers for each clone can be found in Supplementary Data). Where two rice PMs map to one Lolium/Festuca linkage group the BPs from the different rice groups are shown in red or blue. BPs in bold and underlined map out of order as compared with rice. The chequered area on linkage group 3 represents the NOR (King et al., 2002a).
DNA extraction, PCR and purification
Genomic DNA was extracted using an AutoGen 740 (AutoGen, Holliston, MA, USA).
All PCR amplifications carried out in this work were performed using Faststart TAQ DNA polymerase and the manufacturer's buffer system (Roche, Lewes, UK) in ABI9700 thermocyclers (Applied Biosystems, Warrington, UK). Thermo cycling was performed beginning with 5 min at 96 °C; followed by 10 cycles of 1 min at 95 °C, 1 min at 60 °C (with the temperature reduced by 0.5 °C per cycle), 3 min at 72 °C; followed by 30 cycles of 1 min at 95 °C, 1 min at 55 ° and 3 min at 72 °C; and a final extension step of 7 min at 72 °C. For a small number of primers the initial annealing temperature was either raised to 65 ° (dropping to 60 °C over the course of the first 10 cycles) or dropped to 55 °C (dropping to 50 °C over the course of the first 10 cycles).
DNA was purified either directly from the PCR amplification using the QIAquick PCR Purification Kit (Qiagen, Crawley, UK) or from an agarose gel using the QIAquick Gel Extraction Kit (Qiagen).
Single nucleotide polymorphism marker (SNP) development
Primers were developed from conserved orthologous gene sequences of rice. Primers were designed using the Primer 3 software (http://primer3.sourceforge.net) following the strategies outlined in; (King et al., 2007) and incorporating Festuca expressed sequence tag sequences available in public databases. The latter were used in BLAST searches against rice coding sequences to identify potential high confidence orthologs between Lolium/Festuca and rice.
All primers were initially tested by the PCR amplification of genomic DNA from the plants involved in the production of the monosomic substitution lines (F. pratensis, L. perenne-diploid, L. perenne-tetraploid, F. pratensis/L. perenne/L. perenne triploid) and the monosomic substitution lines themselves with the amplification products run on a 1% agarose gel. The amplified bands were cut from the gel, cleaned and sequenced using an ABI 3100 (Applied Biosystems). The sequences obtained were aligned using Genedoc (http://www.psc.edu/biomed/genedoc). The aligned sequences were then characterised for polymorphisms between the parental F. pratensis genotype and the two L. perenne genotypes. The presence of SNP markers between Festuca and Lolium were confirmed by their presence in the sequences of the triploid hybrid and the relevant substitution line. Primers flanking SNPs distinguishing the Lolium and Festuca backgrounds on different linkage groups were used to assign the SNP markers to a bin on the relevant linkage group (Figure 1).
The BPs on each rice PM were numbered 1 to x according to the MTP from the MSU Rice Genome Annotation Project at the start of the project, that is, April 2007. This numbering is used in all figures. Supplementary Information Supplementary Table 1–12 give the GenBank accession number for each BP, the locus identifier of the functionally annotated gene model used and full primer details.
Introgression mapping of SNPs
Having established the presence of a SNP between F. pratensis and L. perenne in any specific substitution line, the primers were used to amplify the genomic DNA of the derived BC2 genotypes. Screening for the presence or absence of these SNPs in the BC2 genotypes allowed each SNP to be assigned to a bin on the appropriate introgression map. The sequence data from F. pratensis was BLASTN searched back against the rice genome to confirm that the products were orthologous to the original sequence obtained from rice.
Determination of the distribution of recombination frequency throughout the genome relative to gene content
In order to determine the relationship between physical distance (as determined via introgression mapping) and genetic distance, bin mapped SNPs were used to genetically map a L. perenne × L. perenne mapping population. The F2 mapping population used was derived from a cross between the varieties Perma and Aurora which has already been extensively mapped with a variety of other genetic markers, for example, restriction fragment length polymorphisms (RFLPs), simple sequence repeats, etc. (Turner et al., 2006). The selection of primer pairs from the bin mapped SNPs for use in the genetic mapping was achieved as follows. Primer pairs of each bin mapped SNP were used to generate amplicons using pooled DNA from 96 genotypes of the F2 mapping family. Where sequencing of these amplicons identified potential SNPs, the surrounding sequence was used for primer design for the KASPar SNP genotyping platform (KBioscience, unit 7 Maple Park, Hoddesdon, Herts, England; http://www.kbioscience.co.uk); 96 genotypes from the F2 mapping family were subsequently scored for segregation of SNP markers. (The genetic mapping data for the bin mapped SNPs segregating in the Aurora × Perma L. perenne × L. perenne mapping population is available in Supplementary Table 13 of the Supplementary Information.)
Comparative mapping
For the comparisons with rice, Brachypodium distachyon and Sorghum bicolor, where L. perenne/F. pratensis sequences had been generated for mapped markers in this study (for example, the SNP markers) or were already published (many restriction fragment length polymorphism markers), these sequences were aligned with the complete PM assemblies for rice (Rice Genome Annotation Project; http://rice.plantbiology.msu.edu/; version_6.1; all.con.gz), Brachypodium distachyon (Phytozome; http://www.phytozome.net/, B.distachyon_114.fa) and Sorghum (Phytozome; S.bicolor_79.fa) using BLAST-generated sequence alignments (cut-off threshold e=<1e-005). In each case, only the most significant BLAST alignment was used to assign putative orthology between the L. perenne/F. pratensis marker sequences and the PM sequences for rice, Brachypodium and Sorghum. The comparative relationships were described as plots of the genetic positions (cM) of sequenced markers on the L. perenne/F. pratensis linkage groups against the physical positions (bp) of the putative orthologues on the rice, Brachypodium and Sorghum PMs. In all cases, the genetic and physical positions were expressed as percentages of the total genetic distance of the linkage group or the complete PM length.
