Background: S-Nitrosylation may regulate 2-Cys peroxiredoxin systems to affect cellular metabolism of hydrogen peroxide.
Results: Nitrosylation impeded the peroxiredoxin-1 catalytic cycle by directly inhibiting the activity of peroxiredoxin-1 and by interfering with the recycling of oxidized peroxiredoxin-1 by the thioredoxin system.
Conclusion: Nitrosylation exerts multilevel control of the thioredoxin-peroxiredoxin system.
Significance: Through its multiple effects on the thioredoxin-peroxiredoxin system, nitrosylation may influence cellular redox homeostasis.
Keywords: Hydrogen Peroxide, Nitrosylation, Peroxiredoxin, Thioredoxin, Thioredoxin Reductase
Abstract
S-Nitrosothiols (SNOs), formed by nitric oxide (NO)-mediated S-nitrosylation, and hydrogen peroxide (H2O2), a prominent reactive oxygen species, are implicated in diverse physiological and pathological processes. Recent research has shown that the cellular action and metabolism of SNOs and H2O2 involve overlapping, thiol-based mechanisms, but how these reactive species may affect each other's fate and function is not well understood. In this study we investigated how NO/SNO may affect the redox cycle of mammalian peroxiredoxin-1 (Prx1), a representative of the 2-Cys Prxs, a group of thioredoxin (Trx)-dependent peroxidases. We found that, both in a cell-free system and in cells, NO/SNO donors such as S-nitrosocysteine and S-nitrosoglutathione readily induced the S-nitrosylation of Prx1, causing structural and functional alterations. In particular, nitrosylation promoted disulfide formation involving the pair of catalytic cysteines (Cys-52 and Cys-173) and disrupted the oligomeric structure of Prx1, leading to loss of peroxidase activity. A highly potent inhibition of the peroxidase catalytic reaction by NO/SNO was seen in assays employing the coupled Prx-Trx system. In this setting, S-nitrosocysteine (10 μm) effectively blocked the Trx-mediated regeneration of oxidized Prx1. This effect appeared to be due to both competition between S-nitrosocysteine and Prx1 for the Trx system and direct modulation by S-nitrosocysteine of Trx reductase activity. Our findings that NO/SNO target both Prx and Trx reductase may have implications for understanding the impact of nitrosylation on cellular redox homeostasis.
Introduction
It is well appreciated that reactive oxygen species and reactive nitrogen species participate in a wide array of biological processes that include cell growth, differentiation, and survival and other specialized functions. Beyond their roles in normal physiology, abnormal formation or metabolism of reactive oxygen species/reactive nitrogen species is implicated in an ever-growing number of human disorders, including metabolic, cardiovascular, and autoimmune diseases, neurodegeneration, and cancer (1–4). There is mounting evidence that biologically relevant reactive oxygen species/reactive nitrogen species, particularly hydrogen peroxide (H2O2) and nitric oxide (NO), affect cellular behavior in part through reversible modifications of cysteine residues in proteins. Specifically, H2O2 can react with a cysteine thiol to form a sulfenic acid (SOH),2 whereas NO promotes the conversion of a thiol to a nitrosothiol (SNO), a process known as S-nitrosylation (or S-nitrosation) (5–8). Formation of SOH or SNO can in turn give rise to other thiol modifications, including S-glutathionylation or intra/intermolecular disulfide formation. By regulating the function of various proteins, these reversible thiol modifications play diverse roles in signal transduction and other cellular processes (5, 7, 9).
To date, much research has focused on specific cellular and molecular effects elicited either by H2O2 or SNOs. Less attention has been given to the question of how H2O2 might affect SNO signaling and metabolism and, conversely, how SNO might influence H2O2 homeostasis and consequent cellular responses. Given the ubiquitous distribution and diverse functions of these reactive species, such cross-talk might be common and have potentially important implications for normal and pathological cellular functions. Pertinent to this notion are recent studies, which established that common enzymatic mechanisms are involved in the cellular degradation of both H2O2 and SNO, thus supporting the premise that the fate of these reactive species is intertwined. Specifically, thioredoxin (Trx) proteins and their reducing systems or dependent pathways have come to light as important components in the cellular metabolism and signaling pathways related not only to H2O2 but also to SNO (10–13).
Central components in the cellular redox network are the peroxiredoxins (Prxs), an abundant family of thiol-based peroxidases that detoxify various peroxides, particularly H2O2 (14, 15). The mammalian Prx family is composed of six members (Prx1–6) that are divided into several groups based on their number of conserved cysteine residues and catalytic mechanism: 2-Cys Prxs (Prx1–4), atypical 2-Cys Prx (Prx5), and 1-Cys Prx (Prx6). The structure and catalytic mechanism of 2-Cys Prxs have been extensively studied. These proteins function as obligate dimers, which are further assembled into multimeric structures, typically decameric rings (15, 16). The peroxidase cycle of typical 2-Cys Prxs involves several steps. In the first step a conserved peroxidatic cysteine (CP, Cys-52 in human Prx1) reacts with H2O2 (or alkyl peroxides) to generate Cys-SOH. A resolving cysteine (CR, Cys-173) located on the adjacent monomer next attacks the nascent SOH moiety to generate an intermonomer disulfide. This oxidized dimeric form of Prx (referred to as Ox-Prx) is subsequently reduced by the Trx system, thus completing the peroxidase cycle. This series of processes are linked to transitions in the oligomeric state of Prx, specifically, oxidation by H2O2 promotes, via CP-CR dimerization, the dissociation of the Prx decamer into dimers (15, 16).
The 2-Cys Prx-mediated cellular catabolism of H2O2 may be regulated by several mechanisms. First, endogenous peroxidase activity may be determined by changes in the intracellular levels of specific Prx proteins or their reducing systems even though they are typically found at high levels in most tissues and cell types (17). Prx activity can also be directly modulated by post-translational modifications, including phosphorylation and glutathionylation (15). Notably, under conditions of elevated production of H2O2, 2-Cys Prxs may become inactivated by over-oxidation of CP to sulfinic acid (-SO2H), a reaction that requires catalytic turnover and thus depends on Trx and Trx reductase (TrxR). Over-oxidation of Prxs is associated with structural-functional changes such as formation of higher order multimers that display chaperone activity; however, the physiological significance of this modification is still debated (18). Given the essential role of Trx/TrxR in the peroxidase cycle of 2-Cys Prxs as well as in their over-oxidation, it is expected that post-translational modifications or other cellular controls of Trx or TrxR will also influence 2-Cys Prx function and cellular H2O2 homeostasis.
Several members of the Prx family have been found to undergo S-nitrosylation in multiple cell types and under varying conditions. In neuronal cells, nitrosylation of Prx2 has been shown to inhibit its activity and thereby to contribute to oxidative stress and cellular damage (19). Nitrosylation likewise inhibits the activity of plant Prx II E (20). Prx1 was found to be nitrosylated in endotoxin-stimulated macrophages (21, 22), and recent studies indicate that nitrosylation protects Prx1 from over-oxidation (23). Despite these reports, it remains to be established exactly how S-nitrosylation controls the structure and function of distinct Prx proteins and, more generally, how and to what extent SNOs and H2O2 cross-regulate each other's function. To address these issues, we undertook a detailed analysis of S-nitrosylation of Prx1. In addition and of particular importance to the present study, we investigated the effects of SNO on the entire Prx/Trx/TrxR system. Our studies reveal that nitrosylation exerts complex multilevel control of the 2-Cys Prx reaction cycle. Both the peroxidase reaction itself and, most notably, the regeneration of oxidized Prx by the Trx/TrxR system are shown to be negatively regulated by SNO. These findings may have broad relevance to understanding the biochemical cross-talk between SNO and H2O2.
EXPERIMENTAL PROCEDURES
Cell Culture and Materials
HeLa and RAW264.7 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37 °C under 5% CO2. Tissue culture media and reagents were from Biological Industries (Beit Haemek, Israel). Rabbit polyclonal antibodies for Prx1 were obtained from Abcam (catalogue no. ab41906). Hydrogen peroxide (H2O2) was obtained from Carlo Erba Reagents, Amplex Red was from Cayman Chemical, and NADPH was from Roche Diagnostics. S-Nitrosocysteine (CysNO) was synthesized by combining an equimolar concentration of l-cysteine with sodium nitrite in 0.2 n HCl and used within 1 h. The Prx1(WT)/pET-17b, Prx1(C52S)/pET-17b, Prx1(C173S)/pET-17b, and Prx1(C52/173S)/pET-17b vectors for expression of the recombinant human proteins in bacteria were generously provided by Dr. Hyun Ae (Ewha Womans University, Seoul, Korea). The pET17b-yTrx1 and pET17b-yTrxR vectors for expression of recombinant yeast Trx and yeast TrxR in bacteria, only used here for measuring 2-Cys Prx activity in cell lysates, were generously provided by Dr. Sang Won Kang (Ewha Womans University). Recombinant rat TrxR1 was produced as previously described (24) and had specific activity of 28 units/mg. Other materials were obtained from Sigma unless otherwise indicated.
Site-directed Mutagenesis
Mutant Prx1 plasmids were constructed by using the pET17b-Prx1 plasmid as a template and employing the QuikChange II kit (Stratagene). The following sets of forward and reverse primers were used to introduce mutations at Cys-83: CTGTGGATTCTCACTTCTCTCATCTAGCATGGGTCAATAC (forward) and GTATTGACCCATGCTAGATGAGAGAAGTGAGAATCCACAG (reverse). The double mutants C52S/C83S and C173S/C83S were constructed by a second round of site-directed mutagenesis on C52S and C173S mutants using the above-mentioned primers. All mutants were verified by DNA sequencing.
