Background: Many invertebrates contain ceramide phosphoethanolamine (CPE) rather than sphingomyelin as key membrane component.
Results: Insect-specific CPE synthase belongs to a novel branch of CDP-alcohol phosphotransferases with unique membrane topology.
Conclusion: CPE production is catalyzed by a CDP-ethanolamine:ceramide ethanolamine phosphotransferase in the Golgi lumen.
Significance: Identification of CPE synthase provides a novel opportunity to elucidate the biological role of an enigmatic but widespread sphingolipid.
Keywords: Bioinformatics, Ceramide, Drosophila, Enzyme Mechanisms, Phospholipid Metabolism, Sphingolipid, CDP-Alcohol Phosphotransferase, Kennedy Pathway, Ceramide Phosphoethanolamine, Phylogenetic Profiling
Abstract
Sphingomyelin (SM) is a vital component of mammalian membranes, providing mechanical stability and a structural framework for plasma membrane organization. Its production involves the transfer of phosphocholine from phosphatidylcholine onto ceramide, a reaction catalyzed by SM synthase in the Golgi lumen. Drosophila lacks SM and instead synthesizes the SM analogue ceramide phosphoethanolamine (CPE) as the principal membrane sphingolipid. The corresponding CPE synthase shares mechanistic features with enzymes mediating phospholipid biosynthesis via the Kennedy pathway. Using a functional cloning strategy, we here identified a CDP-ethanolamine:ceramide ethanolamine phosphotransferase as the enzyme responsible for CPE production in Drosophila. CPE synthase constitutes a new branch within the CDP-alcohol phosphotransferase superfamily with homologues in Arthropoda (insects, spiders, mites, scorpions), Cnidaria (Hydra, sea anemones), and Mollusca (oysters) but not in most other animal phyla. The enzyme resides in the Golgi complex with its active site facing the lumen, contrary to the membrane topology of other CDP-alcohol phosphotransferases. Our findings open up an important new avenue to address the biological role of CPE, an enigmatic membrane constituent of a wide variety of invertebrate and marine organisms.
Introduction
Sphingolipids are essential components of the plasma membrane. They are primarily concentrated in the exoplasmic leaflet, providing an important structural framework for plasma membrane organization and function (1). As in mammals, Drosophila sphingolipids are critical for developmental processes such as embryogenesis, neurogenesis, and gametogenesis, whereas intermediates of sphingolipid metabolism have been associated with signal transduction cascades, cell death, and phagocytosis (2, 3).
Nevertheless, there are some remarkable differences between sphingolipids of Drosophila and mammals. The major sphingoid bases in Drosophila and other dipterans are tetradecasphingenine (C14) and hexadecasphingenine (C16) as compared with octadecasphingenine (C18) in mammals (4, 5). Also, the fatty acids that are amino-linked to the sphingoid bases to create ceramides are shorter in Drosophila sphingolipids in comparison with mammals. These characteristics predict that membranes would remain fluid even at lower temperature, which correlates well with the requirement of lower ambient temperatures for Drosophila survival. Moreover, Drosophila lacks the phosphocholine-containing sphingomyelin (SM)4 found in mammalian membranes and instead synthesizes ceramide phosphoethanolamine (CPE) (4, 6, 7). The smaller cross-sectional area of the phosphoethanolamine headgroup in CPE allows a closer contact between these molecules in comparison with SM, promoting membrane viscosity. Contrary to SM, CPE does not interact favorably with cholesterol and fails to form sterol-rich domains in model bilayers (8). Addressing how each organism evolved functional membranes based on such highly divergent membrane components is an important topic in lipid biology.
SM biosynthesis in mammals is catalyzed by a PC:ceramide cholinephosphotransferase (EC 2.7.8.27) or SM synthase (SMS) (9). This enzyme catalyzes the transfer of phosphocholine from phosphatidylcholine (PC) onto ceramide, yielding SM and diacylglycerol. Mammalian cells contain two SM synthase isoforms, namely SMS1 responsible for bulk production of SM in the Golgi lumen and SMS2 serving a role in regenerating SM from ceramides liberated by sphingomyelin phosphodiesterase on the exoplasmic surface of the plasma membrane (10, 11). Both SMS1 and SMS2 are required for cell growth, at least in certain types of cancer cells (12, 13). Together with a closely related enzyme, SMSr, they form the SMS protein family (10). Mammalian cells also produce CPE, although its concentration in membranes is very low and its biological role is unknown. Two CPE synthase activities have been described in mammalian cells, one enriched in a microsomal fraction (presumably ER) and the other one associated with the plasma membrane (14–16). As PE serves as the headgroup donor for both activities, the enzyme(s) involved can be classified as PE:ceramide ethanolamine phosphotransferases analogous to SM synthase. We previously demonstrated that SMS2 is a bifunctional enzyme that produces both SM and CPE (17). Thus, SMS2 likely accounts for the plasma membrane-resident CPE synthase activity reported previously (14, 16). The function of SMSr had so far been unknown, but we recently identified it to be a monofunctional CPE synthase that resides in the ER (17, 18). SMSr thus qualifies for the microsomal CPE synthase activity first described by Malgat et al. (14).
Drosophila lacks SMS1 and SMS2 homologues, but contains a homologue of SMSr, which we named dSMSr. Although dSMSr possesses CPE synthase activity, its removal had no impact on bulk production of CPE in Drosophila S2 cells (18). In vitro enzyme assays revealed that these cells contain a second, dSMSr-independent CPE synthase that uses CDP-ethanolamine rather then PE as headgroup donor in CPE biosynthesis. This implied that the latter enzyme uses a reaction mechanism different from the one used by SMS family members, but similar to that of the enzymes producing phosphatidyl-ethanolamine via the Kennedy pathway. We here set out to identify the enzyme responsible for bulk production of CPE in Drosophila.
EXPERIMENTAL PROCEDURES
Chemicals
C6-7-nitro-2,1,3-benzoxadiazole (NBD)-ceramide (NBD-Cer) was from Molecular Probes, and NBD-sphingomyelin (NBD-SM), NBD-phosphatidylcholine (NBD-PC), and NBD-phosphatidylethanolamine (NBD-PE) were from Avanti Polar Lipids. NBD-ceramide phosphoethanolamine (NBD-CPE) was a generous gift from Philippe Devaux (Institut de Biologie Physico-chimique, Paris, France). [2-14C]Ethan-1-ol-2-amine hydrochloride and CDP-[14C]ethan-1-ol-2-amino hydrochloride were from Amersham Biosciences. All other lipids and chemicals were from Sigma-Aldrich.
Antibodies
Rabbit polyclonal and mouse monoclonal anti-V5 antibodies were from Sigma and Invitrogen, respectively. The mouse monoclonal anti-GM130 antibody was from BD Biosciences, and the rabbit polyclonal anti-calnexin antibody was from Santa Cruz Biotechnology. The rabbit polyclonal anti-dGolgin245 antibody was a generous gift from Sean Munro (Cambridge, UK). The rabbit polyclonal anti-dGMAP antibody was described by Kondylis et al. (19). Horseradish peroxidase-conjugated secondary antibodies were from PerBio, whereas antibodies conjugated to FITC and Texas Red or Alexa dyes were purchased from Jackson ImmunoResearch Laboratories or Molecular Probes, respectively. The antibody against dSMSr was obtained as described (18).
Selection, Cloning, and Expression of dCCS Sequences
Selection of candidate CPE synthases (CCS) from the National Center for Biotechnology Information (NCBI) database involved the following steps: 1) selection of proteins containing a CDP-alcohol phosphatidyltransferase motif (NCBI accession number cI00453); 4982 RefSeq proteins; 2) restriction of results to Drosophila melanogaster; 14 RefSeq proteins; 3) restriction of results to one isoform per gene; 8 RefSeq proteins; 4) removal of one incomplete (CG40928) and one misannotated sequence (CG6921); 6 RefSeq proteins; 5) removal of phosphatidylinositol synthase (PIS) (CG9245) and cardiolipin synthase (CLS) (CG4774) proteins because their biochemical function is certain; 4 RefSeq proteins. This procedure yielded four CCS proteins in Drosophila, namely dCCS1 (CG33116), dCCS2 (CG6016), dCCS3 (CG7149), and dCCS4 (CG4585). The open reading frames (ORFs) of the corresponding dCSS sequences were amplified by RT-PCR (Titan One, Roche Applied Science) from mRNA isolated from Drosophila S2 cells (TRIzol, Invitrogen) using the primers listed in Table 1. PCR products were cloned into mammalian expression vector pcDNA3.1/V5-His-TOPO (Invitrogen), and the resulting plasmids dCCS1-V5, dCCS2-V5, dCCS3-V5, and dCCS4-V5 were used to transfect HeLa cells. For expression studies in S2 cells, the cDNAs were subcloned into the copper-inducible pMT/V5-His B (Invitrogen) vector using the restriction sites KpnI and XhoI (for dCCS1 and dCCS3) or KpnI and NotI (for dCCS2 and dCCS4). PE-methyltransferase-GFP plasmid was obtained as described in Ref. 18.
