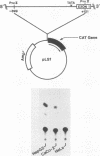
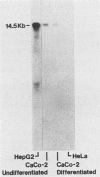
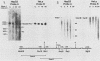
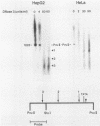
Abstract
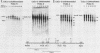
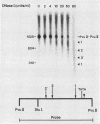
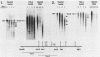
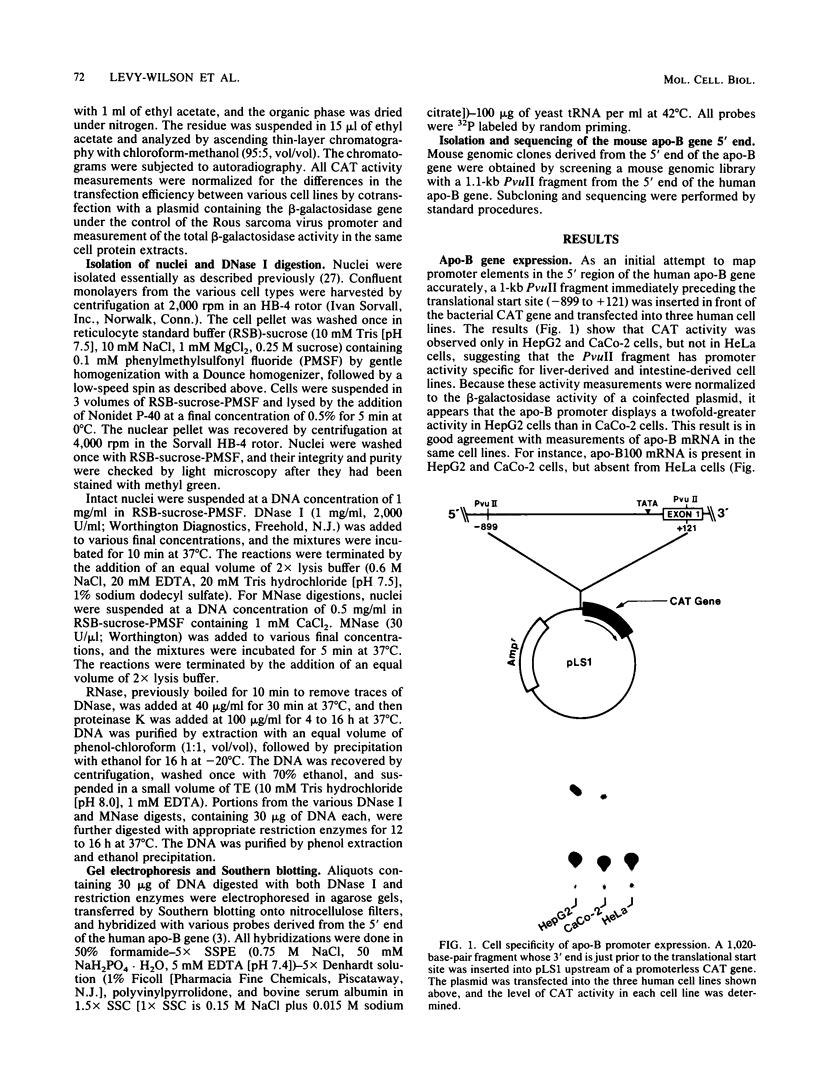
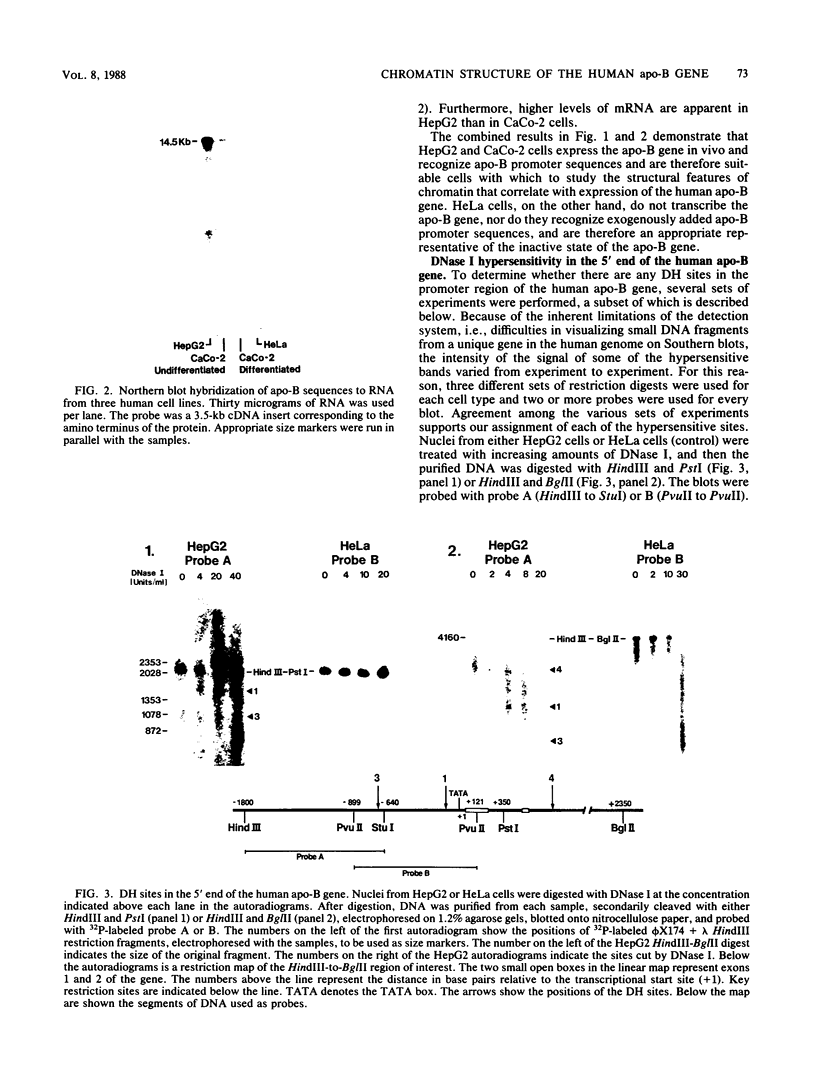
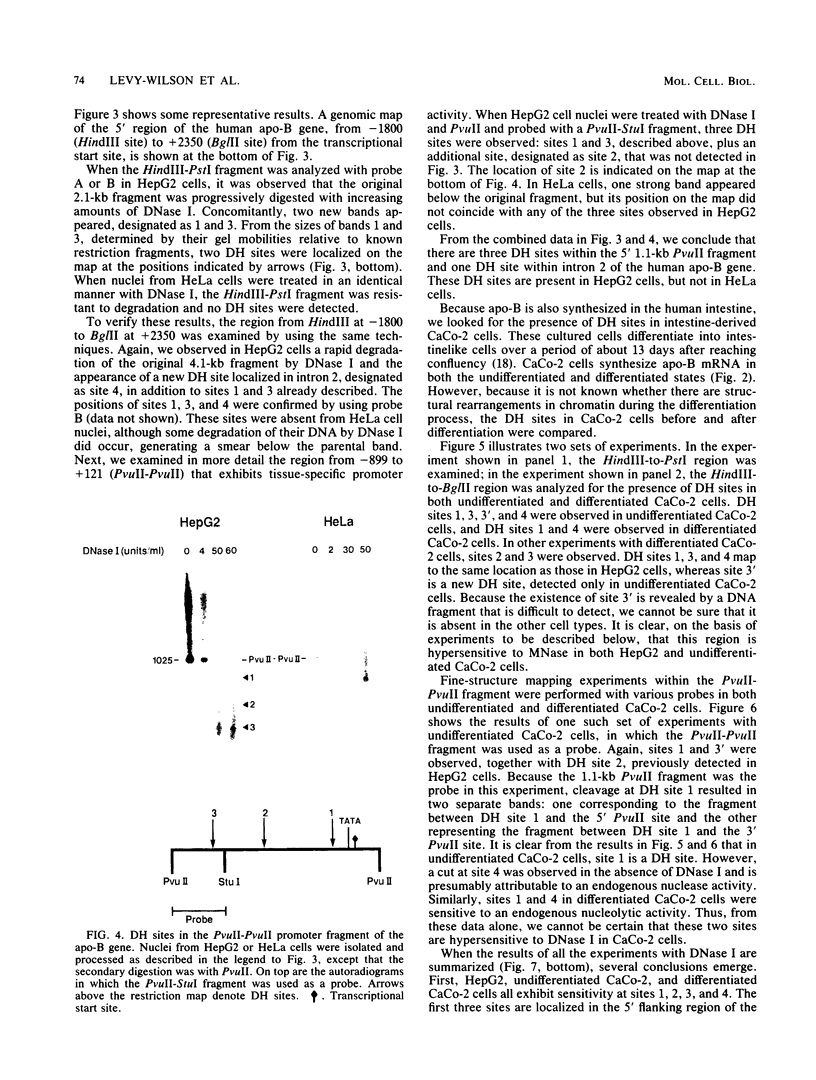
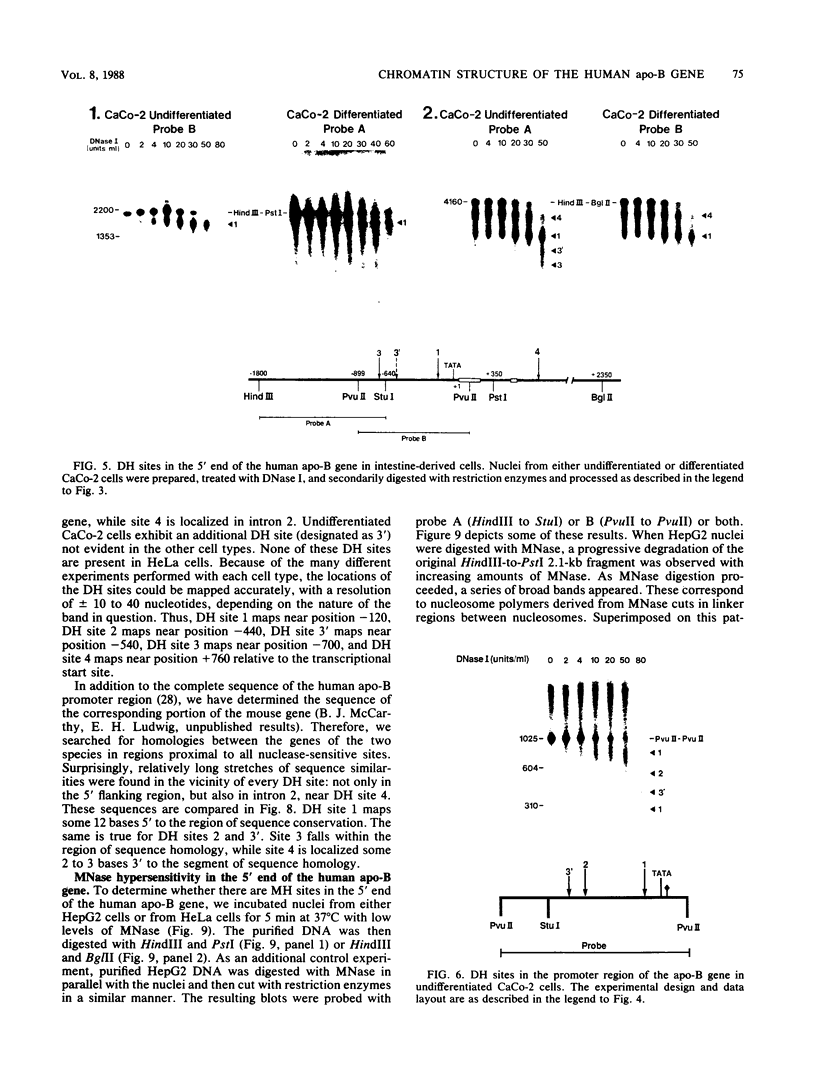
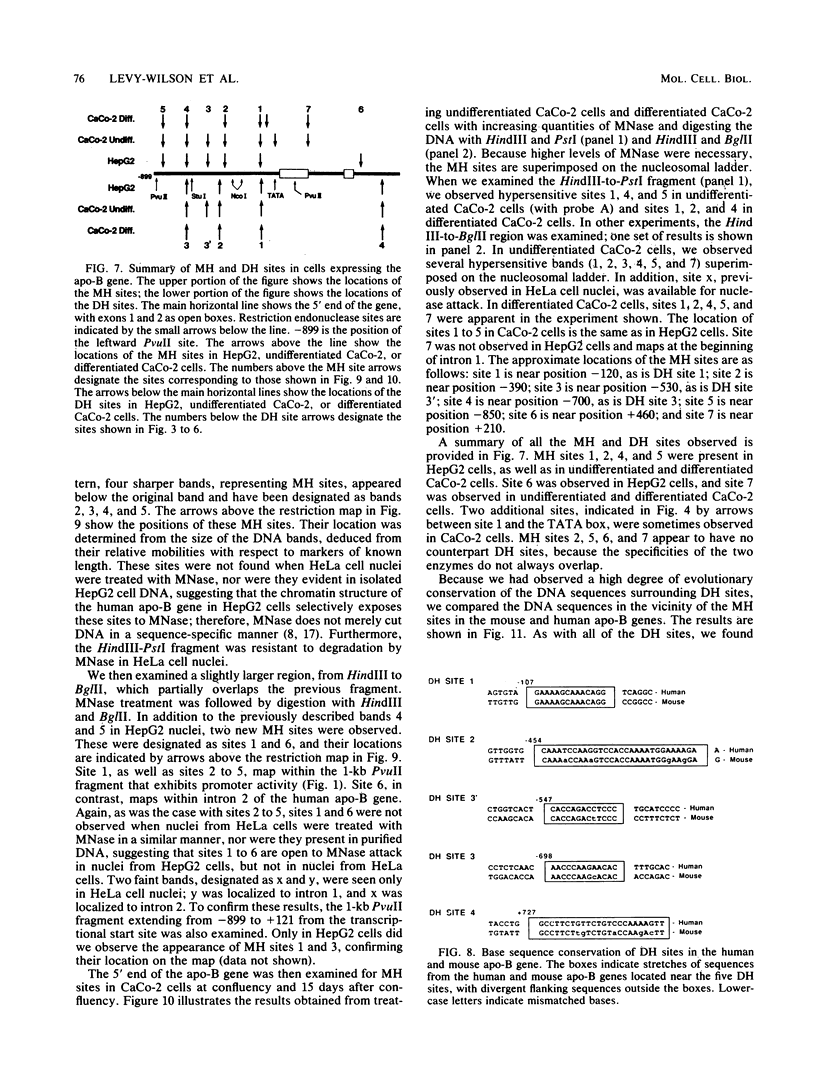
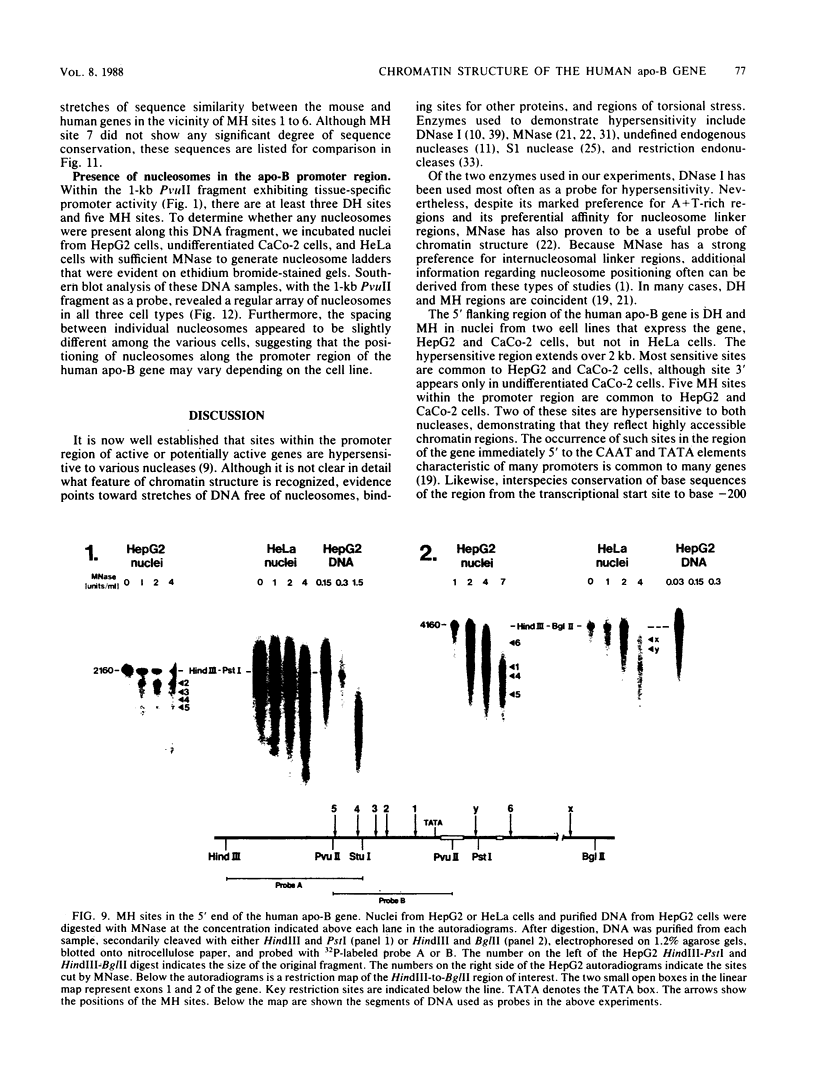
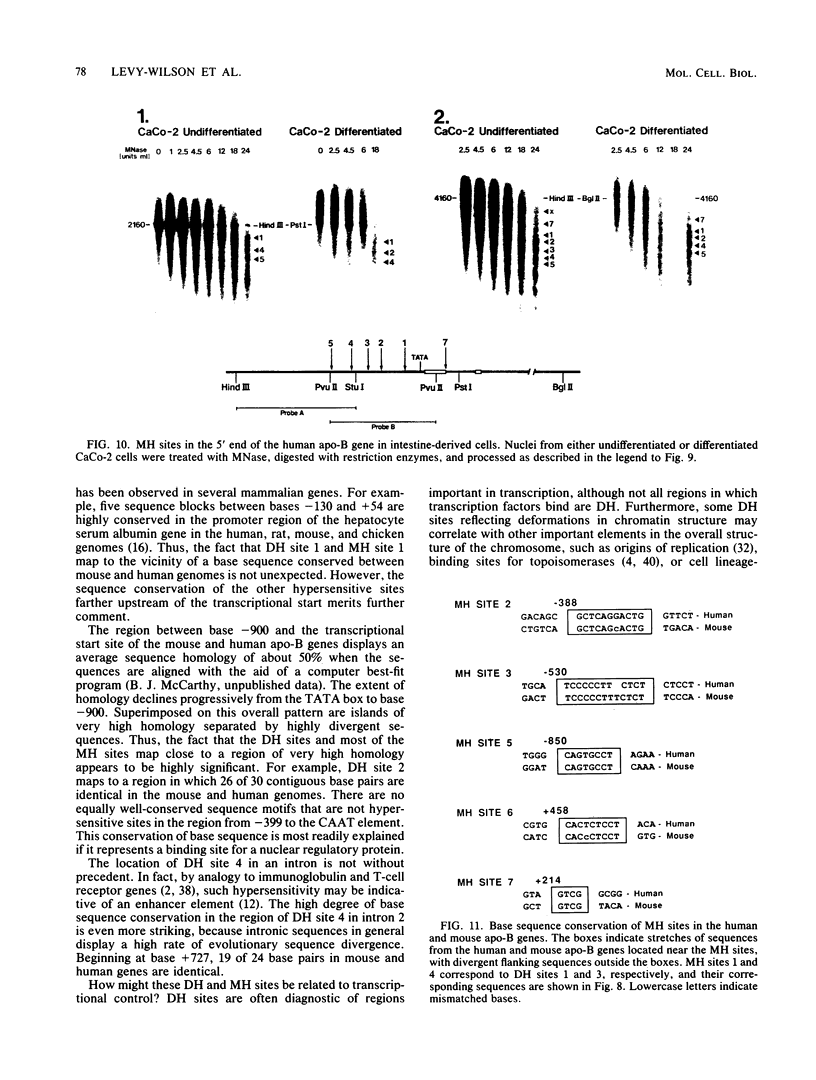
We have mapped the DNase I- and micrococcal nuclease-hypersensitive sites present in the 5' end of the human apolipoprotein B (apo-B) gene in nuclei from cells expressing or not expressing the gene. Four DNase I-hypersensitive sites were found in nuclei from liver-derived HepG2 cells and intestine-derived CaCo-2 cells, which express the apo-B gene, but not in HeLa cells, which do not. These sites are located near positions -120, -440, -700, and +760 base pairs relative to the transcriptional start site. Undifferentiated CaCo-2 cells exhibited another site, near position -540. Six micrococcal nuclease-hypersensitive sites were found in nuclei from HepG2 and CaCo-2 