Background: Mechanisms that redirect androgen receptor signaling to primarily support prostate tumor growth are poorly understood.
Results: Prostate cancer cells were addicted to ELK1, which tethered AR to activate growth genes in hormone-dependent and castration-recurrent PC without ELK1 phosphorylation.
Conclusion: ELK1 directs a critical arm of transcriptional growth signaling by AR that is preserved in CRPC.
Significance: The ELK1-AR interaction offers a functionally tumor-selective drug target.
Keywords: Androgen Receptor, Gene Regulation, Nuclear Receptors, Prostate Cancer, Transcription
Abstract
The androgen receptor (AR) is essential for diverse aspects of prostate development and function. Molecular mechanisms by which prostate cancer (PC) cells redirect AR signaling to genes that primarily support growth are unclear. A systematic search for critical AR-tethering proteins led to ELK1, an ETS transcription factor of the ternary complex factor subfamily. Although genetically redundant, ELK1 was obligatory for AR-dependent growth and clonogenic survival in both hormone-dependent PC and castration-recurrent PC cells but not for AR-negative cell growth. AR required ELK1 to up-regulate a major subset of its target genes that was strongly and primarily enriched for cell growth functions. AR functioned as a coactivator of ELK1 by association through its A/B domain, bypassing the classical mechanism of ELK1 activation by phosphorylation and without inducing ternary complex target genes. The ELK1-AR synergy per se was ligand-independent, although it required ligand for nuclear localization of AR as targeting the AR A/B domain to the nucleus recapitulated the action of hormone; accordingly, Casodex was a poor antagonist of the synergy. ELK3, the closest substitute for ELK1 in structure/function and genome recognition, did not interact with AR. ELK1 thus directs selective and sustained gene induction that is a substantial and critical component of growth signaling by AR in PC cells. The ELK1-AR interaction offers a functionally tumor-selective drug target.
Introduction
Both androgen-responsive and advanced prostate tumors are generally dependent on the androgen receptor (AR)2 for growth (1–4). The growth could be driven by circulating androgen, postablation synthesis of intratumoral androgen, or by AR acting completely independently of androgen (5–10). In all cases, the current clinical paradigm for adjuvant therapy is total and ubiquitous attenuation of AR signaling by androgen ablation and the use of AR ligands that antagonize, sequester, or deplete AR (11). However, currently available antiandrogens have limited efficacy in blocking disease progression; furthermore, androgen ablation is associated with undesirable side effects in a variety of non-target tissues and organ systems (12). Therefore, it is desirable to identify and effectively disrupt a functional aspect of AR that is critical for tumor growth but not for the physiological role of androgen in differentiated normal tissues.
Specific mechanisms by which prostate cancer cells reprogram androgen/AR signaling to primarily support tumor growth are not well understood. In the classical model of gene regulation by AR, the receptor requires bound ligand to homodimerize, enter the nucleus, and bind to well characterized androgen response elements (AREs) associated with target genes (13–17). Androgen binding also enables phosphorylation of AR that is required for its stabilization and activity (18, 19). When the bound ligand is an agonist, AR then recruits coactivators; in contrast, when AR is bound to antagonists, co-repressors are preferentially recruited (15, 16). AR contains sites of coregulator binding that are either ligand-dependent or -independent. However, in prostate cancer cells that are adapted to grow in the absence of hormone, the AR apoprotein is localized in an active form in the nucleus. In such cells, the AR apoprotein activates a gene set that is distinct from genes that require androgen for activation in the same cells (9, 20). This set of genes strikingly overlaps the signature gene overexpression profile of clinical castration-recurrent prostate tumors and is enriched for gene clusters primarily supporting mitotic cell division (9, 20). Furthermore, we have demonstrated that in those cells the AR apoprotein can support growth through gene activation that occurs without the direct binding of AR to AREs and likely through tethered associations of the receptor with its target genes (9). Our previous detailed studies of the interaction of AR with CCAAT/enhancer-binding protein α (involved in terminal tissue differentiation) suggested that tethered associations of AR with DNA may not require hormone except in cell contexts in which androgen is needed for nuclear import of AR (21). Therefore, it may be possible to identify one or a few critical AR-tethering proteins whose interaction with the receptor may be necessary, although not sufficient, for androgen/AR-dependent growth of both early stage and advanced prostate cancer cells. If such interactions do not occur in, or are not critical for, the normal physiological actions of AR, then they could be targeted for functionally selective and tumor-specific intervention in prostate cancer.
In prostate cancer cells in which AR is localized in the nucleus in the absence of hormone, the AR apoprotein has profound genotropic effects. However, in genome-wide chromatin binding studies using cell line models of advanced prostate cancer in the absence of hormone, putative tethered associations of AR with chromatin generally give relatively weak signals at best. This is presumably due to the poor efficiency of immunoprecipitation of AR in such complexes (9, 20). Nevertheless, the association of AR with DNA-bound transcription factors has been reported by screening a synthetic cis-element array of transcription factor binding sites for AR recruitment from a nuclear extract of LNCaP cells (22). After narrowing this list on the basis of the cis-elements that could be functionally validated as mediators of transactivation by AR, we undertook a detailed investigation of ELK1. ELK1 belongs to the ternary complex factor (TCF) subgroup of the ETS family transcription factors.
