Background: 53BP1 counteracts BRCA1 in DNA repair.
Results: RIF1 acts downstream of 53BP1 and counteracts BRCA1 in DNA end resection. It also has a 53BP1-independent role in regulating BLM chromatin association.
Conclusion: RIF1 is the major downstream effector of 53BP1.
Significance: These results reveal that RIF1 antagonizes BRCA1, functions in DNA end protection, and prevents homologous recombination repair.
Keywords: BRCA1, Cell Cycle, Cell Division, DNA Damage, DNA Recombination, DNA Repair, Homologous Recombination
Abstract
BRCA1 promotes homologous recombination repair and antagonizes 53BP1-dependent nonhomologous end joining (NHEJ) pathway. However, the molecular basis of the competition between BRCA1 and 53BP1 pathways remains elusive. Here we report that RIF1 protein translocates to damage sites via ATM-dependent 53BP1 phosphorylation. Strikingly, loss of RIF1 rescues initial DNA end resection and checkpoint activation in BRCA1-depleted cells. Interestingly RIF1 accumulation at damage sites is antagonized by BRCA1 in S and G2 phases. Conversely, the translocation of BRCA1 to damage sites is inhibited by RIF1 in G1 phase. However, loss of RIF1 differs from that of 53BP1 deficiency, as it cannot fully rescue RAD51 foci formation, homologous recombination defect, and radio-hypersensitivity in BRCA1-deficient cells. This is likely because RIF1, but not 53BP1, also regulates the foci formation and chromatin loading of BLM (the Bloom syndrome helicase). Thus, RIF1 not only acts downstream of 53BP1 and counteracts BRCA1-mediated end resection but also has a secondary role in promoting BLM function in DNA repair.
Introduction
Chromosomal double-strand breaks (DSBs)2 are among the most severe lesions that occur in eukaryotic cells. Proper repair of chromosomal DSBs is critical for maintaining genome stability and preventing tumorigenesis. Two major repair pathways, homologous recombination (HR) and nonhomologous end joining (NHEJ), allow the repair of DSBs. The HR repair pathway requires a homology template and is initiated by DNA end resection, which is mediated by MRN complex and facilitated by CtIP (1, 2). A more extensive end resection is carried out by Dna2, EXO1, and BLM to generate longer ssDNA stretch (3–5). Replication protein A (RPA) then binds to ssDNA, protects ssDNA from nuclease cleavage, and prevents hairpin formation. After that, the BRCA2/PALB2 complex removes RPA and loads the recombinase RAD51 onto ssDNA to form a nucleoprotein filament that catalyzes homologous search and strand invasion, which leads to strand exchange (6–9). In contrast, DSB repair by NHEJ does not need any homology template; instead, it promotes a direct ligation of two broken ends. Besides the different requirement for homology template, HR and NHEJ also differ at the initiation steps. Resection at the 5′ end of DSBs is not only a requirement for the initiation of HR repair, but it simultaneously blocks KU70/80 complex-mediated classic NHEJ, as KU cannot bind to resected ssDNA. Thus, DNA end resection is the key step that controls DSB repair pathway choice (10, 11).
Different DNA damage response proteins control these two repair pathways. For example, 53BP1 plays a key role in NHEJ-dependent rejoining of distant DSBs, including class switch recombination (12, 13), V(D)J recombination (14), and telomere fusion (15). A critical HR repair protein is BRCA1, which is encoded by a tumor suppressor gene frequently mutated in familial breast and ovarian cancers. Deficiency of BRCA1 leads to impaired HR (16), which may result from inefficient DNA end resection (2, 17) and the impaired loading of BRCA2-PALB2 to DSBs (7). BRCA1 and 53BP1 were previously believed to work independently. However, recent studies using mouse models demonstrated that BRCA1 and 53BP1 are involved in a competition between HR and NHEJ repair pathways. Genetic deletion of 53BP1 rescued embryonic lethality, HR deficiency, and genome instability associated with BRCA1 loss (18–20). The detailed mechanism as how 53BP1 participates in this competition remains largely unexplored.
RIF1 was first identified in yeast. It localizes at the telomere via its interaction with RAP1 (21) and negatively regulates telomere length (22). Unlike its yeast ortholog, mammalian RIF1 is not a telomeric protein and does not associate with telomere protein RAP1. Instead, mammalian RIF1 translocates to double-strand break sites in response to DNA damage via a 53BP1- and ATM-dependent manner (23). Here we report that RIF1 acts downstream of phosphorylated 53BP1 to limit initial DNA end resection, which is promoted by BRCA1. In addition, RIF1 also has a 53BP1-independent function in facilitating BLM chromatin association and localization at DSB sites.
EXPERIMENTAL PROCEDURES
Antibodies
The following antibodies were used in this study: anti-FLAG (F3165, Sigma), anti-HA (H9658, Sigma), anti-RPA32 (ab2175, Abcam,), anti-phospho-RPA32 (S4/S8) (A300-245, Bethyl), anti-GAPDH (MAB374, Millipore), anti-Histone H3(05-928, Millipore), anti-BRCA1 (D-9, Santa Cruz), anti-BLM (A310-029A, Bethyl), anti-RIF1(A300-569A, Bethyl), anti-phospho-CHK1 (S345) (2348S, Cell Signaling), and anti-phospho-KAP1 (S824) (4127, Cell Signaling). Rabbit antibodies against BRCA1, RAD51, γH2AX, and 53BP1 were described previously (1).
Cell Culture, Transfection, and Small Interfering RNAs (siRNAs)
The culture of human cells has been described previously (24). The sequence for BRCA1 siRNA is CAGCUACCCUUCCAUCAUAdTdT. All siRNAs were synthesized by Dharmacon, Inc. Oligofectamine was used for transfection. U2OS or HeLa cells were transfected with siRNAs 2 times at 24-h intervals. 24 h after the last transfection, cells were irradiated or collected.
Cell Cycle Synchronization
To enrich HeLa cells in G1 phase, cells were first arrested in M phase with nocodazole (100 ng ml−1) treatment for 12 h and then released for 8 h. To enrich cells in S and G2/M phases, cells were synchronized with the use of a double thymidine block as previously described (1). Cells were blocked in S or G2/M phase upon release from thymidine block for 4 or 8 h, respectively. 2 h before the indicated time points, cells were treated with 5 Gy of γ-IR to induce IRIF formation.
shRNAs
shRNAs targeting RIF1, 53BP1, and BRCA1 were cloned in pLKO.1 vector (Sigma). Sequences of non-targeting shRNA: 5′-CCTAAGGTTAAGTCGCClCTCG-3′; three nonoverlapping RIF1 targeting shRNAs (3#, 5′-GCTCTATTGTTAGGTCCCATTCT-3′; 4#. 5′-GCTATCTGGAAGGAGCTAATT-3′; 5#, 5′-CGCATTCTGCTGTTGTTGATT-3′); 53BP1 targeting shRNA, 5′-GATACTCCTTGCCTGATAATT-3′; CtIP targeting shRNA, 5′-CAGAAGGATGAAGGACAGTTT-3′; BRCA1 targeting shRNA, 5′-ATTCATGCCAGAGGTCTTATA-3′. Knockdown cells are generated as previously described (25).
