Abstract
Purpose.
Single nucleotide polymorphisms (SNPs) located near or within the COL5A1 gene, at 9q34.2-q34.3 chromosomal region have been reported in association with central corneal thickness (CCT). Using family linkage analysis, we identified a keratoconus susceptibility locus at 9q34. These findings led us to perform an association study between COL5A1 variation and keratoconus susceptibility.
Methods.
A Caucasian case–control cohort of 222 keratoconus patients and 3324 controls was selected as the discovery panel. An independent case–control panel of 304 cases and 518 controls and a family panel of 186 subjects were replicated for genotyping and association. Forty-four SNPs (21 for discovery and 23 for fine-mapping) spanning 300 kilobases in and around COL5A1 were genotyped and tested for genetic association. Logistic regression models implemented in PLINK were used to test for association in case controls. Generalized estimating equation models accounting for familial correlations implemented in genome-wide interaction analyses with family data were used for association testing in families.
Results.
Two CCT associated SNPs (rs1536482 and rs7044529 near and within COL5A1) were identified in the keratoconus discovery cohort (P values of 6.5 × 10−3 and 7.4 × 10−3). SNP rs1536482 was replicated in the second case–control sample (P = 0.02), and SNP rs7044529 was replicated in a keratoconus family panel (P = 0.03). Meta P values of rs1536482 and rs7044529 in the keratoconus cohorts were 1.5 × 10−4 (odds ratio [OR] = 1.30) and 2.9 × 10−3 (OR = 1.39). After Bonferroni correction, the association of SNP rs1536482 remained significant (P = 6.5 × 10−3).
Conclusions.
SNPs in the COL5A1 region, which regulate normal variation in CCT, may play a role in the thinning associated with keratoconus.
Keywords: keratoconus, association, COL5A1
Variants in the COL5A1 gene may contribute to genetic susceptibility to corneal thinning associated with keratoconus, in addition to their role in genetic regulation of normal variation in central corneal thickness.
Introduction
Keratoconus (KC, MIM #148300) is a noninflammatory corneal dystrophy characterized by progressive corneal thinning resulting in moderate to marked visual impairment.1 The prevalence of keratoconus has been estimated between approximately 1 in 12002 to 1 in 20003 in the Caucasian population. Although spectacles or contact lenses may be used for vision correction in early and moderate disease, corneal transplantation is often required for visual rehabilitation in severe cases, making it one the most common indications for corneal transplantation.4
One of the hallmarks of keratoconus is marked corneal thinning. There is an average difference of 75 μm in central corneal thickness (CCT) in keratoconus patients when compared with normal controls.5–8 Extreme corneal thinning is also a dramatic clinical feature for rare congenital connective tissue disorders with gross corneal abnormalities, such as Brittle cornea syndrome and several types of osteogenesis imperfecta.9,10 Yet mildly reduced CCT is associated with more common eye conditions, including keratoconus11,12 and primary open-angle glaucoma.13 Variation in CCT is one of the most highly heritable human traits.14,15 Several recently performed genome-wide association studies (GWAS) identified a number of genomic loci associated with differences in CCT and revealed differences in genetic determinants of CCT between ethnic groups.16–18 Since the corneal stroma is composed of collagen fibrils, it is not surprising that a number of genomic loci associated with CCT contain genes that code for various forms of collagen, such as COL1A1 and COL1A2,19 COL8A2,17 and COL5A1.16–18 Interestingly, COL5A1 is located under a keratoconus linkage peak at 9q34 we identified through a prior linkage analysis study, using sib-pair families, with the maximum logarithm of odds of 2.7.20
In the present study, we selected SNPs located within or near COL5A1, including those associated with CCT, for genotyping and testing for association with keratoconus in a Caucasian population. We identified genetic evidence of an association between SNPs within or near COL5A1 and keratoconus, possibly independent of the association with CCT, supporting a potential involvement of COL5A1 in both familial and sporadic keratoconus.
Materials and Methods
Study Subjects
Clinically affected keratoconus patients were recruited at three major sites: the Cornea Genetic Eye Institute (n = 551) at Cedars-Sinai Medical Center, Los Angeles, CA; the Jules Stein Eye Institute (n = 26) at UCLA, Los Angeles, CA; and the University Hospitals Eye Institute, Case Western Reserve University, Cleveland, OH (n = 46). Institutional Review Board (IRB) approval was obtained at all clinic sites. Written informed consent was obtained from all subjects after explanation of the nature and possible consequences of the study. The study was conducted in concordance with the provisions of the Declaration of Helsinki.
Discovery Cohort.
Clinically affected Caucasian individuals with keratoconus (n = 240) were enrolled into the GWAS as a part of the longitudinal videokeratography and genetic study at the Cornea Genetic Eye Institute at Cedars-Sinai Medical Center in Los Angeles.21 After removing samples with poor quality of genotyping, 222 samples were included in the analysis. Caucasian controls (n = 3324) were obtained from the Cardiovascular Health Study (CHS), a population-based cohort study of risk factors for cardiovascular disease and stroke in adults 65 years of age or older, recruited at four field centers.22,23 In all, 5201 predominantly Caucasian individuals were recruited in 1989 to 1990 from random samples of Medicare eligibility lists, followed by an additional 687 African-Americans recruited in 1992–1993 (total n = 5888). All these patients underwent ocular evaluations. CHS was approved by the IRB at each recruitment site, and subjects provided informed consent for the use of their genetic information. African-American CHS participants were excluded from analysis due to an insufficient number of ethnically matched cases.
Replication Cohort.
An independent group of keratoconus cases and controls was recruited as a part of the collaborative replication study, consisting of 304 keratoconus cases: 232 independent keratoconus cases recruited at the Cornea Genetic Eye Institute, 26 cases recruited at the Jules Stein Eye Institute, and 46 cases recruited at University Hospitals Eye Institute; 518 normal controls were also recruited at Cedars-Sinai Medical Center in Los Angeles.
Keratoconus Families.