ETS family proteins share a conserved winged helix-turn-helix DNA binding domain of about 85 amino acids and bind to a core GGA sequence. The many ETS proteins demonstrate both commonalities and differences in tissue specificity and binding site (target gene) selectivity (23, 24). The TCF subfamily proteins have the additional capability of forming a ternary complex with the serum response factor (SRF) and serum response elements (SREs) to induce immediate early genes (25, 26). TCF proteins are activated by MAPK signaling to control growth or to respond to stress (27). ELK1 ordinarily represses genes in its SUMOylated form (28); its activation through MAPK occurs by phosphorylation that is accompanied by loss of SUMOylation (28), resulting in a loss of its ability to repress genes. Although ELK1 regulates a broad network of genes (29), deletion of the ELK1 gene does not result in significant abnormalities in phenotype (30). This is presumably due to functional redundancy within the TCF subfamily (23, 24).
ELK1 is redundant for normal mammalian development but shows consistent expression in the epithelial cells of clinical prostate tumors (31). ELK1 also appears to support transcriptional signaling by AR. It was therefore of interest to further examine the nature and significance of its interactions with AR in prostate cancer.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
Normal primary prostate epithelial cells from two donors aged 17 and 29 years were purchased from Lifeline Cell Technology (Oceanside, CA). LNCaP, VCaP, DU145, PC-3, and HeLa cell lines were from the American Type Culture Collection (Manassas, VA). C4-2 cells were kindly provided by Dr. Edwin Sanchez (University of Toledo). 293FT cells were from Invitrogen. LNCaP and C4-2 cells were routinely grown at 37 °C in 5% CO2 in RPMI 1640 medium supplemented with 10% FBS (Invitrogen); 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mm l-glutamine mixture (Invitrogen); and sodium pyruvate (1 mm) (Invitrogen). VCaP, HeLa, and DU145 cells were grown in DMEM supplemented with 10% FBS and 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mm l-glutamine mixture. PC-3 cells were grown in RPMI 1640 medium supplemented with 10% FBS and 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mm l-glutamine mixture. 293FT cells were grown in DMEM supplemented with 10% FBS, non-essential amino acids (Invitrogen), 500 μg/ml Geneticin (Invitrogen), and 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mm l-glutamine mixture. Affinity-purified rabbit anti-human antibodies to AR (sc-816) and ELK1 (sc-355) and mouse anti-human antibodies to AR (sc-7305), ELK1 (sc-65986), and GAPDH (sc-47724) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit monoclonal anti-human antibody to ELK1 (ab 32106) was from Abcam (Cambridge, MA). Phospho-ELK1 (Ser-383) antibody (catalogue number 9181) was purchased from Cell Signaling Technology (Danvers, MA). R1881 and Casodex were kindly provided by Dr. Lirim Shemshedini (University of Toledo). Cisplatin used for the Annexin V assay was a gift from Dr. Steve Patrick (University of Toledo). LipofectamineTM 2000 was purchased from Invitrogen. Protease inhibitor mixture was purchased from Thermo Scientific (product number 78410). Phosphatase inhibitor mixture (catalogue number P-5726) and phorbol 12-myristate 13-acetate were purchased from Sigma-Aldrich. For hormone depletion, cells were grown in either phenol-red free RPMI 1640 medium or phenol red-free DMEM supplemented with 10% charcoal stripped FBS (Invitrogen) and 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mm l-glutamine mixture for 48 h before the experiments.
Plasmids
GAL4-TATA-Luc plasmid (pG5luc) and expression plasmid for VP16 and Gal4 were purchased from Promega (Madison, WI) (CheckMate Mammalian Two-hybrid System). The (ELK1)2-TATA-Luc plasmid was constructed using an EMSA-validated oligonucleotide sequence representing a tandem repeat of the optimal binding site for ELK1 (5′-GAGCCGGAAGATCGGAGCCGGAAG-3′) that was custom synthesized. The complementary oligonucleotides were annealed to obtain double-stranded DNA. The synthetic DNA was designed with the addition of 5′ KpnI and 3′ NheI sites and substituted for the Gal4 element in the pG5luc vector (Promega) upstream of the TATA box. The ISRE-TATA-Luc and ARE-TATA-Luc plasmids were similarly constructed but with the insertion of the ISRE (5′-GATCGGGAAAGGGAAACCGAAACTGAAGCC-3′) or a consensus ARE (5′-AGTACGTGATGTTCT-3′), respectively, instead of the ELK1 element. The pRL plasmid encoding Renilla luciferase was purchased from Promega. The PSA-Luc plasmid containing a 6.1-kb DNA fragment encompassing the promoter and distal enhancer regions of the PSA gene was a kind gift from Dr. Lirim Shemshedini. The AR expression plasmid (pSG5 vector) was a kind gift from Dr. Lirim Shemshedini. The expression plasmids for human full-length ELK1 and ELK3 in the pCMV plasmid were purchased from OriGene (Rockville, MD). The Gal4 fusion with ELK1 in which the DNA binding domain of ELK1 (amino acids 1–86) was deleted was constructed by PCR using the ELK1 expression plasmid as the template and the appropriate primers and subcloned at BamHI (upstream) and NotI (downstream) sites in a vector expressing Gal4 fusions (pBind). VP16 fusion constructs for the various domains of AR were constructed using the VP16 expression plasmid from Promega. The AR(A/B)-NLS construct was generated by PCR amplification of the A/B domain (residues 1–555) from the full-length AR plasmid and cloning into the pCDH vector with an in-frame insertion of tandem repeats of a nuclear localization sequence (NLS) at its C terminus. pCDH-ELK3 was generated by cloning the full-length ELK3 cDNA into the pCDH vector. The TIPARP promoter-Luc construct was generated by PCR amplification of the nucleotide sequence −536 to +60 of the TIPARP gene from human genomic DNA. The ELK1 binding element within this sequence spans nucleotides −357 to −348. This PCR-amplified product was cloned into pGL3-Basic vector. TIPARP (ΔELK1) promoter-Luc construct was made by deleting the ELK1 binding element from the TIPARP promoter-Luc construct and cloned into pGL3-Basic vector. shRNAs targeting AR, ELK1, and SRF and non-targeting control shRNA in the lentiviral expression vector pLKO.1-puro were purchased from Sigma-Aldrich. All shRNA sequences are in Supplement 8.