Immunostaining
Immunostaining was performed similar to that described previously (25). Of note, pre-extraction was performed in all RPA2 and BLM immunostaining.
Cell Fractionation and Immunoprecipitation
To separate soluble and chromatin fractions, cells were lysed in NETN buffer supplemented with protease and phosphatase inhibitors (100 mm NaCl, 20 mm Tris-HCl (pH 8.0), 1 mm EDTA, 0.5 mm DTT, 0.5% Nonidet P-40, 1 μg ml−1 aprotinin, pepstatin A, 25 mm NaF, and 50 mm β-glycerophosphate), and the supernatant was saved as “soluble fraction.” The pellet was washed twice with NETN and boiled in 1× SDS loading buffer.
For co-immunoprecipitation experiments cells were harvested and resuspended in low salt buffer (10 mm HEPES (pH 7.4), 10 mm KCl, 1.0 mm CaCl2, 1.5 mm MgCl2, 1 mm DTT, 1% digitonin, and 10% glycerol) with protease and phosphatases inhibitor and 50 units/ml TurboNuclease (Abnova) and homogenized with a Dounce homogenizer (20 strokes). After high speed centrifugation, the supernatant was collected, and the concentration of NaCl was adjusted to 100 mm. The lysates were then incubated with streptavidin-conjugated beads (Amersham Biosciences) for 1 h at 4 °C and washed 3 times with NETN buffer.
HR and NHEJ Assay
U2OS cells containing a single copy of the DR-GFP reporter (U2OS-DR) were employed using experimental procedures that were described previously (6). NHEJ repair was assessed by a cell-based plasmid integration as previously described (25). IR sensitivity assay was performed as described previously (24) except 1000 cells were seeded into 10-cm dishes.
RESULTS
ATM-dependent 53BP1 Phosphorylation Recruits RIF1 to DNA Damage Sites
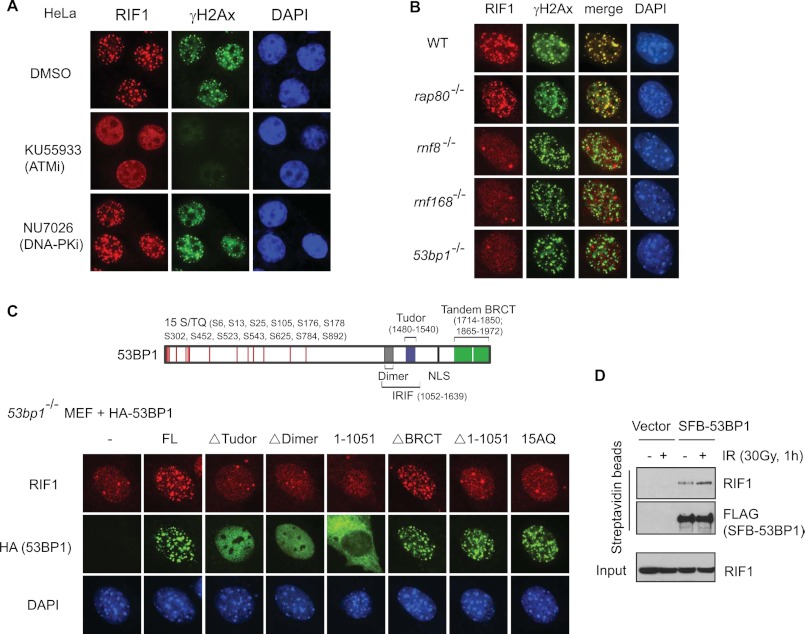
We and others have shown previously that the DNA repair function of 53BP1 requires both its localization to DSB sites and its phosphorylation by ATM in response to DNA damage (26, 27); however, how 53BP1 phosphorylation affects DNA repair is unclear. RIF1 is one of the few proteins that has been shown to act downstream of 53BP1 (23), and consistent with previous report, the accumulation of RIF1 at DNA damage sites is ATM-dependent, as the ATM inhibitor abolished IR-induced foci formation (IRIF) of RIF1 (Fig. 1A). Further genetic studies using murine knock-out embryonic fibroblasts (MEFs) revealed that RIF1 IRIF depends on the RNF8-RNF168–53BP1 pathway but not the RAP80-mediated BRCA1 pathway (28) (Fig. 1B).
FIGURE 1.
Localization of mammalian RIF1 in response to DNA double-strand breaks. A, ATM, but not DNA-PK, mediates IRIF of RIF1. HeLa cells were pretreated with DMSO, ATM inhibitor KU55933 (10 μm), or DNA-PK inhibitor NU7026 (2.5 μm) and then irradiated (10 Gy). 2 h after irradiation, cells were fixed and immunostained with anti-RIF1 and anti-γH2AX antibodies. B, RIF1 re-localization after DNA damage in wild-type (WT), rnf8−/− (25), rnf168−/− (49), 53bp1−/− (50), and rap80−/− (51) MEF cells is shown. Immunostaining experiments were performed using anti-RIF1 and anti-γH2AX antibodies. C, 53BP1 DSB localization and N-terminal ATM phosphorylation are both required for targeting RIF1 to DNA damage sites. Top, shown is schematic diagram of domain organization of human 53BP1. Bottom, shown are 53bp1−/− MEF cells transfected with HA-tagged wild-type or various mutants of human 53BP1. 48 h later cells were irradiated (10 Gy) and immunostained with anti-RIF1 and anti-HA antibodies. 15AQ is a mutant that abolishes all 15 ATM-phosphorylation sites in 53BP1. FL, full-length. IR induced RIF1–53BP1 interaction. 293T cells were transiently transfected with empty vector or plasmids encoding S-FLAG-SBP (SFB)-tagged 53BP1. 48 h later cells were irradiated (30 Gy) or left untreated, and cell lysates were subjected to pulldown using streptavidin-conjugated beads and blotted with anti-RIF1 antibody.
To understand exactly how RIF1 is recruited by 53BP1, we examined RIF1 IRIF in 53BP1-deficient MEF cells that had been reconstituted with wild type or various mutants of 53BP1. Cells reconstituted with wild type or the BRCT deletion mutant of 53BP1 fully restored IRIF of RIF1, but deletion of the 53BP1 N terminus (Δ1–1051), which still form foci, failed to support RIF1 IRIF formation. 53BP1 N terminus contains a cluster of potential ATM target sites (15 consensus (S/T)Q) (26) (Fig. 1C, top panel). Thus, we used a 15AQ mutant of 53BP1 in which all of the 15 conserved (S/T)Q sites were mutated to Ala. This mutant of 53BP1 also failed to recruit RIF1 to DNA damage sites (Fig. 1C, bottom panel). Together, these data suggest that ATM-dependent phosphorylation of 53BP1 facilitates the relocalization of RIF1 to DNA damage sites.
These results raised the possibility that phosphorylated 53BP1 might bind to RIF1. Indeed a small amount of RIF1 was detected in 53BP1 immunocomplex, and this interaction was increased by 2-fold in response to irradiation (Fig. 1D). Therefore, RIF1 binds to 53BP1 in a DNA damage-induced, phosphorylation-dependent manner.