Family members of keratoconus cases diagnosed at the Cornea Genetic Eye Institute were recruited to perform family-based studies. A total of 186 individuals from 41 Caucasian families consisting of 79 keratoconus patients and 107 unaffected family members were obtained for this study. This panel was ascertained through a clinically affected keratoconus proband as a part of the longitudinal videokeratography and genetic study of keratoconus at the Cedars-Sinai Medical Center in Los Angeles, CA.20
Clinical Diagnosis
The diagnosis of keratoconus was based on clinical examination and confirmed with videokeratography. All family members underwent clinical examinations including slit-lamp biomicroscopy, cycloplegic retinoscopy, and fundus evaluations. The slit-lamp biomicroscope was used to identify corneal stromal thinning, Vogt striae, or a Fleischer ring. The retinoscopy examination was performed with a fully dilated pupil (20 minutes after phenylephrine 2.5% and cyclopentolate 1% drops had been instilled in the eye) to determine the presence or absence of retro illumination signs of keratoconus, such as the oil droplet sign and scissoring of the red reflex. Videokeratography evaluation was also performed on each eye using the Topographic Modeling System (TMS-4). Subjects were assigned as having keratoconus if they had at least one clinical sign of keratoconus and a confirmatory videokeratography map with an asymmetric bowtie with skewed radial axis above and below the horizontal meridian (AB/SRAX) pattern.24
CCT Measurements
CCT was measured using a Fourier domain optical coherence tomography (OCT) device (Optovue, Inc., Fremont, CA). The OCT system operated at a 1.3-μm wavelength with a scan rate of 2000 axial scans per second. A pachymetry scan pattern (8 radials, 128 axial scans each; 10-mm diameter) centered at the corneal vertex was used to map the corneal thickness. The pachymetry map was divided into zones by octants and annular rings. Five pachymetric parameters were calculated from the region inside the 5-mm diameter: minimum, minimum–median, inferior–superior (I-S), inferotemporal–superonasal (IT-SN), and the vertical location of the thinnest cornea. The 1-percentile value of the normal group was used to define the diagnostic cutoff. This method has been shown to be highly reproducible and as accurate as ultrasonic pachymetry and scanning slit tomography for CCT assessment.8,25
DNA Extraction
DNA was extracted from Epstein–Barr virus transformed lymphoblastoid cell lines established from peripheral whole blood of each study participant (NucleoSpin Tissue kit; Macherey-Nagel Inc., Bethlehem, PA). The salt extraction protocol was used to extract DNA from buffy coats whenever cell lines were not available.
Genotyping
Discovery.
All genotyping was performed at the genotyping laboratory of the General Clinical Research Center and the Medical Genetics Institute at Cedars-Sinai Medical Center. Discovery genotyping was performed using a whole genome genotyping beadchip (Illumina Human CNV370 Quad; Illumina, Inc., San Diego, CA), following the manufacturer's protocol (Illumina Inc.).26,27 The details of GWAS genotyping and quality control were performed as previously described.28 In all, 21 SNPs located within and near COL5A1 were genotyped and analyzed in the discovery cohort.
Replication.
Using a commercial software package (Tagger [http://www.broad.mit.edu/mpg/tagger] as implemented in Haploview29,30; Broad Institute of MIT and Harvard University, Cambridge, MA) and data from the CEPH population in the International HapMap project, release 2,31,32 we designed a panel of 44 SNPs spanning 300 kilobases in and around COL5A1 (including 21 SNPs genotyped in the discovery cohort and 23 fine-mapping SNPs in the replication cohort) in COL5A1 and genotyped them using custom beadchip technology33 (iSelect Infinium; Illumina Inc.) in both case–control and family samples. The average genotyping rate for samples genotyped on the iSelect platform and passing quality control was 99.98%. Genotyping concordance among 20 pairs of blind duplicates was 100%.
One SNP (rs7044529, genotyped in the discovery cohort) failed for genotyping in the iSelect panel and was regenotyped using predesigned genotyping assay (TaqMan; Applied Biosystems/Life Technologies, Carlsbad, CA) with allelic discrimination on a real-time PCR system (ABI 7900; Applied Biosystems/Life Technologies).
Imputation
Since 23 fine-mapping SNPs were genotyped in replication cohorts but not in the original discovery cohort, multilevel imputation software (IMPUTE version 2.1.0; SAS Institute, Cary, NC)34 was used to perform imputation for the Caucasian participants in the discovery cohort on chromosome 9 (HapMap Phase I and II, CEU as the reference panel, release #24; National Center for Biotechnology Information Build 36 [dbSNP 126], Bethesda, MD).
Statistical Analyses
Discovery and Replication Study.
Under the additive genetic model, we examined the association between keratoconus and each SNP individually, adjusted for potential covariates, using the logistic regression model. Odds ratios (ORs) and their SE values associated with each additional minor allele for each SNP were calculated. Association tests by the logistic regression model for case–control samples were performed with the use of the PLINK program (v1.07) (http://pngu.mgh.harvard.edu/purcell/plink/).35 Principal component analysis (PCA) for population stratification was performed in the discovery GWAS panel using EIGENSTRAT.36 Sex and three significant principal components variables identified from PCA were used as covariates. Because of the large age difference between patients and controls in the discovery panel, age was not used as a covariate. We also adjusted for the sample collection site in the replication case–control samples. Association tests controlling for familial correlations by a generalized estimating equation (GEE) model were performed in families using the GWAF package.37
Association Testing After Conditioning on Significant SNPs.
Additional association tests in the discovery cohort were performed after adjustment for each of the top two significant SNPs, and both of them together by logistic regression.
Meta-Analysis.
Meta-analyses of 44 SNPs obtained from the three cohorts were calculated using an inverse-variance weighting by PLINK.35 We also calculated and present the adjusted P values based on Bonferroni correction for the number of SNPs tested. Given that a number of 44 SNPs tested were in linkage disequilibrium (LD), that is a rather conservative approach. Regional association plots were generated by LocusZoom38 (http://cgs.sph.umich.edu/locuszoom/).
Analysis of CCT in Patients Carrying Different Alleles of COL5A1 SNPs.
To identify whether the association between identified COL5A1 SNPs and keratoconus was independent of CCT, we tested the difference in CCT among individuals carrying different genotypes of SNPs rs1536482 and rs7044529 using a regression model in a subset of 197 keratoconus patients (all cases from the replication case–control cohort).