TABLE 1.
Primer pairs used in this study
ACC. No., accession number.
| Protein | Acc. No. | Primer pairs for cloning | Primer pairs for creation of dsRNA |
|---|---|---|---|
| dCCS1 | Q8T9G2 | CAATAATCATGAGGCAACTATTCAGACGC/ACTATATCTTTCTTCAATGGCTCC | T7-ATGAGGCAACTATTCAGACGC/T7-GACTAGTGGAAGATTCGCC |
| dCCS2 | A1Z9D9/A1Z9E0 | CAATAATCATGGCGCTGCTCGCCTAC/ACGTGCGGTTTGCTCTTCGAC | T7-ATGGCGCTGCTCGCCTAC/T7-CAGGACCCAGTGCAGCCG |
| dCCS3 | Q8T0S3 | CAATAATCATGGGTTGCATGCGCTACTTG/ACATGGCTTTTCAACGGTTTGATT | T7-ATGGGTTGCATGCGCTACTTG/T7-CGCCATGGTGAAACCATATA |
| dCCS4 | CG4585 | CACGGTACCATAATCATGATCGGACCCAGTTCGC/CACCTCGAGACCAGTGAAATATTACTGCACGC | T7-ATGATCGGACCCAGTTCGC/T7-CAGACAGGCCACGCCCAC |
Cell Culture and RNA Interference
Human HeLa cells were grown in DMEM with 10% FCS. Transfections with dCCS1-V5, dCCS2-V5, dCCS3-V5, and dCCS4-V5/pcDNA3.1 constructs were performed using Lipofectamine reagent (Invitrogen) following the manufacturer's instructions. Drosophila S2 cells were grown in Schneider's insect medium with 10% FBS (Cambrex) at 27 °C in a humidified atmosphere. Cells were transfected with dCCS1, dCCS2, dCCS3, or dCCS4/pMT/V5-HisB constructs using Effectene (Qiagen). Expression of recombinant dCCS proteins was induced by the addition of 1 mm CuSO4 for 3 h followed by a 2-h chase in the presence of 150 μg/ml cycloheximide. RNAi on Drosophila S2 cells was performed by treatment with double-stranded RNA (dsRNA) synthesized by in vitro transcription of PCR products flanked by T7 RNA polymerase binding sites (TTAATACGACTCACTATAGGGAGA) using the MEGASCRIPT T7 transcription kit (Ambion). PCR products of 753, 604, 651, and 530 bp were amplified from dCCS1, dCCS2, dCCS3, and dCCS4 cDNAs, respectively, using primer sets listed in Table 1. dsRNAs targeting dSMSr and green fluorescent protein (GFP) were obtained as described in Ref. 18, and dsRNA treatment was performed as described in Ref. 20. On day 1, 106 cells were plated in a 35-mm dish and incubated with 30 μg of dsRNA in 1 ml of serum-free medium for 1 h at room temperature followed by the addition of 2 ml of complete medium. After 3 days, cells were either harvested for enzyme assays and metabolic labeling or fixed for immunofluorescence.
In Vitro Enzyme Assay
HeLa and S2 cells were lysed in ice-cold reaction buffer (0.3 m sucrose, 15 mm KCl, 5 mm NaCl, 1 mm EDTA, 20 mm Hepes-KOH, pH 7.0) containing freshly added protease inhibitors by passing 20 times through a 23-gauge 3/4 needle. 200 μl of postnuclear supernatant (700 × g, 10 min, 4 °C) was combined with 200 μl of reaction buffer containing 50 μm C6-NBD-ceramide or 1 μCi of CDP-[14C] ethanolamine and incubated for 2 h at 27 °C in the presence or absence of 10 mm MnCl2 and 500 μm CDP-ethanolamine. Reactions were stopped by adding 1 ml of MeOH and 0.5 ml of CHCl3, and lipids were extracted according to Bligh and Dyer (21). The lower phase was evaporated under N2, and the reaction products were analyzed by TLC using CHCl3/acetone/MeOH/acetic acid/H2O (50/20/10/10/5, v/v/v/v/v; reactions containing NBD-Cer) or CHCl3/MeOH/25% NH4OH (50/25/6, v/v/v; reactions containing CDP-[14C] ethanolamine). Fluorescent lipids were visualized on a STORM 860 image analysis system (GE Healthcare) and quantified with Quantity One software (Bio-Rad). Radiolabeled lipids were detected by exposure to BAS-MS imaging screens (Fuji Photo Film), scanned on a Bio-Rad personal molecular imager, and quantified with Quantity One software.
Cell Surface Enzyme Assay
This assay was performed essentially as described in Ref. 17. In brief, HeLa cells transfected with dCCS4 or empty vector were grown to 80–90% confluence in a 10-cm dish. The cells were washed in HBSS and preincubated with HBSS containing 1% fatty acid-free BSA at 5 °C for 30 min. To permeabilize the plasma membrane, cells were treated with 1 μg/ml streptolysin in HBSS for 15 min at 37 °C prior to preincubation in BSA-supplemented HBSS at 5 °C. Next, NBD-Cer dissolved in ethanol was added to a final concentration of 2 μm (0.2% ethanol in the medium), and the cells were incubated at 5 °C for 3 h in the presence or absence of 10 mm MnCl2 and 500 μm CDP-ethanolamine. The incubation medium was saved, and the cells were washed by incubating with HBSS containing 1% fatty acid-free BSA at 5 °C for 30 min. Incubation medium and wash were combined and subjected to lipid extraction according to Bligh and Dyer (21). Fluorescent lipids were analyzed by TLC using CHCl3/acetone/MeOH/acetic acid/H2O (50/20/10/10/5, v/v/v/v/v) and visualized as described above.
Metabolic Labeling
S2 cells (2–5 × 106) were labeled in 0.5 ml of complete Schneider's insect medium with 10 nmol NBD-Cer or with 1 μCi of [14C]ethanolamine at 27 °C for 2 h. Lipids were extracted in CHCl3/MeOH/10 mm acetic acid (1/4.4/0.2, v/v/v) and then processed according to Bligh and Dyer (21). Half of the extract was subjected to mild alkaline hydrolysis. Lipids were analyzed by TLC in CHCl3/MeOH/25% NH4OH (50/25/6, v/v/v; [14C]ethanolamine) or CHCl3/acetone/MeOH/acetic acid/H2O (50/20/10/10/5, v/v/v/v/v; NBD-Cer) and visualized as described above.
Microscopy and Image Analysis
Cells were fixed in 4% paraformaldehyde/PBS and processed for immunofluorescence after permeabilization with Triton (S2 cells) as in Ref. 22 or saponin (HeLa cells) as in Ref. 13. Images were captured using a confocal microscope D-eclipse C1, Nikon with 60× 1.40 NA Plan Apo oil objective (Nikon). Images presented are confocal sections.