Transfection and Reporter Luciferase Assays
Transient transfections of LNCaP and C4-2 cells were performed using Cell Line Nucleofector kit R from Amaxa Biosystems (Germany) following the manufacturer-optimized protocol for LNCaP cells. 2 × 106 cells were used for each nucleofection. After nucleofection with appropriate plasmids and shRNA, the cells were plated in 12-well poly-d-lysine-coated plates from BD Discovery Labware. In all cases, the appropriate empty vector plasmids were used to equalize total DNA for transfection. For promoter analysis, 2 μg of each promoter-luciferase reporter construct was transfected. In the AR or ELK1 knockdown experiments, 1.5 μg of each shRNA or non-targeting control shRNA plasmid was transfected. HeLa cells were transfected using Lipofectamine 2000 transfection reagent. The cells were lysed with Passive Lysis Buffer (Promega), and the luciferase activities were measured using substrates for either firefly luciferase or Renilla luciferase provided in the Luciferase Assay System (Promega) in a luminometer (Lumat LB9501, Berthold, Wildbad, Germany). In all cases, uniformity of transfection was confirmed using the pRL-null plasmid expressing Renilla luciferase.
Lentivirus-mediated Transduction
For lentivirus-mediated gene knockdown, shRNAs for AR, ELK1, and SRF and non-targeting control were packaged in 293FT cells using lentiviral packaging plasmids as described previously (9). The virus-containing supernatant was harvested 48 and 72 h after transfection, filtered, and stored at −80 °C until the time of infection. 24 h before infection, 5 × 105–6 × 105 cells were plated in poly-d-lysine-coated 6-well plates (for LNCaP or C4-2 cells) in phenol red-free medium supplemented with 10% heat-inactivated charcoal-stripped FBS and 2 mm l-glutamine. The next day, cells were infected with either non-targeting control shRNA lentivirus, AR shRNA lentivirus, ELK1 shRNA lentivirus, a combination of AR shRNA and ELK1 shRNA lentiviruses, or SRF shRNA lentivirus with Polybrene (8 μg/ml) for a duration of 5 h followed by a similar second lentiviral infection for an additional 5 h. 10 h after the infection, the virus was replaced with fresh phenol red-free medium containing 10% charcoal-stripped FBS. For the AR-negative cell lines DU145 and PC-3, after the lentiviral infection, the virus was replaced with fresh phenol red-free medium containing 10% FBS.
VCaP cells are sensitive to Polybrene. Therefore, to increase the lentiviral transduction efficiency in the absence of Polybrene, the MISSIONTM ExpressMag Super Magnetic kit from Sigma-Aldrich was used. Briefly, 24 h before infection, 5 × 105–6 × 105 cells were plated in poly-d-lysine-coated 6-well plates in phenol red-free DMEM supplemented with 10% heat-inactivated charcoal-stripped FBS and 2 mm l-glutamine. The next day, cells were infected with either non-targeting control shRNA or ELK1 shRNA lentivirus using MISSION ExpressMag Beads according to the manufacturer's protocol. 18–20 h after the infection, the virus was replaced with fresh phenol red-free DMEM containing 10% FBS.
Cell Proliferation Assay
Cells were trypsinized and 4000–6000 cells/well were seeded in 96-well plates in phenol red-free medium supplemented with 10% charcoal-stripped FBS and grown at 37 °C in 5% CO2 for different time periods. For LNCaP, VCaP, and C4-2 cells, it was necessary to use plates coated with poly-d-lysine. For LNCaP and VCaP cells, 24 h after seeding in 96-well plates, the cells were treated with vehicle (ethanol) or R1881 (1 nm). The culture medium was not changed during the entire time course. At the end of each time point, cell viability was determined using the MTT assay. Briefly, 10 μl of MTT (5 mg/ml) was added to each well and incubated for 2 h at 37 °C. The formazan crystal sediments were dissolved in 100 μl of DMSO, and the absorbance at 570 nm was measured using a SpectraMax Plus spectrophotometer (Molecular Devices Corp., Sunnyvale, CA). The assay was conducted in sextuplicate wells, and all values were normalized to day 0.