For the comparisons with barley and wheat, wheat expressed sequence tag and bin map positions were obtained from GrainGenes (http://wheat.pw.usda.gov/cgi-bin/westsql/map_locus.cgi) and barley sequence and genetic map information was obtained from (Close et al. 2009). The comparative relationships were described in terms of the introgression (L. perenne/F. pratensis), genetic (L. perenne and barley) or deletion bin (wheat) positions on each chromosome of mapped, sequenced, markers relative to the position (bp) of each marker's most significant BLAST alignment on the rice PMs.
Results
Introgression map development
131 polymorphic simple sequence repeats were used to screen the seven BC2 populations derived by inter-crossing the seven monosomic substitutions with the diploid L. perenne parent. In total, 2784 BC2 individuals were screened, that is, 204, 218,126, 548,188, 1335 and 165 from monosomic substitution lines 1, 2, 3, 4, 5, 6 and 7, respectively (this does not include the BC2 plants used to generate the initial introgression map for L. perenne/F. pratensis linkage group 3 (King et al., 2007)). Cytogenetic analysis enabled introgression maps to be generated for the remaining six linkage groups, that is, linkage groups 1, 2, 4, 5, 6 and 7, which were divided into 12 (11 BC2 genotypes), 18 (16 BC2 genotypes), 19 (18 BC2 genotypes), 16 (15 BC2 genotypes), 15 (14 BC2 genotypes) and 27 bins (25 BC2 genotypes), respectively. The previously published introgression map of linkage group 3 (King et al., 2002a, 2007) was increased in resolution from 18–26 bins (using a total of 22 BC2 genotypes) (Figure 1).
SNP development and bin mapping
Four thousand three hundred and thirty three new primer pairs were designed and tested for the production of amplification products from all three parental genotypes (primers designed and tested for the initial introgression mapping of L. perenne/F. pratensis linkage group 3 (King et al., 2007) are not included). The frequency of successful amplification varied slightly between rice PMs but, in all cases, at least 90% (usually greater than 95%) could be taken forward to the sequencing stage (Table 1). Thirty five per cent of the primers designed were successful in mapping a SNP to a bin on the introgression maps. However, the success rate varied depending on the rice PM origin of the target sequences (rice PMs 1–12 gave success rates of 28%, 31%, 45%, 38%, 39%, 35%, 42%, 35%, 32%, 26%, 31% and 33%, respectively). Hence, the lowest success rate was with the primers designed from PM 10 (26%) and the highest success rate was with primers designed from PM 3 (45%) (Table 1). Rice PM 1 showed a relatively low level of success (28%). This could be directly attributed to the difficulty in designing primers which spanned Lolium/Festuca polymorphisms from two specific regions of this PM. Between BACs 108 and 191 on rice PM 1, 27 pairs of primers were designed of which only three were successful in bin mapping a SNP; a success rate of 11%. In a second region (between BACs 275 and 301) 25 pairs of primers were designed, of which again, only three were successful; a success rate of 12%. Although the success rate of mapping a SNP marker to a bin was lowest with primers designed from rice PM 10, the lowest percentage of BPs represented on the bin maps were from rice PMs 10 (27% of total BPs mapped), 11 (27% of total BPs mapped) and 12 (29% of total BPs mapped) (Table 1). This result is consistent with our previously published results on heat maps between Lolium/Festuca and rice (Armstead et al., 2007b) which showed a lower level of synteny between rice PMs 10, 11 and 12 and Lolium/Festuca.
Table 1. The number of primers designed from sequences from each of the rice linkage groups and their success rate in amplifying products in all the parental genotypes, bin mapping SNPs and bin mapping individual rice BPs on the seven Lolium/Festuca introgression maps.
| Rice linkage group | Number of primers designed | Number of primers giving amplification products (%) | Number of SNPs mapped (%) | Number of BACs mapped (% total BACs mapped) |
|---|---|---|---|---|
| 1 | 314 | 307 (98) | 87 (28) | 76 (19) |
| 2 | 426 | 398 (94) | 131 (31) | 119 (33) |
| 3 | 603 | 590 (98) | 274 (45) | 221 (68) |
| 4 | 407 | 397 (98) | 156 (38) | 123 (42) |
| 5 | 364 | 357 (98) | 140 (39) | 127 (44) |
| 6 | 377 | 338 (90) | 131 (35) | 109 (39) |
| 7 | 373 | 366 (98) | 155 (42) | 141 (49) |
| 8 | 341 | 331 (97) | 120 (35) | 105 (38) |
| 9 | 296 | 282 (95) | 94 (32) | 83 (37) |
| 10 | 239 | 230 (96) | 63 (26) | 55 (27) |
| 11 | 273 | 264 (97) | 85 (31) | 69 (27) |
| 12 | 320 | 318 (99) | 107 (33) | 79 (29) |
Abbreviations: BACs, bacterial artificial chromosome; SNPs, single-nucleotide polymorphisms. (The numbers shown for rice PM 1 do not include primers and mapped SNPs already published—King et al., 2007).
Over the course of the programme, a number of the primers were tested in other systems, for example, Lolium temulentum/Festuca pratensis hybrids, Lolium multiflorum/Festuca glaucescens hybrids, bread wheat (Triticum aestivum) and some wheat wild relatives including Thinopyrum bessarabicum, Aegilops sp., Triticum urartu, rye (Secale cereale), etc. In the other grass hybrids, generally over 90% of the primers tested, produced amplification products and in L. temulentum/F. pratensis hybrids 95% gave SNP polymorphisms. In wheat and its wild relatives 75% and 63%, respectively, of the primers tested produced amplification products. Circa 45% of the primers tested identified polymorphisms between T. aestivum and the wild species with which it was being compared (data not shown).