RESULTS AND DISCUSSION
Drosophila Contains a Unique CPE Synthase Unrelated to dSMSr
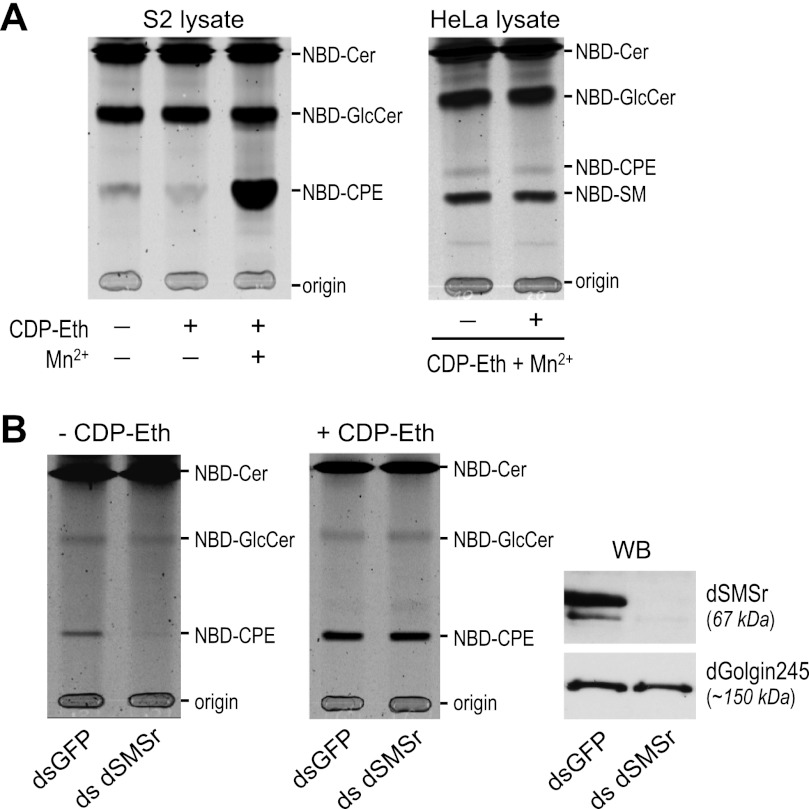
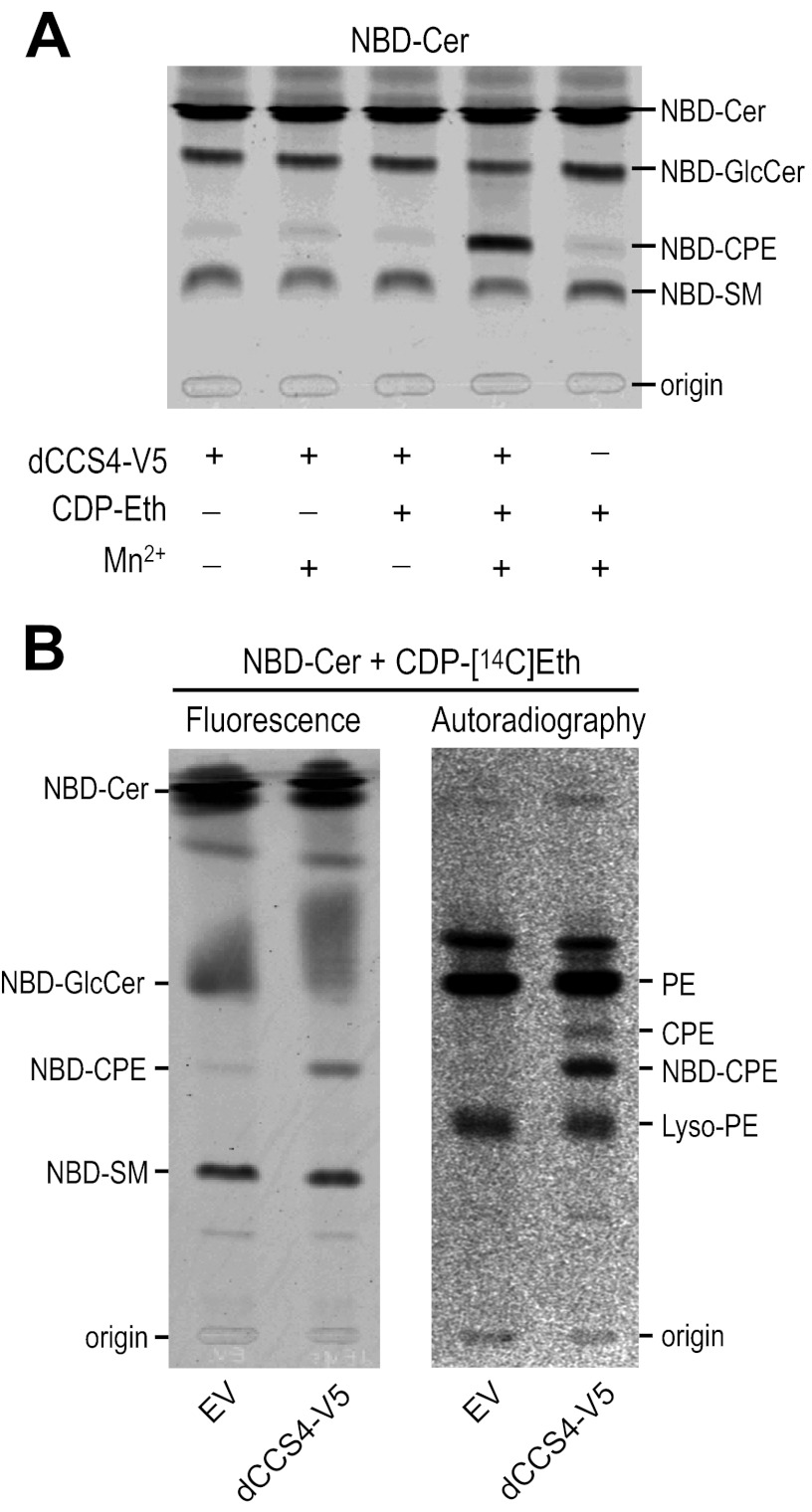
Drosophila S2 cell lysates incubated with fluorescent NBD-Cer form NBD-CPE (Fig. 1A, left panel). Depletion of CPE synthase dSMSr abolished NBD-CPE formation in S2 cell lysates (Fig. 1B, left panel) but had no effect on NBD-CPE formation in intact S2 cells (18). This indicated that S2 cells contain a second, dSMSr-independent CPE synthase. We reasoned that this second enzyme might not be detectable in cell lysates if it would require a soluble substrate that is continuously regenerated in living cells. CDP-ethanolamine (CDP-Eth) is an attractive candidate for such substrate given its role as headgroup donor in PE biosynthesis (23). Indeed, the addition of CDP-Eth dramatically enhanced NBD-CPE formation in S2 cell lysates when Mn2+ ions were present (Fig. 1A, right panel) (18). CDP-Eth-dependent CPE synthase activity was unaffected by dSMSr depletion (Fig. 1B, right panels). Together, these results indicate that Drosophila S2 cells contain two distinct CPE synthases, namely a PE:ceramide ethanolamine phosphotransferase corresponding to dSMSr and a CDP-Eth:ceramide Eth-phosphotransferase of unknown identity (Fig. 2A). The latter enzyme appears unique for insect cells as the addition of CDP-Eth and Mn2+ to lysates of human HeLa cells did not enhance NBD-CPE formation from NBD-Cer (Fig. 1A, right panel). The insect-specific CPE synthase shares two important features with ethanolamine phosphotransferases of the Kennedy pathway, i.e. the use of CDP-Eth as headgroup donor and a requirement for Mn2+ ions for proper catalysis (24, 25). One may therefore anticipate that the enzymes share a certain degree of structural similarity. This provided the starting point of a bioinformatics-based cloning strategy to identify the insect-specific CPE synthase. From now on, we will refer to this enzyme as CPES.
FIGURE 1.
Drosophila S2 cells contain a CDP-Eth-dependent CPE synthase. A, TLC analysis of reaction products from lysates of Drosophila S2 cells (left) or human HeLa cells (right) incubated with NBD-Cer in the presence or absence of CDP-Eth and MnCl2 (Mn2+). B, TLC separation of reaction products from lysates of dSMSr-depleted (dsSMSr) or mock-depleted (dsGFP) Drosophila S2 cells incubated with NBD-Cer and MnCl2 in the absence (left) or presence (middle) of CDP-Eth. Right, immunoblots (WB) of dSMSr- or mock-depleted Drosophila S2 cells stained with anti-dSMSr and anti-dGolgi245 antibodies.
