Background: The flavoenzyme quinone reductase 2 (NQO2) has an unknown cellular function and binds many drugs and bioactive molecules.
Results: Reduction of FAD and binding of chloroquine cause a change in NQO2 conformation.
Conclusion: NQO2 functions as a chloroquine-dependent flavin redox switch.
Significance: A flavin redox switch function of NQO2 could explain off-target effects of chloroquine and other bioactive molecules.
Keywords: Enzyme Inhibitors, Flavin, Protein Conformation, Protein Structure, Reductase, Chloroquine, Flavin Redox Switch, Malaria, Off-target Drug Interaction, Primaquine
Abstract
Quinone reductase 2 (NQO2) is an FAD-linked enzyme and the only known human target of two antimalarial drugs, primaquine (PQ) and chloroquine (CQ). The structural differences between oxidized and reduced NQO2 and the structural basis for inhibition by PQ and CQ were investigated by x-ray crystallography. Structures of oxidized NQO2 in complex with PQ and CQ were solved at 1.4 Å resolution. CQ binds preferentially to reduced NQO2, and upon reduction of NQO2-CQ crystals, the space group changed from P212121 to P21, with 1-Å decreases in all three unit cell dimensions. The change in crystal packing originated in the negative charge and 4–5º bend in the reduced isoalloxazine ring of FAD, which resulted in a new mode of CQ binding and closure of a flexible loop (Phe126–Leu136) over the active site. This first structure of a reduced quinone reductase shows that reduction of the FAD cofactor and binding of a specific inhibitor lead to global changes in NQO2 structure and is consistent with a functional role for NQO2 as a flavin redox switch.
Introduction
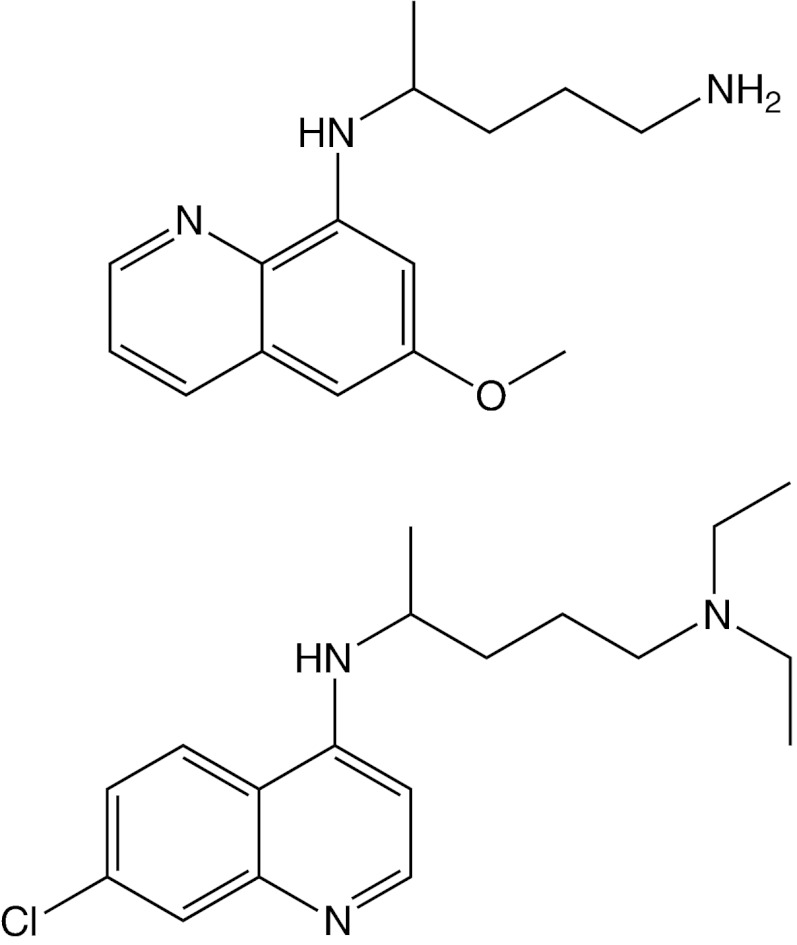
Despite 6 decades of routine use in treating malaria, the pharmacologic mechanisms of primaquine (PQ)3 and chloroquine (CQ) are not completely understood, although it is known that the two antimalarial drugs work at different stages of the Plasmodium life cycle. Malaria begins when Plasmodium sporozoites gain entry to the bloodstream via a bite from an infected mosquito, after which the parasite infects liver cells. During this liver stage infection, the 8-aminoquinoline, PQ (Fig. 1), acts as a hepatic schizontocide to clear infection (1). If left untreated, the parasites mature into merozoites and are released into the bloodstream to infect erythrocytes. In erythrocytes, the parasites either spawn into more merozoites or mature into gametocytes capable of infecting mosquitoes through additional bites. During the erythrocytic stage of infection, the 4-aminoquinoline, CQ (Fig. 1), acts as a blood schizontocide to clear infection (2). As such, PQ and CQ are routine medications for treating malaria. In recent years, however, the emergence of chloroquine-resistant Plasmodium falciparum has rendered CQ ineffective in most endemic areas. PQ, on the other hand, is still widely used as a prophylaxis of malaria because parasites rarely exhibit resistance against this drug.
FIGURE 1.
Structures of primaquine and chloroquine. Primaquine (top) and chloroquine (bottom) are two antimalarial drugs that act at different stages of the Plasmodium life cycle.
In terms of therapeutic mechanisms, PQ is thought to work by generating toxic oxygen species via its reactive metabolites (3). CQ, on the other hand, is thought to prevent a detoxification of excess heme in the parasite, which is generated as a result of its metabolism of hemoglobin (3). Despite these findings, no protein target has been identified for either drug in the parasite, causing some controversy about the mode of action of the two drugs (4, 5). This led Graves et al. (6) to search for target proteins of CQ and PQ using a proteomics approach. Surprisingly, no parasitic protein targets were identified; instead, two human proteins present in infected erythrocytes, quinone reductase 2 (NQO2) and aldehyde dehydrogenase 1, were identified. Of the two proteins, NQO2 was potently inhibited by concentrations of the quinoline drugs used in clinical treatment, whereas aldehyde dehydrogenase 1 was not. It is not known whether the inhibition of NQO2 by PQ and/or CQ plays a role in the antimalarial effects of the drugs.
The identification of NQO2 as a human target of PQ and CQ is intriguing because both drugs are used to treat non-malarial diseases. For instance, PQ is used to treat pneumocystis pneumonia and is active against leishmaniasis and trypanosomiasis in vivo (7–9). Similarly, CQ is used to treat rheumatoid arthritis, lupus erthosyma, and amoebic hepatitis; is under clinical trials for use in HIV-1/AIDS; and is being investigated for use in cancer chemotherapy (10–12). Thus, the inhibition of NQO2 may contribute to the therapeutic effects of PQ and CQ against diseases other than malaria.
NQO2 is a cytosolic and ubiquitously expressed metalloflavoprotein that catalyzes the two-electron reduction of quinone substrates. It has been the subject of extensive study over the last 15 years and binds a number of bioactive compounds, including imatinib (13, 14), inhibitors against CKII and PKC (15, 16), resveratrol (17), and melatonin (18, 19); however, the only natural substrate identified is ubiquinone, and the physiological function of NQO2 is not understood (20). NQO2 is a member of the thioredoxin family of enzymes but is unique in that it uses dihydronicotinamide riboside (NRH) as a reducing coenzyme rather than NADH or NADPH. The oxidized form of the co-enzyme, nicotinamide riboside, is involved in NAD metabolism (21), but the cellular source of the reduced form, NRH, is not known, and it is not clear why NQO2 has evolved to use NRH. In addition to an enzymatic role in quinone reduction, NQO2 stabilizes the p53 tumor suppressor against 20S proteasomal degradation in the presence of NRH (22, 23). Furthermore, NQO2 is capable of generating reactive oxygen species (24). Yet the current understanding of NQO2 and its inhibition does not adequately explain the effects of PQ and CQ on malaria and non-malarial diseases.
PQ and CQ inhibit NQO2 with different mechanisms. Kinetic studies show that PQ and CQ both inhibit NQO2 in the micromolar range, with KI values of 1.0 and 0.6 μm, respectively (25), but PQ exhibits competitive inhibition against the reducing co-factor (NRH), whereas CQ competes with the quinone substrate. This pattern of inhibition can be explained by the ping-pong kinetic mechanism that NQO2 uses. The tightly bound FAD co-enzyme is first reduced by NRH to FADH2; nicotinamide riboside is replaced by the quinone substrate, which is then reduced by FADH2. As such, NQO2 can exist as either the oxidized (NQO2ox) or reduced (NQO2red) form, and inhibitors may have a higher affinity for one or the other.