Expression and Purification of Recombinant Proteins
Escherichia coli BL21 cells expressing human Prx1, either the wild-type or various mutants, were cultured in 500 ml of LB and induced by the addition of 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h. Cells were harvested by centrifugation at 3000 rpm for 10 min at 4 °C, resuspended in 10 ml of extraction buffer (25 mm Tris, 1 mm EDTA, 2 mm DTT, with protease inhibitors, pH 8.8), and disrupted by ultrasonication. The lysate was treated with streptomycin sulfate (1% final concentration) overnight at 4 °C to precipitate nucleic acids. After centrifugation at 12,000 × g for 35 min at 4 °C, the supernatant was loaded on a DE52 anion exchange column (Whatman, Maidstone, England) equilibrated in 25 mm Tris, 1 mm EDTA, 2 mm DTT, pH 8.8. Under these conditions, Prx1 was present in the unbound material. Column fractions were analyzed by SDS-PAGE, pooled, and stored at −80 °C. Protein purity was at least 95% as judged by SDS-PAGE.
E. coli BL21 cells expressing His-tagged human Trx were cultured in 500 ml LB and induced by the addition of 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 6 h. Cells were harvested by centrifugation at 3000 rpm for 10 min at 4 °C, resuspended in 10 ml of extraction buffer (300 mm KCl, 50 mm KH2PO4, 5 mm imidazole, with protease inhibitors, pH 7.4), and disrupted by ultrasonication. After centrifugation at 12,000 × g for 30 min at 4 °C, the supernatant was applied to nickel-nitrilotriacetic acid beads (Thermo Scientific) overnight at 4 °C. The beads were washed with extraction buffer containing imidazole (10 mm), and finally, the protein was eluted with 250 mm imidazole. Fractions were analyzed by SDS-PAGE, pooled, and stored at −80 °C. Protein purity was at least 95% as judged by SDS-PAGE.
Yeast Trx and TrxR proteins were prepared using the same conditions described above for the preparation of human Prx1, with one exception; for yeast TrxR, the pH of the extraction buffer and anion-exchange equilibration buffer was 7.5. These yeast proteins were only used for determination of Prx activity in cell lysates (see below), whereas all other experiments with Trx1 or TrxR1 were performed with the mammalian enzymes.
Nitrosylation and Oxidation of Prx1
Prx1 protein (100 μm) in assay buffer (25 mm HEPES, 0.1 mm EDTA, 0.2 mm diethylenetriaminepentaacetate, 10 μm neocuproine, 0.1% CHAPS, 100 mm NaCl, pH 7.5) was reduced by incubation with 50 mm DTT for 30 min at 37 °C. Excess reducing agent was removed by gel filtration using Sephadex G-25 columns (GE Healthcare) equilibrated in the same buffer. Reduced Prx1 was then incubated with NO/SNO donors at 37 °C as indicated in the legends of Figs. 1 and 2. After excess donor was removed by Sephadex G-25 chromatography, the number of nitrosylated thiols was determined as described below. For oxidation of Prx1, the reduced protein (100 μm) was incubated in the same assay buffer described above and exposed to 100 μm H2O2 for different times at 37 °C. Excess H2O2 was then removed by Sephadex G-25 gel chromatography.
FIGURE 1.
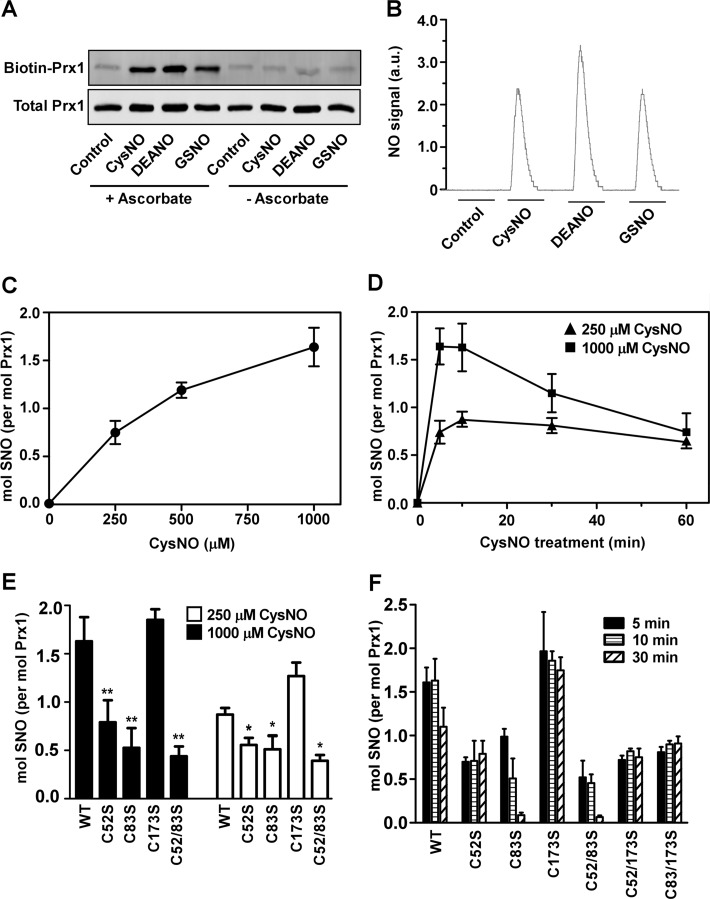
S-Nitrosylation of Prx1 in vitro. A, fully reduced Prx1 was incubated for 10 min at 37 °C with CysNO (250 μm), DEANO (500 μm), or GSNO (250 μm). SNO-Prx1 was assessed by the biotin switch assay in the presence or absence of ascorbate. B, after treatment with different NO/SNO donors as in A, nitrosylation of Prx1 was assessed by chemical reductive chemiluminescence. a.u., arbitrary units. C, after incubation of fully reduced Prx1 with different concentrations of CysNO for 5 min at 37 °C, SNO content was measured by the Saville-Griess assay. D, Prx1 was incubated with 250 or 1000 μm CysNO for different times at 37 °C. SNO content was determined as in C. E, reduced Prx1, WT, or Cys to Ser mutants were incubated with 250 or 1000 μm CysNO for 10 min. SNO content was determined as in C. F, reduced Prx1, WT, or Cys to Ser mutants were incubated with CysNO (1000 μm) for the indicated times. SNO content was determined as in C. Results shown represent the mean ± S.D. n ≥ 3. *, p < 0.05; **, p < 0.01 for mutant versus WT by one-way analysis of variance with post hoc Tukey test.
FIGURE 2.
SNO-induced changes in the thiol-disulfide and oligomerization state of Prx1. A, reduced Prx1 was incubated with CysNO (250 μm) for different times at 37 °C, and its redox state was then assessed by non-reducing SDS-PAGE. The Prx1 monomer is indicated by an asterisk, the dimer is indicated by double asterisk, and the high molecular weight species are indicated by triple asterisks. B, reduced Prx1, WT, or Cys to Ser mutants were incubated with or without CysNO (250 μm) for 60 min at 37 °C. The samples were then analyzed by non-reducing SDS-PAGE. C, shown are gel filtration chromatograms of reduced, nitrosylated, and oxidized Prx1. Reduced Prx1 was either left untreated or exposed to CysNO (1 mm, 60 min) or to H2O2 (100 μm, 60 min). Protein samples (0.66 mg) were fractionated using Superdex 200 size exclusion chromatography as described under “Experimental Procedures.” D, reduced Prx1 (either WT or C52S) were left untreated or exposed to CysNO (250 μm, 60 min). Protein samples (0.66 mg) were analyzed as in C. The elution profile of standard proteins was as follows: ferritin, 10.7 ml; aldolase, 12.8 ml; conalbumin, 14.5 ml; ovalbumin, 15 ml; ribonuclease, 18 ml.
Determination of Free Thiol Groups
Protein thiols were measured spectrophotometrically after reaction with 5,5′-dithio-bis(2-nitrobenzoic acid) (DTNB, Ellman's reagent). In brief, samples were diluted into a 200-μl reaction mix (1 mm DTNB, 6 m guanidine hydrochloride, 90 mm HEPES, pH 7.5) for 10 min, and the absorbance readings were taken at 412 nm. Values were derived by comparison with glutathione standards and were normalized to protein concentration.
Assessment of Nitrosylation of Prx1 under Cell-free Conditions
Quantification of nitrosylated thiols in Prx1 was determined by the Saville-Griess assay. Briefly, the nitrosylated protein was incubated in a final volume of 200 μl of assay buffer (1% sulfanilamide, 0.1% N-(1-naphthyl)ethylenediamine dehydrochloride, 1% HCl) in the absence or presence of 1 mm HgCl2 for 30 min, and absorbance readings were measured at 540 nm. Mercury-dependent absorbance was converted to SNO concentrations using S-nitrosoglutathione (GSNO) standards treated identically. The concentration of GSNO was determined by UV-visible spectrophotometer by measuring absorbance at 334 nm (extinction coefficient of 920 m−1cm−1).
Nitrosylation of Prx1 was also determined by chemical reductive chemiluminescence (25) or using a modified version of biotin switch method (26). In brief, after the various treatments (as indicated in the legend of Fig. 1), the blocking step was performed at 50 °C for 30 min in the presence of 50 mm N-ethyl maleimide (NEM) with frequent vortexing. Excess NEM was removed by Sephadex G-25 chromatography. This material was added to 50 mm sodium ascorbate and 1 mm N-(3-maleimidylpropionyl)biocytin (Invitrogen) for 1 h at room temperature. Excess labeling reagent was removed by Sephadex G-25 chromatography. The samples were analyzed by Western blotting using 5-FAM-streptavidin conjugate (AnaSpec).