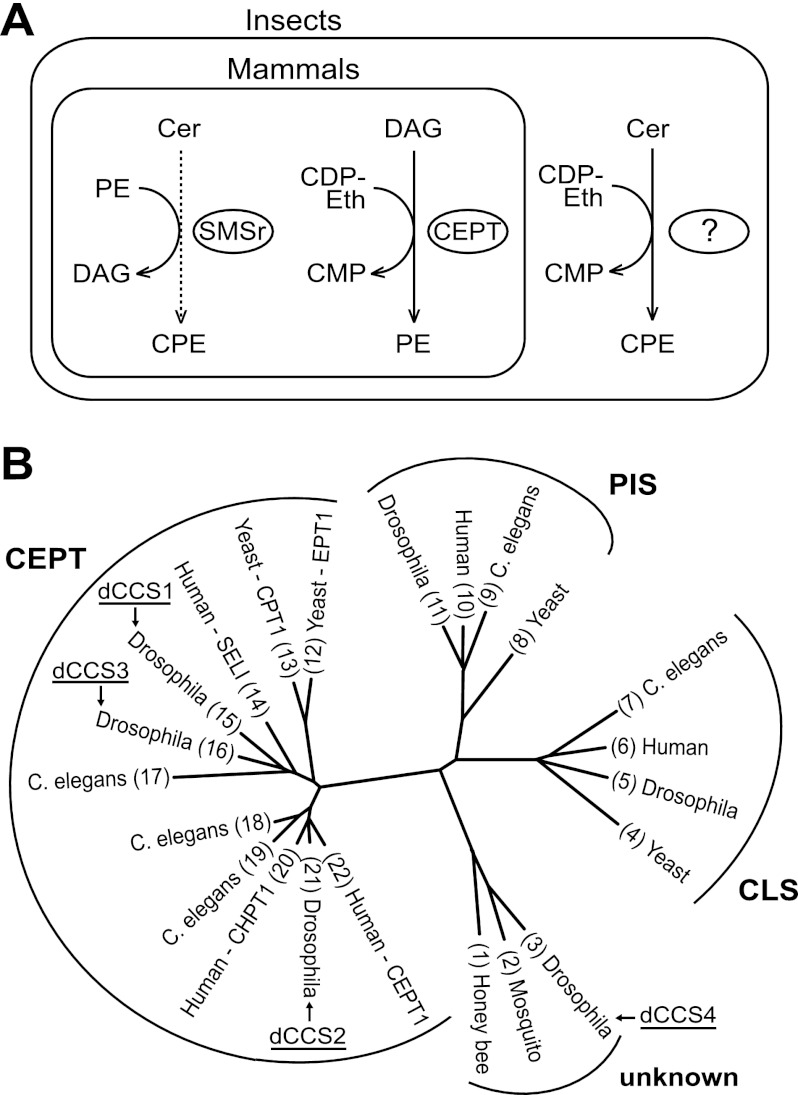
FIGURE 2.
Selection and phylogenetic analysis of candidate CPE synthases. A, reaction chemistry of PE and CPE biosynthesis in mammals and insects. See “Results and Discussion” for further details. DAG, diacylglycerol. B, phylogenetic tree of proteins in insects, Caenorhabditis elegans, human, and yeast containing a CAPT motif. The four candidate CPE synthases in Drosophila are marked dCCS1–4. The protein sequences were aligned with DIALIGN 2 (33) with manual editing to ensure correct alignment of the conserved pattern, D(X)2DG(X)2(A/Y)R(X)8–16G(X)3D(X)3D. Alignment columns with a quality score of 4 or better were used to draw a phylogenetic tree with Protdist and Fitch from the PHYLIP package. The tree was displayed and edited with TreeIllustrator (34). NCBI GI numbers are: 1) 110750730 (translation of bases 327125–328027); 2) 158033532 (translation of bases 2339192–2338023); 3) O77475; 4) 6320059; 5) 23172318; 6) 10092647; 7) 71984834; 8) 6325370; 9) 17537129; 10) 5453906; 11) 24642243; 12) 6321915; 13) 42742307; 14) 50083289; 15) 28574275; 16) 28574587; 17) 71986977; 18) 193209951; 19) 115534720; 20) 50726996; 21) 24653393; 22) 5174415.
Selection of CPES Candidates from the Insect Database
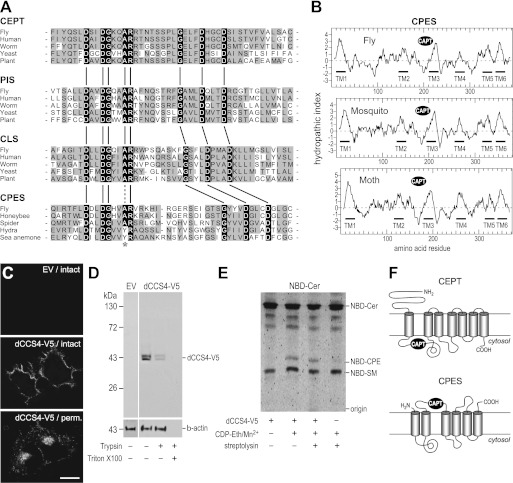
The reaction catalyzed by CPES is very similar to the one catalyzed by CDP-Eth:diacylglycerol Eth-phosphotransferase (EPT; EC 2.7.8.1) during PE formation via the Kennedy pathway, except that ceramide instead of diacylglycerol serves as acceptor of the phosphoethanolamine headgroup (Fig. 2A). EPT belongs to the superfamily of CDP-alcohol phosphotransferases (NCBI accession number cl00453). Members of this superfamily share the CDP-alcohol phosphotransferase (CAPT) sequence motif D(X)2DG(X)2(A/Y)R(X)8–16G(X)3D(X)3D. In human choline/ethanolamine phosphotransferase CEPT1, the final two aspartates of this motif are essential for catalysis, whereas the remainder of the conserved residues serves a role in substrate affinity or steric stability (26, 27).
The CDP-alcohol phosphotransferase superfamily includes six Drosophila proteins, with three of them showing high similarity to CEPT (CG33116, CG6016, and CG7149), one showing high similarity to PIS proteins (CG9245), and one showing high similarity to CLS proteins (CG4774), whereas one does not show similarity to any protein of known function (CG4585). We considered the possibility that one of the three CEPT-related proteins might have evolved into a CPES by a change of the acceptor substrate. This hypothesis appeared especially attractive because CG33116 and CG7149 (46% identity) have only one human orthologue and must therefore have originated from a gene duplication specific to the fly lineage (Fig. 2B, sequences 15 and 16). The protein of unknown function (CG4585) also represents an interesting CPES candidate as its homologues are present in insects and other arthropods, but not in most other animal phyla (Fig. 2B, sequence 3). Hence, we considered four CCS in Drosophila, namely three proteins from the CEPT subfamily (dCCS1/CG33116, dCCS2/CG6016 and dCCS3/CG7149) and one protein (dCCS4/CG4585) belonging to a novel protein family with a phylogenetic distribution mainly restricted to Arthropoda (Fig. 2B). These four proteins were next subjected to a detailed functional analysis.
Subcellular Distribution of CPES Candidates
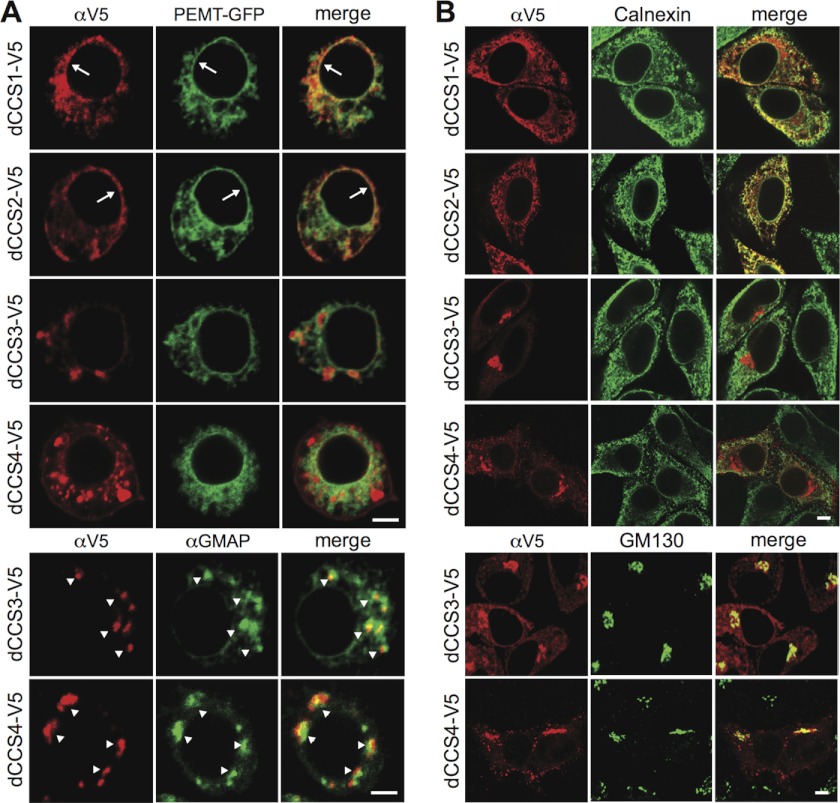
Because bulk production of CPE in Drosophila S2 cells occurs independently of dSMSr (18), it is likely mediated by CPES. Indeed, this would explain why mammalian cells, which lack the latter enzyme, contain only trace amounts of CPE. Efficient CPE production in Drosophila requires the ceramide transfer protein CERT (28), which mediates ER-to-Golgi transport of newly synthesized ceramides (29). This implies that CPES resides in the Golgi, so we first mapped the subcellular distributions of the four dCCS proteins. To this end, V5-tagged versions of these proteins were expressed in Drosophila S2 cells and localized by immunofluorescence microscopy. Both dCCS1-V5 and dCCS2-V5 localized exclusively to the ER, as evidenced by a reticular and nuclear envelope staining that overlapped extensively with the ER marker PE-methyltransferase (Fig. 2A, top). In contrast, the bulk of dCCS3-V5 and dCCS4-V5 was found in punctate structures containing the Golgi marker dGMAP, suggesting that these two proteins are at least partially associated with the Golgi (Fig. 3A, bottom). Some dCCS4-V5 was occasionally found at the plasma membrane, which is not unusual for Golgi proteins expressed at a high level. Localization studies in human HeLa cells produced very similar results; dCCS1 and dCCS2 localized exclusively to the ER, whereas dCCS3 and dCCS4 partially co-localized with the Golgi marker GM130 (Fig. 3B). Hence, unlike dCCS1 and dCCS2, dCCS3 and dCCS4 each meet at least one additional characteristic of CPES.