RIF1 Inhibits Homologous Recombination
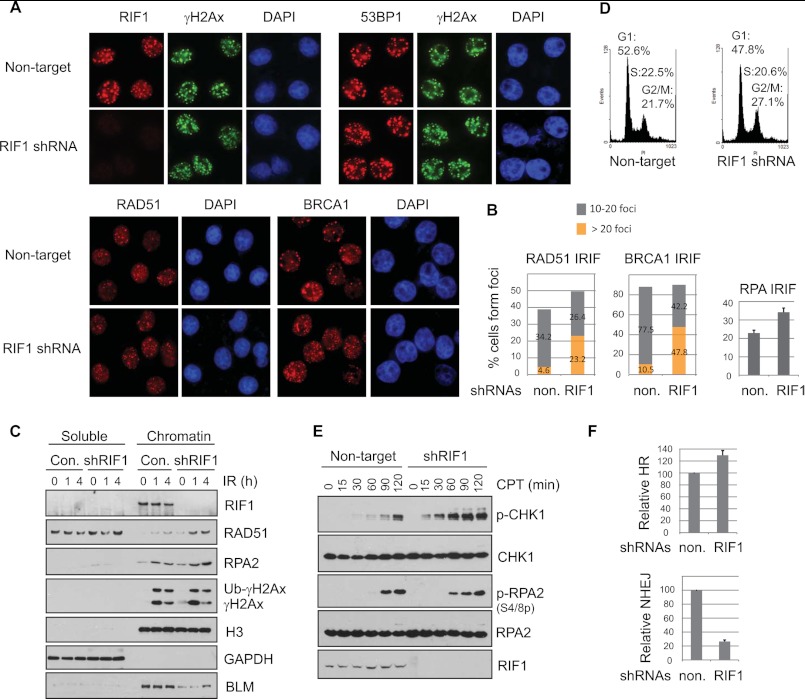
Recent evidence from yeast implicated a role of yeast RIF1 in inhibiting checkpoint activation and RPA loading at dysfunctional telomeres (29, 30). The anti-checkpoint function of yeast RIF1 is independent of its binding to RAP1 (29, 30). We asked whether this function of yeast RIF1 is conserved in human RIF1. We depleted RIF1 expression using lentiviral shRNAs and confirmed RIF1 depletion by immunofluorescent staining and Western blotting analysis (Fig. 2, A and C). RIF1 depletion did not result in any detectable change in 53BP1 IRIF (Fig. 2A, top panel); however, the number of damage-induced HR repair protein foci, such as BRCA1 and RAD51 foci, increased in RIF1-deficient cells when compared with those in RIF1-proficient cells (Fig. 2, A, bottom panel, and B, left panel). Besides, the percentage of cells forming IR-induced RPA foci, a marker of resected DSBs (31), was also significantly elevated by the knockdown of RIF1 (Fig. 2B, right panel). Analysis of soluble and chromatin-bound fractions also showed that more RAD51 and RPA proteins were enriched in chromatin fractions in response to DNA damage in the absence of RIF1 (Fig. 2C). Fractionation results also revealed that RIF1 tightly associated with chromatin regardless of DNA damage (Fig. 2C). Loss of RIF1 did not have any major impact on cell cycle distribution. The percentages of cells in G1 or S phases were comparable in control and RIF1-depleted cells. The only noticeable change was the percentage of cells in G2/M phase, which was modestly increased in the absence of RIF1 (Fig. 2D). This minor change in cell cycle profile suggests that RIF1 depletion did not inhibit the loading of HR proteins to DNA damage sites by diminishing the population of cells in S and G2 phases.
FIGURE 2.
Impact of RIF1 on accumulation of DNA repair proteins at sites of DNA damage. A, HeLa cells depleted of endogenous RIF1 were irradiated (5 Gy) and recovered for 4 h before fixation and permeabilization. Immunostaining experiments were performed as described under “Experimental Procedures.” Three different, non-overlapping lentiviral shRNAs for RIF1 were used, and similar results were obtained. B, left, a histogram shows the percentage of cells shown in A containing low (<20) or high (>20) levels of RAD51 or BRCA1 foci. Right, shown is quantification of cells that show RPA foci. At least 300 cells were counted in each experiment. C, HeLa cells as described in A were irradiated or left untreated and harvested at the indicated time points. Cells were lysed, and soluble and chromatin fractions were prepared as described under “Experimental Procedures.” Samples were immunoblotted with antibodies as indicated. D, a cell cycle profile of RIF1-proficient and -deficient HeLa cells is shown. E, HeLa cells were infected with lentivirus carrying non-target or RIF1-specific shRNA. Cells were either left untreated or treated with 0.5 μm camptothecin (CPT) for the indicated times and then harvested and immunoblotted with the indicated antibodies. F, top, increased HR repair efficiency was observed in RIF1-depleted cells. DR-U2OS cells were infected with non-target or RIF1-specific lentiviral shRNAs and then transfected with I-SceI expression plasmid (pCBASce); the latter induced double-strand breaks. Successful repair by HR resulted in the appearance of GFP+ cells. The relative HR frequencies in RIF1-depleted cells are shown in comparison to those in control cells. Results are the means (±S.D.) of three independent experiments. Bottom, loss of RIF1 decreased NHEJ repair. HeLa cells infected with non-targeting shRNA or RIF1-specific lentiviral shRNA were transfected with linearized plasmid pcDNA3.1/hygro. Cells were incubated in selective media containing 100 μg ml−1 hygromycin for 14 days, and the numbers of colonies were determined. Results are the means (±S.D.) of three independent experiments.
As RIF1 inhibits RPA foci formation (Fig. 2B), we further explored the role of RIF1 in DNA end resection. We treated cells with camptothecin, a DNA topoisomerase I inhibitor that generates single-strand breaks that can be converted to DSBs during DNA replication to induce robust activation of ATR pathway (32). Checkpoint kinase-1 (CHK1) and RPA2 are two well known substrates for ATR. Although the short ssDNA-dsDNA junction rapidly activates ATR-CHK1, RPA2 phosphorylation is a relatively slow process and requires the presence of long stretches of ssDNA (31, 33). We found that RIF1 depletion led to a robust hyperactivation of CHK1 in all time points after camptothecin treatment. RPA2 phosphorylation was also increased at early time points in RIF1-depleted cells; however, at late time points, RIF1 depletion did not further increase RPA2 phosphorylation (Fig. 2E). These results indicate that RIF1 only protects DNA ends from initial processing. Once the initial end resection starts, RIF1 is probably displaced and, therefore, is no longer involved in this process.
To confirm a role of RIF1 in inhibiting HR repair, we used U2OS-DR cells, which have a single, stably integrated copy of HR reporter (6). Depletion of RIF1 in U2OS-DR cells resulted in a 30∼40% increase in GFP-positive cells (Fig. 2F, top panel), implying RIF1 inhibited HR in this setting. Conversely, loss of RIF1 led to a significant reduction in NHEJ repair (Fig. 2F, bottom panel), supporting a role of RIF1 in favoring NHEJ and repressing HR.