Analysis of Modifications of Prx1 by Mass Spectrometry
After treatment of reduced Prx1 with vehicle or CysNO (250 μm, 10 min) sample buffer was replaced with 8 m urea (in 65 mm ammonium bicarbonate) using a 10-kDa cutoff filter (Millipore). The proteins in the samples were digested in 2 m urea with modified trypsin (Promega) at a 1:50 enzyme-to-substrate ratio overnight at 37 °C without reduction/alkylation of cysteines. The resulting tryptic peptides were resolved by reverse-phase chromatography on 0.075 X 200-mm fused silica capillaries (J&W) packed with Reprosil reversed phase material (Dr Maisch GmbH, Germany). The peptides were eluted with 45-min linear gradients of 5 to 45% and 10 min at 95% acetonitrile with 0.1% formic acid in water at flow rates of 0.25 μl/min. Mass spectrometry (MS) was performed by an ion-trap mass spectrometer (Orbitrap XL, Thermo) in a positive mode using repetitively full MS scan followed by collision induces dissociation of the seven most dominant ion selected from the first MS scan. Data were processed using Thermo Scientific Proteome Discoverer software (Version 1.3). MS peptide spectra were searched by the SEQUEST against the human UniProt-SwissProt protein database and decoy database (to determine the false discovery rate) and against the specific database. The mass tolerances for precursor and fragment ions were set to 10 ppm and 0.5 Da, respectively. Variable modifications selected for searching included oxidation of methionine and cysteine, dioxidation and trioxidation of cysteine, and nitrosylation of cysteines, tryptophan, and phenylalanine.
Assessment of Nitrosylation of Cellular Prx1
Detection of nitrosylated Prx1 was performed using the SNO-RAC (resin-assisted capture) method (21) with minor modifications as follows. After different treatments as indicated in the legend of Fig. 3, cells were incubated with phosphate-buffered saline containing NEM (100 mm) for 10 min at room temperature and then lysed in lysis buffer (50 mm HEPES, 1% Nonidet P-40, 150 mm NaCl, 1 mm EDTA, 0.1 mm diethylenetriaminepentaacetate, 50 mm NEM with protease inhibitors, pH 7.5). Cell debris was removed by centrifugation at 10,000 × g for 10 min at 4 °C. A total of 3 mg of protein was used for each experimental condition. The blocking step was performed at 50 °C in the presence of 20 mm NEM with frequent vortexing. To remove excess NEM, proteins were precipitated with 3 volumes of acetone at −20 °C for 30 min. The proteins were recovered by centrifugation at 5000 × g for 5 min, and the pellets were resuspended in HENS buffer (250 mm HEPES, 1 mm EDTA, 0.1 mm neocuproine, 1% SDS, pH 7.7). This material was added to 40 μl of thiopropyl-Sepharose resin in the presence of 20 mm sodium ascorbate. After rotation in the dark overnight at 4 °C, the resin was washed with 4 × 1 ml of HENS buffer, then 2 × 1 ml of HENS/10 buffer (HENS diluted 1:10). Captured proteins were eluted with 30 μl of HENS/10 containing 100 mm 2-mercaptoethanol for 20 min at room temperature and analyzed by Western blotting using an anti-Prx1 antibody.
FIGURE 3.
Nitrosylation and resultant redox structural changes in cellular Prx1. A, RAW264.7 cells were treated with CysNO (500 μm) for the indicated times, and S-nitrosylation of Prx1 was then assessed by SNO-RAC. The redox status of Prx1 was further assessed by non-reducing Western blotting. B, RAW264.7 cells were treated for 10 min with GSNO or DEANO (500 μm each), and S-nitrosylation of Prx1 was then assessed by SNO-RAC. C, HeLa cells were treated for 10 min with the indicated concentrations of CysNO. S-Nitrosylation and dimerization of Prx1 were assessed as in A. D, RAW264.7 cells were either left untreated or treated with CysNO, DEANO, or GSNO (500 μm each). Cell lysates were fractionated by gel filtration chromatography as described under “Experimental Procedures.” Each fraction was separated by SDS-PAGE and immunoblotted (IB) for Prx1. The positions where the peaks of protein standards eluted (440, 158, 43, and 13 kDa) are indicated.
SDS-PAGE and Western Blot Analysis
Reduced, nitrosylated, or oxidized recombinant Prx1 (2 μg) was analyzed by SDS-PAGE using reducing or non-reducing conditions. The gels were stained with Krypton Infrared Protein Stain (Pierce) and visualized using the Odyssey infrared imaging system (LI-COR Biosciences). For immunodetection, aliquots containing 200 ng of recombinant Prx1 or 20 μg of total cell lysate were subjected to SDS-PAGE and Western blotting with anti-Prx1 antibody. Protein bands were detected and quantified with the Odyssey system.
Gel Filtration Chromatography
A total of 0.66 mg of reduced, nitrosylated, or oxidized Prx1 was loaded onto a Superdex 200 column (10 × 300 mm) equilibrated with buffer containing 25 mm HEPES, 0.1 mm EDTA, and 100 mm NaCl, pH 7.5. Proteins were eluted at a flow rate of 0.5 ml/min, and 0.5 ml fractions were collected. Ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa), ovalbumin (43 kDa), and ribonuclease (13.7 kDa) were used as molecular weight standards. Gel filtration analysis of endogenous Prx1 was performed using similar conditions. In brief, cells were treated with vehicle, CysNO (500 μm, 10 min), diethylamine NONOate (DEANO; 500 μm, 10 min), or GSNO (500 μm, 60 min). Superdex 200 fractionation was carried out using 1.5 mg of cell lysates. Twenty-microliter aliquots were analyzed by SDS-PAGE and Western blotting with anti-Prx1 antibody.
Determination of Peroxidase Activity
Peroxidase activity was measured based on a previously reported method (27) and as follows. Untreated or nitrosylated Prx1 (10 μm) diluted in 25 mm potassium phosphate, 1 mm EDTA, pH 7.0, was incubated with the Trx system (5 μm Trx, 0.1 μm TrxR, and 450 μm NADPH) and 500 μm H2O2 at 37 °C for 10 min. Twenty-microliter aliquots were taken every minute and transferred into 980 μl of Fox reagent (100 mm sorbitol, 125 μm xylenol orange, 250 μm ferrous ammonium sulfate, and 25 mm H2SO4). The absorbance at 560 nm was read after 30 min and compared with a H2O2 standard curve. Peroxidase activity was derived from the linear regression of absorbance versus time. Prx activity in whole cell lysates was measured according to the method of Kim et al. (28), employing yeast Trx and yeast TrxR.
For measurement of cell-dependent peroxide reduction, cells incubated in serum-free medium were treated with H2O2 and/or CysNO as indicated in the legend of Fig. 5, and the amount of H2O2 remaining in the medium over time was then determined using an Amplex Red-based assay as described elsewhere (29).
FIGURE 5.
Influence of CysNO and H2O2 on the coupled Prx1/Trx/TrxR system; potent inhibition by CysNO of peroxidase activity and mild inhibition by H2O2 of CysNO reduction. A, reduced Prx1 (10 μm) was incubated with the Trx system (5 μm Trx, 0.1 μm TrxR, and 450 μm NADPH), H2O2 (500 μm) and with different concentrations of CysNO for 2 min at 37 °C. H2O2 levels were determined using the Fox assay and are compared with controls. The activity of reduced (unmodified) Prx1 was considered as 100%. B, the Trx/TrxR system was incubated with reduced Prx1 (10 μm) and CysNO (100 μm) in the presence of different concentrations of H2O2 for 30 min at 37 °C. SNO content was determined using the Saville-Griess assay. Denitrosylase activity in the absence of H2O2 was considered as 100%. C, HeLa cells were treated with or without CysNO (50 μm) for 10 min followed by the addition of H2O2 (200 μm). Remaining H2O2 levels at the indicated times were measured by Amplex Red assay. Results shown represent the mean ± S.D. (n ≥ 3).
Denitrosylation Assay
The Trx system (5 μm Trx, 0.1 μm TrxR, and 450 μm NADPH) was incubated in assay buffer (25 mm potassium phosphate, 1 mm EDTA, pH 7.0) with reduced Prx1 (10 μm), CysNO (100 μm) and different concentrations of H2O2 at 37 °C for different times. SNO content was determined using the Saville-Griess reaction as described above. Denitrosylation activity was derived from the slope of linear regression of absorbance versus time plots.
Determination of Trx/TrxR Activity
The activities of Trx and TrxR were determined using the Trx-linked insulin reduction assay (30). In brief, the Trx system (5 μm Trx, 0.1 μm TrxR, and 250 μm NADPH) diluted in assay buffer (50 mm HEPES, 2.6 mm EDTA, pH 7.5) was incubated with 1.5 mg/ml insulin, and the rate of NADPH oxidation was monitored at 340 nm. To test the effect of SNO on Trx and TrxR activities, a slightly modified procedure was employed as follows. Trx (100 μm) was reduced by incubation with 10 mm DTT for 30 min at 37 °C, and the reduced protein (20 μm) was then exposed to different CysNO concentrations (0–200 μm) for 10 min at 37 °C. After each of these steps, excess modifying reagent (DTT or CysNO) was removed by gel filtration. Finally, the protein was adjusted to 5 μm, to which the remainder of the insulin assay system was added. Alternatively, TrxR (0.4 μm) was incubated with CysNO (0–200 μm) in the presence of 1 mm NADPH for 10 min at 37 °C. The reaction mixture was then diluted 1:4 and combined with the rest of the insulin assay mixture.