cells, but not in HeLa cells or free DNA. These sites are located near positions -120, -390, -530, -700, -850, and +210. HepG2 cells exhibited another site, near position +460. Comparison of the DNA sequence of the 5' flanking regions of the human and mouse apo-B genes revealed a high degree of evolutionary conservation of short stretches of sequences in the immediate vicinity of each of the DNase I- and most of the micrococcal nuclease-hypersensitive sites.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almer A., Hörz W. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J. 1986 Oct;5(10):2681–2687. doi: 10.1002/j.1460-2075.1986.tb04551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E., Hashimoto Y., Greene M. I., Maxam A. M. Active T-cell receptor genes have intron deoxyribonuclease hypersensitive sites. Science. 1985 Aug 9;229(4713):528–534. doi: 10.1126/science.3927483. [DOI] [PubMed] [Google Scholar]
- Blackhart B. D., Ludwig E. M., Pierotti V. R., Caiati L., Onasch M. A., Wallis S. C., Powell L., Pease R., Knott T. J., Chu M. L. Structure of the human apolipoprotein B gene. J Biol Chem. 1986 Nov 25;261(33):15364–15367. [PubMed] [Google Scholar]
- Bonven B. J., Gocke E., Westergaard O. A high affinity topoisomerase I binding sequence is clustered at DNAase I hypersensitive sites in Tetrahymena R-chromatin. Cell. 1985 Jun;41(2):541–551. doi: 10.1016/s0092-8674(85)80027-1. [DOI] [PubMed] [Google Scholar]
- Chen S. H., Yang C. Y., Chen P. F., Setzer D., Tanimura M., Li W. H., Gotto A. M., Jr, Chan L. The complete cDNA and amino acid sequence of human apolipoprotein B-100. J Biol Chem. 1986 Oct 5;261(28):12918–12921. [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Cladaras C., Hadzopoulou-Cladaras M., Nolte R. T., Atkinson D., Zannis V. I. The complete sequence and structural analysis of human apolipoprotein B-100: relationship between apoB-100 and apoB-48 forms. EMBO J. 1986 Dec 20;5(13):3495–3507. doi: 10.1002/j.1460-2075.1986.tb04675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C., Lomonossoff G. P., Laskey R. A. High sequence specificity of micrococcal nuclease. Nucleic Acids Res. 1981 Jun 25;9(12):2659–2673. doi: 10.1093/nar/9.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J. C., Cartwright I. L., Thomas G. H., Elgin S. C. Selected topics in chromatin structure. Annu Rev Genet. 1985;19:485–536. doi: 10.1146/annurev.ge.19.120185.002413. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. DNAase I-hypersensitive sites of chromatin. Cell. 1981 Dec;27(3 Pt 2):413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- Fritton H. P., Sippel A. E., Igo-Kemenes T. Nuclease-hypersensitive sites in the chromatin domain of the chicken lysozyme gene. Nucleic Acids Res. 1983 Jun 11;11(11):3467–3485. doi: 10.1093/nar/11.11.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies S. D., Morrison S. L., Oi V. T., Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983 Jul;33(3):717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard J. M., Herbomel P., Ott M. O., Mottura-Rollier A., Weiss M., Yaniv M. Determinants of rat albumin promoter tissue specificity analyzed by an improved transient expression system. Mol Cell Biol. 1987 Jul;7(7):2425–2434. doi: 10.1128/mcb.7.7.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T. E., Sasak W. V., Ordovas J. M., Forte T. M., Lamon-Fava S., Schaefer E. J. A novel cell line (Caco-2) for the study of intestinal lipoprotein synthesis. J Biol Chem. 1987 Mar 15;262(8):3762–3767. [PubMed] [Google Scholar]
- Hörz W., Altenburger W. Sequence specific cleavage of DNA by micrococcal nuclease. Nucleic Acids Res. 1981 Jun 25;9(12):2643–2658. doi: 10.1093/nar/9.12.