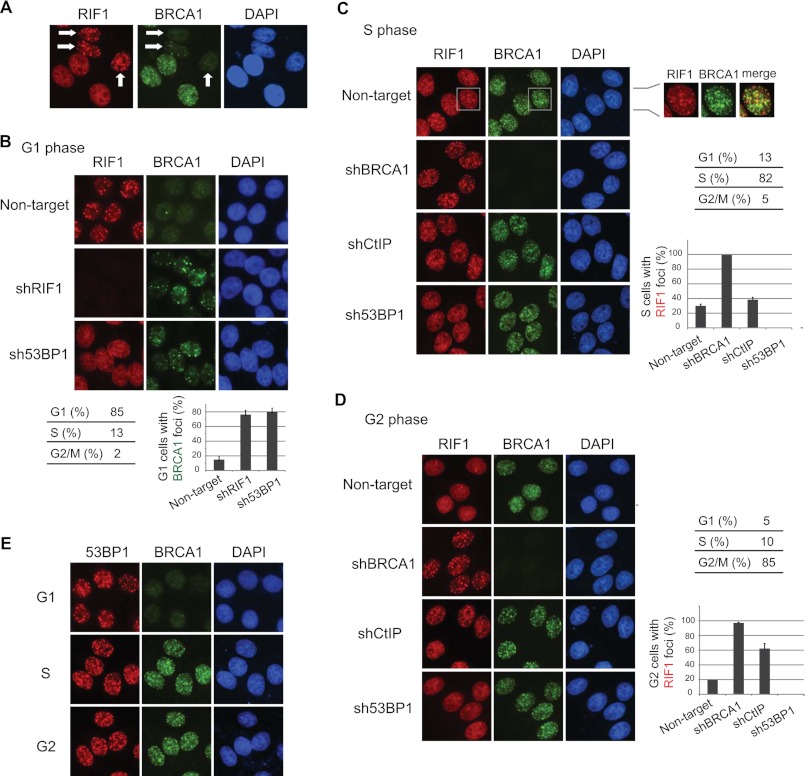
RIF1 and BRCA1 Accumulate at DNA Damage Sites in a Mutually Exclusive Manner
We noticed that RIF1 formed IR-induced foci in a subpopulation of asynchronous cells in which the BRCA1 foci were absent (Fig. 3A). To further dissect the localization of RIF1 in response to double-strand breaks in different cell cycle phases, we synchronized cells in G1, S, and G2 phases. The focus formation of RIF1 mainly took place in G1 phase (Fig. 3B), was diminished to a few dots in S phase, which did not co-localized with BRCA1 (Fig. 3C) and were not induced by DNA damage (data not shown), and were absent in G2 phase (Fig. 3D). This damage-induced, cell cycle-dependent regulation of RIF1 foci formation is completely contrary to the pattern of damage-induced BRCA1 foci formation (Fig. 3, B–D). More strikingly, depletion of RIF1 as well as that of 53BP1 enabled BRCA1 to translocate to DNA damage sites in G1 phase (Fig. 3B). Conversely, knockdown of BRCA1 led to RIF1 IRIF formation in S and G2 cells (Fig. 3, C and D). This cell cycle-dependent antagonistic effect is quite specific to RIF1 and BRCA1 because 53BP1, the upstream regulator of RIF1, formed damage-induced foci throughout the cell cycle (Fig. 3E), and CtIP, a binding partner of BRCA1, only had minimal inhibitory effect on RIF1 IRIF in S phase (Fig. 3C) and showed modest inhibition on RIF1 foci formation in G2 phase (Fig. 3D), which agrees with our previous observation that CtIP-BRCA1 complex mainly acts in G2 phase (1). These data provide the molecular basis for the competition between RIF1 and BRCA1 in regulating the balance of NHEJ and HR; the former is a dominant repair pathway in G1 cells, and the latter is mainly active in S and G2 phases.
FIGURE 3.
RIF1 and BRCA1 accumulate at DNA damage sites in a mutually exclusive manner. A, shown is IR-induced RIF1 and BRCA1 foci formation in asynchronous cells. Arrows indicate the cells with RIF1 but no BRCA1 foci. B, depletion of RIF1 or 53BP1 promoted BRCA1 localization to DNA damage sites in G1 cells. HeLa cells infected with lentiviral particles carrying the indicated shRNAs were enriched in G1 phase by treating cells with nocodazole and then released for 8 h. 2 h before the desired time point, cells were irradiated with 5 Gy of IR. Quantification of BRCA1 foci formation is shown in the bottom panel, and the data are represented as the mean ± S.E. (n = 3). C, BRCA1 deficiency enabled RIF1 foci formation in S phase cells. HeLa cells were manipulated as described in B except that they were enriched in S phase by double thymidine block and released for 4 h. Quantification of RIF1 foci formation is shown in the right panel, and data are represented as the mean ± S.E. (n = 3). Please note that the majority of S phase RIF1 foci did not co-localize with BRCA1, and they form independent of DNA damage. D, HeLa cells were manipulated as described in C except they were enriched in G2 phase by a double thymidine block and released for 8 h. Quantification of RIF foci formation was shown in the bottom panel, and results are presented as the means (±S.D.) of three independent experiments. Cell cycle distributions were analyzed by flow cytometry and summarized in B–D. E, 53BP1 IRIF in G1, S, and G2 phases are shown. Cell synchronization was carried out as described in B–D.
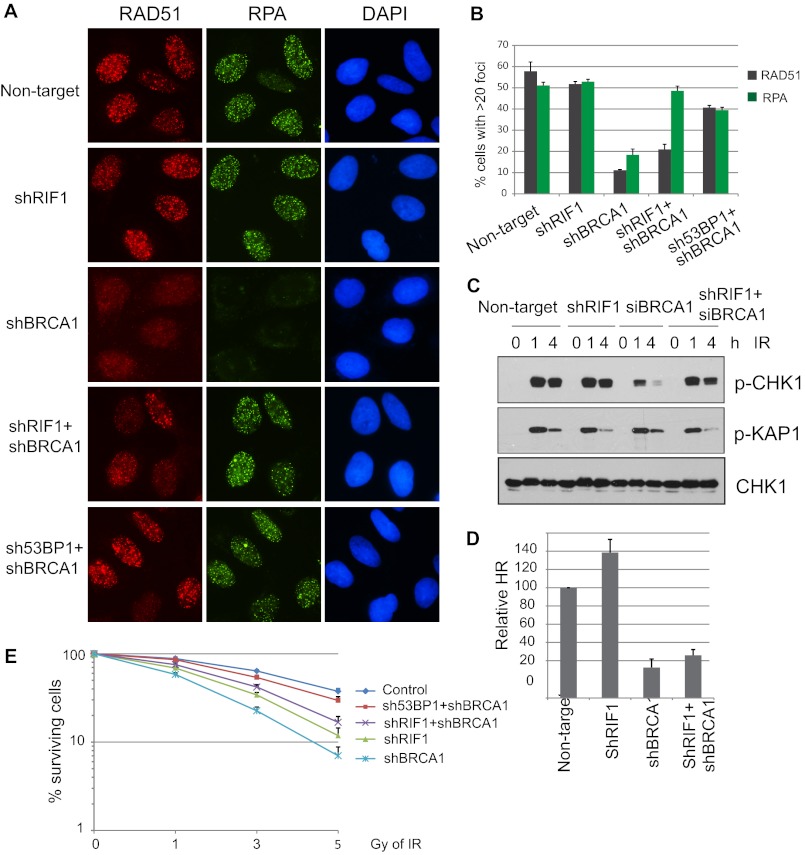
Loss of RIF1 Rescues an End Resection Defect Caused by BRCA1 Deficiency
It has been shown recently that loss of 53BP1 reverses the HR defects in brca1-null cells (19). As RIF1 works downstream of 53BP1 (Fig. 1), we would like to further explore whether or not RIF1 depletion could also restore HR defects in BRCA1-deficient cells. As previously reported, abrogating BRCA1 diminished RAD51 foci formation after IR (Fig. 4, A and B). In addition, RPA foci formation, a marker for DNA end resection and ssDNA region, was also greatly reduced in BRCA1-depleted cells (Fig. 4, A and B), confirming that BRCA1 acts by promoting end resection at early stages of DSB repair. Strikingly, co-depletion of RIF1 and BRCA1 restored RPA foci formation, comparable with that of control-depleted cells (Fig. 4, A and B). However, it only partially rescued RAD51 loading at DSB sites (Fig. 4, A and B). In contrary, co-depletion of 53BP1 and BRCA1 largely recovered both RPA and RAD51 foci formation (Fig. 4, A and B).