TrxR activity was also measured using the direct DTNB reduction assay (30). TrxR was diluted in assay buffer (25 mm potassium phosphate, 1 mm EDTA, pH 7.0) to 15 nm and incubated with 250 μm NADPH and 2.5 mm DTNB. The rate of DTNB reduction in the absence or presence of CysNO was monitored at 412 nm.
Determination of Trx and TrxR Activities in Cell Lysates
Activities of TrxR and Trx in cell lysates were measured in 96-well microtiter plates using an end point insulin assay (30). For TrxR activity measurement, 25 μg of protein cell lysate was incubated in a final volume of 50 μl containing 0.3 mm insulin, 660 μm NADPH, 2.5 mm EDTA, and 5 μm human Trx in 85 mm HEPES, pH 7.5, for 20 min at room temperature. Control reactions excluding human Trx were also set up. Then, 250 μl of 1 mm DTNB, 240 μm NADPH, and 200 mm Tris-HCl, pH 8, in 6 m guanidine hydrochloride was added, and the absorbance was monitored at 412 nm. For determination of Trx activity, procedures were similar as for TrxR activity assay; 10 μg of protein cell lysate was incubated in reaction solution but with the addition of 600 nm TrxR instead of 5 μm Trx (except for control wells).
RESULTS
Nitrosylation of Prx1 in Vitro
To investigate potential cross-talk between SNO and H2O2 metabolism, we first set out to characterize nitrosylation of Prx1 in vitro using several NO/SNO donors. Chemically reduced human Prx1, which contains four thiols groups, was reacted with 2–5 equivalent excess of CysNO, DEANO, or GSNO. After removal of unreacted donors by gel filtration, nitrosylation was assessed by three different methods, namely the biotin switch assay, chemical reductive chemiluminescence, and the Saville-Griess assay, as described under “Experimental Procedures.” Brief exposure (5–10 min) to CysNO, DEANO, or GSNO led to appreciable nitrosylation of Prx1 (Fig. 1, A and B, and Table 1). The biotin switch assay specificity was confirmed by the observation that omission of ascorbate in the assay system abolished the detection of SNO-Prx1 (Fig. 1A). Likewise, SNO-Prx1 was not detected after treatment of Prx1 with cysteine, glutathione, or diethylamine (data not shown). A quantitative dose-response analysis using CysNO as a donor (250–1000 μm; 5 min exposure) showed that the number of SNO moieties was between ∼0.75 and 1.65 mol of SNO/mol Prx1 (Fig. 1C and Table 1). We examined the kinetics of Prx1 nitrosylation at two doses of CysNO, 250 and 1000 μm. This analysis revealed that Prx1 was maximally nitrosylated already at 5–10 min after the addition of CysNO; after 10 min, Prx1 progressively denitrosylated, with the time-dependent effect more evident at the higher CysNO dose (Fig. 1D). Of note, thiol quantification of the samples indicated that treatment of Prx1 with the different NO/SNO donors did not induce significant cysteine oxidation (distinct from nitrosylation; see Table 1).
TABLE 1.
The characteristics of reduced and nitrosylated Prx1
The number of thiols in the control sample (vehicle treated) is slightly below the expected value of 4 due to small degree of oxidation during gel filtration. The values are shown as the means ± S.D. (n ≥ 3).
| Treatment | Number of free SH groups | Number of SNO groups |
|---|---|---|
| Vehicle | 3.25 ± 0.20 | 0.00 ± 0.00 |
| CysNO (250 μm) | 2.28 ± 0.11 | 0.74 ± 0.12 |
| CysNO (500 μm) | 2.11 ± 0.30 | 1.19 ± 0.02 |
| CysNO (1000 μm) | 1.58 ± 0.32 | 1.64 ± 0.19 |
| DEANO (500 μm) | 2.00 ± 0.15 | 1.00 ± 0.16 |
| GSNO (250 μm) | 2.63 ± 0.23 | 0.48 ± 0.03 |
To localize the nitrosylation sites within Prx1 we employed several mutant constructs in which key cysteine residues were replaced by serine. As shown in Fig. 1, E and F, a Prx1 mutant lacking the peroxidatic Cys-52 (C52S) displayed significant reduction (up to 50%) in nitrosylation compared with wild type (WT). Cys-83 carries another redox-active thiol that may play a role in stabilizing the Prx1 decamer by increasing the affinity between dimers and under certain conditions may form disulfide bonds (Cys-83–Cys-83) at the dimer-dimer interface (31). Similar to Prx1(C52S), the C83S mutant was also less nitrosylated. Contrasting with these results, nitrosylation of a Prx1 mutant lacking resolving Cys-173 was somewhat elevated compared with WT, particularly at the lower dose of CysNO. Among the Prx1 variants tested, a mutant lacking both Cys-52 and Cys-83 (C52S/C83S) appeared to be the least nitrosylated (Fig. 1, E and F). These results were confirmed using another quantitative method (chemical reductive chemiluminescence; data not shown). These findings suggest that under the current experimental conditions, nitrosylation of Prx1 substantially occurs at Cys-52 and also Cys-83.
To directly identify cysteine residues of Prx1 that are nitrosylated, we performed mass spectrometric analyses. Untreated and CysNO-treated Prx1 were submitted to tryptic digestion, and peptides were analyzed by LC-MS/MS (see “Experimental Procedures”). As summarized in Table 2, the MS analysis revealed the presence of several peptides containing a cysteine residue with an additional mass of 29 Da, equivalent to a NO moiety. Consistent with the mutant analysis, the MS/MS data indicated the presence of Cys-52-SNO and Cys-83-SNO adducts in the nitrosylated sample (Table 2). In addition, formation of SNO adducts with Cys-71 and Cys-173 could also be inferred from the MS/MS spectra; however, the data did not allow a quantitative estimate of the relative abundance of the different nitrosopeptides. As such, the MS results may be taken as qualitative confirmation of nitrosylation. In addition to nitrosylation, several other nitroso-oxidative modifications were identified, specifically, sulfenic acid formation (at Cys-173), sulfonic acid formation (at Cys-71), and tryptophan nitrosation (Trp-177) (Table 2). As noted above, however, other forms of thiol oxidation were quantitatively very small compared with nitrosylation (see Table 1).
TABLE 2.
Nitrosopeptides identified by mass spectrometry analysis of CysNO-treated Prx1
The nitrosopeptides were identified as described under “Experimental Procedures.” Mass accuracy was less than 5 ppm for all selected fragment ions. The q-value is the minimal false discovery rate at which the identification is considered correct. No nitrosopeptides were identified in the control, untreated Prx1 sample. c, Cys; w, Trp.
| Residues | Sequence | Modifications | MH+ | q-Value | Charge |
|---|---|---|---|---|---|
| 46–78 | LDFTFVcPTEIIAFSDRAEEFKKLNcQVIGASV | Cys-52-SNO, Cys-71-SO3H | 3767.82 | 0.004848 | 5 |
| 169–190 | HGEVcPAGWKPGSDTIKPDVQK | Cys-173-SNO | 2378.16 | 0.000463 | 4 |
| 69–92 | LNcQVIGASVDSHFCHLAWVNTPK | Cys-71-SNO | 2668.28 | <10−5 | 4 |
| 68–92 | KLNCQVIGASVDSHFcHLAWVNTPK | Cys-83-SNO | 2796.35 | <10−5 | 4 |
| 169–190 | HGEVcPAGwKPGSDTIKPDVQK | Cys-173-SOH, Trp-177-NNO | 2394.16 | <10−5 | 3 |
Effect of Nitrosylation on the Thiol-Disulfide and Oligomerization State of Prx1
We next asked whether S-nitrosylation might influence the disulfide redox state of Prx1. To address this question, we analyzed the untreated and nitrosylated protein on non-reducing SDS-PAGE. Whereas reduced Prx1 almost entirely migrated as a monomer at 22 kDa, CysNO-treated Prx1 gradually accumulated in the dimeric form (about 44 kDa, Fig. 2A). Thus, nitrosylation induces the transition toward the oxidized (disulfide-linked) form of Prx1 (Ox-Prx1), an effect similar to that induced by H2O2 (14), albeit at a slower rate. In addition to dimers, higher molecular weight (MW) species were also observed in the nitrosylated sample (Fig. 2A). Formation of disulfides paralleled the kinetics of auto-denitrosylation of Prx1 noted above (Fig. 1D). SNO-dependent dimer formation was virtually abolished in the C52S Prx1 mutant (Fig. 2B). In contrast, the C83S mutant displayed elevated dimer content, a finding consistent with its faster denitrosylation (see also Fig. 1F); however, little accumulation of the higher MW species was detected in this mutant (Fig. 2B). The double mutants, C52S/C83S, C52S/C173S, and C83S/C173S, were affected little by CysNO, essentially remaining in their reduced, non-disulfide-linked form. These data suggest that nitrosylation of Prx1 facilitates the formation of intermolecular disulfides, most likely through Cys-52–Cys-173 and Cys-83–Cys-83 couples, which thereby also appears to denitrosylate the protein. Although less likely from our data, we cannot rule out the possibility that the CysNO-induced dimerization may to some extent occur through oxidative-based mechanisms.
To assess possible effects of nitrosylation on Prx1 oligomerization, we employed gel filtration chromatography. Consistent with previous reports (31–33), we observed that reduced Prx1 in solution existed mainly in the decameric state (Fig. 2C). Treatment of Prx1 with CysNO, however, resulted in marked changes of its oligomeric state. In particular, a decrease of the decamer peak with the concomitant increase of the dimer peak was conspicuous (Fig. 2C). Additional species of intermediate size (larger than dimer but smaller than decamer) were also apparent in the nitrosylated Prx1 sample. These structural changes were similar to those induced by H2O2, although some differences were noted. In particular, H2O2, but not CysNO, induced the formation of very large molecular species (>500 kDa), which likely represent the formation of dodecamers or higher order multimers (Fig. 2C) (16). Notably, CysNO failed to induce decamer breakdown in the C52S mutant (Fig. 2D). Thus, it appears that nitrosylation of Cys-52 promotes disulfide bond formation with Cys-173, thereby disrupting the decameric structure (34).