One thousand six hundred and twelve SNPs were bin mapped on the seven L. perenne/F. pratensis introgression lines derived from 1376 BPs from the rice MTP (Figure 1). Table 1 shows the rice-anchored markers which map to each L. perenne/F. pratensis linkage group. Only L. perenne/F. pratensis linkage groups 3 and 6 were mapped with SNPs anchored to just one rice PM (PMs 1 and 2, respectively). All other L. perenne/F. pratensis linkage groups were mapped with SNPs anchored to two rice PMs. Where SNPs anchored to two different rice PMs were bin mapped to the same L. perenne/F. pratensis linkage group, it was common to find the SNPs anchored to one of the rice PMs inserted into the middle of the SNPs anchored to the other rice PM. The exception to this is Lolium/Festuca linkage group 5 where one end is syntenic to rice PM 9 and the other end to rice PM 12.
Comparative relationship between the forage grasses and rice
The data obtained enabled the determination of the macro-syntenic relationships between the rice PMs and the L. perenne/F. pratensis genomes. It should be noted that bin mapping only places a gene from a BP within a bin; the relative order within each bin is derived according to the BP order in the rice MTP for that rice PM (Figure 1). For simplicity, the order and identity of orthologous genes from rice, which have been mapped into Lolium/Festuca bins is described in terms of the BP order in the MTP; that is, BP 1, a rice gene(s) present on the first BP in the MTP for that rice PM.
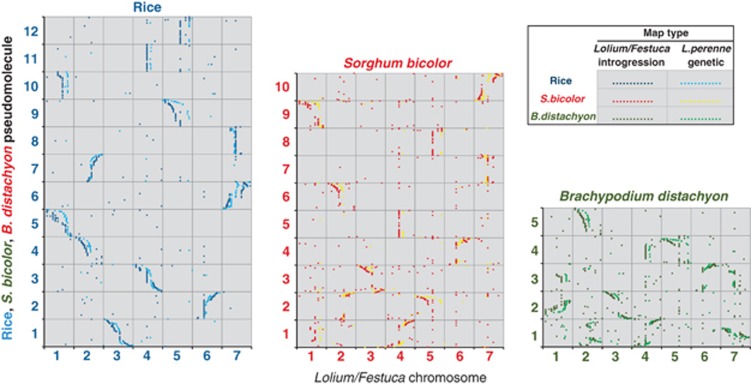
The results show that five of the Lolium/Festuca linkage groups are composed of regions syntenic to two rice PMs (thus accommodating the difference in the number of linkage groups between the two species while maintaining a high level of synteny at the macro level). In the four cases, one rice PM has been translocated into another rice PM, that is, L. perenne/F. pratensis linkage groups 1, 2, 4, 7, and in a further case one rice PM has been translocated on to the end of another linkage group, that is, L. perenne/F. pratensis linkage group 5 (Figures 1 and 2).
Figure 2.
Conserved syntenic relationships between Lolium/Festuca, L. perenne and rice, S. bicolor and B. distachyon. The x axes represent the introgression (Lolium/Festuca) or genetic (L. perenne) positions on each chromosome of mapped, sequenced, markers relative to the position (bp) of each marker's most significant BLAST alignment on the rice, S. bicolor or B. distachyon PMs (y axes).
The bin map order of genes on L. perenne/F. pratensis linkage groups 4 and 6 was consistent with the order of their orthologues on the rice PMs that is, there were no obvious chromosomal inversions at this level of resolution. In contrast, when compared with their order on the syntenic rice PMs, the bin map order of genes on the introgression maps of L. perenne/F. pratensis linkage groups 1, 2, 3, 5 and 7 showed at least one disruption in macro-colinearity (Figure 1).
Lolium/Festuca linkage group 1 (rice PMs 5 and 10)
There are three disruptions in macro-colinearity between rice PM 5 and L. perenne/F. pratensis linkage group 1 (as determined by the linear order of the BPs from rice PM 5 relative to their bin mapped position in the L. perenne/F. pratensis introgression map of linkage group 1). No disruptions were found in macro-colinearity between rice PM 10 and L. perenne/F. pratensis linkage group 1.
Lolium/Festuca linkage group 2 (rice PMs 4 and 7)
This linkage group exhibits only one disruption in macro-colinearity relative to rice involving BPs from rice PM 4.
Lolium/Festuca linkage group 3 (rice PM 1)
The two disruptions to the linear order of the rice 1 BPs on L. perenne/F. pratensis linkage group 3 were reported in our last paper (King et al., 2007). However, the first of these linkages was originally thought to comprise only BPs 1–5 in a small inversion. The increased resolution of the linkage group 3 introgression map now shows that this disruption actually involves an inversion of all BPs at the end of the chromosome, that is, BPs 1–21.
Lolium/Festuca linkage group 5 (rice PMs 9, 11 and 12)
The introgression map of L. perenne/F. pratensis linkage group 5 shows no disruption to the linear order of the BPs from rice PM 9 but two disruptions to the linear order of the BPs from rice PM 12.