FIGURE 3.
Subcellular distribution of candidate CPE synthases. A, confocal sections of Drosophila S2 cells double-transfected with V5-tagged dCCS1, dCCS2, dCCS3, or dCCS4 and ER marker PE-methyltransferase-GFP. Cells were stained with anti-V5 antibody (top) or co-stained against the V5 epitope and Golgi marker dGMAP (bottom). Arrows indicate nuclear envelope staining, whereas arrowheads mark immunopositive Golgi structures. B, confocal sections of human HeLa cells transfected with V5-tagged dCCS1, dCCS2, dCCS3, or dCCS4 and stained with anti-V5 and anti-calnexin (ER) or anti-GM30 (Golgi) antibodies. Scale bars: 5 μm.
dCCS4 Corresponds to the Elusive CPES
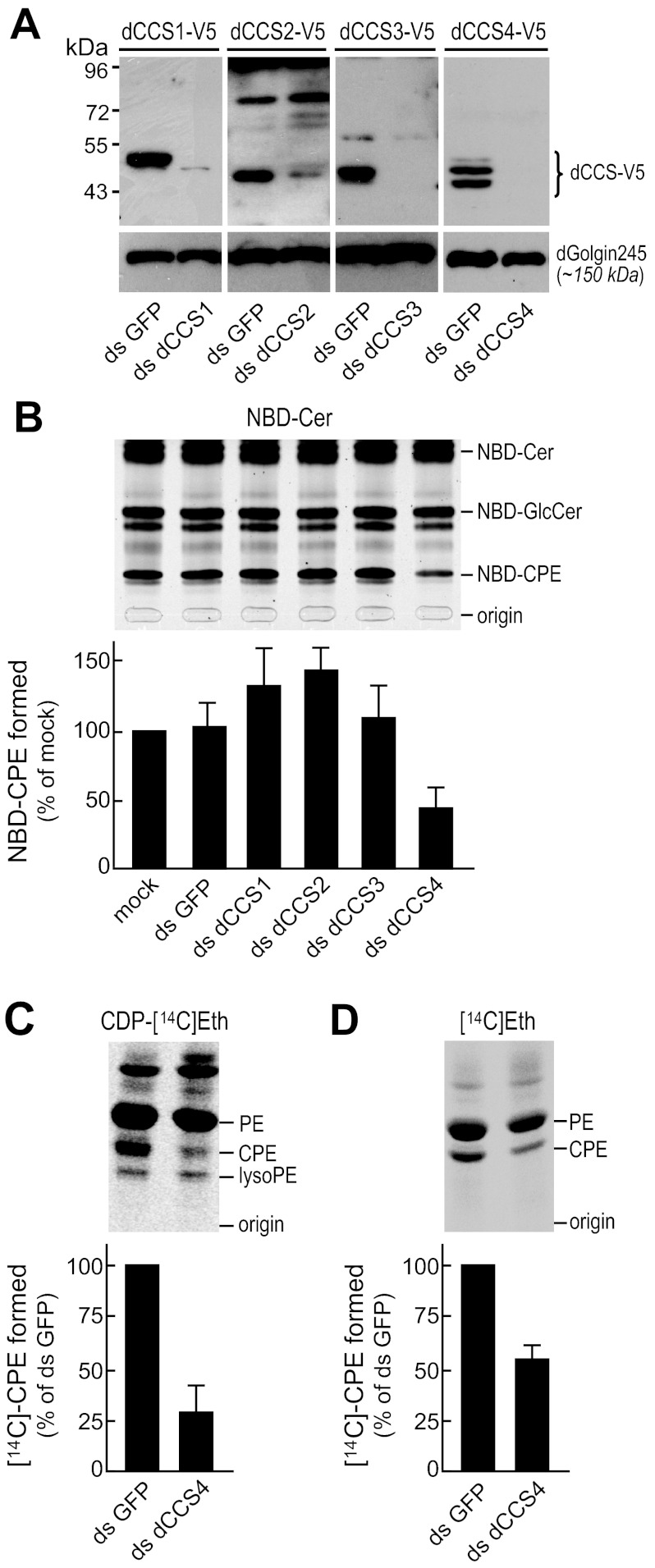
We next screened dCCS proteins for CPES activity. As a first approach, Drosophila S2 cells were treated with dCCS-targeting dsRNAs to deplete individual dCCS proteins, lysed, and then incubated with NBD-Cer in the presence of CDP-Eth and Mn2+ ions. Formation of NBD-CPE was monitored by TLC. dsRNA targeting GFP served as control. The efficiency of depletion was verified by immunoblotting of dsRNA-treated S2 cells expressing individual V5-tagged dCCS proteins (Fig. 4A). Contrary to removal of dCCS1, dCCS2, or dCCS3, depletion of dCCS4 caused a major (∼60%) reduction in CPES activity (Fig. 4B). When incubated with CDP-[14C]Eth in the presence of Mn2+ ions, lysates of dCCS4-depleted cells synthesized only a minor (∼25%) fraction of the radiolabeled CPE formed in lysates of control (dsGFP-treated) cells (Fig. 4C). In addition, loss of dCCS4 caused a substantial drop in de novo synthesis of CPE as monitored by metabolic labeling of S2 cells with [14C]Eth (Fig. 4D). This was accompanied by a defect in cell growth.5 Together, these results indicate that Drosophila S2 cells require dCCS4 for CDP-Eth-dependent CPE production and growth.
FIGURE 4.
dCCS4 is required for CDP-Eth-dependent CPE production in Drosophila S2 cells. A, immunoblots of Drosophila S2 cells transfected with V5-tagged dCCS1, dCCS2, dCCS3, or dCCS4 and then treated with dsRNA targeting individual dCCS proteins (ds dCCS1–4). Cells treated with dsRNA targeting GFP (ds GFP) served as control. Blots were stained with anti-V5 and anti-dGolgin245 antibodies. B, lysates of Drosophila S2 cells treated with dsRNA as in A were incubated with NBD-Cer in the presence of CDP-Eth, MnCl2, and UDP-glucose. The amount of NBD-CPE formed was determined by TLC analysis, normalized against NBD-GlcCer levels, and then expressed as the percentage of control (ds GFP-treated cells). Error bars: S.D., n = 3. C, lysates of Drosophila S2 cells depleted for dCCS4 (ds dCCS4) or mock-depleted for GFP (ds GFP) were incubated with CDP-[14C]ethanolamine and then subjected to lipid extraction, TLC analysis, and autoradiography. Levels of [14C]CPE were normalized against [14C]PE levels and expressed as the percentage of control (ds GFP-treated cells). Error bars: range, n = 2. D, Drosophila S2 cells depleted for dCCS4 (ds dCCS4) or mock-depleted for GFP (ds GFP) were metabolically labeled for 2 h at 27 °C with [14C]ethanolamine and then subjected to lipid extraction, TLC analysis, and autoradiography. Levels of 14C-labeled CPE were normalized against [14C]PE levels and expressed as percentage of control (ds GFP-treated) cells. Error bars: S.D., n = 3.
To investigate whether dCCS4 is not only required, but also directly responsible for CDP-Eth-dependent CPE formation, we next analyzed its ability to synthesize CPE in human HeLa cells. When added to HeLa cell lysates, NBD-Cer is converted to NBD-CPE by the CPE synthase SMSr (18) and the dual specificity SM/CPE synthase SMS2 (17). The addition of CDP-Eth has no effect on NBD-CPE formation because SMSr and SMS2 each use PE as headgroup donor. However, the addition of CDP-Eth to lysates of HeLa cells expressing dCCS4 caused a dramatic increase in NBD-CPE formation (Fig. 5A). This increase was strictly dependent on the presence of Mn2+ ions. Moreover, when incubated with NBD-Cer and CDP-[14C]Eth simultaneously, lysates of dCCS4-expressing HeLa cells, but not of control cells, supported formation of radiolabeled NBD-CPE (Fig. 5B). In sum, these findings indicate that dCCS4 corresponds to the elusive CDP-Eth:ceramide ethanolamine phosphotransferase or CPES in Drosophila.