Nitrosylation of Prx1 in Cells
To investigate S-nitrosylation of endogenous Prx1, we exposed RAW264.7 macrophages to CysNO treatment and assayed for SNO-Prx1 formation with the SNO-RAC method (21). Similar to the in vitro results, endogenous Prx1 was rapidly nitrosylated in response to treatment with CysNO, with maximal nitrosylation observed at the 5-min time point (Fig. 3A). After 5 min, the level of nitrosylation gradually decreased. Concomitant with the denitrosylation phase, a higher proportion of Prx1 accumulated in the disulfide form, as assessed by non-reducing SDS-PAGE (Fig. 3A). However, under these experimental conditions, we did not detect accumulation of higher molecular weight species (larger than dimers) as observed under the cell-free conditions. Nitrosylation of Prx1 was also seen after macrophage treatment with DEANO or GSNO (Fig. 3B) and in a second cell type (HeLa cells, Fig. 3C). Fractionation of whole cell lysates by gel filtration chromatography revealed that in resting cells, Prx1 predominated in the high MW fractions (>200 kDa), whereas in cells exposed to exogenous NO/SNO it was present primarily in the low MW fractions, ranging from ∼20 to ∼40 kDa (Fig. 3D), suggesting that nitrosylation promoted oligomer dissociation. Overall, the results demonstrate that cellular Prx1 is susceptible to S-nitrosylation and that nitrosylation triggers similar redox-coupled structural changes in vitro and in intact cells.
Effect of Nitrosylation on the Peroxidase Activity of Prx1
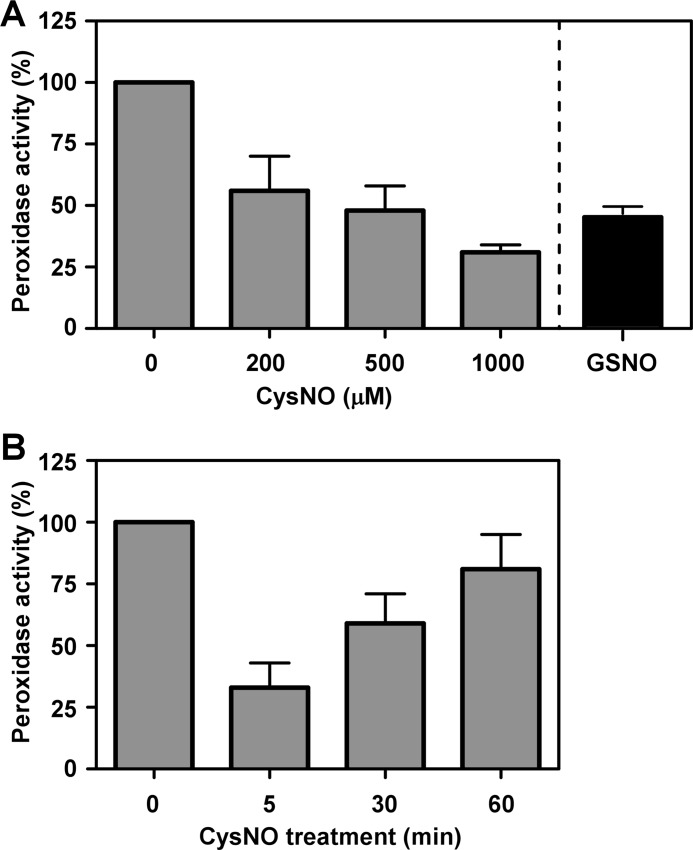
We next examined whether nitrosylation might affect the peroxidase activity of Prx1 in vitro. We used similar experimental conditions to induce nitrosylation of Prx1 as described above (Figs. 1 and 2) and subsequently assayed the H2O2 reduction activity of the untreated or the CysNO-treated protein as described under “Experimental Procedures.” Results of these experiments showed that CysNO rapidly and dose-dependently inhibited Prx1 activity. Specifically, exposure to 1 mm CysNO for 5 min (the time point where nitrosylation was maximal) resulted in a 70% decrease in Prx1 activity in comparison to the non-nitrosylated protein (Fig. 4A). Similarly, Prx1 that was nitrosylated for 10 min with 1 mm GSNO exhibited diminished activity (45 ± 3% versus control, Fig. 4A). A time course analysis revealed that the SNO-dependent Prx1 inhibition was transient and was followed by a reactivation phase (Fig. 4B). Indeed, at the 60-min time point, Prx1 inhibition amounted to only 20% (Fig. 4B). Thus, nitrosylation directly inhibits the peroxidase activity of Prx1, but as the protein undergoes auto-denitrosylation coupled to disulfide formation its activity is restored.
FIGURE 4.
SNO effect on the peroxidase activity of Prx1. A, reduced Prx1 was incubated at 37 °C for 5 min with different concentrations of CysNO or with GSNO (1 mm, 10 min). After SNO removal, thioredoxin-dependent peroxidase activity was determined as described under “Experimental Procedures.” The activity of reduced (unmodified) Prx1 was considered as 100%. B, reduced Prx1 was incubated with 1 mm CysNO at 37 °C for different times. Peroxidase activity was determined as in A. Results shown represent the mean ± S.D. n ≥ 3.
Effect of SNO on the Prx1/Trx/TrxR System
The peroxidase reaction cycle of 2-Cys Prxs (such as Prx1) is dependent upon Trx and TrxR. Therefore, we deemed it important to explore the possible effects of SNO on the entire Prx/Trx/TrxR system. For this purpose, we first employed a cell-free system using reactant concentrations (10 μm Prx1, 5 μm Trx, and 0.1 μm TrxR) that approximate those found in vivo. Interestingly, under these experimental conditions, we found that the peroxidase activity of the coupled system was rapidly and robustly inhibited by low doses of CysNO (Fig. 5A). Indeed, short (2 min) treatment with only 5 μm CysNO (in the presence of 500 μm H2O2) resulted in close to 50% inhibition of peroxidase activity. Treatment with higher CysNO concentrations led up to 80% inhibition (Fig. 5A).
As Trx/TrxR not only support Prx activity but also directly catalyze SNO reduction (11–13, 35, 36), the results depicted in Fig. 5A could be indicative of competition between CysNO and H2O2 for the Prx/Trx/TrxR system (or a component thereof); hence, we assessed the reverse case, namely, if H2O2 can inhibit the CysNO reduction activity of the system. In fact, we found this to be the case; however, the inhibitory effect of H2O2 on Trx/TrxR-dependent denitrosylation activity was relatively mild (up to 40%) and required a 5-fold molar excess of H2O2 relative to CysNO (Fig. 5B). These data show that each of the two substrates in the system (the SNO and H2O2) can slow down the reduction of the other, and they further indicate that the SNO has much greater potency in inhibiting H2O2 reduction compared with the reverse case. In line with this, we also found that CysNO attenuated H2O2 removal in a cellular context (Fig. 5C), but whether this effect was mediated by the Prx-Trx system is not clear and calls for further studies.
SNO Inhibits Trx/TrxR-dependent Regeneration of Oxidized Prx1
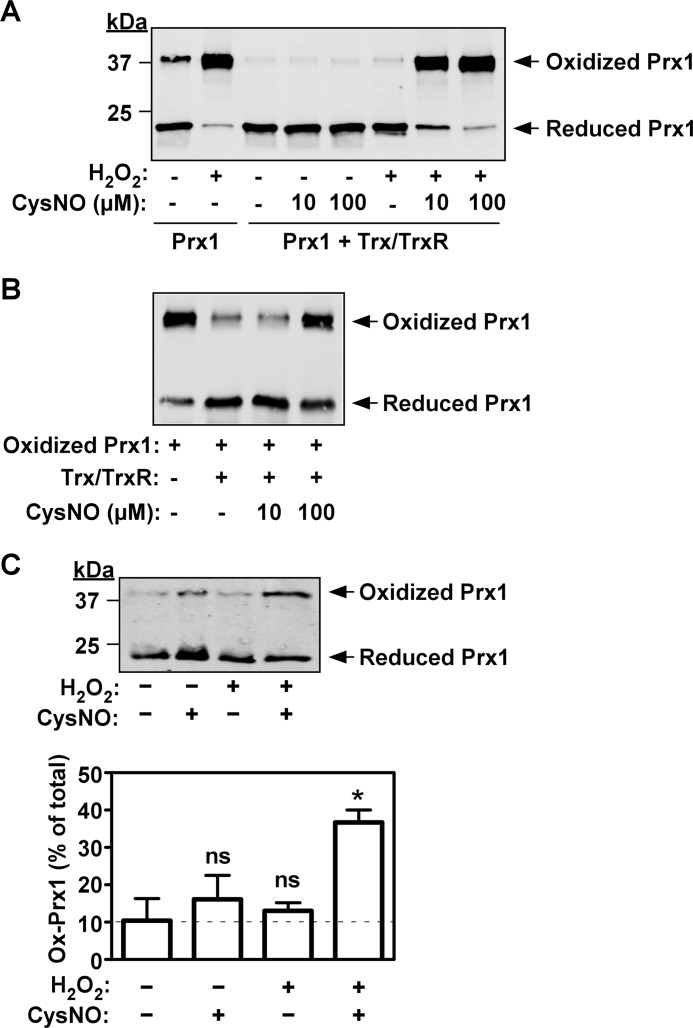
How does SNO exert such potent inhibition on the peroxidase system? 2-Cys Prxs are oxidized very rapidly by H2O2 (rate constants of 1 × 105 to 1 × 107 m−1 s−1) (14, 15, 37); thus, a direct competition for Prx between the SNO and H2O2 is unlikely, and an alternative explanation must be sought. Both in vitro and in vivo Ox-Prx is effectively regenerated by the Trx system, and therefore, under normal physiologically relevant conditions, the population Ox-Prx1 should constitute only a small fraction of total Prx. Thus, it was conceivable that the inhibition of the peroxidase activity could be explained by competition for the Trx system between the SNO and the relatively small pool of Ox-Prx1. To explore this possibility, the complete coupled enzyme system was exposed to CysNO, and the redox status of Prx1 was then analyzed on non-reducing gel. This analysis showed that in the absence of CysNO but in the presence of a relatively high H2O2 concentration (500 μm), the Trx system effectively maintained Prx1 in its reduced form (Fig. 6A). Strikingly, when CysNO (10 μm) was co-administered with H2O2, Prx1 was found to be almost completely in the oxidized (disulfide-linked) form (Fig. 6A), a finding that highlights the capacity of CysNO to interfere with Trx/TrxR-mediated reduction of Ox-Prx1. When the initial concentration of Ox-Prx1 was increased to 50 μm by complete oxidation with H2O2 (followed by peroxide removal), a higher concentration of CysNO (100 μm) was required to effectively inhibit Prx1 regeneration (Fig. 6B). These data are consistent with the notion that by competing for the Trx system, SNO attenuates the recycling of Ox-Prx1.