Results
In this study, we have studied three independent cohorts of patients with keratoconus. Table 1 summarizes the characteristics of study cohorts.
Table 1. .
Characteristics of the Study Samples
|
Variable |
Discovery |
Replication,
Case–Control |
Replication, Family |
||||||
|
Case, n
= 222 |
Control, n
= 3324 |
P |
Case, n
= 304 |
Control, n
= 518 |
P |
Case, n
= 79 |
Control, n
= 107 |
P |
|
| Age (SD)* | 44 (13) | 72 (5) | <0.01 | 43 (16) | 45 (14) | 0.02 | 41 (15) | 55 (18) | <0.01 |
| Male, % | 55 | 39 | <0.01 | 68 | 52 | <0.01 | 55 | 50 | 0.58 |
| CCT (SD)* | — | — | — | 466 (49)† | — | — | — | — | — |
Mean and SD.
197 patients had CCT measurements.
SNPs in COL5A1 Gene Region Are Associated With Keratoconus in the Discovery and Replication Cohorts
Twenty-one SNPs located within and near COL5A1 were genotyped (Illumina Human CNV370 Quad beadchip) in the keratoconus GWAS discovery phase.28 An additional 23 fine-mapping SNPs were further genotyped in replication cohorts. To have genotyping data for all 44 SNPs analyzed in both the discovery and replication cohorts, 23 fine-mapping SNPs were also imputed in the discovery cohort. Four of the 44 SNPs (rs3118516: imputed; rs1536482: genotyped; rs3118520: imputed; and rs7044529: genotyped) were found to be associated with keratoconus in the discovery cohort with nominal P ≤ 0.03 (Table 2). The same two SNPs, rs1536482 and rs7044529, have previously been associated with CCT in three independent GWAS16–18 and in large-scale meta-analysis of CCT.39 Results of the association testing for SNPs rs1536482 and rs7044529 in the discovery cohort were previously published in a collaborative study of keratoconus risk of the CCT-associated loci.39
Table 2. .
Association Testing of SNPs Located in the COL5A1 Gene Associated in GWAS Discovery Cohort*
|
SNP |
Gene Location |
Chromosomal Position |
MA |
Discovery |
Replication, Case–Control |
Replication, Family |
Meta-Analysis |
|||||||||||
|
F_A |
F_U |
OR |
P |
F_A |
F_U |
OR |
P |
F_A |
F_U |
OR |
P |
OR |
P |
P_A† |
||||
| rs3118516 | 5′ to COL5A1 | 136,579,612 | A | 0.39 | 0.28 | 1.37 | 0.01 | 0.42 | 0.36 | 1.29 | 0.02 | 0.36 | 0.31 | 1.26 | 0.23 | 1.31 | 3.6E-4 | 0.02 |
| rs1536482 | 5′ to COL5A1 | 136,580,348 | A | 0.41 | 0.33 | 1.32 | 6.5E-3 | 0.42 | 0.36 | 1.28 | 0.02 | 0.37 | 0.31 | 1.27 | 0.20 | 1.30 | 1.5E-4 | 6.5E-3 |
| rs3118520 | 5′ to COL5A1 | 136,581,415 | G | 0.47 | 0.37 | 1.31 | 0.03 | 0.48 | 0.41 | 1.31 | 0.009 | 0.44 | 0.37 | 1.33 | 0.11 | 1.31 | 2.0E-4 | 8.6E-3 |
| rs7044529 | COL5A1 | 136,707,872 | T | 0.17 | 0.14 | 1.44 | 7.4E-3 | 0.16 | 0.15 | 1.02 | 0.92 | 0.22 | 0.12 | 1.96 | 0.03 | 1.39 | 2.9E-3 | 0.12 |
P value < 0.05 in replication case–control and family cohorts and by meta-analysis.
Bonferroni-corrected P values.
Three of these four SNPs (rs3118516, rs1536482, and rs3118520, clustered in one LD block) also demonstrated significant association with keratoconus in the replication cohort (P = 0.02, 0.02, and 0.009, respectively). However, although the three SNPs demonstrated the same direction of association in the familial cohort, they did not attain individual significance in that cohort. Only one of the four SNPs that were genotyped (rs7044529) demonstrated significant association in the familial cohort (although all four SNPs had an effect in the same direction). Although there were differences in age and sex distribution between cases and controls, the P values remained similar before and after adjustments for age, sex, and other covariates (data not shown).