Two- and Three-dimensional Colony Formation Assays
For the two-dimensional colony formation assay, cells were trypsinized, and 500 cells/well were seeded in poly-d-lysine-coated 6-well plates in phenol red-free medium supplemented with 10% charcoal-stripped FBS. 24 h later, the cells were treated with vehicle or R1881 (1 nm) and grown at 37 °C in 5% CO2 for 2 weeks until colonies had formed. The treatments were replenished every 96 h. Colonies were fixed with methanol and stained with crystal violet. Each treatment was conducted in triplicate, and pictures of individual wells were taken.
For the three-dimensional colony formation assay, 24-well plates were coated with a bottom layer of 0.8% SeaPrep ultralow gelling temperature agarose (BioWhittaker Molecular Applications, Rockland, ME) in phenol red-free medium supplemented with 10% charcoal-stripped FBS. Cells were trypsinized, serially diluted in the same medium, and applied as the top agarose layer. The agarose gel bed was overlaid with phenol red-free medium supplemented with 10% charcoal-stripped FBS containing R1881 (1 nm). The plates were incubated at 37 °C in 5% CO2 for 2 weeks until colonies formed. R1881 was replenished every 96 h. The colonies were stained with MTT by applying 500 μl of MTT (5 mg/ml) to each well and incubated for 30 min at 37 °C.
Apoptosis Assay
Cells were trypsinized and seeded in poly-d-lysine-coated 6-well plates in phenol red-free medium supplemented with 10% charcoal-stripped FBS. Apoptosis was measured by Guava Nexin analysis using the Guava Nexin reagent staining kit according to the manufacturer's instructions.
RNA Isolation, Reverse Transcription, and Quantitative Real Time PCR
Total RNA from cells was isolated using the RNeasy minikit (Qiagen, Georgetown, MD) according to the manufacturer's protocol. Reverse transcription was performed using 500 ng of total RNA and the High-Capacity cDNA Archive kit (Applied Biosystems, Invitrogen) according to the vendor's protocol. cDNA was measured by quantitative real time PCR using the StepOnePlus Real-Time PCR System (Applied Biosystems, Invitrogen) and TaqMan Fast Universal PCR Master Mix (Applied Biosystems, Invitrogen). All primers and TaqMan probes were purchased from the Applied Biosystems inventory (Invitrogen). All samples were measured in triplicate and normalized to the values for GAPDH.
Western Blot Analysis
Cells were lysed with radioimmune precipitation assay buffer (150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mm Tris of pH 8.0) containing protease inhibitor mixture (Pierce, Thermo Fisher Scientific) and incubated on ice for 30 min. Total protein concentration was determined by Bradford assay (Bio-Rad). Cell lysates were heated at 95 °C for 5 min. Protein samples (25–50 μg) were resolved by electrophoresis on 8% SDS-polyacrylamide gels and electrophoretically transferred to PVDF membranes (Millipore, Billerica, MA). The blots were probed with appropriate primary antibody and the appropriate horseradish peroxidase-conjugated secondary antibody. The protein bands were visualized using the HyGLO Chemiluminescent HRP Antibody Detection Reagent (Denville Scientific, Metuchen, NJ).
Detection of Phospho-ELK1
LNCaP cells were washed twice with PBS and incubated for a further 24 h in serum-free medium. The cells were then treated with vehicle, phorbol 12-myristate 13-acetate (10 μm), or R1881 (1 nm) for various durations. After the treatment, the cells were harvested and lysed in radioimmune precipitation assay buffer containing 1× protease inhibitor mixture and 1× phosphatase inhibitor mixture. 60 μg of total protein was heated at 95 °C for 5 min in SDS sample loading buffer and analyzed by Western blot. Phospho-ELK1 was detected using anti-phospho ELK1 (Ser-383) antibody.
Chromatin Immunoprecipitation (ChIP) and Re-ChIP
Cells were treated with either vehicle or R1881 (1 nm) for 2 h and then subjected to ChIP using anti-AR antibody (sc-816) and anti-ELK1 antibody (ab 32106). The ChIP assay was performed using the EZ ChIP chromatin immunoprecipitation kit (catalogue number 17-371) according to the vendor's protocol (Millipore, Temecula, CA), and ChIP signals were measured by quantitative real time PCR analysis of chromatin-immunoprecipitated products. Each sample was tested in triplicate. The primers and TaqMan probes used to amplify genomic sequences by real time quantitative RT-PCR are in Supplement 8.