To provide a structural basis for the difference in specificity between PQ and CQ, we solved high resolution crystal structures of NQO2ox in complex with either PQ or CQ as well as NQO2red in complex with CQ. The NQO2red-CQ complex is particularly interesting because CQ is present in a completely different orientation compared with the NQO2ox-CQ complex, and this difference in binding is accompanied by movement of an active site loop, a change in crystal packing, and a change in space group symmetry. The structure of the NQO2red-CQ complex provides support for the idea that, in common with some other flavin-containing proteins (26), NQO2 acts as a redox-sensitive “switch” that is responsive to the presence of cellular NRH in conjunction with binding of certain bioactive compounds.
MATERIALS AND METHODS
Protein Expression and Purification
Recombinant NQO2 was expressed in Escherichia coli and purified as described previously (27). A critical step in the purification was full reconstitution of the enzyme with the FAD cofactor. When expressed in E. coli, recombinant NQO2 typically contains substoichiometric levels of flavin mononucleotide (FMN) and no FAD. A partial denaturation of NQO2 and reconstitution with FAD was incorporated into the purification procedure, leading to full saturation of the enzyme with FAD, as reported previously (27).
Crystallization of NQO2
NQO2ox was co-crystallized with 1 mm PQ or CQ by hanging drop vapor diffusion against reservoirs containing 0.1 m HEPES, pH 7.5, and 1.3–2.0 m (NH4)2SO4. To prepare NQO2red-CQ crystals, crystals of NQO2-CQ were soaked in 1 μl of reducing-soak solution consisting of 0.1 m HEPES, pH 7.5, 2.0 m (NH4)2SO4, 10 mm 1-(3-sulfonatopropyl)-3-carbamoyl-1,4-dihydropyrimidine (SCDP; an NRH analog), and 1 mm chloroquine, for 2 min; the soak was repeated until crystals turned pale yellow. Whereas larger rod-shaped crystals (0.5 mm × 0.2 mm) cracked when introduced into the reducing soak, medium size crystals (0.3 mm × 0.1 mm) remained intact. The reduced crystals were transferred to a soak of 1 mm chloroquine, 2.0 m (NH4)2SO4, 0.1 m Hepes, pH 7.5, to decrease the of the amount of SCDP, which may be competitive with CQ at high concentrations. Crystals were then briefly passed through a cryoprotectant solution (20% glycerol, 2.0 m (NH4)2SO4, 0.1 m Hepes, pH 7.5) and plunged into liquid nitrogen. To prevent autoxidation of NQO2, the entire crystal mounting process from harvesting to cryocooling was performed under an anoxic atmosphere in a glove bag purged with N2. To prepare for the reduction protocol, the glove bag was initially evacuated and flushed with nitrogen for 5 min. All soaking solutions were then deoxygenated by sparging with nitrogen gas inside the glove bag. In the dry nitrogen atmosphere, crystals and crystal soak solutions quickly evaporated; therefore, to maintain an appropriate level of humidity, the nitrogen used to flush the glove bag was bubbled through a flask of crystallization reservoir solution (0.1 m HEPES, pH 7.5, 2.0 m (NH4)2SO4).
X-ray Data Collection, Refinement, and Analysis
Data were collected at the CMCF-1 beamline of the Canadian Light Source, processed using XDS (28) or MOSFLM (29), and merged using Scala (30). The structures were solved by molecular replacement with Protein Data Bank entry 1QR2 as a starting model (31); refinement was carried out using PHENIX (32). Given the high resolution of the crystallographic data, NCS restraints were not used for any of the refinements.
Topology files for PQ and CQ were generated using PHENIX.ELBOW in conjunction with small molecule crystal structures of the two compounds (33, 34). For refinement of the NQO2red structure, the FAD topology file was modified to allow bending along the N5–N10 axis of FAD: the planarity restraints incorporating N5 and N10 of the isoalloxazine ring were removed so that it was separated into two planes, one incorporating the pyrimidine ring and the second incorporating the benzyl ring. Also, the bond length, bond angle, and dihedral angle restraints involving N5 and N10 were removed. The degree of bending in the isoalloxazine ring was calculated by finding equations for the two planes using principal component analysis incorporating all atoms in each of the two planes. The final structures for NQO2ox-PQ, NQO2ox-CQ, and NQO2red-CQ were deposited in the Protein Data Bank as 4FGJ, 4FGK, and 4FGL respectively.
To validate the observed changes in FAD structure and, in particular, to assess the contribution of the FAD and FADH2 stereochemical restraint files to the structure of the isoalloxazine ring, each model was refined using an identical simulated annealing strategy with the FADH2 stereochemical restraint file to assess the degree of bending along the N5–N10 axis. In each case, the refinement was performed three times using the same starting model with a different random seed to calculate a final average bending angle. To ensure an “active” convergence to an optimal conformation, FAD in the starting model of NQO2 was bent to an angle of ∼30°.
RESULTS
Oxidized NQO2-Primaquine Complex
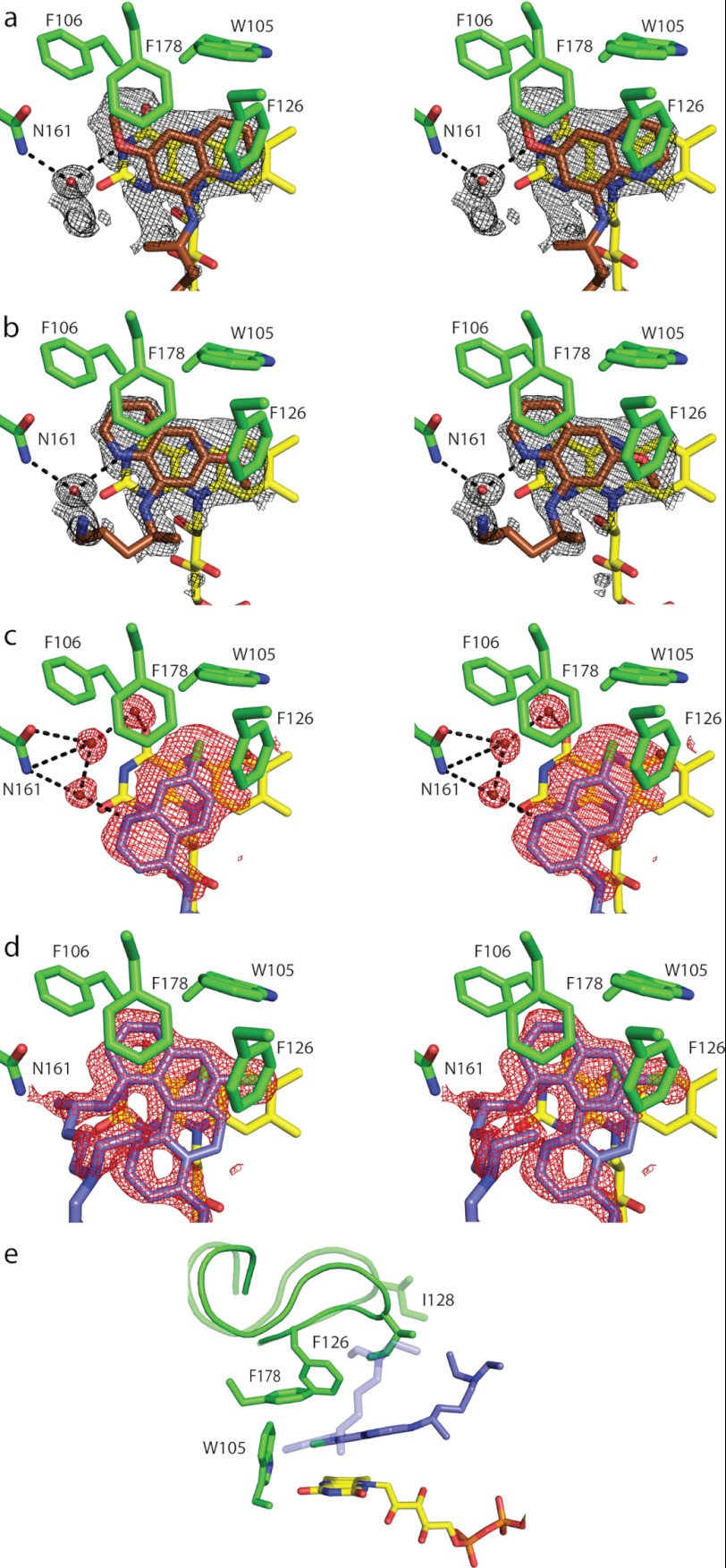
NQO2 was co-crystallized with PQ in space group P212121. Crystals of NQO2-PQ diffracted to 1.35 Å and contained a homodimer in the asymmetric unit; the structure was solved by molecular replacement and refined to an Rfree value of 0.167 (Table 1). Primaquine was initially modeled in a single orientation in both active sites of the homodimer (Fig. 2a), and after several rounds of refinement, a second orientation was incorporated to fully account for the electron density (Fig. 2b). The occupancies for bound PQ molecules refined to values of ∼0.5 for each of the conformations, indicating that PQ binds with roughly equal affinity in each mode. In both orientations, the quinoline ring of PQ is buried in the active site, making planar stacking interactions with the FAD isoalloxazine ring and the phenyl ring of Phe178. PQ also makes contact with additional aromatic residues (Trp105, Phe106, and Phe126), which form the interior wall of the active site. As well, both orientations of PQ make a water-mediated hydrogen bond with the side chain of Asn161. In the first orientation of PQ (Fig. 2a), a water molecule (position 557 in A chain and 450 in B chain) mediates hydrogen bonding between the aromatic nitrogen in the quinoline ring and the side chain of Asn161. In the alternate orientation (Fig. 2b), the quinoline ring is flipped 180°, and the water molecule mediates a hydrogen bond between the 6-methoxy group of PQ and the side chain of Asn161.