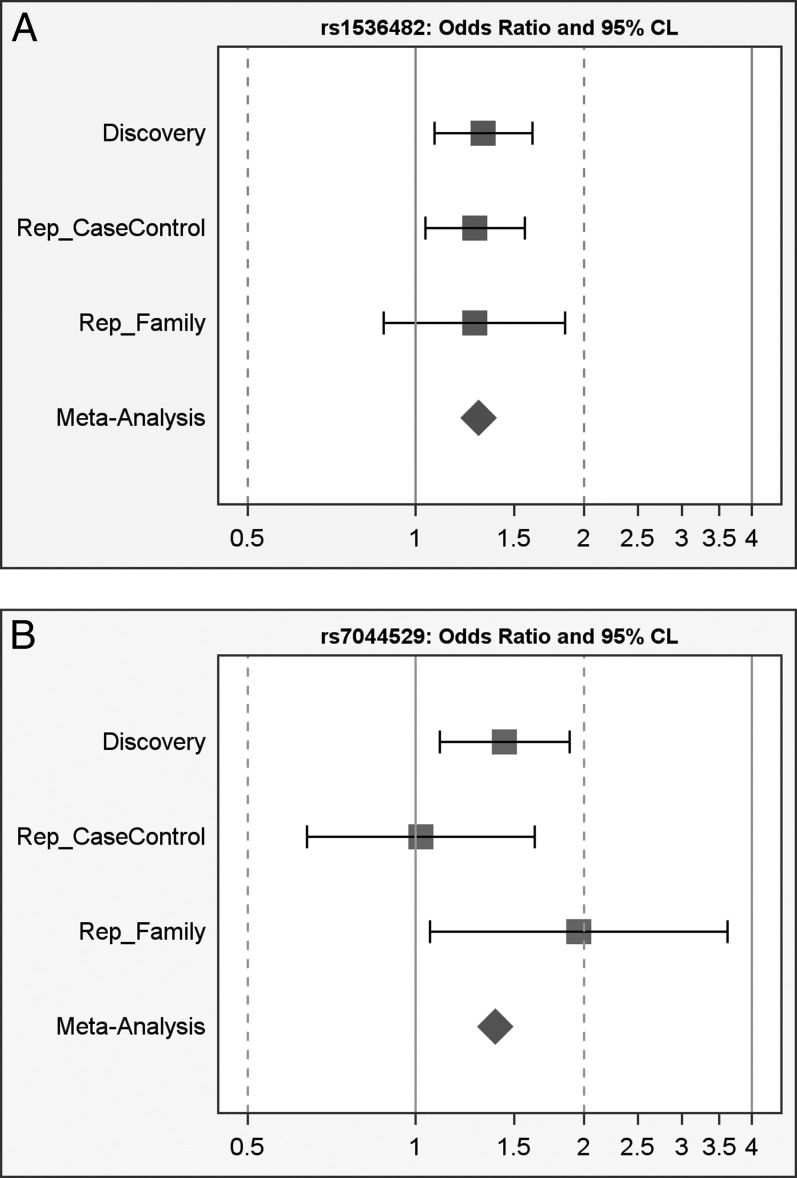
Meta-analysis combing the three cohorts identified rs1536482 as having the greatest association (meta P value = 1.5 × 10−4; Bonferroni corrected P = 6.5 × 10−3, representing the first LD block), followed by rs7044529 (meta P value = 2.9 × 10−3; Bonferroni corrected P = 0.12) (Table 2, Figs. 1A, 1B).
Figure 1. .
Forest plots for top SNPs. (A) rs1536482-A: OR and 95% CI; (B) rs7044529-T: OR and 95% CI.
Variation Across COL5A1 Gene Region and Association With Keratoconus
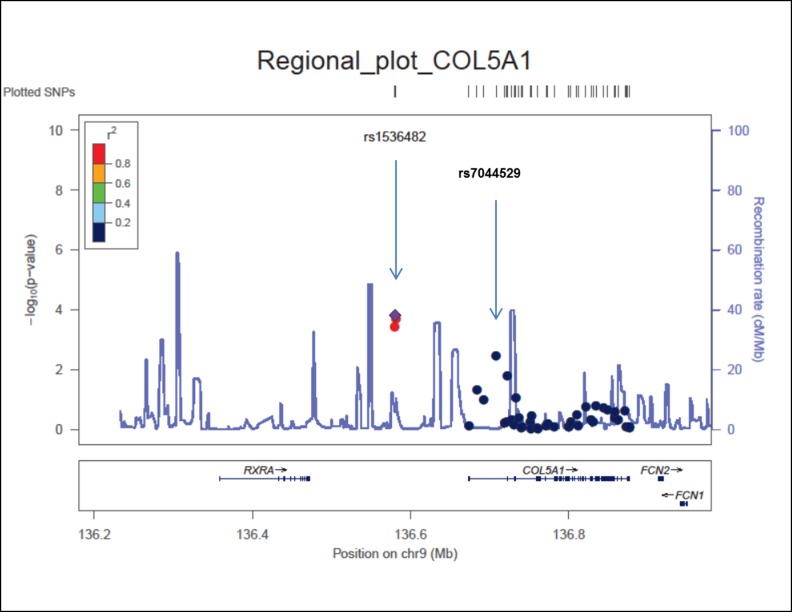
Two independent signals were identified in the region of COL5A1 as evident from the regional plot of meta P values (Fig. 2). Three SNPs, rs3118516, rs1536482, and rs3118520, are clustered in one LD block, located in the intergenic region between RXRA and COL5A1. The other tentative association signal was observed at rs7044529 located in another LD block, in an intron of COL5A1 and located approximately 126 kilobases away from rs1536482. To determine whether these signals represent independent associations, we used genotyping data of the GWAS discovery cohort to perform conditional testing for genetic association after adjusting for the top SNPs using logistic regression. After adjustment for rs7044529, a P value of 8 × 10−3 was calculated for rs1536482. Similarly, after adjustment for rs1536482, a P value of 7 × 10−3 was calculated for rs7044529. After adjusting for both of these SNPs, no other nominally significant associations (P < 0.05) were identified. These results indicate two possibly independent association signals for this region.
Figure 2. .
Regional plot of COL5A1 region.
Analysis of CCT Measurements in Keratoconus Patients With Different Alleles of Associated SNPs
In a subset of keratoconus patients (n = 197) collected for this study, we found the mean CCT measurements to be 466 ± 49 μm (as measured by OCT). These values are much lower than normal values reported for CCT measurements in the literature. This raises the question whether the COL5A1 association with keratoconus is an independent finding or is due to association with corneal thinning in general. Although formal correction for CCT was not performed because these measurements were not available for samples in discovery cohort and also not for the control samples in the replication cohorts, we addressed this issue by assessing CCT values in 197 keratoconus patients from the replication cohort carrying different combination of alleles in the two SNPs that demonstrated the greatest association with keratoconus (rs1536482 and rs7044529). The distribution of CCT between three genotypes of rs1536482 was: GG, 470 ± 49 μm (n = 67); GA, 465 ± 52 μm (n = 93); and AA, 465 ± 42 (n = 35) μm. For rs7044529, the distribution of CCT was: CC, 470 ± 49 μm (n = 149); CT, 453 ± 54 μm (n = 41); and TT, 465 ± 37 μm (n = 7). Although the difference in CCT between the three genotypes was not statistically significant for rs1536482 and rs7044529 (P > 0.05), the effect size of the risk allele was −3 and −10 μm, respectively, which was similar with GWAS in the general population.16–18 In addition, in keeping with data reported in the literature,40 there was no correlation between CCT and age (r2 = 0.002, P = 0.55). No difference of CCT between males and females was identified either (P = 0.09). These results suggest that the association between keratoconus and this gene may be independent of CCT.