FIGURE 6.
SNO-mediated inhibition of the Trx/TrxR-catalyzed reduction of oxidized Prx1. A, reduced Prx1 (10 μm) was incubated with or without H2O2 (500 μm) for 2 min at 37 °C in the absence or presence of the Trx system (5 μm Trx, 0.1 μm TrxR, and 450 μm NADPH) and CysNO as indicated. The redox status of Prx1 was determined by Western blot analysis using non-reducing conditions. B, oxidized Prx1 (50 μm, generated by pretreatment of reduced Prx1 with 500 μm H2O2 for 10 min) was incubated for 2 min at 37 °C in the absence or presence of the Trx system and CysNO as indicated. The redox status of Prx1 was determined as in A. C, top, HeLa cells were treated with or without CysNO (50 μm) for 10 min followed by the addition of H2O2 (200 μm) for 10 min. The redox status of Prx1 in whole cell lysates was determined by Western blot analysis using non-reducing conditions. Bottom, quantitative analysis of the levels of Ox-Prx1 is shown. The dashed line indicates the fraction of Ox-Prx1 in resting cells. Results (mean ± S.D.) are based on three independent experiments. *, p < 0.01 versus untreated cells; ns, non-significant versus untreated cells by one-way analysis of variance with post-hoc Tukey test.
To assess if the cellular Trx/Prx system is similarly affected by SNO, we employed HeLa cells, in which Prx1 accounts for the majority of 2-Cys Prx activity (28). In resting cells, about 10% of Prx1 was found to exist as Ox-Prx1, and this fraction remained essentially unchanged by treatment with H2O2 (Fig. 6C). Treatment with CysNO alone only marginally increased the level of Ox-Prx1. Notably, combined treatment with CysNO and H2O2 significantly and synergistically increased the proportion of Ox-Prx1 relative to control (Fig. 6C). These observations suggest that elevated SNO levels interfere with Trx system-dependent regeneration of Ox-Prx1 also in a cellular context.
Effect of SNO on Trx/TrxR Activities
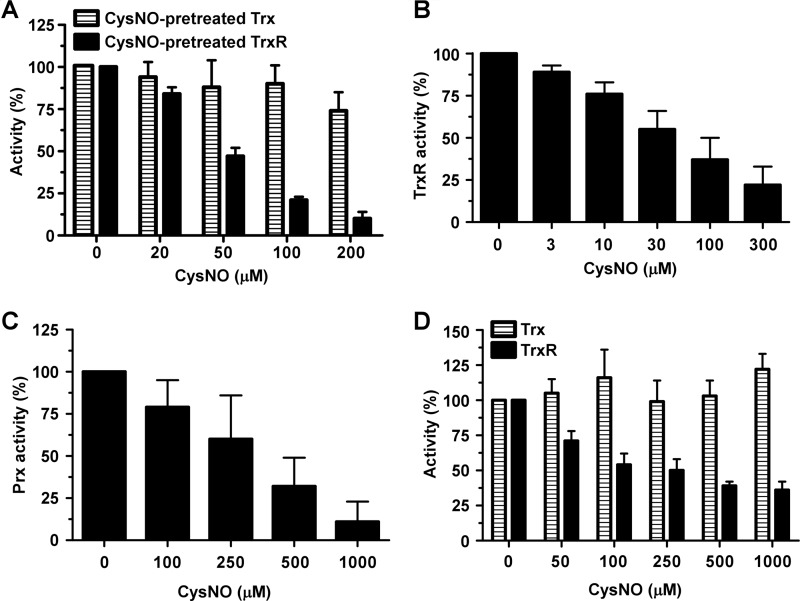
Despite the above findings suggesting that SNO blocks Prx1 recycling by competing for the reducing system (i.e. Trx/TrxR), we suspected that SNO may also directly inhibit (or modulate) the activity of Trx and/or TrxR. To address this possibility we employed the Trx-linked insulin reduction assay to quantitatively evaluate and compare the activities of untreated and SNO-treated Trx and TrxR. Specifically, either Trx or TrxR in reduced forms were preincubated with CysNO, then reconstituted with the complementary, untreated components of the system and finally assayed for insulin reduction. Interestingly, we found that exposure of TrxR to increasing concentrations of CysNO markedly and dose-dependently inhibited insulin reduction, whereas similar treatments of Trx had a minimal effect (Fig. 7A). These data suggest that TrxR activity can be directly modulated by CysNO. Indeed, inhibition of TrxR by CysNO was also confirmed by the use of the direct DTNB reduction assay (Fig. 7B). The in vitro experiments described in Figs. 4 and 7, A and B, collectively suggest that SNOs directly modulate the activities of Prx1 as well as TrxR. To further verify these findings, we measured the activities of Prx, Trx, and TrxR in lysates obtained from control and CysNO-treated HeLa cells. Total Prx activity was found to be inhibited in CysNO-treated cells in a dose-dependent fashion (Fig. 7C). Also, in the same setting, TrxR activity was effectively inhibited (Fig. 7D). In contrast, Trx activity was essentially unaffected by CysNO (Fig. 7D). Put together, these data strongly suggest that the SNO inhibitory effects on the Prx cycle largely occur at the level of Prx and TrxR, with the latter playing a dominant role in the regeneration step.
FIGURE 7.
SNO effects on the activities of recombinant and endogenous Trx and TrxR and endogenous Prx. A, either Trx or TrxR was preincubated for 10 min with the indicated concentrations of CysNO and then reconstituted with the complementary, untreated components of the Trx system. Insulin reduction activity was then measured as described under “Experimental Procedures.” B, TrxR activity using DTNB as substrate was determined in the presence of the indicated concentrations of CysNO. C, HeLa cells were treated for 10 min with different concentrations of CysNO. Total Prx activity in cell lysates was determined as described under “Experimental Procedures.” D, HeLa cells were treated as in C followed by measurement of Trx or TrxR activity in cell lysates. In A–D, the activities of the untreated samples were taken as control (100%). Results shown are represent means ± S.D. n ≥ 3.
DISCUSSION
Both NO and H2O2 are recognized as mediators and/or modulators of a broad range of cellular signaling pathways. The influence of these reactive species on signal transduction and other cellular functions is substantially mediated through nitrosylation/oxidation of critical cysteine residues in proteins (5–8). As such, cross regulation between S(NO) and H2O2 metabolism can potentially affect many cellular processes, yet molecular insights into the nature of this cross-talk remain scarce. Our exploration of the effect of SNO on the 2-Cys Prx cycle advances our understanding of this topic.
Extensive research has shown that Prxs play important roles in the cellular response to peroxides, particularly H2O2 (14, 15). More than a decade ago it was discovered that Prxs have a peroxynitrite reductase activity, thus linking these thiol peroxidases to reactive nitrogen species metabolism (38). Recent reports have provided growing evidence for cross-regulation between Prxs and NO. For example, in a number of experimental models, NO or nitrosative stress induces the expression of specific Prxs, including Prx1 (39, 40). Other recent studies have indicated that S(NO) can directly modulate several members of the Prx family by means of S-nitrosylation (19, 20, 23). In this study we provided additional mechanistic insights into the regulation of a 2-Cys Prx by SNO. A novel and important aspect of the present study was to investigate the effects of SNO on the complete coupled Prx/Trx/TrxR system, thus examining multiple events within the Prx catalytic cycle.
2-Cys Prxs contain multiple redox-sensitive cysteines that may be modified by S-nitrosylation. Previous work reported that S-nitrosylation inhibited human Prx2 activity by targeting both the CP and CR residues (19). In another study related to human Prx1, CR (Cys-173) and Cys-83, have been implicated as nitrosylation sites (23). Similar to the findings on Prx2, our analyses of Prx1 suggest that CP (Cys-52) is a major site of nitrosylation. A second thiol, Cys-83, appears to be nitrosylated as well under our experimental conditions. Indeed, nitrosylation of Cys-52 and Cys-83 is supported by our analysis of the various cysteine null mutants; it is also consistent with the MS/MS data. In addition, the MS/MS spectra inferred nitrosylation of Cys-71 and Cys-173; however, when considering all the analyses carried out in this study, it appears that nitrosylation at these thiols occurs at low SNO occupancy. In fact, our analyses suggest a model in which Cys-173, through formation of a disulfide bond with the nitrosylated Cys-52, promotes denitrosylation. 2-Cys Prxs are characterized by an intricate redox chemistry involving various thiol reactions and interactions. Differences in rates of nitrosylation/denitrosylation among the different thiols of Prx1 as well as possible transfer of NO moieties between thiols (trans-nitrosylation) may contribute to a complex nitrosylation profile. Here, we reported that SNO affected the dimerization and oligomeric state of Prx1. Reciprocally, the thiol-disulfide redox and oligomeric state of Prx1 may influence nitrosylation/denitrosylation events (41). Further research is needed to fully characterize the various aspects of nitrosylation of Prx1 and related Prxs.