TABLE 1.
Crystallographic data collection and refinement statistics
| Oxidized NQO2 crystal |
Reduced NQO2 crystal (chloroquine ligand) | ||
|---|---|---|---|
| Primaquine ligand | Chloroquine ligand | ||
| Wavelength (Å) | 1.033217 | 1.033217 | 0.97949 |
| Space group | P212121 | P212121 | P21 |
| Unit cell dimensions (Å) | 56.5, 83.0, 106.5 | 56.37, 83.11, 106.6 | 54.31, 105.71, 82.00 β = 90.17° |
| Resolution | 30.1-1.35 | 32.7-1.40 | 48.3-1.20 |
| Rsyma | 0.066 (0.63) | 0.065 (0.50) | 0.041 (0.288) |
| I/σ(I)a | 10.4 (2.4) | 14.5 (3.7) | 19.3 (4.4) |
| Completenessa | 99.5 (98.8) | 99.6 (99.0) | 99.8 (99.9) |
| Redundancy | 4.8 | 7.3 | 3.7 |
| Unique reflections | 110,793 | 98,537 | 287,298 |
| Rwork/Rfree | 0.1413/0.1666 | 0.1325/0.1745 | 0.1059/0.1362 |
| Ramachandran plotb (%) | |||
| Most favored | 90.7 | 89.9 | 89.9 |
| Additionally allowed | 8.8 | 9.5 | 9.6 |
| Generously allowed | 0.5 | 0.5 | 0.4 |
| Disallowed | 0 | 0 | 0.1 |
| Root mean square deviations | |||
| Bond lengths (Å) | 0.008 | 0.007 | 0.009 |
| Bond angles (degrees) | 1.255 | 1.223 | 1.174 |
| Dihedral angles (degrees) | 14.594 | 14.471 | 14.188 |
| Mean atomic displacement parameter values (Å2) | |||
| Protein | |||
| All atoms | 12.815 | 18.083 | 12.691 |
| Main chain | 11.659 | 16.253 | 11.336 |
| Side chain | 13.790 | 19.642 | 13.829 |
| Solvent | 24.975 | 28.55 | 25.312 |
| FAD | 12.315 | 17.265 | 11.447 |
| Inhibitor | 17.453 | 26.280 | 22.062 |
a Values in parentheses refer to the highest resolution shell.
b Ramachandran plot statistics were calculated using PROCHECK (53).
FIGURE 2.
Binding of primaquine and chloroquine to oxidized NQO2. a–d, stereodiagrams illustrating the binding of PQ and CQ to NQO2ox. The electron density represents the final 2Fo − Fc maps contoured at 1σ around the inhibitors. a and b show PQ bound to the A subunit in two alternate conformations. The B subunit of the NQO2ox-PQ homodimer contained similar electron density and was modeled in the same way as the A subunit. c and d show the active sites for the NQO2ox-CQ structure. Electron density in the NQO2ox-CQ subunit A (c) was different from that observed in the B subunit (d), and CQ was modeled in two alternate conformations in the B subunit. E, the two positions of CQ in the B subunit correspond to two different conformations of the active site loop, comprising residues 126–136. The active site loop in the “open” conformation (light shading) accommodates CQ when it is deeply buried in the active site, whereas the “closed” conformation (dark shading) is adopted when CQ is bound in a more peripheral location. One-letter amino acid codes are used.
Oxidized NQO2-Chloroquine Complex
NQO2 catalysis proceeds by a ping-pong mechanism. The reducing co-factor, NRH, binds in the active site, reduces the FAD, and then dissociates; the quinone substrate then enters the active site and undergoes a two-electron reduction. Thus, NQO2 can exist in either an oxidized (NQO2ox) or reduced (NQO2red) state, and inhibitors of NOQ2 will exhibit a preference for one or the other. Kinetic studies indicate that CQ exhibits competitive inhibition against the quinone substrate and therefore binds preferentially to NQO2red (25). However, high concentrations of CQ will also compete against the reducing co-enzyme, NRH, indicating that CQ also has affinity with NQO2ox. To fully characterize CQ binding to NQO2, we co-crystallized NQO2ox in complex with CQ. CQ-NQO2ox crystals diffracted to 1.4 Å, and the structure was refined to an Rfree of 0.175 (Table 1).
Electron density for the bound CQ was initially quite poor, but a large peak (greater than 10σ) in the difference (Fo − Fc) electron density map was used to position the CQ chlorine atom, after which the rest of the CQ molecule was placed in the remaining electron density. For the “A” subunit of the homodimer, CQ is present in one position, with the quinoline ring less deeply buried than primaquine and the aminopentane arm projecting toward the solvent (Fig. 2c). In the active site of the “B” subunit, additional difference density was observed after several rounds of manual adjustment and refinement, and an alternate orientation of CQ was modeled into the structure (Fig. 2d). The occupancies for the two modes of CQ binding to the B subunit refined to 0.52 and 0.48. In conjunction with these two positions for CQ, an active site loop consisting of residues 126–136 was modeled in two different conformations (Fig. 2e). In the position of CQ unique to the B subunit, in which the quinoline ring is more deeply buried in the active site, the bulky aminopentane arm of CQ interferes with the ability of the 126–136 loop to close over the active site, and therefore the loop adopts an “open” conformation (Fig. 2e). There is no evidence of cooperative interactions or nonequivalence between the two subunits of NQO2, and therefore this minor difference in CQ binding between the A and B subunits probably originates from different crystal packing environments.
Reduced NQO2-Chloroquine Complex
The reduced form of NQO2 (NQO2red) was not sufficiently stable to allow for co-crystallization with CQ. To obtain crystals of NQO2red in complex with CQ, crystals of NQO2ox-CQ were treated with SCDP (an NRH analog) in an anoxic environment until the crystal was bleached, indicating reduction of FAD to FADH2, and then frozen and stored in liquid nitrogen for data collection. Data from three such crystals of NQO2red-CQ were collected and processed (Table 2). In all three cases, the crystallographic data for NQO2red could be processed using the P212121 space group, with unit cell dimensions ∼1 Å shorter in all three directions compared with those for NQO2ox-CQ. However, data processed in P212121 yielded unacceptably high Rsym values, making it clear that the crystals had undergone a structural transition to a lower symmetry space group with four rather than two protomers in the asymmetric unit. Data processed and merged in P21 yielded acceptable Rsym values (Table 2), and the highest resolution data set was used for refinement and analysis (Table 1). The NQO2 structure underwent several rounds of manual rebuilding and refinement to yield an Rfree of 0.17. Analysis of the crystallographic data indicated the presence of pseudomerohedral twinning, and further refinement in Phenix using the twin operator (h, −k, −l) yielded an estimated twin fraction of 0.15, with a final Rfree value of 0.136 and improved electron density maps. This is the first structure of a reduced quinone reductase, and the combined effects of FAD reduction and chloroquine binding are described below.
TABLE 2.
Space group change upon reduction of NQO2-chloroquine crystals
| Crystal 1a (1.24 Å resolution) |
Crystal 2a (1.35 Å resolution) |
Crystal 3a,b (1.15 Å resolution) |
||||
|---|---|---|---|---|---|---|
| P212121c | P21c | P212121c | P21c | P212121c | P21c | |
| Unit cell | ||||||
| a | 53.75 | 53.67 | 53.90 | 53.68 | 54.24 | 54.29 |
| b | 81.68 | 105.61 | 81.84 | 105.56 | 82.02 | 105.71 |
| c | 105.67 | 81.68 | 105.66 | 81.66 | 105.71 | 82.01 |
| β | 90 | 90.14 | 90 | 90.35 | 90 | 90.16 |
| Rsym | 0.171 | 0.070 | 0.327 | 0.061 | 0.347 | 0.061 |
a Crystals of NQO2ox in complex with chloroquine were soaked with SCDP, an NRH analogue, to effect reduction of the FAD cofactor.
b Crystal 3 was used for structure solution, refinement, and detailed analysis.
c Data were indexed, processed, and merged in either the P212121 or P21 space group using Mosflm and Scala.