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordano J., Perucho M. Chromatin structure of the promoter region of the human c-K-ras gene. Nucleic Acids Res. 1986 Sep 25;14(18):7361–7378. doi: 10.1093/nar/14.18.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane J. P. Apolipoprotein B: structural and metabolic heterogeneity. Annu Rev Physiol. 1983;45:637–650. doi: 10.1146/annurev.ph.45.030183.003225. [DOI] [PubMed] [Google Scholar]
- Kaye J. S., Bellard M., Dretzen G., Bellard F., Chambon P. A close association between sites of DNase I hypersensitivity and sites of enhanced cleavage by micrococcal nuclease in the 5'-flanking region of the actively transcribed ovalbumin gene. EMBO J. 1984 May;3(5):1137–1144. doi: 10.1002/j.1460-2075.1984.tb01942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene M. A., Elgin S. C. Micrococcal nuclease as a probe of DNA sequence organization and chromatin structure. Cell. 1981 Nov;27(1 Pt 2):57–64. doi: 10.1016/0092-8674(81)90360-3. [DOI] [PubMed] [Google Scholar]
- Knott T. J., Rall S. C., Jr, Innerarity T. L., Jacobson S. F., Urdea M. S., Levy-Wilson B., Powell L. M., Pease R. J., Eddy R., Nakai H. Human apolipoprotein B: structure of carboxyl-terminal domains, sites of gene expression, and chromosomal localization. Science. 1985 Oct 4;230(4721):37–43. doi: 10.1126/science.2994225. [DOI] [PubMed] [Google Scholar]