Lolium/Festuca linkage group 7 (rice PMs 6 and 8)
The linear order of the BPs from rice PM 6 is disrupted three times while that from rice PM 8 is disrupted once. The first disruption to the BPs from rice PM 6 is the only disruption to clearly precede the fusion of the two rice PMs making up Lolium/Festuca linkage group 7. Rice 6 BPs 121–156 have been inverted and rice PM 8 has then inserted into the middle of this inversion between BPs 133 and 143. (Supplementary Table 14 in the supplementary Information gives a summary of the disruptions to macro-colinearity found between rice PMs 1–12 and L. perenne/F. pratensis linkage groups 1–7.)
As stated, the linear order of the BPs from rice PM 2 remain in the expected order on L. perenne/F. pratensis linkage group 6. It is interesting to note, however, that no BPs from rice PM 2 mapped to either end of the chromosome (15% of the physical distance of the chromosome at one end of the chromosome and 17% of the physical distance of the chromosome at the other end). These bins remain ‘empty' of SNP markers as no BPs from any of the other rice PMs are physically mapped to them. We did, however, map one microsatellite marker to the end bin covering 15% of the physical distance and two microsatellite markers plus five DArT markers to the other end bin (data not shown). Hence, all the markers we have mapped to the end two bins were from genomic libraries.
Comparative relationships between L. perenne/F. pratensis, B. distachyon, S. bicolor, barley and wheat
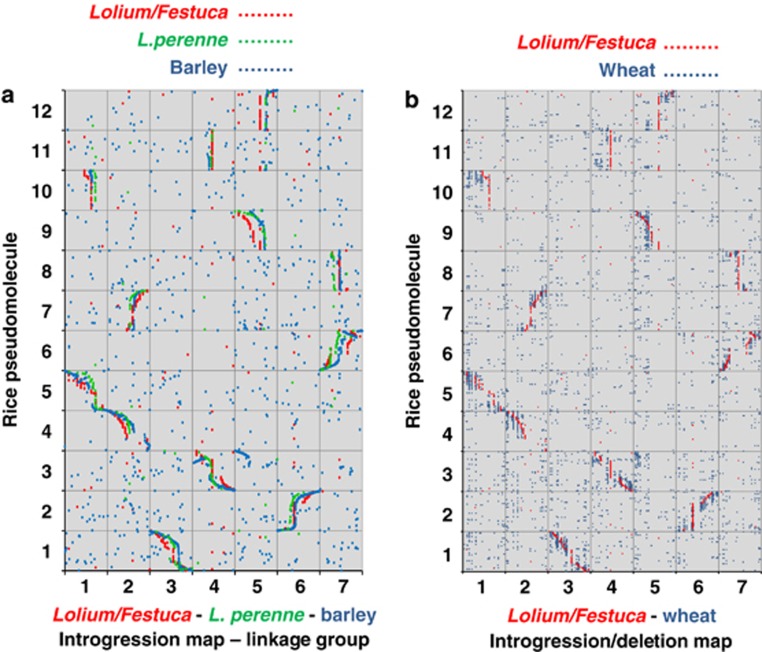
Initial analyses were also made of the syntenic relationships between L. perenne/F. pratensis and S. bicolor, B. distachyon, wheat and barley (Figures 2 and 3). For both S. bicolor and B. distachyon there are clearly strongly conserved blocks of genomic synteny with L. perenne/F. pratensis distributed across the 10 (S. bicolor) and 5 (B. distachyon) linkage groups of the two species. The patterns of conserved synteny observed between L. perenne/F. pratensis, S. bicolor and B. distachyon are very similar to those reported by (Mayer et al. 2011) for the conserved syntenic relationships between barley, S. bicolor and B. distachyon, indicating the close evolutionary distance between L. perenne/F. pratensis and barley. This close relationship is also illustrated by the comparisons of L. perenne/F. pratensis with wheat and barley (Figure 3). The one major exception to this conserved synteny occurs on linkage groups 4 and 5. L. perenne/F. pratensis linkage group 4 contains all the mapped BPs from rice PM 3. However, in barley and wheat, one end of rice PM 3 has been translocated to the end of linkage group 5 in these species. Thus, wheat and barley show a chromosome 4/5 translocation relative to L. perenne/F. pratensis.
Figure 3.
Conserved syntenic relationships between (a) Lolium/Festuca, L. perenne, barley and rice, and (b) Lolium/Festuca, wheat and rice. The x axes represent the introgression (Lolium/Festuca), genetic (L. perenne and barley) or deletion bin (wheat) positions on each chromosome of mapped, sequenced, markers relative to the position (bp) of each marker's most significant BLAST alignment on the rice, S. bicolor or B. distachyon PMs (y axes).
Determination of the frequency of meiotic recombination throughout the genome relative to gene content
Two hundred and eighty-three bin mapped primer pairs spanned SNPs that segregated in the Perma × Aurora mapping population. With very few exceptions, the position of SNPs determined by genetic mapping was the same as that determined via bin mapping. (Supplementary Information Supplementary Table 13 shows the genetic mapping data for the 283 bin mapped SNPs that segregate in the Perma × Aurora mapping population.) The main exceptions observed were as follows: SNPs 364 and 392 from rice PM 1 both bin mapped to L. perenne/F. pratensis linkage group 3. On the genetic map, however, SNP 364 was located on L. perenne/F. pratensis linkage group 6 and SNP 392 on linkage group 5. SNP 41 from rice PM 6 bin mapped to L. perenne/F. pratensis linkage group 7 but on the genetic map was located on linkage group 1. SNP 138 from rice PM 11 also genetically mapped to L. perenne/F. pratensis linkage group 1 but bin mapped to linkage group 4. Finally, we bin mapped two SNPs from the same putative gene locus on rice PM 9. One of the SNPs bin mapped to L. perenne/F. pratensis linkage group 5 as expected but the other SNP bin mapped to linkage group 6. Only the second SNP segregated in the Perma × Aurora mapping population. In this case the genetic mapping confirmed the bin mapping by placing this SNP on linkage group 6.