Chloroquine Binding to Reduced NQO2
The reduced isoalloxazine ring is expected to be deprotonated and carry a negative charge when it is bound to NQO2. In solution, N1 of FADH2 exhibits a pKa of 6.8 (35); when FAD is bound to NQO2, N1 accepts a hydrogen bond from the amide nitrogen of Gly149, making it highly unlikely that N1 is protonated in NQO2red. Instead, the negative charge, which would be delocalized between N1 and O2, is neutralized by two hydrogen bonds donated to O2, one from the ring hydroxyl of Tyr155 and a second from the amide of Gly150.
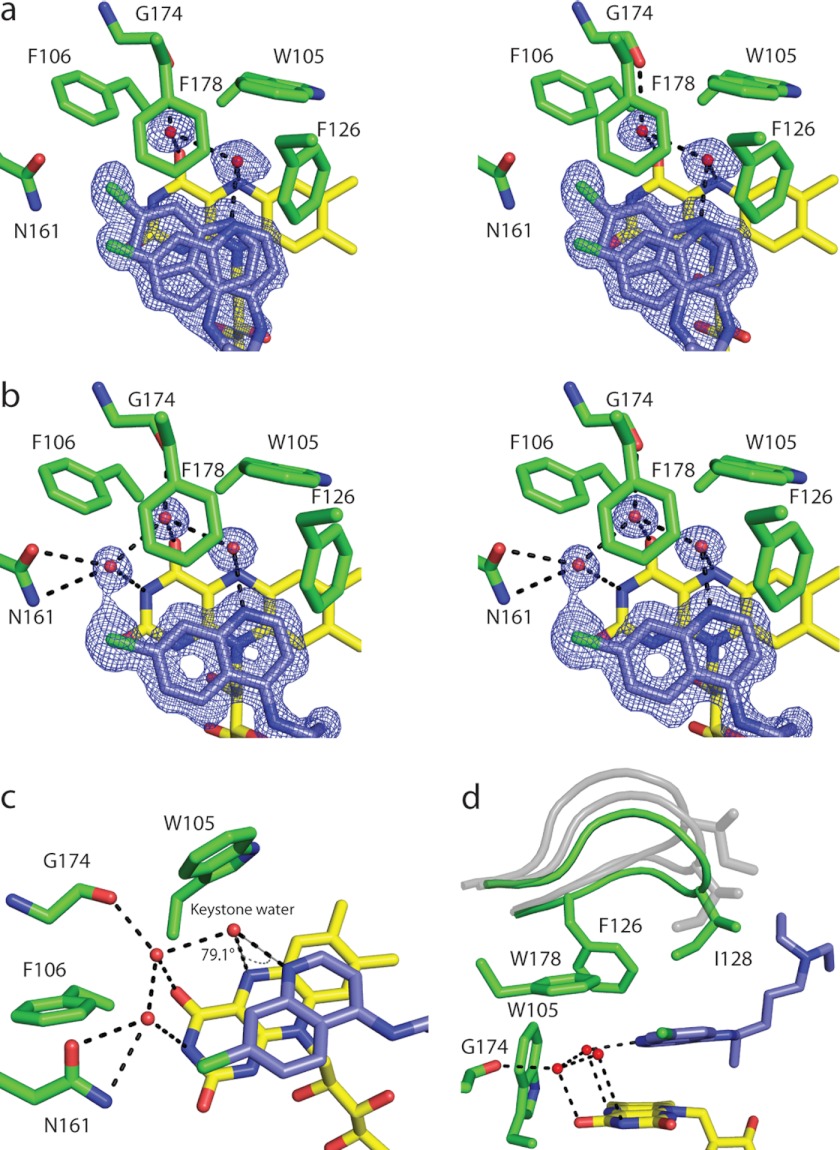
Reduction of FAD drastically changes the nature of its interaction with CQ. In NQO2ox, CQ tends to be rather deeply buried and able to make direct contact with aromatic residues that line the back wall of the active site (Fig. 2c), with the chlorine atom positioned above C5A of the isoalloxazine ring. In the NQO2red structure, the quinoline ring of CQ has flipped so that the opposite face interacts with the surface of reduced FAD, and the chlorine atom is positioned roughly above O2 on the periphery of the isoalloxazine ring (Fig. 3, a and b). The mode of CQ binding in NQO2red allows CQ to participate in a water-mediated hydrogen-bonding network that is centered on the proton on N5, a characteristic feature of reduced FAD (Fig. 3c). The keystone for this network is a water molecule (HOH 32, 312, 204, and 92 in chains A, B, C, and D, respectively) that accepts hydrogen bonds from the N5 proton of FAD and also from the proton on N4 of the CQ quinoline ring. The keystone water donates a hydrogen bond to a second water molecule (HOH 149, 154, 157, and 63 in chains A, B, C, and D, respectively) that in turn donates hydrogen bonds to O4 of the FAD and to the carbonyl oxygen of Gly174. The relative positions of these water molecules are conserved in all four subunits of the asymmetric unit. The geometry of the interaction between the keystone water and the FAD N5 atom deviates from what would be expected for a good hydrogen bond; the angle between CQ-N4, the keystone water, and FAD-N5 is 79 ± 1° (mean ± S.D. for four subunits) rather than a more typical angle of ∼110°. In addition to these hydrogen bonds, the aromatic rings of Trp105, Phe106, Phe126, and Phe178 enclose the two water molecules; indeed, the keystone water appears to donate a hydrogen bond to the indole ring of Trp105, whereas the second water may donate a hydrogen bond to the benzene ring of Phe178 (36).
FIGURE 3.
Binding of chloroquine to reduced NQO2. a and b, stereodiagrams illustrating the binding of CQ to the NQO2red active sites in the two dimers of the asymmetric unit; the electron density corresponds to 2Fo − Fc maps contoured at 1σ around CQ. a, the electron density for AB dimer. Shown is the active site of the A subunit, with CQ bound in two alternate positions, one of which corresponds to the position observed in the C and D subunits, whereas the second is shifted slightly. The electron density and positions of bound CQ were similar for the B subunit. b, the electron density and position of bound CQ in the CD dimer. Shown is the active site of the C subunit; the electron density in the D subunit was similar, and for both subunits, CQ was modeled in the same single position. c, the hydrogen bonding network, with the “keystone water” bridging N4 of CQ and N5 of the FAD isoalloxazine ring. d, the active site loop (residues 126–136) is shown in the fully closed conformation (green) observed in NQO2red-CQ; in this state, residue Ile128 makes van der Waals contact with CQ. For reference, the conformations of the loop observed in crystals of oxidized NQO2 are shown in gray. One-letter amino acid codes are used.
The 2Fo − Fc electron density for bound CQ is extremely well defined in one of the NQO2 dimers (C and D chains) of the asymmetric unit, and there is clear density for a third water molecule in the active sites of the CD dimer (Fig. 3b). This third water molecule donates a hydrogen bond to the second water and to the chlorine atom on CQ, and it accepts a hydrogen bond from the side-chain amide nitrogen of Asn161. In the second dimer (A and B chains) of the asymmetric unit, there was additional electron density for the bound CQ, and therefore a second CQ molecule was included in a slightly different orientation from the first (Fig. 3a). For the A and B subunits, the third water molecule was not present. This minor difference in CQ binding between the two reduced NQO2 dimers in the asymmetric unit may be a product of their interaction with each other or may be due to differences in the surrounding crystal packing environment.
Regarding the stereochemistry of the bound inhibitors, it is noteworthy that both CQ and PQ have a single stereocenter and exist as racemic mixtures. For the structures of NQO2ox with bound PQ or CQ, the electron density for the aminopentane arm was generally weak, and it was not possible to discern whether a single enantiomer was selectively bound by the protein. In the case of NQO2red, however, the (R)-CQ enantiomer was built into the structure, and there was enough electron density for the aminopentane arm to exclude binding of the (S)-CQ enantiomer. In this regard, binding of CQ to reduced NQO2 may be stereoselective for the (R)-enantiomer.