For the re-ChIP assay, the first immunoprecipitation was performed with either anti-ELK1 antibody (ab 32106), anti-AR antibody (sc-816), or normal rabbit IgG (sc-2027). The chromatin complexes from the first round of immunoprecipitation were eluted with 10 mm DTT at 37 °C for 30 min, diluted 20 times with ChIP dilution buffer (1% Triton X-100, 2 mm EDTA, 150 mm NaCl, 20 mm Tris HCl, pH 8.1), and subjected to the second round of immunoprecipitation with anti-AR antibody (sc-816) or anti-ELK1 antibody (ab 32106). The chromatin eluted from the second immunoprecipitation was detected by PCR amplification of a 407-bp fragment of the promoter region of the TIPARP gene. A 478-bp genomic fragment within the GAPDH gene was amplified as the non-targeting negative control. The forward and reverse primer sequences for the PCR amplifications are in Supplement 8. The PCR products were analyzed by electrophoresis on a 1% agarose gel. The PCR cycle used to amplify the TIPARP and GAPDH sequences is as follows: initial denaturation at 95 °C for 15 min followed by 35 cycles of 1 min at 94 °C, 1 min at 68.5 °C, and 1 min 30 s at 72 °C and a final extension at 72 °C for 10 min.
mRNA Expression Profiling and Gene Ontology Analysis
The Affymetrix DNA microarray analysis was performed as a full service global gene expression study at the transcriptional profiling core facility of The Cancer Institute of New Jersey (New Brunswick, NJ). The data obtained are from two separate experiments. Total RNA samples were used to generate labeled cRNA, which was hybridized to human U133 Plus 2.0 Affymetrix microarrays. Scanned image files were analyzed using the Gene Chip Operating System version 1.4 software, and standard thresholding and filtering operations were used. The data were normalized using housekeeping genes. Normalization assumes that for a subset of genes (i.e. housekeeping genes) the ratio of measured expression averaged over the set should be 1. All data are minimum information about a microarray experiment-compliant, and the raw data have been deposited in a minimum information about a microarray experiment-compliant database (Gene Expression Omnibus) as detailed by the Microarray Gene Expression Data Society. The Gene Expression Omnibus (GEO) accession number for the data is GSE34589.
Differentially expressed genes were identified by comparing R1881 treatment with vehicle treatment (R1881-activated genes, 1.5-fold cutoff) in control shRNA- versus ELK1 shRNA-infected cells. In cells treated with vehicle, genes repressed or activated by ELK1 alone were identified by comparing samples from cells infected with control shRNA versus ELK1 shRNA (0.5-fold cutoff for repression by ELK1; 2-fold cutoff for activation by ELK1). The adjusted p values for the comparisons, calculated using the Bioconductor limma program, were <0.05. Gene ontology analysis was performed using DAVID Bioinformatics Resources 6.7 (32, 33).
Coimmunoprecipitation
For C4-2 cells, cells plated in hormone-depleted medium were harvested in radioimmune precipitation assay lysis buffer and 1× protease inhibitor mixture. 350 μg of whole cell lysate was precleared using protein A-agarose beads (Calbiochem, Merck KGaA) for 2 h. Immunoprecipitation was performed using 2 μg of rabbit polyclonal AR antibody (sc-816), a 1:80 dilution of rabbit monoclonal ELK1 antibody (ab 32106), and normal rabbit IgG (sc-2027) followed by six washings with radioimmune precipitation assay buffer. The Western blot was probed with mouse monoclonal AR antibody (sc-7305) and mouse monoclonal ELK1 antibody (sc-65986). For LNCaP cells, cells were harvested in Triton X-100 lysis buffer (20 mm Tris, pH 7.4, 137 mm NaCl, 2 mm EDTA, 1% Triton X-100, 10% glycerol, 0.5 mm DTT) and 1× protease inhibitor mixture. 350 μg of lysate was precleared with protein G Plus-agarose beads (Calbiochem, Merck KGaA) for 2 h. Immunoprecipitation was performed using 2 μg of mouse monoclonal AR antibody (sc-7305), mouse monoclonal ELK1 antibody (sc-65986), and normal mouse IgG (sc-2025) followed by six washings with Triton X-100 lysis buffer. The Western blot was probed with rabbit polyclonal AR antibody (sc-816) and rabbit monoclonal ELK1 antibody (ab 32106).
Mammalian Two-hybrid Assay
The CheckMate Mammalian Two-hybrid System (Promega) was used. HeLa cells were plated in 24-well plates in hormone-free phenol red-free DMEM without antibiotics. When the cells were about 90% confluent, they were co-transfected with pG5Luc, pBind vector expressing Gal4 or Gal4-ELK1 fusion proteins, and pACT vector expressing VP16 or VP16 fusion proteins using Lipofectamine 2000 transfection reagent. After 48 h of transfection, the cells were lysed with Passive Lysis Buffer, and the luciferase activity was determined as described above.
Statistical Analysis
All of the experiments were repeated at least three times. Statistical significance was determined using one-way analysis of variance. The error bars in all graphs represent S.D. The p values are indicated in the figure legends.