- Knott T. J., Wallis S. C., Powell L. M., Pease R. J., Lusis A. J., Blackhart B., McCarthy B. J., Mahley R. W., Levy-Wilson B., Scott J. Complete cDNA and derived protein sequence of human apolipoprotein B-100. Nucleic Acids Res. 1986 Sep 25;14(18):7501–7503. doi: 10.1093/nar/14.18.7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Law S. W., Grant S. M., Higuchi K., Hospattankar A., Lackner K., Lee N., Brewer H. B., Jr Human liver apolipoprotein B-100 cDNA: complete nucleic acid and derived amino acid sequence. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8142–8146. doi: 10.1073/pnas.83.21.8142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Wilson B. Modulations of prolactin and growth hormone gene expression and chromatin structure in cultured rat pituitary cells. Nucleic Acids Res. 1983 Feb 11;11(3):823–835. doi: 10.1093/nar/11.3.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig E. H., Blackhart B. D., Pierotti V. R., Caiati L., Fortier C., Knott T., Scott J., Mahley R. W., Levy-Wilson B., McCarthy B. J. DNA sequence of the human apolipoprotein B gene. DNA. 1987 Aug;6(4):363–372. doi: 10.1089/dna.1987.6.363. [DOI] [PubMed] [Google Scholar]