Examples of Relationship Between Low CCT and Keratoconus
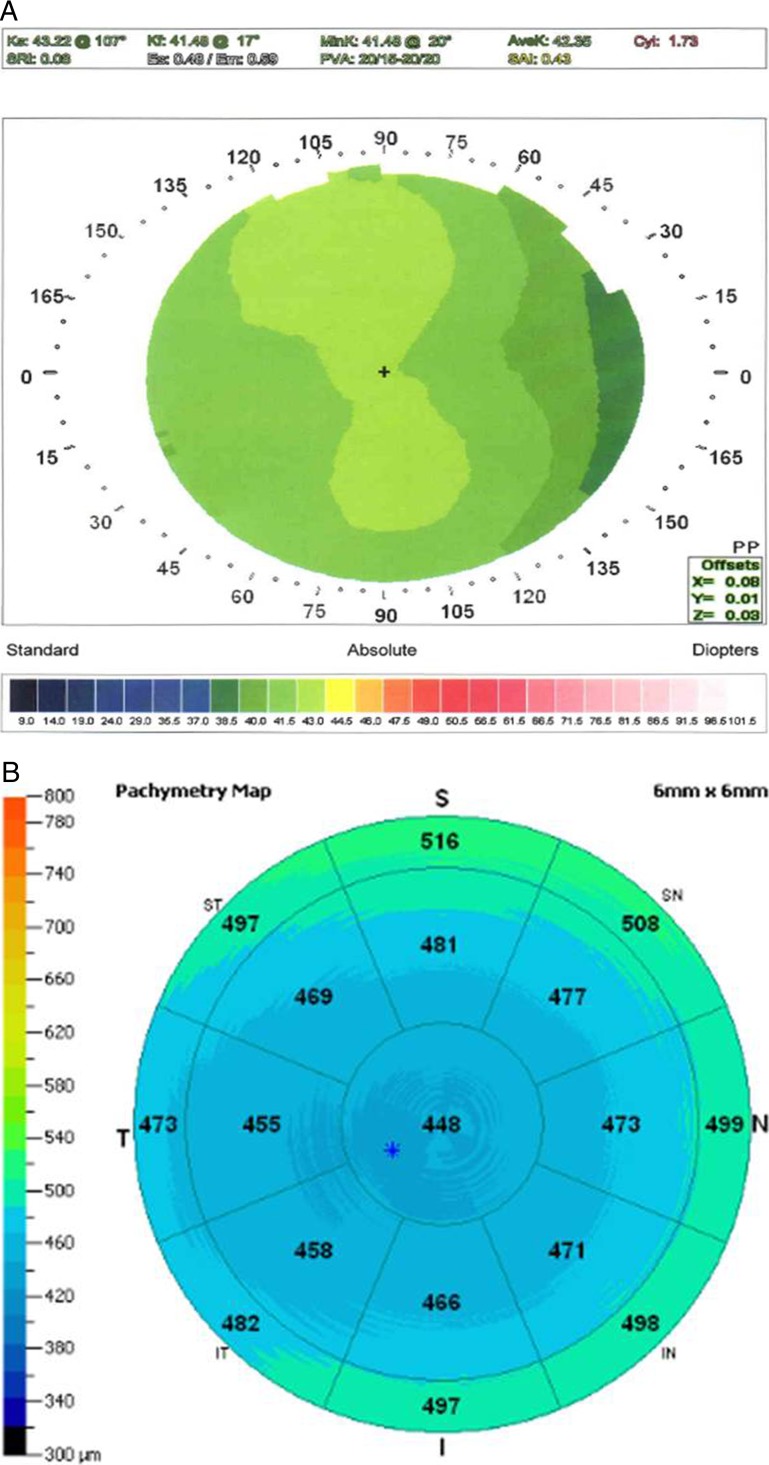
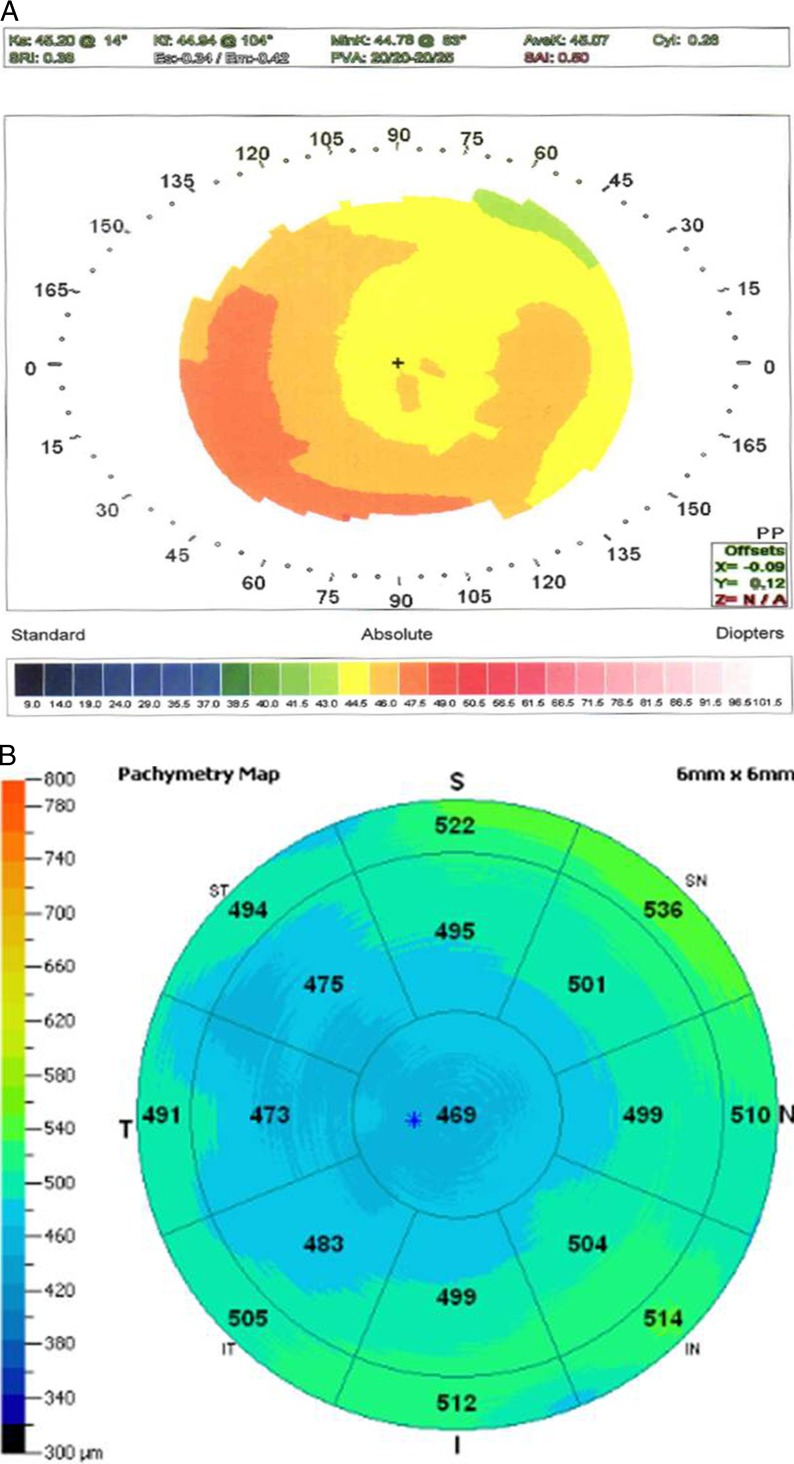
Because CCT measurements are available to us for only a moderate sample size of keratoconus patients, we had limited power to address whether the genetic effect with CCT and keratoconus is independent; however, we have observed many first-degree relatives of patients with keratoconus with abnormally low CCT values without any topographic abnormalities or progression over time. An illustrative example of such a case is an 18-year-old male referred to rule out keratoconus, prior to refractive surgery, who had two cousins with keratoconus on his mother's side. He had no videokeratography signs of keratoconus, but an abnormally low CCT (see Figs. 3A, 3B). His mother also had a low CCT without signs of keratoconus on topography (see Figs. 4A, 4B).
Figure 3. .
(A) Videokeratography right of an 18-year-old male with a family history of keratoconus (two cousins on maternal side). Topography illustrates with the rule astigmatism and no evidence of “forme frusta” keratoconus. (B) OCT pachymetry of the same eye demonstrating low CCT measurements with diffuse corneal thinning and no evidences of focal ectasia.
Figure 4.
(A) Videokeratography of the right eye of the mother of the 18-year-old male described in Figure 3A. Topography demonstrates mild contact lens warpage with no evidence of “forme frusta” keratoconus. (B) OCT pachymetry of this eye demonstrating low CCT measurements with diffuse corneal thinning and no evidence of focal ectasia.
Discussion
An association between SNPs rs1536482 and rs7044529 located near and within COL5A1 and CCT has been documented in both Caucasian and Asian populations,16,17 with the risk alleles of these SNPs associated with reduced CCT. Based on data from HapMap 3 Release 2 data, SNP rs3118520 is in strong linkage disequilibrium with SNP rs1536482 with pairwise r2 of 0.8 (SNAP; http://www.broadinstitute.org/mpg/snap/ldsearchpw.php). However, based on the same data set, r2 between both rs3118520/rs1536482 and rs7044529 is <0.1, confirming the existence of two independent signals in this region. Correspondingly, we observed two independent association peaks in the region of COL5A1, although only SNP rs1536482 remained significant after stringent Bonferroni correction was applied. However, the Bonferroni correction approach with the number of SNPs test is conservative, given that the number of LD blocks in this region is much less than that of SNPs (Fig. 2). This could indicate that the second signal associated with rs7044529 (although not significant after Bonferroni correction), identical to the one repeatedly identified in normal individuals with thinner corneas, is related to the underlying corneal thinning that characterizes keratoconus.