RESULTS
The cell lines chosen for this study include (i) LNCaP, a common model of hormone-dependent PC; (ii) VCaP, a hormone-dependent PC model harboring the common TMPRSS2-ERG fusion as well as recently identified functional splice variants of AR; (iii) C4-2, a model of CRPC developed by xenotransplantation in castrated mice; and (iv) DU145 and PC-3, which are both AR-negative PC cells.
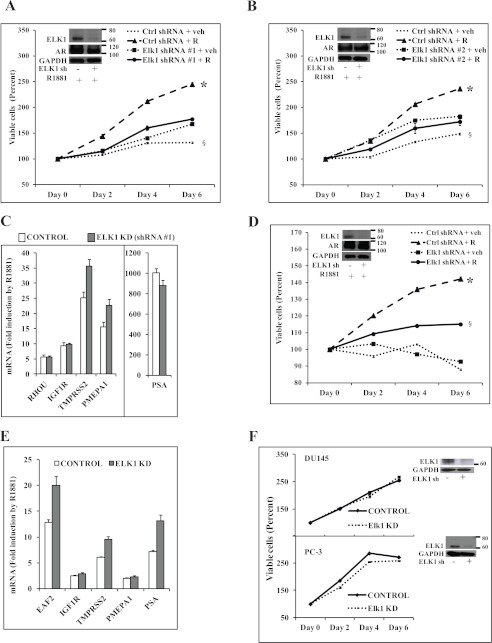
ELK1 Is Essential for Androgen-stimulated Growth but Does Not Regulate the Level of Active AR
In LNCaP cells, in the absence of hormone, depletion of ELK1 using two shRNA lentiviruses with different target site specificities modestly increased the basal level of cell growth (Fig. 1, A and B), consistent with the role of ELK1 in suppressing a number of growth genes when it is not activated (discussed in the next section). However, depletion of ELK1 abrogated the growth response to androgen (Fig. 1, A and B). Depletion of ELK1 did not affect the ability of androgen to induce several classical androgen target genes (Fig. 1C for ELK1 shRNA 1 and supplemental Fig. 1 for ELK1 shRNA 2), indicating that ELK1 was not required for the general transcriptional activity of AR. VCaP cells are dependent on androgen for survival as well as growth as evident from the progressive decline in viable cells in the absence of hormone (Fig. 1D); however, depletion of ELK1 did attenuate androgen-dependent growth (Fig. 1D). Depletion of ELK1 in VCaP cells did not affect the androgen-stabilized level of AR (Fig. 1D, inset) or the ability of AR to activate a number of its classical target genes (Fig. 1E). A modest increase in the androgen-induced activity was observed for its classical target genes in some instances upon depletion of ELK1; this may reflect a modestly repressive association of ELK1 with AR bound to its classical response elements in the chromatin as previously noted for other AR-tethering proteins. Depletion of ELK1 did not affect the growth of DU145 cells and PC-3 cells (Fig. 1F), consistent with a specific role for ELK1 in supporting AR-dependent growth.
FIGURE 1.
Effect of depleting ELK1 on hormone-dependent PC cells. A and B, hormone-depleted LNCaP cells were infected with ELK1 shRNA 1 lentivirus (A), ELK1 shRNA 2 lentivirus (B), or control (Ctrl) shRNA lentivirus. 72 h later (after the knockdown (KD) had occurred), cells were treated with vehicle (veh) or R1881 (1 nm), and cell growth was monitored by the MTT assay. A and B, insets, Western blots showing ELK1 and AR with GAPDH as the loading control. C, LNCaP cells treated as described for A were harvested at 48 h of treatment to measure mRNA levels for the indicated androgen-activated genes by quantitative real time PCR. D, hormone-depleted VCaP cells were infected with ELK1 shRNA lentivirus or control shRNA lentivirus. 72 h later (after the knockdown had occurred), cells were treated with vehicle or R1881 (1 nm), and cell growth was monitored by the MTT assay. D, inset, Western blot showing ELK1 and AR with GAPDH as the loading control. E, VCaP cells treated as described for D were harvested at 48 h of treatment to measure mRNA levels for the indicated androgen-activated genes by quantitative real time PCR. F, DU145 and PC-3 cells were infected with ELK1 shRNA lentivirus or control shRNA lentivirus. 72 h later (after the knockdown had occurred), cell growth was monitored by the MTT assay. F, inset, Western blot showing ELK1 with GAPDH as the loading control. R denotes R1881. * and §, p < 0.001. Error bars represent S.D.