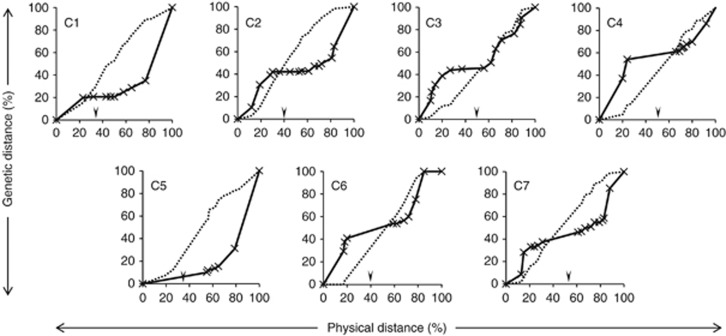
Figure 4 estimates the relationship between the physical positions of bin mapped SNPs relative to their genetic positions as determined from the Perma × Aurora mapping family. The highest levels of recombination were observed at the two distal regions of each chromosome. Low to very low levels of recombination were observed in the proximal regions of the chromosomes. The exception to this was chromosome 5, which showed a peak of recombination in only one of the two distal regions.
Figure 4.
Estimates of the relationship between physical/cytogenetic and genetic distance (black line) for L. perenne/F. pratensis chromosomes (C) 1–7. Physical/cytogenetic distances (horizontal axis) were derived from the L. perenne/F. pratensis introgression mapping population and genetic distances (vertical axis) from a L. perenne F2 genetic mapping population; both types of distances are expressed as % of the total observed for each chromosome. The cumulative % total of rice-anchored markers mapped in each physical bin for L. perenne/F. pratensis chromosomes 1–7 is indicated by the dotted line. The vertical arrowhead above each x axis indicates the approximate positions of the centromere for each chromosome.
Although regions of high and low recombination were observed, the cumulative frequency of SNP markers is reasonably evenly distributed, physically, along the length of each chromosome. Hence, recombination does not reflect SNP (mapped gene) content; that is, while some genes lie in regions of high recombination, a significant proportion of genes in the L. perenne/F. pratensis genome lie in regions of low/very low recombination frequency.
The syntenic relationship between L. perenne/F. pratensis and rice is such that four L. perenne/F. pratensis linkage groups carry a region syntenic to a rice PM in a proximal region (Figures 1 and 2). Three of the proximal regions in L. perenne/F. pratensis that are syntenic to rice PMs 10, 11 and 8 (L perenne/F. pratensis linkage groups 1, 4 and 7, respectively) undergo very low frequencies of recombination. L. perenne/F. pratensis linkage group 2 is composed of two distal regions, which are syntenic to rice PM 4 and a region syntenic to rice PM 7 between the two rice PM 4 regions. Part of the region syntenic to rice PM 7 does undergo a higher frequency of recombination. However, while this region covers the proximal part of L. perenne/F. pratensis linkage group 2, it also extends into the distal region of one arm with only a relatively small region of genes syntenic to rice 4 located terminally to it.
Similar patterns are observed in the comparisons between L. perenne/F. pratensis and S. bicolor (Figure 2). The highest frequencies of recombination occur distally with the lowest occurring proximally. Furthermore, proximal regions of L. perenne/F. pratensis syntenic with complete S. bicolor PMs undergo a very low frequency of recombination. An analysis of the relationship of recombination between L. perenne/F. pratenesis and B. distachyon again revealed that higher recombination is observed in the distal regions of the chromosomes and low frequencies of recombination occur at the proximal regions (Figure 2).
Discussion
This paper describes the transfer of the entire genome of F. pratensis in overlapping chromosome segments into L. perenne and also the development of 1612 bin mapped SNP markers anchored to the rice genome. It has also demonstrated that the primers developed are of value for genetic mapping of SNPs in the Aurora × Perma L. perenne × L. perenne mapping population. In addition, many of the primers developed, amplify and identify SNPs in wheat and its wild and cultivated relatives (data not shown). These are presently being exploited in a wheat/alien introgression programme.
Exploitation of Lolium/Festuca introgression lines
Determination of the genetic control of target traits
The F. pratensis introgressions generated, provide a new and important source of genetic variation for agronomically important target traits within the grasses for the development of new varieties and fundamental research. The strategy for utilising this resource for the identification and determination of the genetic control of target traits leading to exploitation in breeding programmes is as follows: (1) The seven monosomic substitution lines (and the four parental genotypes) will be screened for variation for a target trait. (2) Once a substitution line(s) that shows variation for the target trait has been identified the overlapping recombinant series derived from it will be screened for the presence or absence of the trait thus enabling the gene(s) responsible to be mapped to a specific bin. The SNPs that map within the bin will then be used to transfer the gene into elite breeding material and also provide a platform from which the gene can be isolated (for example, Armstead et al., 2006a, 2007a).
Comparative analysis between Lolium/Festuca and other monocots
The bin mapping of the BPs from rice showed that L. perenne/F. pratensis linkage groups 3 and 6 were collinear with rice PMs1 and 2, respectively. The remaining L. perenne/F. pratensis linkage groups, that is, 1, 2, 4, 5 and 7, were each colinear with two rice PMs (LF1, R5+R10, LF2, R4+R7, LF4, R3+R11, LF5, R9+R12, LF7, R6+R8). Analogous macro-syntenic relationships have previously been reported between rice and the Triticeae (initially by Moore, 1995; Moore et al., 1995). L. perenne/F. pratensis linkage groups 1, 2, 4 and 7 are composed of two colinear rice PMs, where one rice PM has been inserted into another PM. The insertion of one rice chromosome into the middle of the second, leading to a reduction in chromosome number, is also consistent with previous findings from other Poaceae species, for example barley—(Korzun and Künzel, 1996); finger millet—(Srinivasachary et al., 2007); wheat—(Salse et al., 2008); (Luo et al., 2009).