FIGURE 4.
Depletion of RIF1 alleviates DNA repair defect in BRCA1-knockdown cells. A, U2OS cells were infected with lentiviral particles carrying the indicated shRNAs. 48 h post-infection, cells were irradiated with 10 Gy IR and recovered for 4 h. Immunostaining was performed using antibodies as indicated. B, quantification of RAD51 and RPA foci formation in cells was as described in A. More than 150 cells were counted to determine the percentages of foci forming cells in each sample. C, checkpoint activation in single- or double-knockdown cells is shown. Control or RIF1 stable knockdown cells were transfected with control or BRCA1-specific siRNAs. 48 h later cells were exposed to 0 or 10 Gy IR and harvested at the indicated time points. Total lysates were subjected to Western blot analysis with the indicated antibodies. D, DR-GFP reporter (U2OS-DR) cells were infected with indicated lentivival shRNAs for 48 h and then electroporated with I-SceI expression plasmid (pCBASce). The percentage of GFP-positive cells was determined by flow cytometry 48 h after electroporation. The data were normalized to those obtained from cells infected with non-targeting shRNA. Data represent the means (±S.D.) of three independent experiments. E, shown is clonogenic survival of cells as described in A after exposed to the indicated doses of IR. Results are the means (±S.D.) of three independent experiments.
Double knockdown of RIF1 and BRCA1 also rescued CHK1 phosphorylation to a level similar to those in control cells (Fig. 4C). Because CHK1 phosphorylation requires RPA-ssDNA for the activation of its upstream kinase ATR (31, 34), this result further supports that RIF1 inhibits BRCA1-dependent DNA end processing. Because RIF1 and BRCA1 have opposing effects on HR, we then tested the efficiency of HR in RIF1 and BRCA1 double knockdown cells. Single depletion of RIF1 increased HR as shown in Fig. 2E; however, double depletion of RIF1 and BRCA1 only slightly rescues HR deficiency caused by BRCA1 loss (Fig. 4D), which agrees with the partial restoration of RAD51 IRIF observed in RIF1 and BRCA1 double knockdown cells (Fig. 4, A and B). This rescue of end resection defect by RIF1 loss is specific to BRCA1 deficiency, as knockdown of RIF1 or 53BP1 was unable to rescue impaired RPA loading in CtIP-depleted cells (data not shown). Finally we tested the effect of RIF1 in IR sensitivity. Single depletion of either RIF1 or BRCA1 resulted in increased sensitivity to IR. However, consistent with RAD51 foci formation and HR activity shown in Fig. 4A, B and D, depletion of RIF1 only slightly alleviated hypersensitivity of BRCA1-deficient cells to IR (Fig. 4E) while abrogating 53BP1 in BRCA1-deficient cells desensitized cells to IR treatment, which is consistent with previous reports (18–20). These data indicate that although knockdown of RIF1 could rescue the loading of RPA to ssDNAs, it could not efficiently restore the subsequent HR events in BRCA1-deficient cells.
RIF1 Regulates Nuclear Foci Formation and Chromatin Association of BLM
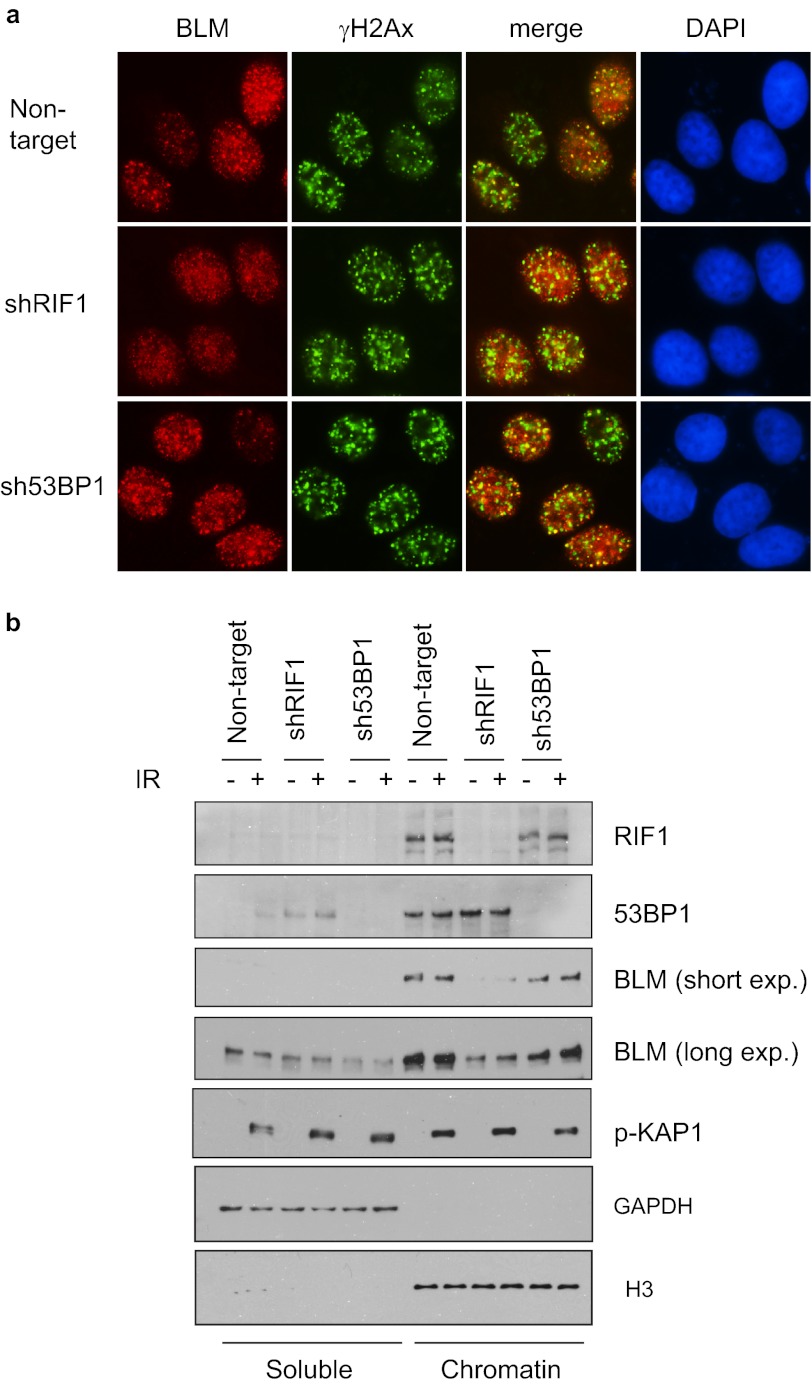
Unlike 53BP1, loss of RIF1 could not fully restore HR repair in BRCA1-deficient cells (Fig. 4). This observation suggests that RIF1 may have a role in HR repair that is independent of 53BP1. Vertebrate RIF1 was shown to interact with BLM helicase (35). We thus explored whether RIF1 would affect BLM function in response to DSBs. BLM translocalizes to DSB sites after IR (36). We found that the formation of BLM foci was severely impaired after RIF1 depletion but not so in 53BP1-depleted cells (Fig. 5a). Moreover, knockdown RIF1 also led to impaired loading of BLM onto chromatin as shown by fractionation assay (Fig. 5b), indicating that compromised BLM foci formation may reflect reduced presence of BLM on chromatin. Consistently, 53BP1 depletion did not have any impact on BLM chromatin loading (Fig. 5b).