COL5A1 encodes an alpha chain for type V collagen, one of the low-abundance fibrillar collagens. It is a constituent of type I collagen-rich fibrils in many connective tissues including the cornea41 and appears to regulate the diameter of heterotypic fibers composed of both type I and type V collagen.42 Several studies have been performed to compare composition and distribution of different collagen types in normal and keratoconus corneas with conflicting results.43–46 Preliminary studies of corneas in our laboratory indicate a strong presence of COL5A1 transcripts. We are following this up with more detailed expression studies of this gene, comparing keratoconus corneal postkeratoplasty buttons to normal.
In addition to our molecular findings, there is clinical evidence to support findings suggested by our genetic studies: that the variation in CCT may be an independent genetic risk factor that predisposes individuals to developing keratoconus. Corneal thinning associated with keratoconus more often than not commences paracentrally and inferotemporally and may be progressive in nature.1,47 This phenomenon frequently occurs in clinically normal family members of patients with keratoconus with mild topographic abnormalities and in the clinically normal fellow eyes of patients with unilateral keratoconus as first shown by our group48,49 and further confirmed by the work of Colin et al.11 and Salabert et al.50
Phenotypically subclinical CCT is different from that associated with keratoconus. It is central in position, nonprogressive in nature, and it occurs in the absence of clinical or topographic features of keratoconus.14 In a retrospective review of 171 patients who developed ectasia after LASIK surgery (an adverse outcome indicating a high probability of underlying undiagnosed keratoconus), Randleman et al.51 found that decreased CCT measurements constituted a major risk factor predisposing LASIK patients to developing ectasia.
One of the limitations of our study is the unmatched design of cases and controls, especially the large difference in age between cases and controls in the discovery panel. We could not adjust for age as the covariate in the association tests. However, keratoconus typically commences in the early teens and progresses to the mid-30s, when it typically arrests. Therefore, using an elderly population as controls, the misclassification probability of disease due to age is small, although age-related genes could potentially be detected as false positives. For replication cohorts, we did comparisons of association tests before and after adjustments for age and sex, and did not identify significant age and/or sex effect.