FIGURE 5.
dCCS4 shows CDP-Eth:ceramide ethanolamine phosphotransferase activity. A, lysates of human HeLa cells transfected with V5-tagged dCCS4 or empty vector (EV) were incubated with NBD-Cer in the absence or presence of CDP-Eth and MnCl2 and then subjected to lipid extraction and TLC analysis. B, lysates of HeLa cells transfected with V5-tagged dCCS4 or empty vector (EV) were incubated with NBD-Cer in the presence of CDP-[14C]Eth and MnCl2. Lipids were extracted, separated by TLC, and then analyzed for fluorescence (left) and radioactivity (right). Note that only dCCS4p-expressing cells synthesized NBD-CPE that was labeled with 14C.
CPES Structure and Topology
Homologues of CPES occur in a variety of Arthropoda, including flies, mosquitoes, bees, spiders, and mites. In addition, CPES homologues are present in at least two species of Cnidaria: i.e. Hydra and sea anemones. All CPES homologues contain a highly conserved CAPT motif (Fig. 6A). However, what distinguishes the CAPT motif in CPES from those present in CEPT, PIS, and CLS proteins is the insertion of 7–8 additional amino acid residues between the invariant Arg and the second invariant Gly residue. Although no tertiary structure is available for any of these enzymes, it is conceivable that this change in spacing corresponds to a change in specificity for the acceptor substrate, namely from diacylglycerol (for CEPT, PIS, and CLS) to ceramide (for CPES).
FIGURE 6.
Structure and membrane topology of CPES and related enzymes. A, alignment showing the conserved amino acid motif D(X)2DG(X)2(A/Y)R(X)8–16G(X)3D(X)3D in the CAPT domain of various members of the CDP-alcohol phosphotransferase superfamily. Note the change in spacing between the two halves of the motif in CLS and CPES. The conserved Ala residue in the CAPT motif of CDP-alcohol phosphotransferases is substituted by a Tyr residue in CPES from Hydra and sea anemone (asterisk), which might have to do with the use of a phosphonate analogue of CDP-Eth as substrate in Cnidaria. Protein sequences were aligned with T-Coffee (35) for each family separately (CEPT, PIS, CLS, CPES). The alignment was trimmed manually and displayed with prettyplot from the EMBOSS package (36). NCBI GI numbers or UniProtKB identifiers are 24653393, 5174415, 193209951, 6321915, and 15222885 for CEPT; 24642243, 5453906, 6325370, 17537129, and 15220618 for PIS; 23172318, 10092647, 71984834, 6320059, and 18412722 for CLS; and O77475, 110750730 (translation of bases 327125–328027), B5M6F6, 204837661 (translation of bases 20637–21278), and A7S4T3 for CPES. B, hydrophobicity plots of CPES homologues from D. melanogaster (fly), Aedes aegypti (mosquito), and Bombyx mori (moth) created according to Kyte and Doolittle (37). Positions of putative transmembrane domains (TM1–6) and the CAPT motif are indicated. C, confocal sections of HeLa cells transfected with V5-tagged dCCS4 or empty vector (EV) and stained with anti-V5 antibodies before (intact) or after saponin-mediated membrane permeabilization (perm.). Scale bar: 15 μm. D, HeLa cells transfected with V5-tagged dCCS4 or empty vector (EV) were treated with trypsin in the presence or absence of Triton X-100 and then subjected to immunoblot analysis using anti-V5 and anti-β-actin antibodies. E, HeLa cells transfected with V5-tagged dCCS4 or empty vector were incubated at 15 °C with NBD-Cer in the presence or absence of CDP-Eth, MgCl2, and streptolysin. NBD-labeled lipids formed on the cell surface were extracted by BSA, separated by TLC, and analyzed for fluorescence. F, schematic view of the predicted membrane topologies of CEPT (27) and CPES.
Apart from the CAPT motif, CPES does not display any obvious sequence similarity with other members of the CDP-alcohol phosphotransferase superfamily. Hydrophobicity analysis using a combination of different methods (Octopus (Stockholm Bioinformatics Center); TMHMM Server v. 2.0; Phobius (Stockholm Bioinformatics Center)) predicted six membrane-spanning α helices connected by hydrophilic regions that would form extramembrane loops (Fig. 6B). The hydrophilic CAPT motif is situated between the second and third membrane span. The amino and carboxyl termini of CPES are predicted to face the luminal or exoplasmic space. This would position the CAPT motif in the Golgi lumen (Fig. 6F). When heterologously expressed in HeLa cells, a significant portion of dCCS4-V5 reaches the plasma membrane (Fig. 3B). According to the model (Fig. 6F), this would result in exposure of the C-terminal V5 epitope on the cell surface. To test this prediction, HeLa cells expressing dCCS4-V5 were immunostained with anti-V5 antibodies either before (Fig. 6C, intact) or after fixation and permeabilization (Fig. 6C, perm.). In line with the model, intact cells displayed a cell surface staining, whereas in permeabilized cells, both the plasma membrane and the Golgi were stained (Fig. 6C). Moreover, trypsinization of intact cells caused a substantial loss of V5-tagged dCCS4 (Fig. 6D), consistent with an exoplasmic orientation of the C-terminal V5 epitope. These results suggest that the active site of CPES is facing the luminal or exoplasmic space. To verify this directly, HeLa cells expressing dCCS4-V5 were analyzed for their ability to catalyze CDP-Eth-dependent CPE synthesis on the cell surface. For this approach, cells were incubated at 5 °C to block endocytosis, and NBD-Cer was added to the medium, which was supplemented with BSA to extract any newly formed NBD-labeled lipid from the cell surface. Under these conditions, cells synthesized NBD-SM and trace amounts of NBD-CPE because of the plasma membrane-associated and bifunctional SM/CPE synthase, SMS2 (17). However, the external addition of CDP-Eth and Mn2+ ions greatly stimulated NBD-CPE formation (Fig. 6E). This stimulation was strictly dependent on dCCS4. Permeabilization of the plasma membrane by streptolysin did not result in any further stimulation of CDP-Eth-dependent NBD-CPE formation (Fig. 6E). From these results, we conclude that dCCS4/CPES catalyzes CPE production on the exoplasmic side of the membrane, which is thus consistent with the model depicted in Fig. 6F.
Phylogenetic Distribution of CPES and SMS Enzymes
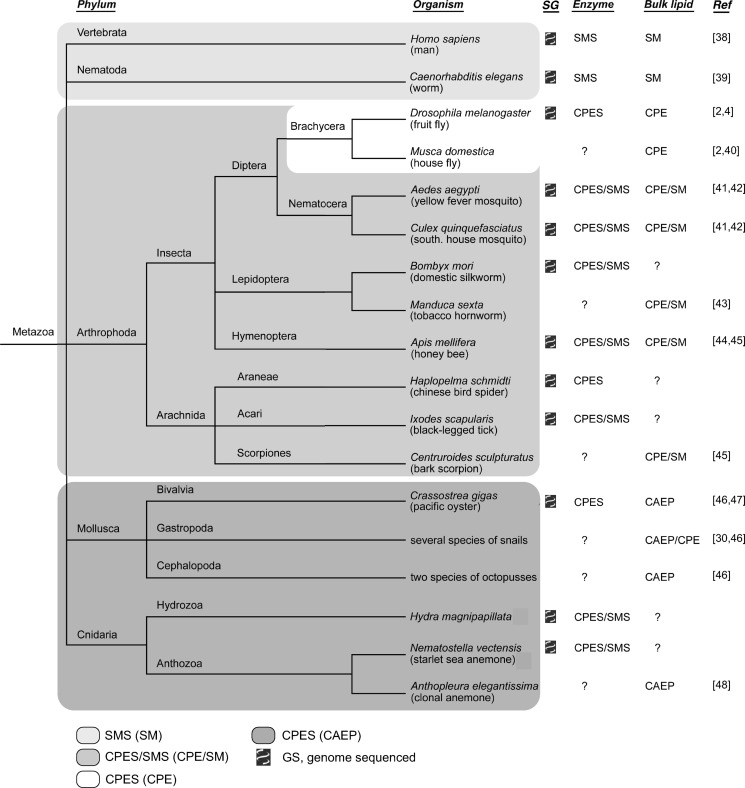
Our data predict that organisms containing a CPES homologue would produce bulk amounts of CPE, analogous to SM being an abundant membrane component in organisms harboring SMS. Indeed, as illustrated in Fig. 7, the presence of these enzymes in different organisms correlates well with their membrane lipid composition. Vertebrates and nematodes lack CPES homologues but contain multiple SMS enzymes (11). Accordingly, they have SM as their principal phosphosphingolipid and produce only trace amounts of CPE through SMSr and SMS2 (17, 18, 38, 39). Arthropods in general have CPES in addition to SMS and therefore produce bulk amounts of both CPE and SM (2, 4, 40–45). However, within the Arthropoda, the fly lineage (Brachycera) has lost SMS and thus the ability to synthesize SM. Flies therefore have CPE as their only phosphosphingolipid.