FIGURE 5.
RIF1 regulates BLM DSB recruitment and chromatin loading. a, defective BLM IRIF formation in RIF1-depleted cells is shown. HeLa cells were infected with shRNAs as indicated. 4 h after irradiation, cells were fixed and immunostained with anti-BLM and anti-γH2Ax antibodies. Three different, nonoverlapping lentiviral shRNAs for RIF1 were used, and the same results were obtained. b, depletion of RIF1 reduces the amount of chromatin-associated BLM. RIF1 and 53BP1 knockdown cells were mocked-treated or treated with ionizing radiation. 1 h after IR, the cells were harvested, and soluble and chromatin fractions of cell lysates were analyzed by immunoblotting using the indicated antibodies.
As shown in Fig. 2C, RIF1 tightly associates with chromatin regardless of DNA damage, which is similar to that of 53BP1 and BLM. Although RIF1 IRIF formation depends on the presence of 53BP1 (Fig. 1), its chromatin loading was not affected by 53BP1 depletion (Fig. 5b). These results suggest that RIF1 is important for maintaining chromatin association of BLM. This function of RIF1 does not require its recruitment of DSB sites, which is mediated by 53BP1.
DISCUSSION
In this study we demonstrate that RIF1 plays a role in early processing of double-strand beaks. RIF1 acts downstream of 53BP1 and contributes to the capacity of 53BP1 in limiting DNA end resection after DNA damage. Correspondingly, depletion of RIF1 rescues DNA end resection defects caused by BRCA1 deficiency. On the other hand, RIF1 fine-tunes HR by promoting BLM localization to chromatin and to DSB sites. Together, our results provide a new player, RIF1, that participates in the competition between BRCA1 and 53BP1 and suggests a complex regulation of NHEJ and HR in DSB repair.
The requirement of ATM-dependent phosphorylation of 53BP1 in DNA damage response and DNA repair was established a while ago (10, 11). Interestingly, we showed here that RIF1 is recruited to DNA damage sites via ATM-dependent phosphorylation of 53BP1 (Fig. 1). Mammalian RIF1 contains HEAT repeats at its N terminus and a DNA binding domain at its C terminus (23, 35), but RIF1 does not have any known phosphopeptide binding domains. Therefore, we assume that either RIF1 has a previously unrecognized phosphoprotein binding motif or a yet-to-be-identified bridging protein that may mediate the phosphorylated 53BP1-dependent recruitment of RIF1 to DSB sites. Among all known 53BP1-interacting proteins, PTIP is the only protein that contains BRCT domains and associates with the ATM-phosphorylated form of 53BP1 (37). However, using PTIP-deficient cells, we showed that PTIP is not required for RIF1 IRIF formation (data not shown).
Loss of RIF1 phenocopies 53BP1 deficiency in terms of heightened DNA end resection (27, 38), increased RPA loading (39), and the recovery of BRCA1 IRIF in G1 phase (Fig. 3). RIF1 inhibits the loading of HR proteins to DNA damage sites (Fig. 2) and antagonizes BRCA1 and BRCA1-mediated resection of DNA ends (Figs. 3 and 4). The molecular basis for the end processing-inhibitory role of RIF1 may be the existence of its C-terminal DNA binding domain, which strongly binds to dsDNA but not ssDNA in vitro (35). Upon DNA damage, the short ssDNA-dsDNA junctions generated at the newly formed DSBs may provide loading sites for RIF1 and, therefore, prevents RPA loading and the following excessive end resection. However, when the ssDNA stretch becomes longer, RIF1 can no longer inhibit RPA loading because its ssDNA binding activity is weak and cannot compete with RPA. Therefore, it is conceivable that RIF1 functions as a shield against initial DNA end resection at DSBs and thus commits cells to undergo NHEJ-mediated, but not HR-mediated, DNA repair. This notion is supported by studies in yeast. Chromatin immunoprecipitation assay revealed that yeast Rif1 caps at short, but not long, ssDNAs to block accumulation of RPA (29, 30). In this study performed in mammalian cells, we found that RIF1 affects the initial, but not the sustained, RPA phosphorylation (Fig. 2E), suggesting that it constrains DNA ends from initial processing rather than inhibits further excessive DNA end resection. It may also explain why RIF1 IRIF only occurs in G1 cells (Fig. 3), in which the DNA end resection activity is low. When extensive end processing occurred in S and G2 phases, RIF1 is released from DNA break sites due to its inability to bind to ssDNA (35); as a result more BRCA1 translocate to DSB sites to facilitate HR repair. During the preparation of this manuscript, several studies reported the function of RIF1 in DNA end protection (40–43). Our findings largely agree with these very recent reports. Although RIF1 works downstream of phosphorylated 53BP1 to prevent DNA end resection, the function of RIF1 and 53BP1 may not completely overlap. For example, the class switching defect is much milder in rif1−/− B cells than that observed in 53bp1−/− cells (41). RIF1 is also not required for the mobility of dysfunctional telomeres, as does 53BP1 (40). Furthermore, unlike 53BP1 depletion, loss of RIF1 only partially suppressed sensitivity of BRCA1-deficient cells to PARP inhibitor (40, 43).