Structural Changes in FAD
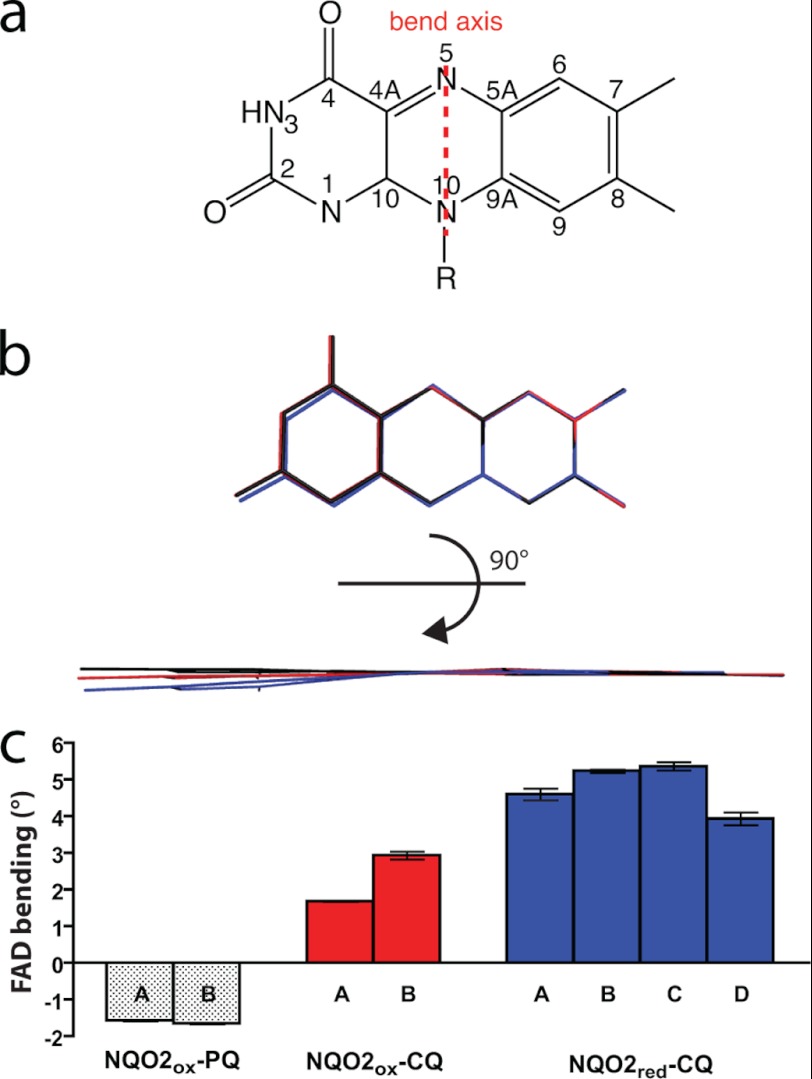
Crystals of NQO2ox in complex with CQ were consistently isomorphous with other oxidized forms, indicating that the CQ itself is not sufficient for the observed structural change in crystals of the NQO2red-CQ complex. Instead, the origin of the change in structure is the reduction of the FAD cofactor. The FAD isoalloxazine ring is planar, but reduction should lead to a “butterfly bend” along the N5–N10 axis (37) (Fig. 4). The electron density of the isoalloxazine ring in NQO2red-CQ indicated that the bending upon reduction is relatively minor. Refinement using a modified stereochemical restraint set for FAD, in which planar restraints and associated bond and angle restraints around N5 and N10 were removed, indicated an angle of ∼4–5° between the two planes of the “butterfly wings” (Fig. 4).
FIGURE 4.
Structural changes in the FAD isoalloxazine ring. a, chemical structure and atom designations in the FAD isoalloxazine ring, and the axis for the “butterfly bend” that is brought about by reduction of FAD to FADH2. b, an edge-on view of the structure of the isoalloxazine ring for the following: NQO2ox with PQ (black), NQO2ox with CQ (red), and NQO2red with CQ (blue). c, binding of PQ induces a slight (−1.6°) concave bend in the isoalloxazine system, whereas binding of CQ induces a convex bend of ∼2.3°. Reduction of FAD further increases this bend to 4.8°. To determine the bend in the isoalloxazine rings, the crystal structures were subjected to simulated annealing without stereochemical restraints at the N5 and N10 positions. FAD bending along the N5–N10 axis (butterfly bend) was calculated using the atomic positions of the dimethylbenzene and pyrimidine “wings” and principal component analysis to find the angle between the two best fit planes. Error bars, S.D.
To validate that the small change in the FAD isoalloxazine structure was due to reduction of the FAD, and not simply removal of stereochemical restraints, the refined NQO2 structures were subjected to simulated annealing refinement against data from both oxidized and reduced crystals. In the absence of stereochemical restraints around N5 and N10, the isoalloxazine ring of NQO2ox in complex with PQ refined to an average bend of −1.6°; for NQO2ox in complex with CQ, the average bend was 2.3°; whereas for NQO2red in complex with CQ, the bend was 4.8° (Fig. 4c). It is noteworthy that for NQO2ox, PQ brings about a small “upward” bend in the ring, forming a very slight concave binding surface, whereas with CQ, the isoalloxazine system bends in the opposite direction, with CQ binding to a slightly convex surface. Reduction of the FAD further enhances this bend by ∼2–3°. Other notable changes occur around N5, which moves toward a tetrahedral configuration in NQO2red; the C4A-N5-C5A bond angle was decreased from ∼116–117° in NQO2ox-CQ and NQO2ox-PQ to 112° in NQO2red-CQ (Table 3). In contrast, there was no significant difference in the C10-N10-C9A angle between the oxidized and reduced forms. Thus, binding of CQ to NQO2ox appears to push the isoalloxazine ring toward a slightly bent conformation, and reduction of FAD to FADH2 results in a further increase in the bend as well as the transition of N5 toward a tetrahedral configuration.
TABLE 3.
Conformational changes in the isoalloxazine ring upon reduction
| NQO2 | Inhibitor | Subunit | N5-N10 benda,b | C4A-N5-C5A anglea | C10-N10-C9A anglea |
|---|---|---|---|---|---|
| Oxidized | Primaquine | A | −1.56 ± 0.01 | 117.1 ± 0.2 | 120.0 ± 0.1 |
| B | −1.64 ± 0.00 | 116.9 ± 0.1 | 118.6 ± 0.1 | ||
| Average | −1.60 | 117.0 | 119.3 | ||
| Chloroquine | A | 1.69 ± 0.00 | 118.2 ± 0.2 | 118.9 ± 0.1 | |
| B | 2.94 ± 0.11 | 113.8 ± 0.2 | 116.5 ± 0.2 | ||
| Average | 2.32 | 116.0 | 117.7 | ||
| Reduced | Chloroquine | A | 4.61 ± 0.28 | 112.2 ± 0.5 | 117.6 ± 0.3 |
| B | 5.24 ± 0.05 | 112.1 ± 0.4 | 118.8 ± 0.5 | ||
| C | 5.36 ± 0.19 | 112.3 ± 1.2 | 116.1 ± 0.2 | ||
| D | 3.94 ± 0.30 | 113.1 ± 1.1 | 117.9 ± 0.2 | ||
| Average | 4.79 | 112.4 | 117.6 |
a Both oxidized and reduced structures were subjected to three separate rounds of simulated annealing refinement in the absence of planar restraints for N5-N10 axis and angle restraints for the C4A-N5-C5A/C10-N10-C9A angles.
b The “butterfly” bend along the N5-N10 axis was calculated using the atomic positions of the dimethylbenzene and pyrimidine “wings” and principal component analysis to find the angle between the two best fit planes.
Conformational Changes in Reduced NQO2-Chloroquine
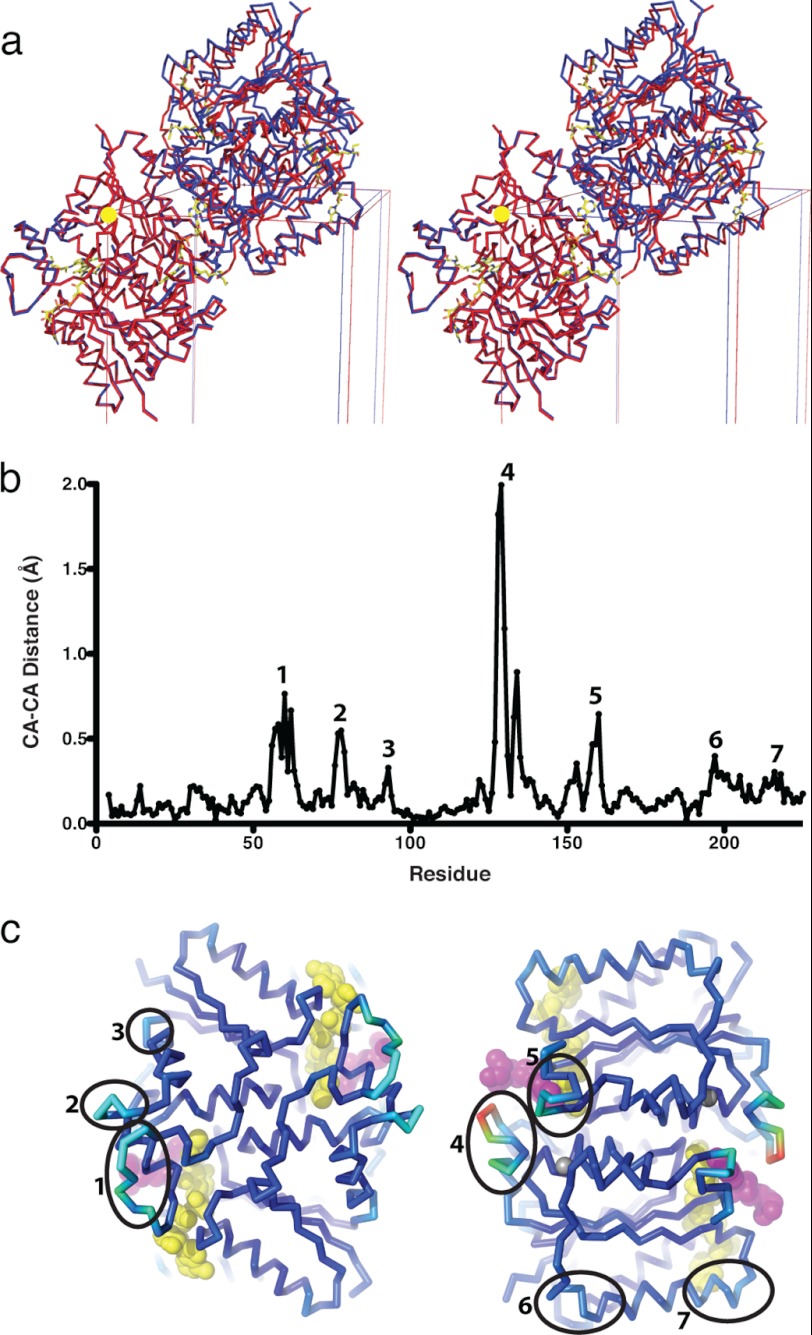
The monoclinic crystals of NQO2red-CQ contained four NQO2 protomers, two functional dimers, in the asymmetric unit, rather than the single functional dimer observed in the asymmetric unit of orthorhombic crystals of NQO2ox. Thus, reduction and CQ binding caused a reorientation of the two dimers, removing the crystallographic symmetry that related them in orthorhombic crystals of NQO2ox (Fig. 5a).