A novel finding of the present study was that nitrosylation converted the decameric Prx1 to its dimeric structure, a process that was associated with formation of disulfide-linked dimers. The observed redox structural changes triggered by nitrosylation of Prx1 show similarities to those induced by H2O2 (albeit at a slower rate) as well as oxidized glutathione (GSSG) (42). However, whereas H2O2 can also promote the formation of higher-order Prx assemblies such as dodecamers (a process favored by over-oxidation) SNOs appear unable to do so.
How is the dynamic equilibrium between Prx1 dimers and decamers affected by S-nitrosylation? Structural studies suggest that disulfide bond formation between CP and CR is accompanied by stabilization of a locally unfolded conformation of the active site, which leads to destabilization of the decamer (for review, see Ref. 34). Nitrosylation-mediated disulfide formation between CP and CR may similarly destabilize the decamer. In line with this, we found that SNO treatment of Prx(C52S) did not induce decamer breakdown, reemphasizing the importance of CP in mediating the effects of SNO on Prx1 structure and function. In addition, it has been suggested that Cys-83, which is localized at the dimer-dimer interface, may play a role in stabilizing the oligomeric structure (31). In agreement with this proposal, we found that, unlike reduced WT Prx1, reduced Prx(C83S) did not form oligomers (data now shown). It is thus possible that nitrosylation of Cys-83 may hinder the stabilization of the Prx1 oligomer. Overall, our data suggest that nitrosylation of CP (and possibly Cys-83 also) promotes disulfide formation leading to the dissociation of the decameric Prx1 assembly into dimers.
How is the peroxidase activity of Prx1 controlled by SNO? Congruent with past research on mammalian Prx2 (19) and plant Prx II E (20), we found that S-nitrosylation inhibited the peroxidase activity of Prx1. Nitrosylation of CP followed by disruption of the Prx1 decamer, as shown by our data, may explain at least in part the inhibitory effect of SNO on Prx1 activity. Our results further show that this inhibition is transient, and concomitant with the progression of denitrosylation (promoted by CR), Prx1 activity is restored. Denitrosylation is frequently initiated by a thiol attack on the sulfur atom of the SNO resulting in formation of a disulfide (11, 12, 43). In the case of SNO-Prx1, auto-denitrosylation (involving CP and CR) enables reactivation, contingent on the action of a disulfide reduction system (i.e. Trx/TrxR). The rate of reactivation is thus dependent on the rate of denitrosylation. For Prx1, condensation of CR with the nitrosylated form of CP appears to be a slow process compared with the condensation promoted by the sulfenic acid form of CP. As such, nitrosylation temporarily withdraws Prx1 from the peroxidase cycle, resulting in reduced activity. Similar kinetic considerations could likely apply to other SNO-regulated thiol-disulfide-based systems. We note that exposure of Prx1 to S(NO) was associated with only minor thiol oxidation; however, we cannot rule out completely the possibility that the S(NO)-induced effects involve, to some extent, oxidative-based mechanisms (41).
In addition to the direct effects of SNO on Prx1-mediated peroxide reduction, we found that SNO exerted a marked inhibitory effect on the regeneration of Ox-Prx1 by the Trx/TrxR system. Indeed, both in vitro and in cells, the recycling step in the Prx cycle seemed most susceptible to inhibition by relatively low concentrations of SNO. These findings point to Trx/TrxR as key elements in the SNO-based regulation of the Prx cycle. In principle, two mechanisms may explain the aforementioned inhibition of Prx1 recycling, namely, 1) competition between Ox-Prx1 and SNO for Trx/TrxR and 2) direct inhibition/modulation of Trx/TrxR by SNO. Our data suggest that both mechanisms contribute to the inhibition of Prx recycling. With regard to the second mechanism, it should be noted that S-nitrosylation of Trx has been reported in multiple studies; however, the identity of the modified thiols and the functional consequences of the modification remain controversial as reviewed in Ref. 13. Our studies highlight TrxR, rather than Trx, as the relevant SNO target in the inhibitory effects reported herein. Indeed, experiments both in vitro and in vivo revealed that TrxR activity was significantly and potently inhibited by SNO. In contrast, under the same conditions, Trx activity was little affected. These data suggest that SNO-based modulation/inhibition of TrxR is responsible at least in part for the inhibition of the recycling of Prx1. These findings are consistent with and expand on early in vitro data (36), and they provide strong evidence and support for the concept that nitrosylation functionally regulates the Prx/Trx/TrxR system. We also note the attenuation by H2O2 of Trx/TrxR-mediated denitrosylation, but overall this inhibitory effect was less pronounced than that of SNO on the peroxidase reaction (Fig. 5). These observations indicate a functional asymmetry between SNO and H2O2, in which SNO affects H2O2 metabolism to a greater extent as compared with the converse case.
Distinct from the inhibitory effects on 2-Cys Prx function, SNOs have also been found to protect Prx1 and 2 from over-oxidation (19, 23). Much the same, a protective effect of NO has been observed in immunostimulated macrophages (39). In that context, NO-related attenuation of over-oxidation was accounted for, at least in part, by the up-regulation of sulfiredoxin, an enzyme that reduces over-oxidized 2-Cys Prxs (15, 44). Taken together, the present findings and previous observations suggest two non-exclusive alternative mechanisms by which SNO may safeguard 2-Cys Prxs from over-oxidation: 1) nitrosylation of the CP renders it inaccessible to subsequent attack by H2O2, and 2) competition with or direct modulation of Trx/TrxR slows down the required turnover of the catalytic sites.
In summary, the present work demonstrates that SNOs may exert multiple regulatory controls on the Prx1/Trx/TrxR system to affect H2O2 metabolism. Whereas relatively high SNO levels (>100 μm) inhibit peroxide removal by directly modifying Prx1, intermediate to low levels inhibit the complete peroxidase system mainly by affecting TrxR. The physiological ramifications of these SNO-based regulatory mechanisms remain to be fully explored. In this context, cooperativity in the action of S(NO) and H2O2 has long been recognized (45–47), but the underlying mechanisms remained obscure. Our present analysis of the Prx/Trx/TrxR system may provide a mechanistic explanation for these observations. Finally and more generally, our results suggest that SNO, through its effects on TrxR, may affect additional effector functions of the Trx/TrxR system that were not explored in this study, such as reduction of ribonucleotides or oxidized methionine in proteins. Accordingly, the influence of S-nitrosylation on redox-dependent processes may be greater than currently recognized.
Acknowledgments
We thank Professors Mickey Fry and Herman Wolosker for critically reading the manuscript and helpful discussions.
This work was supported by grants from the Israel Science Foundation (1336/10), the FP7 European Commission (Marie Curie) grant program (256438), and the Mallat family foundation (to M. B.). This work was also supported by grants from the Karolinska Institutet, the Swedish Research Council (Medicine), and the Swedish Cancer Society (to E. S. J. A.).
- SOH
- sulfenic acid
- SNO
- S-nitrosothiol
- Trx
- thioredoxin
- TrxR
- thioredoxin reductase
- CysNO
- S-nitrosocysteine
- GSNO
- S-nitrosoglutathione
- Prx
- peroxiredoxin
- DTNB
- 5,5′-dithio-bis(2-nitrobenzoic acid)
- RAC
- resin-assisted capture
- DEANO
- diethylamine NONOate
- CP
- peroxidatic cysteine
- CR
- resolving cysteine
- NEM
- N-ethyl maleimide.