In this study we provide additional evidence to reveal the differences between RIF1 and 53BP1. First, the localization of RIF1 to DSB sites is restricted in G1 phase and is suppressed by BRCA1 in S and G2 phases, whereas 53BP1 accumulates at DSB sites in all stages of cell cycle regardless of BRCA1 status (Fig. 3). It is unlikely that the phosphorylation of 53BP1 by ATM is absent in S and G2 phases because the activation of ATM kinase is not cell cycle-dependent (44). These cell cycle-dependent regulations of BRCA1 and RIF1 foci formation indicate that RIF1 and BRCA1 may directly counteract each other at DSBs. Exactly how BRCA1 competes with RIF1 for accumulation at DSB sites is currently unknown. One possibility is that the BRCT domain of BRCA1 may interact with cell cycle-regulated phosphorylation of 53BP1 to mask the binding surface for RIF1. Another possibility is that RIF1 may not be able to recognize ubiquitinated chromatin structure modified by BRCA1. Second, we found that depletion of RIF1 recovers RPA-ssDNA filament formation in BRCA1-deficient cells but cannot rescue the entire HR repair defects conferred by BRCA1 loss, which are different from a recent report (43). On the contrary, 53BP1 depletion rescues both RPA and RAD51 IRIF in BRCA1-deficient cells (Fig. 4). These different phenotypes observed after RIF1 or 53BP1 depletion may be explained by the fact that a substantial portion of RIF1 associates with BLM helicase (35) and regulates BLM stability at chromatin in a 53BP1-independent manner (Fig. 5). Because BLM works together with EXO1 and Dna2 to promote extensive DNA end processing (3–5), it is conceivable that RIF1 deficiency on one hand facilitates BRCA1-mediated initial DNA end processing and on the other hand impairs the following extensive DNA end resection carried out by BLM-EXO1 complex, although the latter is less prominent as a significant portion of BLM still associates with chromatin in the absence of RIF1 (Fig. 5b). As a result, knockdown of RIF1 promotes the initial, but not the extensive, DNA end resection (Fig. 2E) despite the fact that the initial DNA end resection stimulates extensive end resection (10). We hope that future studies using purified RIF1 and resection machinery may lead to a better understanding of the roles of RIF1 in end resection and DSB repair. As for 53BP1, it may also have other downstream factors besides RIF1 that contribute to its competition with BRCA1 in HR repair, in particular, the displacement of RPA-ssDNA filament and the conversion to RAD51 filament catalyzed by BRCA1-PLAB2-BRCA2 complex (6–9).
Although CtIP is the only known BRCA1-binding protein implicated in DNA end processing (1, 2, 45), depletion of RIF1 is unable to rescue end resection defect in CtIP-deficient cells (data not shown), suggesting that the role of CtIP in DNA end resection is not completely dependent on its association with BRCA1, an idea that is supported by fact that the chicken DT cells expressing CtIPS332 mutant that fail to bind to BRCA1 are still proficient in HR repair (46). On the other hand, unlike BRCA1, which is implicated in both CHK1 (47) and RPA phosphorylation (19), CtIP only controls RPA phosphorylation but has little impact on ATR-CHK1 activation (48).
RIF1 inhibits CHK1 activation and initial RPA phosphorylation (Fig. 2E); this feature is distinct from CtIP loss but partly reconcile with BRCA1 deficiency. In addition, contrary to a recent study (43), we found that depletion of BRCA1, but not CtIP, enables RIF1 forming IRIF in S phase (Fig. 3C), further supporting the idea that RIF1 specifically opposes BRCA1 at DSBs. However, exactly how BRCA1 functions in DNA end resection beyond its association with CtIP, especially in S phase cells, remains a mystery that needs to be solved in near future.
Acknowledgments
We thank all members of the Chen laboratory for discussions, Weidong Wang (National Institutes of Health) for rabbit hRIF1 polyclonal antibodies, Michael S. Y. Huen (The University of Hong Kong) and Ja-Eun Kim (Kyung Hee University) for human 53BP1 expression constructs, Razqallah Hakem (University of Toronto) for rnf168−/− MEF cells, Jo Morris (University of Birmingham) for suggestions on BRCA1 siRNAs, and Selleck Chemicals for ATM and DNA-PK inhibitors.
This work was supported, in whole or in part, by National Institutes of Health Grants CA089239, CA092312, and CA100109 (to J. C.).
- DSB
- double-strand break
- HR
- homologous recombination
- NHEJ
- nonhomologous end joining
- Gy
- gray
- IR
- ionizing radiation
- IRIF
- IR-induced foci formation
- MEF
- mouse embryonic fibroblast
- CHK1
- Checkpoint kinase-1
- ATM
- ataxia telangiectasia mutated
- RPA
- replication protein A.
REFERENCES
- 1. Yu X., Chen J. (2004) DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell Biol. 24, 9478–9486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yun M. H., Hiom K. (2009) CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 459, 460–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gravel S., Chapman J. R., Magill C., Jackson S. P. (2008) DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 22, 2767–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nimonkar A. V., Ozsoy A. Z., Genschel J., Modrich P., Kowalczykowski S. C. (2008) Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc. Natl. Acad. Sci. U.S.A. 105, 16906–16911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nimonkar A. V., Genschel J., Kinoshita E., Polaczek P., Campbell J. L., Wyman C., Modrich P., Kowalczykowski S. C. (2011) BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 25, 350–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xia B., Sheng Q., Nakanishi K., Ohashi A., Wu J., Christ N., Liu X., Jasin M., Couch F. J., Livingston D. M. (2006) Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell 22, 719–729 [DOI] [PubMed] [Google Scholar]
- 7. Sy S. M., Huen M. S., Chen J. (2009) PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc. Natl. Acad. Sci. U.S.A. 106, 7155–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu J., Doty T., Gibson B., Heyer W. D. (2010) Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 17, 1260–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Buisson R., Dion-Côté A. M., Coulombe Y., Launay H., Cai H., Stasiak A. Z., Stasiak A., Xia B., Masson J. Y. (2010) Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat. Struct. Mol. Biol. 17, 1247–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Symington L. S., Gautier J. (2011) Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45, 247–271 [DOI] [PubMed] [Google Scholar]
- 11. Misteli T., Soutoglou E. (2009) The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat. Rev. Mol. Cell Biol. 10, 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Manis J. P., Morales J. C., Xia Z., Kutok J. L., Alt F. W., Carpenter P. B. (2004) 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat. Immunol. 5, 481–487 [DOI] [PubMed] [Google Scholar]
- 13. Ward I. M., Reina-San-Martin B., Olaru A., Minn K., Tamada K., Lau J. S., Cascalho M., Chen L., Nussenzweig A., Livak F., Nussenzweig M. C., Chen J. (2004) 53BP1 is required for class switch recombination. J. Cell Biol. 165, 459–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Difilippantonio S., Gapud E., Wong N., Huang C. Y., Mahowald G., Chen H. T., Kruhlak M. J., Callen E., Livak F., Nussenzweig M. C., Sleckman B. P., Nussenzweig A. (2008) 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature 456, 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dimitrova N., Chen Y. C., Spector D. L., de Lange T. (2008) 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456, 524–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jasin M. (2002) Homologous repair of DNA damage and tumorigenesis. The BRCA connection. Oncogene 21, 8981–8993 [DOI] [PubMed] [Google Scholar]