FIGURE 5.
Conformational changes in NQO2 upon reduction and binding of CQ. a, stereodiagram illustrating the relationship between NQO2 in the oxidized (red) and reduced (blue) states. CA traces of two dimers are shown, with the origin of the unit cells indicated by a yellow sphere; the FAD molecules are drawn as stick representations. To show the changes that take place in the crystal, the dimer from the orthorhombic (P212121) NQO2ox-CQ crystal has been superimposed on one of the dimers from the monoclinic (P21) crystal of NQO2red-CQ. In orthorhombic crystals of NQO2ox, there is a single dimer in the asymmetric unit, and the second dimer shown is related by crystallographic symmetry. When NQO2-CQ is reduced (blue), the relationship between the two dimers changes, and the crystallographic symmetry is broken, resulting in a monoclinic space group with two dimers in the asymmetric unit. b, a plot of differences in average CA positions between protomers of oxidized NQO2 (NQO2ox-PQ and NQO2ox-CQ) and protomers of NQO2red-CQ. c, two views of the NQO2 dimer (CA trace) colored from blue to red according to the magnitude of the difference in average CA position between reduced and oxidized NQO2-CQ. The views are related by 180° rotation around a vertical axis; the FADH− cofactor and bound CQ are represented as yellow and magenta CPK models. Regions of NQO2 (numbered 1–7) that demonstrate a significant shift upon reduction and binding of CQ (b) are indicated.
The active sites in the NQO2 dimer are at the interface of the two protomers. One of the protomers is largely responsible for binding FAD, whereas the second protomer contributes elements that cover the surface of the isoalloxazine ring. One such element is an irregular loop formed by residues 126–136 (Fig. 3d). In NQO2red-CQ, the loop closes over the active site, with Ile128 coming into direct contact with the CQ quinoline ring, which becomes sandwiched between the side chain of Ile128 and the isoalloxazine ring of FAD. This conformation is seen in the B, C, and D chains of the NQO2red-CQ structure, with the loop in a slightly more open conformation in the A chain.
The loop movement outlined above, as well as other changes in the structure that could explain the repacking of the crystal upon reduction of FAD and binding of CQ, are relatively small and difficult to distinguish from crystal packing artifacts. To assess the structural changes that take place upon reduction and binding of CQ, we made use of the four NQO2 protomers in the asymmetric unit of NQO2red-CQ to eliminate random changes in atomic positions and find regions that show a concerted difference from protomers in crystals of oxidized NQO2. All of the individual subunits from reduced and oxidized NQO2-CQ were superimposed, and average CA positions were calculated for each of the four oxidized protomers (two protomers from each of the NQO2ox-PQ and NQO2ox-CQ structures) and the four reduced protomers (contained in the NQO2red-CQ asymmetric unit). Differences in the average CA positions for the oxidized and reduced protomers were calculated, plotted, and mapped to the structure (Fig. 5, b and c). There were seven regions where consistent differences between the oxidized and reduced NQO2 subunits were observed. Two of these regions (regions 4 and 5 in Fig. 5) make direct contact with the bound chloroquine and comprise the 126–136 active site loop and an oddly structured connecting loop that includes N161; three other regions (regions 1, 2, and 3) form a surface patch that is not in direct contact with either the FAD cofactor or bound chloroquine; the last two regions (regions 6 and 7) are at the N- and C-terminal ends, respectively, of a surface-exposed helix comprising residues 196–212. Region 7 is in direct contact with the adenosine moiety of FAD but is still far removed from the active site. In summary, only two of seven regions that exhibit structural changes between NQO2ox and NQO2red-CQ are in direct contact with CQ and the isoalloxazine ring. On this basis, the combination of reduction and CQ binding appears to exert a global change in structure, consistent with the observed reorientation of NQO2 dimers within the crystal lattice.
DISCUSSION
NQO2 was identified as a human target of both CQ and PQ; kinetic studies showed that PQ binds preferentially to NQO2ox, whereas chloroquine binds to NQO2red (6, 25). Binding of inhibitors to NQO2red has not been studied, and the goal of the current work was to understand the structural basis for the difference in binding specificity. The NQO2red-CQ complex is the first structure of reduced NQO2, and it shows that the mode of CQ binding to NQO2red is completely different from what is observed for CQ binding to NQO2ox. The ring nitrogen of 4-aminoquinoline has a pKa of 9.2 (38), and therefore CQ will be protonated and positively charged at physiological pH; in contrast, the reduced form of the NQO2-bound isoalloxazine ring will have a negative charge, whereas the oxidized form will be neutral. The negative charge on the reduced isoalloxazine ring explains the preference of CQ for NQO2red and will contribute to the striking difference in the mode of CQ binding to NQO2red and NQO2ox.
The difference in CQ binding to NQO2ox and NQO2red is noteworthy but not as surprising and interesting as the effect of reduction and CQ binding on the overall structure of NQO2. There are 40 NQO2ox structures in the Protein Data Bank, most of which were crystallized as complexes with different inhibitors. 38 were crystallized in the same orthorhombic space group observed for the NQO2ox-PQ and NQO2ox-CQ complexes; one structure was crystallized in I422 with a single protomer in the asymmetric unit; and one structure was crystallized in P1 with two dimers in the asymmetric unit. The monoclinic space group and close packing of the two dimers observed for NQO2red-CQ are therefore unique and arise due to the reduction of FAD and chloroquine binding rather than random crystal packing effects. The unexpected conformational change in NQO2 provides a structural basis for the proposed role of quinone reductases as flavin redox switches (39, 40).
Flavin redox switches are FAD- or FMN-containing proteins in which the oxidation state of the cofactor regulates interactions with other proteins, nucleic acids, or membranes (26). The oxidation state of the cofactor is coupled to the conformation of the switch protein through a hydrogen-bonding network that incorporates the N5 atom of the isoalloxazine ring. For example, in the LOV family of light-sensing domains, light-induced reduction of the flavin involves reaction of a cysteine thiolate with C4A of the isoalloxazine ring; the ensuing protonation of N5 causes the side chain amide of a conserved glutamine residue to flip, and this change in hydrogen bonding in the vicinity of N5 is communicated to other parts of the protein (41, 42). In the case of NQO2, reduction of FAD leads to a unique mode of CQ binding; protonation of the isoalloxazine N5 allows it to donate a hydrogen bond to the “keystone” water that is held in place by a second hydrogen bond from CQ. Binding of CQ to NQO2red in this fashion stabilizes an active site loop in a “closed” conformation. Reduction and CQ binding also cause the isoalloxazine ring of FAD to bend by 4.8°. Because the active site of NQO2 is at the interface between the two protomers, the “local” changes in the active site affect the overall structure of the dimer. The reorientation of the dimers in the crystal lattice indicates global changes in structure and/or dynamics. Thus, the flavin switch function of NQO2 requires both reduction of FAD and binding of an appropriate ligand to the reduced isoalloxazine ring to stabilize the protein in an alternate conformation.