The presence of a chromosome 4/5 translocation (confirming the previous observation by Alm et al., 2003) in wheat and barley demonstrated one example where synteny at the macro level had broken down between these species and L. perenne/F. pratensis. A possible further break down in synteny between these species has been reported, that is, a putative insertion of wheat/barley linkage group 2 at one end of Lolium/Festuca linkage group 6 (Alm et al., 2003). However, it was not possible to confirm this as no SNPs were bin mapped to the ends of Lolium/Festuca linkage group 6.
It has been postulated that the progenitor of the monocots had five chromosomes, which then underwent an increase in chromosome number to 12, as seen in rice, followed by a reduction in chromosome number to seven in the Poeae, Triticeae and Aveneae grasses (Murat et al., 2010). The fact that: (1) with one exception, all the inversions observed in Lolium/Festuca relative to rice were observed in a single rice PM and (2) that the macro synteny in L. perenne, F. pratensis, wheat and barley has largely been maintained indicates that the inversions observed relative to rice occurred in the 12 chromosome progenitor of the Poaceae.
The centromeric positions have been established for the 12 rice PMs (web site) (Rice Genome Annotation Project—Ouyang et al., 2007, rice.plantbiology.msu.edu/) and with the resolution of the introgression maps it is possible to compare the location of the 12 rice centromeres with those of the seven Lolium/Festuca centromeres. In L. perenne/F. pratensis, linkage groups 3 and 6, which showed synteny to only one rice PM, the bin containing the physical location of the Lolium/Festuca centromere, also contains BPs surrounding the relevant rice centromere. For example, the rice genome annotation project gives the centromeric position of rice PM 1 centromere as BP B1061G08 (BP 157 as described in this paper). BPs on either side of 157, that is, 147 and 181, have been mapped to the bin located between 47 and 53% of the physical distance along the chromosome, which also contains the L. perenne/F. pratensis linkage group 1 centromere. The same occurs in L. perenne/F. pratensis linkage group 6 although the bin containing the centromere covers a much larger physical distance, that is, from 41–80%.
In the L. perenne/F. pratensis linkage groups showing synteny to two rice PMs, the ‘functional' rice centromeric location maps to the bin containing the L. perenne/F. pratensis centromere. In L. perenne/F. pratensis linkage group 1 the rice 10 centromeric position is located in the same bin as the L. perenne/F. pratensis centromere with the centromeric position of rice PM 5 being located two bins away. The rice PM 7 centromere is located in the same bin as the L. perenne/F. pratensis linkage group 2 centromere. In L. perenne/F. pratensis linkage group 7 the position of the centromere from rice PM 8 is located to the same bin as the L. perenne/F. pratensis centromere. However, in L. perenne/F. pratensis linkage groups 4 and 5 the centromeric position of both syntenic rice PMs is located to the bins containing the L. perenne/F. pratensis centromeres.
The distribution of recombination along the L. perenne/F. pratensis chromosomes
A comparison of recombination along the length of each of the seven L. perenne/F. pratensis chromosomes revealed that the distal regions showed the highest frequencies of recombination while the proximal regions showed the lowest frequencies of recombination. SNP analysis indicated that genes were distributed both proximally and distally along each of the seven L. perenne/F. pratensis chromosomes, that is, in areas of both high and low recombination frequency. This data is in agreement with previous data that showed (1) that genes were distributed throughout the length of L. perenne/F. pratensis chromosome 3 and (2) that a large proportion of genes on chromosome 3 (c. 75%) were located in regions of the genome that showed low/very low frequencies of recombination (King et al., 2007). Furthermore, L. perenne/F. pratensis linkage groups 1, 4 and 7 (and to a lesser extent linkage group 2) carry regions of DNA syntenic to whole rice linkage groups. Although in rice these linkage groups recombine normally, in the Poaceae they are proximally located, and thus undergo a reduced frequency of recombination. It has previously been postulated that recombination was localised to the distal regions of the genome because these were the regions that were thought to be gene rich while proximal regions of chromosomes were thought to be gene poor (Akhunov et al., 2003). However, it appears more likely that the reduction in recombination may result from physical restraints of the proximal location of DNA in large monocot chromosomes as seen in wheat, L. perenne and F. pratensis, barley and rye as compared with smaller genome monocots such as rice.
Plant breeders require novel genetic variation to develop new superior plant varieties. This variation is derived from new allelic combinations that are generated via recombination at meiosis in breeding material. In Lolium/Festuca, where only very low frequencies of recombination occur at proximal regions of the chromosomes, the majority of new allelic combinations will be generated from the distal regions of the chromosomes. Thus, due to very low frequencies of recombination among a large proportion of the genes that are located in the proximal regions of the genomes of Lolium/Festuca and are being under-utilised to generate novel allelic combinations. Therefore, plant breeders are not able to fully exploit a large proportion of the potential genetic variation available for variety development. Low frequencies of recombination in proximal regions in chromosomes are not restricted to L. perenne and F. pratensis. Wheat and barley have a similar syntenic arrangement relative to L. perenne and F. pratensis at the macro level, with the exception of the 4/5 translocation described. Both of these species have also been shown to have distally located chiasmata (Lukaszewski and Curtis, 1993; Künzel et al., 2000; Akhunov et al., 2003).