- 17. Schlegel B. P., Jodelka F. M., Nunez R. (2006) BRCA1 promotes induction of ssDNA by ionizing radiation. Cancer Res. 66, 5181–5189 [DOI] [PubMed] [Google Scholar]
- 18. Cao L., Xu X., Bunting S. F., Liu J., Wang R. H., Cao L. L., Wu J. J., Peng T. N., Chen J., Nussenzweig A., Deng C. X., Finkel T. (2009) A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol. Cell 35, 534–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bunting S. F., Callén E., Wong N., Chen H. T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L., Xu X., Deng C. X., Finkel T., Nussenzweig M., Stark J. M., Nussenzweig A. (2010) 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bouwman P., Aly A., Escandell J. M., Pieterse M., Bartkova J., van der Gulden H., Hiddingh S., Thanasoula M., Kulkarni A., Yang Q., Haffty B. G., Tommiska J., Blomqvist C., Drapkin R., Adams D. J., Nevanlinna H., Bartek J., Tarsounas M., Ganesan S., Jonkers J. (2010) 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 17, 688–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hardy C. F., Sussel L., Shore D. (1992) A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 6, 801–814 [DOI] [PubMed] [Google Scholar]
- 22. Marcand S., Wotton D., Gilson E., Shore D. (1997) Rap1p and telomere length regulation in yeast. Ciba Found. Symp. 211, 76–93; discussion 93–103 [DOI] [PubMed] [Google Scholar]
- 23. Silverman J., Takai H., Buonomo S. B., Eisenhaber F., de Lange T. (2004) Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint. Genes Dev. 18, 2108–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Feng L., Huang J., Chen J. (2009) MERIT40 facilitates BRCA1 localization and DNA damage repair. Genes Dev. 23, 719–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Feng L., Chen J. (2012) The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat. Struct. Mol. Biol. 19, 201–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ward I., Kim J. E., Minn K., Chini C. C., Mer G., Chen J. (2006) The tandem BRCT domain of 53BP1 is not required for its repair function. J. Biol. Chem. 281, 38472–38477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bothmer A., Robbiani D. F., Di Virgilio M., Bunting S. F., Klein I. A., Feldhahn N., Barlow J., Chen H. T., Bosque D., Callen E., Nussenzweig A., Nussenzweig M. C. (2011) Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol. Cell 42, 319–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huen M. S., Sy S. M., Chen J. (2010) BRCA1 and its toolbox for the maintenance of genome integrity. Nat. Rev. Mol. Cell Biol. 11, 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xue Y., Rushton M. D., Maringele L. (2011) A novel checkpoint and RPA inhibitory pathway regulated by Rif1. PLoS Genet. 7, e1002417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ribeyre C., Shore D. (2012) Anticheckpoint pathways at telomeres in yeast. Nat. Struct. Mol. Biol. 19, 307–313 [DOI] [PubMed] [Google Scholar]
- 31. Zou L., Elledge S. J. (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300, 1542–1548 [DOI] [PubMed] [Google Scholar]
- 32. Pommier Y. (2009) DNA topoisomerase I inhibitors. Chemistry, biology, and interfacial inhibition. Chem Rev. 109, 2894–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu J. S., Kuo S. R., Melendy T. (2006) Phosphorylation of replication protein A by S-phase checkpoint kinases. DNA Repair 5, 369–380 [DOI] [PubMed] [Google Scholar]
- 34. Choi J. H., Lindsey-Boltz L. A., Kemp M., Mason A. C., Wold M. S., Sancar A. (2010) Reconstitution of RPA-covered single-stranded DNA-activated ATR-Chk1 signaling. Proc. Natl. Acad. Sci. U.S.A. 107, 13660–13665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu D., Muniandy P., Leo E., Yin J., Thangavel S., Shen X., Ii M., Agama K., Guo R., Fox D., 3rd, Meetei A. R., Wilson L., Nguyen H., Weng N. P., Brill S. J., Li L., Vindigni A., Pommier Y., Seidman M., Wang W. (2010) Rif1 provides a new DNA-binding interface for the Bloom syndrome complex to maintain normal replication. EMBO J. 29, 3140–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu L., Davies S. L., Levitt N. C., Hickson I. D. (2001) Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J. Biol. Chem. 276, 19375–19381 [DOI] [PubMed] [Google Scholar]
- 37. Jowsey P. A., Doherty A. J., Rouse J. (2004) Human PTIP facilitates ATM-mediated activation of p53 and promotes cellular resistance to ionizing radiation. J. Biol. Chem. 279, 55562–55569 [DOI] [PubMed] [Google Scholar]
- 38. Bothmer A., Robbiani D. F., Feldhahn N., Gazumyan A., Nussenzweig A., Nussenzweig M. C. (2010) 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J. Exp. Med. 207, 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Martínez P., Flores J. M., Blasco M. A. (2012) 53BP1 deficiency combined with telomere dysfunction activates ATR-dependent DNA damage response. J. Cell Biol. 197, 283–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zimmermann M., Lottersberger F., Buonomo S. B., Sfeir A., de Lange T. (2013) 53BP1 regulates DSB repair using Rif1 to control 5' end resection. Science 339, 700–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Di Virgilio M., Callen E., Yamane A., Zhang W., Jankovic M., Gitlin A. D., Feldhahn N., Resch W., Oliveira T. Y., Chait B. T., Nussenzweig A., Casellas R., Robbiani D. F., Nussenzweig M. C. (2013) Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science 339, 711–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chapman J. R., Barral P., Vannier J. B., Borel V., Steger M., Tomas-Loba A., Sartori A. A., Adams I. R., Batista F. D., Boulton S. J. (2013) RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol. Cell 49, 858–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Escribano-Díaz C., Orthwein A., Fradet-Turcotte A., Xing M., Young J. T., Tkáč J., Cook M. A., Rosebrock A. P., Munro M., Canny M. D., Xu D., Durocher D. (2013) A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell 49, 872–883 [DOI] [PubMed] [Google Scholar]
- 44. Pandita T. K., Lieberman H. B., Lim D. S., Dhar S., Zheng W., Taya Y., Kastan M. B. (2000) Ionizing radiation activates the ATM kinase throughout the cell cycle. Oncogene 19, 1386–1391 [DOI] [PubMed] [Google Scholar]
- 45. Yu X., Wu L. C., Bowcock A. M., Aronheim A., Baer R. (1998) The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J. Biol. Chem. 273, 25388–25392 [DOI] [PubMed] [Google Scholar]
- 46. Nakamura K., Kogame T., Oshiumi H., Shinohara A., Sumitomo Y., Agama K., Pommier Y., Tsutsui K. M., Tsutsui K., Hartsuiker E., Ogi T., Takeda S., Taniguchi Y. (2010) Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS Genet. 6, e1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yarden R. I., Pardo-Reoyo S., Sgagias M., Cowan K. H., Brody L. C. (2002) BRCA1 regulates the G2/M checkpoint by activating Chk1 kinase upon DNA damage. Nat. Genet. 30, 285–289 [DOI] [PubMed] [Google Scholar]
- 48. Kousholt A. N., Fugger K., Hoffmann S., Larsen B. D., Menzel T., Sartori A. A., Sørensen C. S. (2012) CtIP-dependent DNA resection is required for DNA damage checkpoint maintenance but not initiation. J. Cell Biol. 197, 869–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bohgaki T., Bohgaki M., Cardoso R., Panier S., Zeegers D., Li L., Stewart G. S., Sanchez O., Hande M. P., Durocher D., Hakem A., Hakem R. (2011) Genomic instability, defective spermatogenesis, immunodeficiency, and cancer in a mouse model of the RIDDLE syndrome. PLoS Genet. 7, e1001381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ward I. M., Minn K., van Deursen J., Chen J. (2003) p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol. Cell Biol. 23, 2556–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wu J., Liu C., Chen J., Yu X. (2012) RAP80 protein is important for genomic stability and is required for stabilizing BRCA1-A complex at DNA damage sites in vivo. J. Biol. Chem. 287, 22919–22926 [DOI] [PMC free article] [PubMed] [Google Scholar]