ELK1 Enables Induction by Androgen of a Gene Set That Primarily Supports Cell Cycle Progression and Mitosis
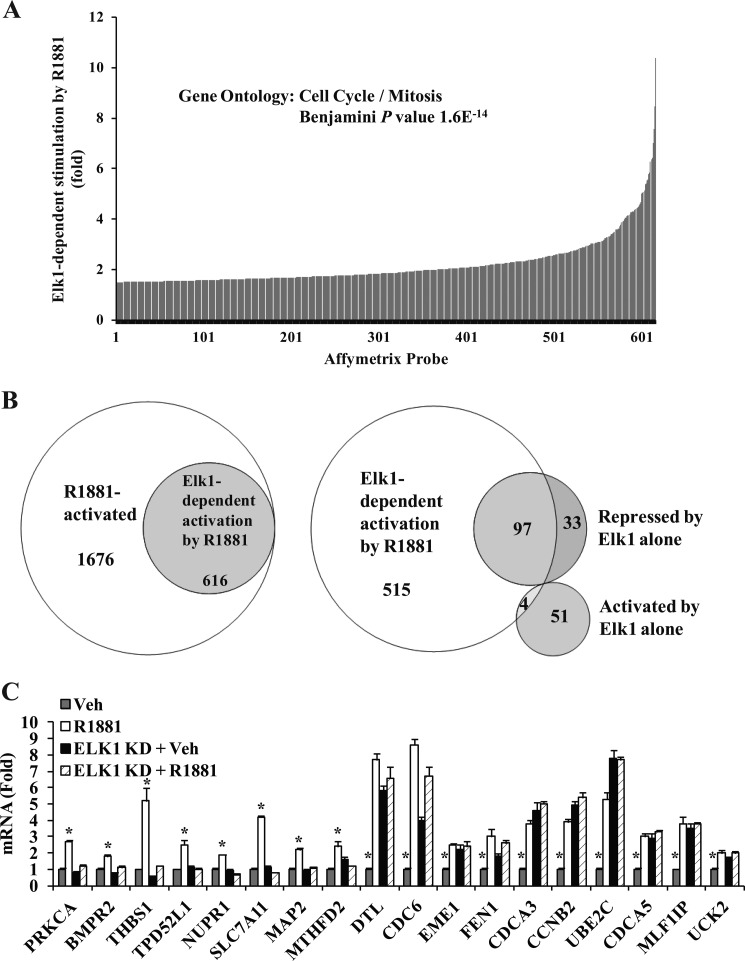
ELK1 is known to both repress and up-regulate genes. The term “activation” of ELK1 refers to regulatory events that either reverse gene repression by ELK1 or cause up-regulation of a target gene by ELK1. mRNA expression profiling of LNCaP cells treated as shown in Fig. 1A using Affymetrix DNA microarray analysis resulted in 616 Affymetrix probe sets showing a ≥50% increase in expression due to androgen treatment in the presence of ELK1 compared with the absence of ELK1 (Fig. 2A and supplemental Table 1). This subset corresponded to 466 unique annotated genes. ELK1 was activated by androgen as evident from both a loss of gene repression by ELK1 and up-regulation of target genes. A substantial proportion (27%) of all probe sets up-regulated by androgen showed fully or partially ELK1-dependent induction by androgen (Fig. 2B); this subset overlapped the majority of a smaller group of probe sets showing ≥50% repression by ELK1 alone (Fig. 2B and supplemental Table 2). A relatively small number of genes showed up-regulation by ELK1 alone (supplemental Table 3), but in contrast to genes repressed by ELK1, only a few of them were further induced by androgen (Fig. 2B). Notably, the list of genes showing induction by androgen in an ELK1-dependent manner did not include well known targets of the ternary complex of ELK1 with SRF and SRE (e.g. c-FOS and EGR1), the activation of which requires phosphorylation of ELK1.
FIGURE 2.
ELK1-dependent genotropic effects of androgen. A, hormone-depleted LNCaP cells were infected with lentivirus expressing ELK1 shRNA or control shRNA. 72 h later (after the knockdown (KD) had occurred), cells were treated with vehicle (Veh) or R1881 (1 nm). 48 h after the treatments, total RNA was extracted and subjected to Affymetrix DNA microarray analysis. The Affymetrix probe profile of ELK1-dependent gene activation by R1881 is plotted. B, the Affymetrix data were used to plot Venn diagrams of sets of probes depicting overlapping subsets of genes coordinately regulated by ELK1 and R1881. C, the results of the Affymetrix microarray analysis are validated for representative genes using quantitative real time PCR. *, p < 0.001. Error bars represent S.D.
Ontology analysis for the gene set showing ELK1-dependent activation by androgen in Fig. 2A was performed using the DAVID Bioinformatics Resources (32, 33). This gene set primarily supported cell cycle and mitosis as represented by the top four clusters with enrichment scores of 14, 11, 5, and 5 and corresponding Benjamini adjusted p values of 1.6e−14, 4.3e−14, 7.1e−8 and 8.6e−6 as well as clusters 6 and 7. This was in contrast to the group of genes that was induced by androgen completely independently of ELK1 (represented by the 1676 probes in Fig. 2B); for this gene set, it was only cluster 19 that showed any enrichment for cell cycle and mitosis genes (enrichment score, 2.21; Benjamini adjusted p value, 2.7e−3).
The Affymetrix data were validated by quantitative RT-PCR for representative genes, most of which are known to support cell cycle progression and mitosis (Fig. 2C). As discussed above, many of the genes were repressed by ELK1 as evident from the fact that depletion of ELK1 increased their expression (e.g. the 10 genes from the right in Fig. 2C); androgen treatment essentially prevented this repression by ELK1 but did not further induce the genes in the absence of ELK1. Many genes (e.g. the eight genes from the left in Fig. 2C) were relatively unaffected by depletion of ELK1 but were activated by androgen; this activation did not occur in the absence of ELK1. The collective patterns of response of target genes to depletion of ELK1 and/or androgen treatment are illustrated in a heat map (supplemental Fig. 2). Thus, androgen increases the expression of ELK1 target genes either by reversing their repression or by simply activating them.