REFERENCES
- 1. Forman H. J., Maiorino M., Ursini F. (2010) Signaling functions of reactive oxygen species. Biochemistry 49, 835–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Finkel T. (2011) Signal transduction by reactive oxygen species. J. Cell Biol. 194, 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rhee S. G. (2006) Cell signaling. H2O2, a necessary evil for cell signaling. Science 312, 1882–1883 [DOI] [PubMed] [Google Scholar]
- 4. Pacher P., Beckman J. S., Liaudet L. (2007) Nitric oxide and peroxynitrite in health and disease. Physiol. Rev. 87, 315–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Janssen-Heininger Y. M., Mossman B. T., Heintz N. H., Forman H. J., Kalyanaraman B., Finkel T., Stamler J. S., Rhee S. G., van der Vliet A. (2008) Redox-based regulation of signal transduction. Principles, pitfalls, and promises. Free Radic. Biol. Med. 45, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Poole L. B., Nelson K. J. (2008) Discovering mechanisms of signaling-mediated cysteine oxidation. Curr. Opin. Chem. Biol. 12, 18–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Paulsen C. E., Carroll K. S. (2010) Orchestrating redox signaling networks through regulatory cysteine switches. ACS Chem. Biol. 5, 47–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hess D. T., Matsumoto A., Kim S. O., Marshall H. E., Stamler J. S. (2005) Protein S-nitrosylation. Purview and parameters. Nat. Rev. Mol. Cell Biol. 6, 150–166 [DOI] [PubMed] [Google Scholar]
- 9. Foster M. W., Hess D. T., Stamler J. S. (2009) Protein S-nitrosylation in health and disease. A current perspective. Trends Mol. Med. 15, 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fourquet S., Huang M. E., D'Autreaux B., Toledano M. B. (2008) The dual functions of thiol-based peroxidases in H2O2 scavenging and signaling. Antioxid. Redox Signal. 10, 1565–1576 [DOI] [PubMed] [Google Scholar]
- 11. Benhar M., Forrester M. T., Hess D. T., Stamler J. S. (2008) Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science 320, 1050–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Benhar M., Forrester M. T., Stamler J. S. (2009) Protein denitrosylation. Enzymatic mechanisms and cellular functions. Nat. Rev. Mol. Cell Biol. 10, 721–732 [DOI] [PubMed] [Google Scholar]
- 13. Sengupta R., Holmgren A. (2012) The role of thioredoxin in the regulation of cellular processes by S-nitrosylation. Biochim. Biophys. Acta 1820, 689–700 [DOI] [PubMed] [Google Scholar]
- 14. Wood Z. A., Schröder E., Robin Harris J., Poole L. B. (2003) Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 28, 32–40 [DOI] [PubMed] [Google Scholar]
- 15. Rhee S. G., Woo H. A. (2011) Multiple functions of peroxiredoxins. Peroxidases, sensors, and regulators of the intracellular messenger H2O2 and protein chaperones. Antioxid. Redox Signal. 15, 781–794 [DOI] [PubMed] [Google Scholar]
- 16. Barranco-Medina S., Lázaro J. J., Dietz K. J. (2009) The oligomeric conformation of peroxiredoxins links redox state to function. FEBS Lett. 583, 1809–1816 [DOI] [PubMed] [Google Scholar]
- 17. Dammeyer P., Arnér E. S. (2011) Human protein atlas of redox systems. What can be learned? Biochim. Biophys. Acta 1810, 111–138 [DOI] [PubMed] [Google Scholar]
- 18. Karplus P. A., Poole L. B. (2012) Peroxiredoxins as molecular triage agents, sacrificing themselves to enhance cell survival during a peroxide attack. Mol. Cell 45, 275–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fang J., Nakamura T., Cho D. H., Gu Z., Lipton S. A. (2007) S-Nitrosylation of peroxiredoxin 2 promotes oxidative stress-induced neuronal cell death in Parkinson's disease. Proc. Natl. Acad. Sci. U.S.A. 104, 18742–18747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Romero-Puertas M. C., Laxa M., Mattè A., Zaninotto F., Finkemeier I., Jones A. M., Perazzolli M., Vandelle E., Dietz K. J., Delledonne M. (2007) S-Nitrosylation of peroxiredoxin II E promotes peroxynitrite-mediated tyrosine nitration. Plant Cell 19, 4120–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Forrester M. T., Thompson J. W., Foster M. W., Nogueira L., Moseley M. A., Stamler J. S. (2009) Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat. Biotechnol. 27, 557–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Benhar M., Thompson J. W., Moseley M. A., Stamler J. S. (2010) Identification of S-nitrosylated targets of thioredoxin using a quantitative proteomic approach. Biochemistry 49, 6963–6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu C., Liu T., Chen W., Oka S., Fu C., Jain M. R., Parrott A. M., Baykal A. T., Sadoshima J., Li H. (2010) Redox regulatory mechanism of trans-nitrosylation by thioredoxin. Mol. Cell. Proteomics 9, 2262–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rengby O., Johansson L., Carlson L. A., Serini E., Vlamis-Gardikas A., Kårsnäs P., Arnér E. S. (2004) Assessment of production conditions for efficient use of Escherichia coli in high-yield heterologous recombinant selenoprotein synthesis. Appl. Environ. Microbiol. 70, 5159–5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fang K., Ragsdale N. V., Carey R. M., MacDonald T., Gaston B. (1998) Reductive assays for S-nitrosothiols. Implications for measurements in biological systems. Biochem. Biophys. Res. Commun. 252, 535–540 [DOI] [PubMed] [Google Scholar]
- 26. Jaffrey S. R., Erdjument-Bromage H., Ferris C. D., Tempst P., Snyder S. H. (2001) Protein S-nitrosylation. A physiological signal for neuronal nitric oxide. Nat. Cell Biol. 3, 193–197 [DOI] [PubMed] [Google Scholar]
- 27. Nelson K. J., Parsonage D. (2011) Measurement of peroxiredoxin activity. Curr. Protoc. Toxicol. Chapter 7, Unit 7.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim J. A., Park S., Kim K., Rhee S. G., Kang S. W. (2005) Activity assay of mammalian 2-cys peroxiredoxins using yeast thioredoxin reductase system. Anal. Biochem. 338, 216–223 [DOI] [PubMed] [Google Scholar]
- 29. Rhee S. G., Chang T. S., Jeong W., Kang D. (2010) Methods for detection and measurement of hydrogen peroxide inside and outside of cells. Mol. Cells 29, 539–549 [DOI] [PubMed] [Google Scholar]
- 30. Arnér E. S., Holmgren A. (2001) Measurement of thioredoxin and thioredoxin reductase. Curr. Protoc. Toxicol. Chapter 7, Unit 7.4 [DOI] [PubMed] [Google Scholar]
- 31. Lee W., Choi K. S., Riddell J., Ip C., Ghosh D., Park J. H., Park Y. M. (2007) Human peroxiredoxin 1 and 2 are not duplicate proteins. The unique presence of CYS83 in Prx1 underscores the structural and functional differences between Prx1 and Prx2. J. Biol. Chem. 282, 22011–22022 [DOI] [PubMed] [Google Scholar]
- 32. Hirotsu S., Abe Y., Okada K., Nagahara N., Hori H., Nishino T., Hakoshima T. (1999) Crystal structure of a multifunctional 2-Cys peroxiredoxin heme-binding protein 23-kDa/proliferation-associated gene product. Proc. Natl. Acad. Sci. U.S.A. 96, 12333–12338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Matsumura T., Okamoto K., Iwahara S., Hori H., Takahashi Y., Nishino T., Abe Y. (2008) Dimer-oligomer interconversion of wild-type and mutant rat 2-Cys peroxiredoxin. Disulfide formation at dimer-dimer interfaces is not essential for decamerization. J. Biol. Chem. 283, 284–293 [DOI] [PubMed] [Google Scholar]
- 34. Hall A., Nelson K., Poole L. B., Karplus P. A. (2011) Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid. Redox. Signal. 15, 795–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sengupta R., Ryter S. W., Zuckerbraun B. S., Tzeng E., Billiar T. R., Stoyanovsky D. A. (2007) Thioredoxin catalyzes the denitrosation of low-molecular mass and protein S-nitrosothiols. Biochemistry 46, 8472–8483 [DOI] [PubMed] [Google Scholar]
- 36. Nikitovic D., Holmgren A. (1996) S-Nitrosoglutathione is cleaved by the thioredoxin system with liberation of glutathione and redox regulating nitric oxide. J. Biol. Chem. 271, 19180–19185 [DOI] [PubMed] [Google Scholar]
- 37. Winterbourn C. C. (2008) Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 4, 278–286 [DOI] [PubMed] [Google Scholar]
- 38. Bryk R., Griffin P., Nathan C. (2000) Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature 407, 211–215 [DOI] [PubMed] [Google Scholar]
- 39. Diet A., Abbas K., Bouton C., Guillon B., Tomasello F., Fourquet S., Toledano M. B., Drapier J. C. (2007) Regulation of peroxiredoxins by nitric oxide in immunostimulated macrophages. J. Biol. Chem. 282, 36199–36205 [DOI] [PubMed] [Google Scholar]
- 40. Bast A., Erttmann S. F., Walther R., Steinmetz I. (2010) Influence of iNOS and COX on peroxiredoxin gene expression in primary macrophages. Free Radic. Biol. Med. 49, 1881–1891 [DOI] [PubMed] [Google Scholar]
- 41. Barglow K. T., Knutson C. G., Wishnok J. S., Tannenbaum S. R., Marletta M. A. (2011) Site-specific and redox-controlled S-nitrosation of thioredoxin. Proc. Natl. Acad. Sci. U.S.A. 108, E600–E606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Park J. W., Piszczek G., Rhee S. G., Chock P. B. (2011) Glutathionylation of peroxiredoxin I induces decamer to dimers dissociation with concomitant loss of chaperone activity. Biochemistry 50, 3204–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Arnelle D. R., Stamler J. S. (1995) NO+, NO, and NO- donation by S-nitrosothiols. Implications for regulation of physiological functions by S-nitrosylation and acceleration of disulfide formation. Arch. Biochem. Biophys. 318, 279–285 [DOI] [PubMed] [Google Scholar]
- 44. Biteau B., Labarre J., Toledano M. B. (2003) ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature 425, 980–984 [DOI] [PubMed] [Google Scholar]
- 45. Filep J. G., Lapierre C., Lachance S., Chan J. S. (1997) Nitric oxide co-operates with hydrogen peroxide in inducing DNA fragmentation and cell lysis in murine lymphoma cells. Biochem. J. 321, 897–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marcinkiewicz J. (1997) Nitric oxide and antimicrobial activity of reactive oxygen intermediates. Immunopharmacology 37, 35–41 [DOI] [PubMed] [Google Scholar]
- 47. Wang J. Y., Shum A. Y., Ho Y. J., Wang J. Y. (2003) Oxidative neurotoxicity in rat cerebral cortex neurons. Synergistic effects of H2O2 and NO on apoptosis involving activation of p38 mitogen-activated protein kinase and caspase-3. J. Neurosci. Res. 72, 508–519 [DOI] [PubMed] [Google Scholar]