- Mahley R. W., Innerarity T. L. Lipoprotein receptors and cholesterol homeostasis. Biochim Biophys Acta. 1983 May 24;737(2):197–222. doi: 10.1016/0304-4157(83)90001-1. [DOI] [PubMed] [Google Scholar]
- Mahley R. W., Innerarity T. L., Rall S. C., Jr, Weisgraber K. H. Plasma lipoproteins: apolipoprotein structure and function. J Lipid Res. 1984 Dec 1;25(12):1277–1294. [PubMed] [Google Scholar]
- McGhee J. D., Wood W. I., Dolan M., Engel J. D., Felsenfeld G. A 200 base pair region at the 5' end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell. 1981 Nov;27(1 Pt 2):45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- Scott W. A., Wigmore D. J. Sites in simian virus 40 chromatin which are preferentially cleaved by endonucleases. Cell. 1978 Dec;15(4):1511–1518. doi: 10.1016/0092-8674(78)90073-9. [DOI] [PubMed] [Google Scholar]
- Shimada T., Inokuchi K., Nienhuis A. W. Chromatin structure of the human dihydrofolate reductase gene promoter. Multiple protein-binding sites. J Biol Chem. 1986 Jan 25;261(3):1445–1452. [PubMed] [Google Scholar]
- Sive H. L., Roeder R. G. Interaction of a common factor with conserved promoter and enhancer sequences in histone H2B, immunoglobulin, and U2 small nuclear RNA (snRNA) genes. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6382–6386. doi: 10.1073/pnas.83.17.6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck N. A., Baltimore D. Six distinct nuclear factors interact with the 75-base-pair repeat of the Moloney murine leukemia virus enhancer. Mol Cell Biol. 1987 Mar;7(3):1101–1110. doi: 10.1128/mcb.7.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcotte B., Guertin M., Chevrette M., LaRue H., Bélanger L. DNase I hypersensitivity and methylation of the 5'-flanking region of the alpha 1-fetoprotein gene during developmental and glucocorticoid-induced repression of its activity in rat liver. Nucleic Acids Res. 1986 Dec 22;14(24):9827–9841. doi: 10.1093/nar/14.24.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischet W. O., Glotov B. O., Schnell H., Zachau H. G. Differences in the nuclease sensitivity between the two alleles of the immunoglobulin kappa light chain genes in mouse liver and myeloma nuclei. Nucleic Acids Res. 1982 Jun 25;10(12):3627–3645. doi: 10.1093/nar/10.12.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Yang L., Rowe T. C., Nelson E. M., Liu L. F. In vivo mapping of DNA topoisomerase II-specific cleavage sites on SV40 chromatin. Cell. 1985 May;41(1):127–132. doi: 10.1016/0092-8674(85)90067-4. [DOI] [PubMed] [Google Scholar]