There has been considerable interest in the exploitation of genetic variation from distant relatives of crop species for many years. However, only limited use of this enormous source of genetic variation in strategic plant breeding programmes has been possible up to now. In this paper we have described how marker technology can be used to introgress small chromosome segments into crops for exploitation in plant improvement programmes. This approach is now being applied in plant improvement programmes for both cereal and forage grass species.
Data Archiving
Data have been deposited at Dryad: doi:10.5061/dryad.90hc5.
Acknowledgments
We thank Mark Sorrells for the critical reading of this manuscript. This work was supported by the Biotechnology and Biological Sciences Research Council (Grant—BB/E00654X/1).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Akhunov ED, Goodyear AW, Geng S, Qi L-L, Echalier B, Gill BS, et al. The organisation and rate of evolution of the wheat genomes are correlated with recombination rates along chromosome arms. Genome Res. 2003;13:753–763. doi: 10.1101/gr.808603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alm V, Fang C, Busso CS, Devos KM, Vollan K, Grieg Z, et al. A linkage map of meadow fescue (Festuca pratensis Huds.) and comparative mapping with other Poaceae species. Theor Appl Genet. 2003;108:25–40. doi: 10.1007/s00122-003-1399-5. [DOI] [PubMed] [Google Scholar]
- Ammar K, Mergoum M, Rajoram S.2004The history and evolution of triticaleIn Megoum M, Gomez-Macpherson H, (eds).Triticale improvement and production. FAO Plant Production and Protection series NO. 179 FAO: Rome, Italy; 1–11. [Google Scholar]
- Armstead IP, Donnison IS, Aubry S, Harper JA, Hortensteiner S, James CL, et al. From crop to model to crop: identifying the genetic basis of the staygreen mutation in the Lolium/Festuca forage and amenity grasses. New Phytol. 2006a;172:592–597. doi: 10.1111/j.1469-8137.2006.01922.x. [DOI] [PubMed] [Google Scholar]
- Armstead IP, Harper JA, Turner LB, Skot L, King IP, Humphreys MO, et al. Introgression of crown rust (Puccinia coronata) resistance from meadow fescue (Festuca pratensis) into Italian ryegrass (Lolium multiflorum): genetic mapping and identification of associated molecular markers. Plant Pathol. 2006b;55:62–67. [Google Scholar]
- Armstead IP, Donnison IS, Aubry S, Harper JA, Hortensteiner S, James CL, et al. Cross-species identification of Mendel's I locus. Science. 2007a;315:73. doi: 10.1126/science.1132912. [DOI] [PubMed] [Google Scholar]
- Armstead IP, Thomas H, Ougham H, Huang L, King J, King IP. Rice pseudomolecule-anchored cross-species DNA sequence alignments indicate regional genomic variation in expressed sequence conservation. BMC Genomics. 2007b;8:283–329. doi: 10.1186/1471-2164-8-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M, Smith JB. Nuclear DNA amounts in angiosperms. Philos Trans R Soc Lond B Biol Sci. 1976;274:227–274. doi: 10.1098/rstb.1976.0044. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Smith JB, Heslop-Harrison JC. Nuclear DNA amounts in angiosperms. Proc R Soc Lond B. 1982;216:179–199. [Google Scholar]
- Close TJ, Bhat PR, Lonardi S, Wu Y, Rostoks N, Ramsay L, et al. Development and implementation of high-throughput SNP genotyping in barley. Bmc Genomics. 2009;10:582. doi: 10.1186/1471-2164-10-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreisigacker S, Kishii M, Lage J, Warburton M. Use of synthetic hexaploid wheat to increase diversity for CIMMYT bread wheat improvement. Austral J Agri Res. 2008;59:413–420. [Google Scholar]
- Febrer M, Goicoechea JL, Wright J, McKenzie N, Song XA, Lin JK, et al. An integrated physical, genetic and cytogenetic map of Brachypodium distachyon, a model system for grass research. PLoS ONE. 2010;5:e13461. doi: 10.1371/journal.pone.0013461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Olmedo F, Delibes A, Sanchez-Monge R. Transfer of resistance to eyespot disease from Aegilops ventricosa to wheat. Proceedings of the 8th Congress of Eucarpia. 1977. pp. 91–97.