In summary, both our genetic findings and clinical evidence suggest that the variants in CCT-associated gene COL5A1 both contribute to normal variation in central corneal thickness and are associated with clinical corneal thinning in keratoconus. However, the lack of association between COL5A1 variants and CCT in keratoconus patients raises the possibility that abnormal thinning in keratoconus is dependent on the effects of other genetic polymorphisms in other keratoconus-associated genes. This hypothesis is being tested in extended families with keratoconus in our clinic and laboratories.
Acknowledgments
Presented in part at the Annual Meeting of the Association for Research in Vision and Ophthalmology, Fort Lauderdale, Florida, May 2012.
Supported by National Eye Institute/National Institutes of Health (NIH) Grant NEI 09052, the Skirball Foundation for Molecular Ophthalmology, and the Eye Defects Research Foundation Inc. DNA handling and genotyping at Cedars-Sinai Medical Center was supported in part by the National Center for Research Resources/NIH Grant UL1RR033176, and is now at the National Center for Advancing Translational Sciences, Clinical and Translational Science Institute Grant UL1TR000124; and the Board of Governors Chair in Medical Genetics (JIR). Community Health Sciences research was supported by the National Heart, Lung, and Blood Institute (NHLBI) contracts N01-HC-85239, N01-HC-85079 through N01-HC-85086; N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, HHSN268201200036C, and NHLBI Grants HL080295, HL085251, HL087652, and HL105756, with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided by National Institute on Aging Grants AG-023629, AG-15928, AG-20098, and AG-027058. See also http://www.chs-nhlbi.org/pi.htm. The authors alone are responsible for the content and writing of the paper.
Disclosure: X. Li, None; Y. Bykhovskaya, None; A.L.C. Canedo, None; T. Haritunians, None; D. Siscovick, None; A.J. Aldave, None; L. Szczotka-Flynn, None; S.K. Iyengar, None; J.I. Rotter, None; K.D. Taylor, None; Y.S. Rabinowitz, None
References
- 1. Rabinowitz YS. Keratoconus. Surv Ophthalmol. 1998; 42: 297–319 [DOI] [PubMed] [Google Scholar]
- 2. Nielsen K. Hjortdal J,. Aagaard Nohr E, Ehlers N. Incidence and prevalence of keratoconus in Denmark. Acta Ophthalmol Scand. 2007; 85: 890–892 [DOI] [PubMed] [Google Scholar]
- 3. Kennedy RH, Bourne WM, Dyer JA. A. 48-year clinical and epidemiologic study of keratoconus. Am J Ophthalmol. 1986; 101: 267–273 [DOI] [PubMed] [Google Scholar]
- 4. Eye Banking Statistical Report. Eye Bank Association of America. Washington, DC; 2011. [Google Scholar]
- 5. Brautaset RL, Nilsson M, Miller WL, Leach NE, Tukler JH, Bergmanson JP. Central and peripheral corneal thinning in keratoconus. Cornea. 2013; 32: 257–261 [DOI] [PubMed] [Google Scholar]
- 6. Doughty MJ, Zaman ML. Human corneal thickness and its impact on intraocular pressure measures: a review and meta-analysis approach. Surv Ophthalmol. 2000; 44: 367–408 [DOI] [PubMed] [Google Scholar]
- 7. Grewal DS, Brar GS, Grewal SP. Assessment of central corneal thickness in normal, keratoconus, and post-laser in situ keratomileusis eyes using Scheimpflug imaging, spectral domain optical coherence tomography, and ultrasound pachymetry. J Cataract Refract Surg. 2010; 36: 954–964 [DOI] [PubMed] [Google Scholar]
- 8. Li Y, Tang M, Zhang X, Salaroli CH, Ramos JL, Huang D. Pachymetric mapping with Fourier-domain optical coherence tomography. J Cataract Refract Surg. 2010; 36: 826–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Evereklioglu C, Madenci E, Bayazit YA, Yilmaz K, Balat A, Bekir NA. Central corneal thickness is lower in osteogenesis imperfecta and negatively correlates with the presence of blue sclera. Ophthalmic Physiol Opt. 2002; 22: 511–515 [DOI] [PubMed] [Google Scholar]
- 10. Pedersen U, Bramsen T. Central corneal thickness in osteogenesis imperfecta and otosclerosis. ORL J Otorhinolaryngol Relat Spec. 1984; 46: 38–41 [DOI] [PubMed] [Google Scholar]
- 11. Colin J, Sale Y, Malet F, Cochener B. Inferior steepening is associated with thinning of the inferotemporal cornea. J Refract Surg. 1996; 12: 697–699 [DOI] [PubMed] [Google Scholar]
- 12. Steele TM, Fabinyi DC, Couper TA, Loughnan MS. Prevalence of Orbscan II corneal abnormalities in relatives of patients with keratoconus. Clin Exp Ophthalmol. 2008; 36: 824–830 [DOI] [PubMed] [Google Scholar]
- 13. Cohen EJ. Keratoconus and normal-tension glaucoma: a study of the possible association with abnormal biomechanical properties as measured by corneal hysteresis (An AOS Thesis). Trans Am Ophthalmol Soc. 2009; 107: 282–299 [PMC free article] [PubMed] [Google Scholar]
- 14. Dimasi DP, Burdon KP, Craig JE. The genetics of central corneal thickness. Br J Ophthalmol. 2010; 94: 971–976 [DOI] [PubMed] [Google Scholar]
- 15. Toh T, Liew SH, MacKinnon JR, et al. Central corneal thickness is highly heritable: the twin eye studies. Invest Ophthalmol Vis Sci. 2005; 46: 3718–3722 [DOI] [PubMed] [Google Scholar]
- 16. Vitart V, Bencic G, Hayward C, et al. New loci associated with central cornea thickness include COL5A1, AKAP13 and AVGR8. Hum Mol Genet. 2010; 19: 4304–4311 [DOI] [PubMed] [Google Scholar]
- 17. Vithana EN, Aung T, Khor CC, et al. Collagen-related genes influence the glaucoma risk factor, central corneal thickness. Hum Mol Genet. 2011; 20: 649–658 [DOI] [PubMed] [Google Scholar]