Although the cellular function of NQO2 is not understood, it binds many drugs and bioactive compounds and has been implicated in apoptosis and cancer (22, 43–45). A role as a flavin redox switch, the function of which is dependent on cellular redox conditions and the presence of an appropriate ligand, could explain the elusive nature of NQO2 cellular function. CQ is a good example of an NQO2-interacting compound that is used to treat a variety of diseases, but the mechanisms underlying the cellular effects of CQ are poorly understood. As an antimalarial, CQ is thought to interact with undimerized ferriprotoporphyrin and prevent its detoxification and storage as β-hematin (4). On the other hand, the process of hemoglobin digestion and storage of ferriprotoporphyrin by the parasite involves endocytosis and a number of steps in which endosomes must fuse with digestive vacuoles, and CQ and related quinolone drugs may interfere with these processes (4). Along these lines, the accumulation of CQ in lysosomes is thought to be responsible for its inhibition of autophagy (46). Furthermore, CQ has effects on glucocorticoid signaling and has been linked to p53 function (47, 48). It is conceivable that some of the cellular effects of CQ are linked to its binding of NQO2 and the ensuing conformational switch, which would only occur under an appropriate metabolic state where NQO2 is reduced.
Both NQO1 and NQO2 have been shown to protect p53 against 20S proteosomal degradation in the presence of their reduced nicotinamide coenzymes (22, 23, 49, 50). In fact, the stabilization and localization of a transcription factor is a common function of flavin redox switches. For example, the oxidized form of the flavin switch NifL forms an inactive complex with the transcription factor NifA; reduction of NifL leads to dissociation of the complex, allowing NifA to activate genes involved in nitrogen fixation. Lot6p, a yeast homologue of human NQO1 and NQO2 (51), provides an example of a quinone reductase that functions as a flavin redox switch; in this case, reduction of Lot6p helps it to stabilize the Yap4p transcription factor against 20S proteosomal degradation, similar to the way in which NQO1 and NQO2 appear to stabilize p53 (39, 52). Lot6p is actually a poor catalyst (the rate of the oxidative half-reaction is slower than that of non-enzymatic model reactions), and this led Sollner et al. (40) to propose that Lot6p may have evolved primarily as a flavin redox switch to regulate 20S proteosomal degradation. The conformational switch that we have observed with NQO2 provides a structural basis for the proposed role of NQO2 and possibly other quinone reductases in the redox-dependent regulation of p53.
As a flavin redox switch, it is not clear why NQO2 uses NRH as an electron donor instead of NAD(P)H, which is used by NQO1 and all other members of the flavodoxin family. NQO2 is the only enzyme known to use NRH, and although oxidized nicotinamide riboside exists as a metabolite of NAD, the origin of cellular NRH is not known. On this basis, the metabolic signal that facilitates conformational switching of NQO2 remains elusive. CQ has been reported to activate the p53 pathway, but whether this process involves NQO2 was not determined (48). In any case, there may be a number of small molecule “inhibitors” of NQO2, as well as proteins or peptides, that could interact specifically with the reduced form of NQO2 and function in a manner similar to CQ to stabilize NQO2 in an alternate conformation. Thus, although there are many unanswered questions regarding the cellular function of NQO2, a flavin switch function that is dependent on the cellular redox state and the presence of an appropriate ligand provides an intriguing, metabolically regulated, link between NQO2, p53, and the poorly defined cellular effects of the many drugs and bioactive compounds that interact with NQO2.
This work was supported by a Natural Sciences and Engineering Research Council Discovery Grant (to B. H. S.). Beamline CMCF-1 and the Canadian Light Source are supported by the Natural Sciences and Engineering Research Council of Canada, Canadian Institutes of Health Research, and the University of Saskatchewan.
The atomic coordinates and structure factors (codes 4FGJ, 4FGK, and 4FGL) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- PQ
- primaquine
- CQ
- chloroquine
- NQO2
- quinone reductase 2
- NQO2ox
- NQO2 oxidized form
- NQO2red
- NQO2 reduced form
- NRH
- dihydronicotinamide riboside
- SCDP
- 1-(3-sulfonatopropyl)-3-carbamoyl-1,4-dihydropyrimidine
- CA
- α-carbon.
REFERENCES
- 1. Hill D. R., Baird J. K., Parise M. E., Lewis L. S., Ryan E. T., Magill A. J. (2006) Primaquine. Report from CDC expert meeting on malaria chemoprophylaxis I. Am. J. Trop. Med. Hyg. 75, 402–415 [PubMed] [Google Scholar]
- 2. Lalloo D. G., Shingadia D., Pasvol G., Chiodini P. L., Whitty C. J., Beeching N. J., Hill D. R., Warrell D. A., Bannister B. A., and HPA Advisory Committee on Malaria Prevention in UK Travellers (2007) UK malaria treatment guidelines. J. Infect. 54, 111–121 [DOI] [PubMed] [Google Scholar]
- 3. Foley M., Tilley L. (1998) Quinoline antimalarials. Mechanisms of action and resistance and prospects for new agents. Pharmacol. Ther. 79, 55–87 [DOI] [PubMed] [Google Scholar]
- 4. Fitch C. D. (2004) Ferriprotoporphyrin IX, phospholipids, and the antimalarial actions of quinoline drugs. Life Sci. 74, 1957–1972 [DOI] [PubMed] [Google Scholar]
- 5. Vale N., Moreira R., Gomes P. (2009) Primaquine revisited six decades after its discovery. Eur. J. Med. Chem. 44, 937–953 [DOI] [PubMed] [Google Scholar]
- 6. Graves P. R., Kwiek J. J., Fadden P., Ray R., Hardeman K., Coley A. M., Foley M., Haystead T. A. (2002) Discovery of novel targets of quinoline drugs in the human purine binding proteome. Mol. Pharmacol. 62, 1364–1372 [DOI] [PubMed] [Google Scholar]
- 7. Benfield T., Atzori C., Miller R. F., Helweg-Larsen J. (2008) Second-line salvage treatment of AIDS-associated Pneumocystis jirovecii pneumonia. A case series and systematic review. J. Acquir. Immune Defic. Syndr. 48, 63–67 [DOI] [PubMed] [Google Scholar]
- 8. Berman J. D., Lee L. S. (1984) Activity of antileishmanial agents against amastigotes in human monocyte-derived macrophages and in mouse peritoneal macrophages. J. Parasitol. 70, 220–225 [PubMed] [Google Scholar]
- 9. Kinnamon K. E., Poon B. T., Hanson W. L., Waits V. B. (1996) Primaquine analogues that are potent anti-Trypanosoma cruzi agents in a mouse model. Ann. Trop. Med. Parasitol. 90, 467–474 [DOI] [PubMed] [Google Scholar]
- 10. Wallace D. J., Gudsoorkar V. S., Weisman M. H., Venuturupalli S. R. (2012) New insights into mechanisms of therapeutic effects of antimalarial agents in SLE. Nat. Rev. Rheumatol. 8, 522–533 [DOI] [PubMed] [Google Scholar]
- 11. Solomon V. R., Lee H. (2009) Chloroquine and its analogs: a new promise of an old drug for effective and safe cancer therapies. Eur. J. Pharmacol. 625, 220–233 [DOI] [PubMed] [Google Scholar]
- 12. Savarino A., Boelaert J. R., Cassone A., Majori G., Cauda R. (2003) Effects of chloroquine on viral infections. An old drug against today's diseases? Lancet Infect. Dis. 3, 722–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rix U., Hantschel O., Dürnberger G., Remsing Rix L. L., Planyavsky M., Fernbach N. V., Kaupe I., Bennett K. L., Valent P., Colinge J., Köcher T., Superti-Furga G. (2007) Chemical proteomic profiles of the BCR-ABL inhibitors imatinib, nilotinib, and dasatinib reveal novel kinase and nonkinase targets. Blood 110, 4055–4063 [DOI] [PubMed] [Google Scholar]
- 14. Winger J. A., Hantschel O., Superti-Furga G., Kuriyan J. (2009) The structure of the leukemia drug imatinib bound to human quinone reductase 2 (NQO2). BMC Struct. Biol. 9, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Duncan J. S., Gyenis L., Lenehan J., Bretner M., Graves L. M., Haystead T. A., Litchfield D. W. (2008) An unbiased evaluation of CK2 inhibitors by chemoproteomics. Characterization of inhibitor effects on CK2 and identification of novel inhibitor targets. Mol. Cell. Proteomics 7, 1077–1088 [DOI] [PubMed] [Google Scholar]