- Gill GP, Wilcox PL, Whittaker DJ, Winz RA, Bickerstaff P, Echt CE. A framework linkage map of perennial ryegrass based on SSR markers. Genome. 2006;49:354–364. doi: 10.1139/g05-120. [DOI] [PubMed] [Google Scholar]
- Gustafson JP, Borlaug NE, Raven PH. World food supply and biodiversity. World Agriculture. 2010;1:37–41. [Google Scholar]
- Griffiths S, Sharp R, Foote TN, Bertin I, Wanous M, Reader S. Molecular characterisation of PH1 as a major chromosome pairing locus in polyploidy wheat. Nature. 2006;439:749–752. doi: 10.1038/nature04434. [DOI] [PubMed] [Google Scholar]
- Harper JA, Armstead IP, Thomas A, James C, Gasior D, Bisaga M, et al. Alien introgression in the grasses Lolium perenne (perennial rye grass) and Festuca pratensis (meadow fescue)—the development of seven monosomic substitution lines and their molecular and cytological characterisation. Ann Bot. 2011;107:1313–1321. doi: 10.1093/aob/mcr083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys J, Harper JA, Armstead IP, Humphreys MW. Introgression mapping of genes for drought resistance transferred from Festuca arundinacea var. glaucescens into Lolium multiflorum. Theor Appl Genet. 2005;110:579–587. doi: 10.1007/s00122-004-1879-2. [DOI] [PubMed] [Google Scholar]
- King IP, Morgan WG, Armstead IP, Harper JA, Hayward MD, Bollard A, et al. Introgression mapping in the grasses I. Introgression of Festuca pratensis chromosomes and chromosome segments into Lolium perenne. Heredity. 1998;81:462–467. [Google Scholar]
- King IP, Morgan WG, Harper JA, Thomas HM. Introgression mapping in the grasses. II. Meiotic analysis of the Lolium perenne/Festuca pratensis triploid hybrid. Heredity. 1999;82:107–112. [Google Scholar]
- King J, Armstead IP, Donnison IS, Thomas HM, Jones RN, Kearsey MJ, et al. Physical and genetic mapping in the grasses Lolium perenne and Festuca pratensis. Genetics. 2002a;161:315–324. doi: 10.1093/genetics/161.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J, Roberts LA, Kearsey MJ, Thomas HM, Jones RN, Huang L, et al. A demonstration of a 1:1 correspondence between chiasma frequency and recombination using a Lolium perenne/Festuca pratensis substitution. Genetics. 2002b;161:307–314. doi: 10.1093/genetics/161.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J, Armstead IP, Donnison SI, Roberts LA, Harper JA, Skøt K, et al. Comparative analyses between Lolium/Festuca introgression lines and rice reveal the major fraction of functional annotated gene models are located in recombination poor/very poor regions of the genome. Genetics. 2007;177:597–606. doi: 10.1534/genetics.107.075515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J, Thorogood D, Edwards KJ, Armstead IP, Roberts LA, Skøt K, et al. Development of a genomic microsatellite library in perennial ryegrass (Lolium perenne) and its use in trait mapping. Ann Bot. 2008;101:845–853. doi: 10.1093/aob/mcn016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzun L, Künzel G. The physical relationship of barley chromosome 5 (1H) to the linkage groups of rice chromosomes 5 and 10. Mol Gen Genet. 1996;252:225–231. doi: 10.1007/s004380050316. [DOI] [PubMed] [Google Scholar]
- Künzel G, Korzun L, Meister A. Cytologically integrated physical restriction fragment length polymorphism maps of the barley genome based on translocation breakpoints. Genetics. 2000;154:397–412. doi: 10.1093/genetics/154.1.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewski AJ, Curtis CA. Physical distribution of recombination in B-genome chromosomes of tetraploid wheat. Theor Appl Genet. 1993;84:121–127. doi: 10.1007/BF00223816. [DOI] [PubMed] [Google Scholar]
- Luo MC, Deal KR, Akhunov ED, Akhunova A, Anderson OD, Anderson JA, et al. Genome comparisons reveal a dominant mechanism of chromosome number reduction in grasses and accelerated genome evolution in Triticeae. Proc Natl Acad Sci USA. 2009;106:15780–15785. doi: 10.1073/pnas.0908195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KFX, Martis M, Hedley PE, Simkova H, Liu H, Morris JA, et al. Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell. 2011;23:1249–1263. doi: 10.1105/tpc.110.082537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore BJ, Donnison SI, Harper JA, Armstead IP, King J, Thomas H, et al. Molecular tagging of a senescence gene by introgression mapping of a mutant stay-green locus from Festuca pratensis. New Phytol. 2005;165:801–806. doi: 10.1111/j.1469-8137.2004.01269.x. [DOI] [PubMed] [Google Scholar]
- Moore G. Cereal genome evolution—pastoral pursuits with lego genomes. Curr Opin Genet Dev. 1995;5:717–724. doi: 10.1016/0959-437x(95)80003-n. [DOI] [PubMed] [Google Scholar]
- Moore G, Devos KM, Wang Z, Gale MD. Cereal genome evolution—grasses, line up and form a circle. Curr Biol. 1995;5:737–739. doi: 10.1016/s0960-9822(95)00148-5. [DOI] [PubMed] [Google Scholar]
- Murat F, Xu JH, Tannier E, Abrouk M, Guihot N, Pont C, et al. Ancestral grass karyotype reconstruction unravels new mechanisms of genome shuffling as a source of plant evolution. Genome Res. 2010;20:1545–1557. doi: 10.1101/gr.109744.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, et al. The TIGR Rice Genome Annotation resource—improvements and new features. Nucl Acid Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roderick HW, Morgan WG, Harper JA, Thomas HM. Introgression of crown rust (Puccinia coronata) resistance from meadow fescue (Festuca pratensis) into Italian rye grass (Lolium multiflorum) and physical mapping of the locus. Heredity. 2003;91:396–400. doi: 10.1038/sj.hdy.6800344. [DOI] [PubMed] [Google Scholar]
- Salse J, Bolot S, Throude M, Jouffe V, Piegu B, Quraishi UM, et al. Identification and characterisation of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell. 2008;20:11–24. doi: 10.1105/tpc.107.056309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears ER. The transfer of leaf rust resistance from Aegilops umbellulata to wheat. Brookhaven Symposium Biology. 1956;9:1–22. [Google Scholar]
- Sears ER. Chromosome engineering in wheat. Stadler Symposium. 1972;4:23–38. [Google Scholar]
- Srinivasachary, Dida MM, Gale MD, Devos KM. Comparative analyses reveal high levels of conserved colinearity between the finger millet and rice genomes. Theor Appl Genet. 2007;115:489–499. doi: 10.1007/s00122-007-0582-5. [DOI] [PubMed] [Google Scholar]
- Turner LM, Cairns AJ, Armstead IP, Ashton J, Skøt K, Whittaker D, et al. Dissecting the regulation of fructan metabolism in perennial ryegrass (Lolium perenne) with quantitative trait locus mapping. New Phytol. 2006;169:45–57. doi: 10.1111/j.1469-8137.2005.01575.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.