FIGURE 7.
Phylogenetic distribution of SM and CPE synthases in the animal kingdom. The phylogenetic distribution of SMS and CPES homologues in sequenced genomes correlates with the occurrence of their products SM, CPE, and CAEP (see under “Results and Discussion” for details). The phylogenetic tree was drawn with TreeViewX according to the classification in the NCBI/GenBankTM Taxonomy Browser. The presence of SMS and CPES genes was checked by a BLAST search (blastp or tblastn) in the NCBI/GenBank genomic BLAST databases, the NCBI/GenBank nonredundant protein database, UniProtKB, and the Silkworm Genome Database: SilkDB (B. mori).
Most Cnidaria, a phylum comprising sea anemones, corals, jellyfish, and Hydra, contain neither SM nor CPE. Instead, these organisms synthesize ceramide aminoethylphosphonate CAEP (Fig. 7) (48). Mollusca (clams, oysters, snails, octopuses) contain CAEP too, whereas the occurrence of CPE in addition to CAEP is restricted to several species of sea snails (30, 46, 47). CAEP is structurally very similar to CPE except for phosphate being replaced by phosphonate with a direct C–P bond connecting phosphonate and ethanolamine (30, 31). The nonhydrolyzable C–P bond would enhance stability of the lipid, especially with respect to phospholipases. It is conceivable that CPES mediates CAEP production in Cnidaria and Mollusca. Interestingly, the genomes of sea anemone, Hydra, and the pacific oyster Crassostrea gigas contain CPES homologues with Ala replaced by an aromatic amino acid (Tyr or Phe) in an otherwise perfectly conserved CAPT motif (Fig. 6A; data not shown). This amino acid substitution may be linked to the need to transfer aminoethylphosphonate instead of phosphoethanolamine from the donor to the acceptor substrate during CAEP biosynthesis. Free 2-aminoethylphosphonate, a likely precursor of CAEP, has been found in sea anemone (32), supporting this hypothesis. Based on the currently available sequence information, it appears likely that CPES in Mollusca catalyzes production of both CAEP and CPE.
Concluding Remarks
In this study, we identified CPES, a Golgi-resident enzyme responsible for bulk production of the SM analogue CPE in Drosophila. Strikingly, CPES is unrelated to members of the SMS family, which synthesize SM and trace amounts of CPE in vertebrates and nematodes. Instead, CPES shares a similar reaction mechanism with the ethanolamine phosphotransferase that mediates PE biosynthesis via the Kennedy pathway. Common features include: 1) the use of CDP-Eth as headgroup donor in the enzymatic reaction; 2) dependence on Mn2+ ions for catalytic activity; and 3) the presence of a CDP-alcohol phosphotransferase or CAPT motif. However, apart from the CAPT motif, CPES does not share any sequence similarity with other members of the CDP-alcohol phosphotransferase superfamily. CPES homologues occur in Arthropoda (insects, spiders, mites, scorpions), Cnidaria (Hydra, sea anemones), and Mollusca (oysters), but not in most other animal phyla (Fig. 7). Another feature that sets CPES apart from all previously identified CDP-alcohol phosphotransferases is that its active site appears to be situated on the exoplasmic surface of the membrane. This implies that CPE biosynthesis in Arthropoda, Cnidaria, and Mollusca relies on the presence of a membrane transporter involved in moving CDP-ethanolamine from the cytosol into the Golgi lumen. Although the identity of this transporter remains to be established, its presence in organisms containing CPES may explain why functional expression of this enzyme in mammalian cells is not sufficient to allow bulk production of CPE.5 Molecular cloning of the CDP-ethanolamine transporter is the subject of ongoing studies. The present identification of CPES provides a novel opportunity to address the biological role of CPE, an enigmatic lipid with a widespread occurrence in the animal kingdom.
Acknowledgments
We thank our colleagues at Membrane Enzymology for stimulating discussions and critical reading of the manuscript.
This article was selected as a Paper of the Week.
A. M. Vacaru, J. van den Dikkenberg, P. Ternes, and J. C. M. Holthuis, unpublished data.
- SM
- sphingomyelin
- SMS
- sphingomyelin synthase
- SMSr
- SMS-related
- CAEP
- ceramide aminoethylphosphonate
- CAPT
- CDP-alcohol phosphotransferase
- CCS
- candidate CPE synthase(s)
- CDP-Eth
- CDP-ethanolamine
- CEPT
- choline/ethanolamine phosphotransferase
- EPT
- ethanolamine phosphotransferase
- Cer
- ceramide
- CLS
- cardiolipin synthase
- CPE
- ceramide phosphoethanolamine
- CPES
- CPE synthase
- ER
- endoplasmic reticulum
- NBD
- 7-nitrobenz-2-oxa-1,3-diazol-4-yl
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PIS
- phosphatidylinositol synthase
- HBSS
- Hanks' balanced salt solution
- dSMSr
- Drosophila SMSr homologue
- dCCS
- Drosophila CCS homologue.
REFERENCES
- 1. van Meer G., Voelker D. R., Feigenson G. W. (2008) Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Acharya U., Acharya J. K. (2005) Enzymes of sphingolipid metabolism in Drosophila melanogaster. Cell. Mol. Life Sci. 62, 128–142 [DOI] [PubMed] [Google Scholar]
- 3. Kraut R. (2011) Roles of sphingolipids in Drosophila development and disease. J. Neurochem. 116, 764–778 [DOI] [PubMed] [Google Scholar]
- 4. Rietveld A., Neutz S., Simons K., Eaton S. (1999) Association of sterol- and glycosylphosphatidylinositol-linked proteins with Drosophila raft lipid microdomains. J. Biol. Chem. 274, 12049–12054 [DOI] [PubMed] [Google Scholar]
- 5. Holthuis J. C., Pomorski T., Raggers R. J., Sprong H., Van Meer G. (2001) The organizing potential of sphingolipids in intracellular membrane transport. Physiol. Rev. 81, 1689–1723 [DOI] [PubMed] [Google Scholar]
- 6. Guan X. L., Cestra G., Shui G., Kuhrs A., Schittenhelm R. B., Hafen E., van der Goot F. G., Robinett C. C., Gatti M., Gonzalez-Gaitan M., Wenk M. R. (2013) Biochemical membrane lipidomics during Drosophila development. Dev. Cell 24, 98–111 [DOI] [PubMed] [Google Scholar]
- 7. Carvalho M., Sampaio J. L., Palm W., Brankatschk M., Eaton S., Shevchenko A. (2012) Effects of diet and development on the Drosophila lipidome. Mol. Syst. Biol. 8, 600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Térová B., Heczko R., Slotte J. P. (2005) On the importance of the phosphocholine methyl groups for sphingomyelin/cholesterol interactions in membranes: a study with ceramide phosphoethanolamine. Biophys. J. 88, 2661–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Voelker D. R., Kennedy E. P. (1982) Cellular and enzymic synthesis of sphingomyelin. Biochemistry 21, 2753–2759 [DOI] [PubMed] [Google Scholar]
- 10. Huitema K., van den Dikkenberg J., Brouwers J. F., Holthuis J. C. (2004) Identification of a family of animal sphingomyelin synthases. EMBO J. 23, 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tafesse F. G., Ternes P., Holthuis J. C. (2006) The multigenic sphingomyelin synthase family. J. Biol. Chem. 281, 29421–29425 [DOI] [PubMed] [Google Scholar]
- 12. Yamaoka S., Miyaji M., Kitano T., Umehara H., Okazaki T. (2004) Expression cloning of a human cDNA restoring sphingomyelin synthesis and cell growth in sphingomyelin synthase-defective lymphoid cells. J. Biol. Chem. 279, 18688–18693 [DOI] [PubMed] [Google Scholar]
- 13. Tafesse F. G., Huitema K., Hermansson M., van der Poel S., van den Dikkenberg J., Uphoff A., Somerharju P., Holthuis J. C. (2007) Both sphingomyelin synthases SMS1 and SMS2 are required for sphingomyelin homeostasis and growth in human HeLa cells. J. Biol. Chem. 282, 17537–17547 [DOI] [PubMed] [Google Scholar]