- 16. Brehmer D., Godl K., Zech B., Wissing J., Daub H. (2004) Proteome-wide identification of cellular targets affected by bisindolylmaleimide-type protein kinase C inhibitors. Mol. Cell. Proteomics 3, 490–500 [DOI] [PubMed] [Google Scholar]
- 17. Buryanovskyy L., Fu Y., Boyd M., Ma Y., Hsieh T. C., Wu J. M., Zhang Z. (2004) Crystal structure of quinone reductase 2 in complex with resveratrol. Biochemistry 43, 11417–11426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mailliet F., Ferry G., Vella F., Berger S., Cogé F., Chomarat P., Mallet C., Guénin S.-P., Guillaumet G., Viaud-Massuard M.-C., Yous S., Delagrange P., Boutin J. A. (2005) Characterization of the melatoninergic MT3 binding site on the NRH:quinone oxidoreductase 2 enzyme. Biochem. Pharmacol. 71, 74–88 [DOI] [PubMed] [Google Scholar]
- 19. Calamini B., Santarsiero B. D., Boutin J. A., Mesecar A. D. (2008) Kinetic, thermodynamic and X-ray structural insights into the interaction of melatonin and analogues with quinone reductase 2. Biochem. J. 413, 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vella F., Ferry G., Delagrange P., Boutin J. A. (2005) NRH:quinone reductase 2. An enzyme of surprises and mysteries. Biochem. Pharmacol. 71, 1–12 [DOI] [PubMed] [Google Scholar]
- 21. Bogan K. L., Brenner C. (2008) Nicotinic acid, nicotinamide, and nicotinamide riboside. A molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu. Rev. Nutr. 28, 115–130 [DOI] [PubMed] [Google Scholar]
- 22. Gong X., Kole L., Iskander K., Jaiswal A. K. (2007) NRH:quinone oxidoreductase 2 and NAD(P)H:quinone oxidoreductase 1 protect tumor suppressor p53 against 20s proteasomal degradation leading to stabilization and activation of p53. Cancer Res. 67, 5380–5388 [DOI] [PubMed] [Google Scholar]
- 23. Khutornenko A. A., Roudko V. V., Chernyak B. V., Vartapetian A. B., Chumakov P. M., Evstafieva A. G. (2010) Pyrimidine biosynthesis links mitochondrial respiration to the p53 pathway. Proc. Natl. Acad. Sci. U.S.A. 107, 12828–12833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reybier K., Perio P., Ferry G., Bouajila J., Delagrange P., Boutin J. A., Nepveu F. (2011) Insights into the redox cycle of human quinone reductase 2. Free Radic. Res. 45, 1184–1195 [DOI] [PubMed] [Google Scholar]
- 25. Kwiek J. J., Haystead T. A., Rudolph J. (2004) Kinetic mechanism of quinone oxidoreductase 2 and its inhibition by the antimalarial quinolines. Biochemistry 43, 4538–4547 [DOI] [PubMed] [Google Scholar]
- 26. Becker D. F., Zhu W., Moxley M. A. (2011) Flavin redox switching of protein functions. Antioxid. Redox Signal. 14, 1079–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leung K. K., Litchfield D. W., Shilton B. H. (2012) Flavin adenine dinucleotide content of quinone reductase 2. Analysis and optimization for structure-function studies. Anal. Biochem. 420, 84–89 [DOI] [PubMed] [Google Scholar]
- 28. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leslie A. G. W., Powell H. R. (2007) Processing diffraction data with Mosflm. Evolving Methods Macromol. Crystallogr. 245, 41–51 [Google Scholar]
- 30. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 31. Foster C. E., Bianchet M. A., Talalay P., Zhao Q., Amzel L. M. (1999) Crystal structure of human quinone reductase type 2, a metalloflavoprotein. Biochemistry 38, 9881–9886 [DOI] [PubMed] [Google Scholar]
- 32. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rubin J. R., Swaminathan P., Sundaralingam M. (1992) Structure of the anti-malarial drug primaquine diphosphate. Acta Crystallogr. C 48, 379–382 [DOI] [PubMed] [Google Scholar]
- 34. Karle J. M., Karle I. L. (1988) Redetermination of the crystal and molecular structure of the antimalarial chloroquine bis(dihydrogenphosphate) dihydrate. Acta Crystallogr. C 44, 1605–1608 [DOI] [PubMed] [Google Scholar]
- 35. Clark W. M., Lowe H. J. (1956) Studies on oxidation-reduction. XXIV. Oxidation-reduction potentials of flavin adenine dinucleotide. J. Biol. Chem. 221, 983–992 [PubMed] [Google Scholar]
- 36. Levitt M., Perutz M. F. (1988) Aromatic rings act as hydrogen bond acceptors. J. Mol. Biol. 201, 751–754 [DOI] [PubMed] [Google Scholar]
- 37. Hemmerich P., Nagelschneider G., Veeger C. (1970) Chemistry and molecular biology of flavins and flavoproteins. FEBS Lett. 8, 69–83 [DOI] [PubMed] [Google Scholar]
- 38. Lange N. A. (1999) Lange's Handbook of Chemistry, 15th Ed. (Dean J. A., ed), p. 8.29, McGraw-Hill, Inc., New York [Google Scholar]
- 39. Sollner S., Schober M., Wagner A., Prem A., Lorkova L., Palfey B. A., Groll M., Macheroux P. (2009) Quinone reductase acts as a redox switch of the 20S yeast proteasome. EMBO Rep. 10, 65–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sollner S., Deller S., Macheroux P., Palfey B. A. (2009) Mechanism of flavin reduction and oxidation in the redox-sensing quinone reductase Lot6p from Saccharomyces cerevisiae. Biochemistry 48, 8636–8643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zoltowski B. D., Schwerdtfeger C., Widom J., Loros J. J., Bilwes A. M., Dunlap J. C., Crane B. R. (2007) Conformational switching in the fungal light sensor Vivid. Science 316, 1054–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Möglich A., Moffat K. (2007) Structural basis for light-dependent signaling in the dimeric LOV domain of the photosensor YtvA. J. Mol. Biol. 373, 112–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Long D. J., 2nd, Iskander K., Gaikwad A., Arin M., Roop D. R., Knox R., Barrios R., Jaiswal A. K. (2002) Disruption of dihydronicotinamide riboside:quinone oxidoreductase 2 (NQO2) leads to myeloid hyperplasia of bone marrow and decreased sensitivity to menadione toxicity. J. Biol. Chem. 277, 46131–46139 [DOI] [PubMed] [Google Scholar]
- 44. Ahn K. S., Gong X., Sethi G., Chaturvedi M. M., Jaiswal A. K., Aggarwal B. B. (2007) Deficiency of NRH:quinone oxidoreductase 2 differentially regulates TNF signaling in keratinocytes. Up-regulation of apoptosis correlates with down-regulation of cell survival kinases. Cancer Res. 67, 10004–10011 [DOI] [PubMed] [Google Scholar]
- 45. Yu K.-D., Di G.-H., Yuan W.-T., Fan L., Wu J., Hu Z., Shen Z.-Z., Zheng Y., Huang W., Shao Z.-M. (2009) Functional polymorphisms, altered gene expression and genetic association link NRH:quinone oxidoreductase 2 to breast cancer with wild-type p53. Hum. Mol. Genet. 18, 2502–2517 [DOI] [PubMed] [Google Scholar]
- 46. Rubinsztein D. C., Codogno P., Levine B. (2012) Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 11, 709–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. He Y., Xu Y., Zhang C., Gao X., Dykema K. J., Martin K. R., Ke J., Hudson E. A, Khoo S. K., Resau J. H., Alberts A. S., MacKeigan J. P., Furge K. A., Xu H. E. (2011) Identification of a lysosomal pathway that modulates glucocorticoid signaling and the inflammatory response. Sci. Signal. 4, ra44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim E. L., Wüstenberg R., Rübsam A., Schmitz-Salue C., Warnecke G., Bücker E.-M., Pettkus N., Speidel D., Rohde V., Schulz-Schaeffer W., Deppert W., Giese A. (2010) Chloroquine activates the p53 pathway and induces apoptosis in human glioma cells. Neuro-Oncology 12, 389–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Asher G., Lotem J., Cohen B., Sachs L., Shaul Y. (2001) Regulation of p53 stability and p53-dependent apoptosis by NADH quinone oxidoreductase 1. Proc. Natl. Acad. Sci. U.S.A. 98, 1188–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Asher G., Shaul Y. (2005) p53 proteasomal degradation. Poly-ubiquitination is not the whole story. Cell Cycle 4, 1015–1018 [DOI] [PubMed] [Google Scholar]
- 51. Vasiliou V., Ross D., Nebert D. W. (2006) Update of the NAD(P)H:quinone oxidoreductase (NQO) gene family. Hum. Genomics 2, 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sollner S., Macheroux P. (2009) New roles of flavoproteins in molecular cell biology. An unexpected role for quinone reductases as regulators of proteasomal degradation. FEBS J. 276, 4313–4324 [DOI] [PubMed] [Google Scholar]
- 53. Laskowski R. A., MacArthur M., Moss D. S., Thornton J. M. (1993) PROCHECK. A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283–291 [Google Scholar]