- 18. Lu Y, Dimasi DP, Hysi PG, et al. Common genetic variants near the Brittle Cornea Syndrome locus ZNF469 influence the blinding disease risk factor central corneal thickness. PLoS Genet. 2010; 6: e1000947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dimasi DP, Chen JY, Hewitt AW, et al. Novel quantitative trait loci for central corneal thickness identified by candidate gene analysis of osteogenesis imperfecta genes. Hum Genet. 2010; 127: 33–44 [DOI] [PubMed] [Google Scholar]
- 20. Li X, Rabinowitz YS, Tang YG, et al. Two-stage genome-wide linkage scan in keratoconus sib pair families. Invest Ophthalmol Vis Sci. 2006; 47: 3791–3795 [DOI] [PubMed] [Google Scholar]
- 21. Wang Y, Rabinowitz YS, Rotter JI, Yang H. Genetic epidemiological study of keratoconus: evidence for major gene determination. Am J Med Genet. 2000; 93: 403–409 [PubMed] [Google Scholar]
- 22. Fried LP, Borhani NO, Enright P, et al. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991; 1: 263–276 [DOI] [PubMed] [Google Scholar]
- 23. Psaty BM, O'Donnell CJ, Gudnason V, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009; 2: 73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rabinowitz YS. Videokeratographic indices to aid in screening for keratoconus. J Refract Surg. 1995; 11: 371–379 [DOI] [PubMed] [Google Scholar]
- 25. Li Y, Meisler DM, Tang M, et al. Keratoconus diagnosis with optical coherence tomography pachymetry mapping. Ophthalmology. 2008; 115: 2159–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gunderson KL, Steemers FJ, Lee G, Mendoza LG, Chee MS. A genome-wide scalable SNP genotyping assay using microarray technology. Nat Genet. 2005; 37: 549–554 [DOI] [PubMed] [Google Scholar]
- 27. Gunderson KL, Steemers FJ, Ren H, et al. Whole-genome genotyping. Methods Enzymol. 2006; 410: 359–376 [DOI] [PubMed] [Google Scholar]
- 28. Li X, Bykhovskaya Y, Haritunians T, et al. A genome-wide association study identifies a potential novel gene locus for keratoconus, one of the commonest causes for corneal transplantation in developed countries. Hum Mol Genet. 2012; 21: 421–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005; 21: 263–265 [DOI] [PubMed] [Google Scholar]
- 30. de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005; 37: 1217–1223 Available at: http://www.broad.mit.edu/mpg/tagger [DOI] [PubMed] [Google Scholar]
- 31. International HapMap Consortium The International HapMap Project. Nature. 2003; 426: 789–796 [DOI] [PubMed] [Google Scholar]
- 32. Frazer KA, Ballinger DG, Cox DR, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007; 449: 851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gunderson KL, Kuhn KM, Steemers FJ, Ng P, Murray SS, Shen R. Whole-genome genotyping of haplotype tag single nucleotide polymorphisms. Pharmacogenomics. 2006; 7: 641–648 [DOI] [PubMed] [Google Scholar]
- 34. Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007; 39: 906–913 [DOI] [PubMed] [Google Scholar]
- 35. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007; 81: 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006; 38: 904–909 [DOI] [PubMed] [Google Scholar]
- 37. Chen MH, Yang Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics. 2010; 26: 580–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010; 26: 2336–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lu Y, Vitart V, Burdon KP, et al. Genome-wide association analyses identify multiple loci associated with central corneal thickness and keratoconus. Nat Genet. 2013; 45: 155–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Prasad A, Fry K, Hersh PS. Relationship of age and refraction to central corneal thickness. Cornea. 2011; 30: 553–555 [DOI] [PubMed] [Google Scholar]
- 41. Smith SM, Birk DE. Focus on molecules: collagens V and XI. Exp Eye Res. 2012; 98: 105–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Francomano CA. Key role for a minor collagen. Nat Genet. 1995; 9: 6–8 [DOI] [PubMed] [Google Scholar]
- 43. Nakayasu K, Tanaka M, Konomi H, Hayashi T. Distribution of types I, II, III, IV and V collagen in normal and keratoconus corneas. Ophthalmic Res. 1986; 18: 1–10 [DOI] [PubMed] [Google Scholar]
- 44. Radda TM, Menzel EJ, Freyler H, Gnad HD. Collagen types in keratoconus. Graefes Arch Clin Exp Ophthalmol. 1982; 218: 262–264 [DOI] [PubMed] [Google Scholar]
- 45. Tsuchiya S, Tanaka M, Konomi H, Hayashi T. Distribution of specific collagen types and fibronectin in normal and keratoconus corneas. Jpn J Ophthalmol. 1986; 30: 14–31 [PubMed] [Google Scholar]
- 46. Zimmermann DR, Fischer RW, Winterhalter KH, Witmer R, Vaughan L. Comparative studies of collagens in normal and keratoconus corneas. Exp Eye Res. 1988; 46: 431–442 [DOI] [PubMed] [Google Scholar]
- 47. Krachmer JH, Feder RS, Belin MW. Keratoconus and related noninflammatory corneal thinning disorders. Surv Ophthalmol. 1984; 28: 293–322 [DOI] [PubMed] [Google Scholar]
- 48. Rabinowitz YS, Garbus J, McDonnell PJ. Computer-assisted corneal topography in family members of patients with keratoconus. Arch Ophthalmol. 1990; 108: 365–371 [DOI] [PubMed] [Google Scholar]
- 49. Rabinowitz YS, Nesburn AB, McDonnell PJ. Videokeratography of the fellow eye in unilateral keratoconus. Ophthalmology. 1993; 100: 181–186 [DOI] [PubMed] [Google Scholar]
- 50. Salabert D, Cochener B, Mage F, Keratoconus Colin J. and familial topographic corneal anomalies [in French]. J Fr Ophtalmol. 1994; 17: 646–656 [PubMed] [Google Scholar]
- 51. Randleman JB, Woodward M, Lynn MJ, Stulting RD. Risk assessment for ectasia after corneal refractive surgery. Ophthalmology. 2008; 115: 37–50 [DOI] [PubMed] [Google Scholar]