- 14. Malgat M., Maurice A., Baraud J. (1986) Sphingomyelin and ceramide-phosphoethanolamine synthesis by microsomes and plasma membranes from rat liver and brain. J. Lipid Res. 27, 251–260 [PubMed] [Google Scholar]
- 15. Malgat M., Maurice A., Baraud J. (1987) Sidedness of ceramide-phosphoethanolamine synthesis on rat liver and brain microsomal membranes. J. Lipid Res. 28, 138–143 [PubMed] [Google Scholar]
- 16. Maurice A., Malgat M., Baraud J. (1989) Sidedness of ceramide-phosphoethanolamine synthesis on rat liver plasma membrane. Biochimie 71, 373–378 [DOI] [PubMed] [Google Scholar]
- 17. Ternes P., Brouwers J. F., van den Dikkenberg J., Holthuis J. C. (2009) Sphingomyelin synthase SMS2 displays dual activity as ceramide phosphoethanolamine synthase. J. Lipid Res. 50, 2270–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vacaru A. M., Tafesse F. G., Ternes P., Kondylis V., Hermansson M., Browers J. F. H. M., Somerharju P., Rabouille C., Holthuis J. C. (2009) Sphingomyelin synthase-related protein SMSr controls ceramide homeostasis in the ER. J. Cell. Biol. 185, 1013–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kondylis V., van Nispen tot Pannerden H. E., Herpers B., Friggi-Grelin F., Rabouille C. (2007) The Golgi comprises a paired stack that is separated at G2 by modulation of the actin cytoskeleton through Abi and Scar/WAVE. Dev. Cell. 12, 901–915 [DOI] [PubMed] [Google Scholar]
- 20. Clemens J. C., Worby C. A., Simonson-Leff N., Muda M., Maehama T., Hemmings B. A., Dixon J. E. (2000) Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. U.S.A. 97, 6499–6503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bligh E. G., Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 22. Kondylis V., Rabouille C. (2003) A novel role for dp115 in the organization of tER sites in Drosophila. J. Cell. Biol. 162, 185–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vance J. E. (2008) Phosphatidylserine and phosphatidylethanolamine in mammalian cells: two metabolically related aminophospholipids. J. Lipid Res. 49, 1377–1387 [DOI] [PubMed] [Google Scholar]
- 24. Henneberry A. L., McMaster C. R. (1999) Cloning and expression of a human choline/ethanolaminephosphotransferase: synthesis of phosphatidylcholine and phosphatidylethanolamine. Biochem. J. 339, 291–298 [PMC free article] [PubMed] [Google Scholar]
- 25. Horibata Y., Hirabayashi Y. (2007) Identification and characterization of human ethanolaminephosphotransferase1. J. Lipid Res. 48, 503–508 [DOI] [PubMed] [Google Scholar]
- 26. Williams J. G., McMaster C. R. (1998) Scanning alanine mutagenesis of the CDP-alcohol phosphotransferase motif of Saccharomyces cerevisiae cholinephosphotransferase. J. Biol. Chem. 273, 13482–13487 [DOI] [PubMed] [Google Scholar]
- 27. Henneberry A. L., Wright M. M., McMaster C. R. (2002) The major sites of cellular phospholipid synthesis and molecular determinants of Fatty Acid and lipid head group specificity. Mol. Biol. Cell. 13, 3148–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rao R. P., Yuan C., Allegood J. C., Rawat S. S., Edwards M. B., Wang X., Merrill A. H., Jr., Acharya U., Acharya J. K. (2007) Ceramide transfer protein function is essential for normal oxidative stress response and lifespan. Proc. Natl. Acad. Sci. U.S.A. 104, 11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hanada K., Kumagai K., Yasuda S., Miura Y., Kawano M., Fukasawa M., Nishijima M. (2003) Molecular machinery for non-vesicular trafficking of ceramide. Nature 426, 803–809 [DOI] [PubMed] [Google Scholar]
- 30. Hori T., Sugita M., Arakawa I. (1968) Structural elucidation of sphingoethanolamine and its distribution in aquatic animals. Biochim. Biophys. Acta 152, 211–213 [PubMed] [Google Scholar]
- 31. Kariotoglou D. M., Mastronicolis S. K. (2003) Sphingophosphonolipid molecular species from edible mollusks and a jellyfish. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 136, 27–44 [DOI] [PubMed] [Google Scholar]
- 32. Kittredge J. S., Roberts E., Simonsen D. G. (1962) The occurrence of free 2-aminoethylphosphonic acid in the sea anemone, Anthopleura elegantissima. Biochemistry 1, 624–628 [DOI] [PubMed] [Google Scholar]
- 33. Morgenstern B. (1999) DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics 15, 211–218 [DOI] [PubMed] [Google Scholar]
- 34. Trooskens G., De Beule D., Decouttere F., Van Criekinge W. (2005) Phylogenetic trees: visualizing, customizing, and detecting incongruence. Bioinformatics 21, 3801–3802 [DOI] [PubMed] [Google Scholar]
- 35. Notredame C., Higgins D. G., Heringa J. (2000) T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217 [DOI] [PubMed] [Google Scholar]
- 36. Rice P., Longden I., Bleasby A. (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277 [DOI] [PubMed] [Google Scholar]
- 37. Kyte J., Doolittle R. F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
- 38. Barenholz Y., Gatt S. (1982) in: Phospholipids (Hawthorne J. N., Ansell G. B., eds) Vol. 4, pp. 129–177, Elsevier Biomedical, Amsterdam [Google Scholar]
- 39. Satouchi K., Hirano K., Sakaguchi M., Takehara H., Matsuura F. (1993) Phospholipids from the free-living nematode Caenorhabditis elegans. Lipids 28, 837–840 [DOI] [PubMed] [Google Scholar]
- 40. Crone H. D., Bridges R. G. (1963) The phospholipids of the housefly, Musca domestica. Biochem. J. 89, 11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang T. K., Means E., Anderson L. E., Jenkin H. M. (1974) Sphingophospholipids of species of Aedes and Culex mosquito cells cultivated in suspension culture from logarithmic and stationary phases of growth. Lipids 9, 1009–1013 [DOI] [PubMed] [Google Scholar]
- 42. Luukkonen A., Brummer-Korvenkontio M., Renkonen O. (1973) Lipids of cultured mosquito cells (Aedes albopictus). Comparison with cultured mammalian fibroblasts (BHK 21 cells). Biochim. Biophys. Acta. 326, 256–261 [DOI] [PubMed] [Google Scholar]
- 43. Abeytunga D. T., Glick J. J., Gibson N. J., Oland L. A., Somogyi A., Wysocki V. H., Polt R. (2004) Presence of unsaturated sphingomyelins and changes in their composition during the life cycle of the moth Manduca sexta. J. Lipid Res. 45, 1221–1231 [DOI] [PubMed] [Google Scholar]
- 44. Karlander S. G., Karlsson K. A., Leffler H., Lilja A., Samuelsson B. E., Steen G. O. (1972) The structure of sphingomyelin of the honey bee (Apis mellifera). Biochim. Biophys. Acta 270, 117–131 [DOI] [PubMed] [Google Scholar]
- 45. O'Connor J. D., Polito A. J., Monroe R. E., Sweeley C. C., Bieber L. L. (1970) Characterization of invertebrate sphingolipid bases: occurrence of eicosasphinga-4, 11-dienine and eicosasphing-11-enine in scorpion. Biochim. Biophys. Acta 202, 195–197 [DOI] [PubMed] [Google Scholar]
- 46. Hori T., Arakawa I., Sugita M. (1967) Distribution of ceramide 2-aminoethylphosphonate and ceramide aminoethylphosphate (sphingoethanolamine) in some aquatic animals. J. Biochem. 62, 67–70 [DOI] [PubMed] [Google Scholar]
- 47. Le Grand F., Kraffe E., Marty Y., Donaghy L., Soudant P. (2011) Membrane phospholipid composition of hemocytes in the Pacific oyster Crassostrea gigas and the Manila clam Ruditapes philippinarum. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 159, 383–391 [DOI] [PubMed] [Google Scholar]
- 48. Simon G., Rouser G. (1967) Phospholipids of the sea anemone: Quantitative distribution; absence of carbon-phosphorus linkages in glycerol phospholipids; structural elucidation of ceramide aminoethylphosphonate. Lipids 2, 55–59 [DOI] [PubMed] [Google Scholar]