ELK1 Is Also Essential for AR-dependent Clonogenic Survival and Growth of CRPC Cells and Cooperates with the AR Apoprotein to Induce Cell Growth Genes
C4-2 cells are capable of a high frequency of colony formation in soft agar, reflecting their high tumorigenic potential (34). C4-2 cells are capable of robust cell growth in the absence of hormone; however, colony survival of C4-2 cells, particularly in two-dimensional culture (where autocrine factors could dissipate more easily), is optimal in the presence of androgen.
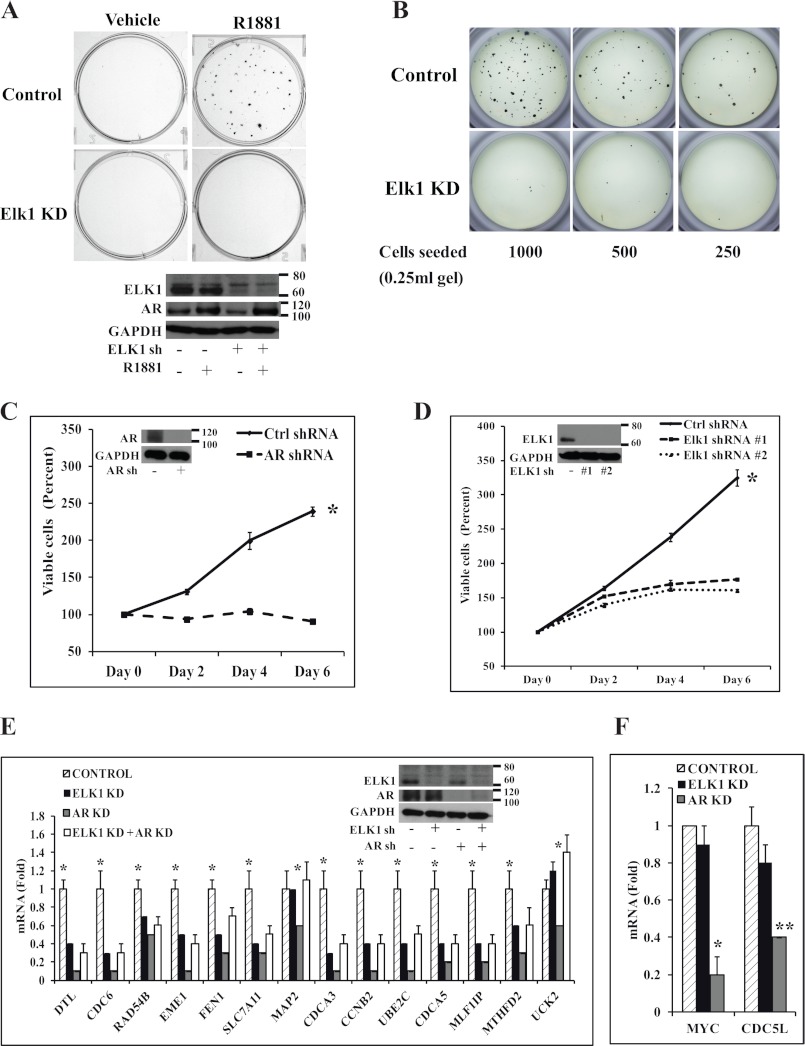
In C4-2 cells, clonogenicity was virtually abrogated by knocking down ELK1 with either one of two lentiviral ELK1 shRNAs when examined by two-dimensional colony formation assay (Fig. 3A, top panel, for ELK1 shRNA 1 and supplemental Fig. 3 for ELK1 shRNA 2). In this experiment, depletion of ELK1 did not appreciably alter the expression of AR (Fig. 3A, lower panel). Similarly, depletion of ELK1 virtually abrogated colony formation using the anchorage-independent clonogenicity assay in soft agar (Fig. 3B).
FIGURE 3.
Effect of depleting ELK1 on clonogenic survival, growth, and AR-regulated gene expression in CRPC cells. A, top, C4-2 cells plated in hormone-depleted medium were infected with ELK1 shRNA lentivirus or control shRNA lentivirus. 72 h later (after the knockdown had occurred), anchorage-dependent colony formation was measured following treatment with vehicle or R1881 (1 nm). At the end of 2 weeks, colonies were stained with crystal violet. A, bottom, at the end of 48 h of treatment, the cells were harvested for Western blot to probe for ELK1 and AR with GAPDH as the loading control. B, C4-2 cells plated in hormone-depleted medium were infected with ELK1 shRNA lentivirus or control shRNA lentivirus. 72 h later (after the knockdown had occurred), anchorage-independent colony formation was measured in 0.8% soft agar containing a serial dilution of cells following treatment with R1881 (1 nm). At the end of 2 weeks, colonies were stained with MTT. C, hormone-depleted C4-2 cells were infected with AR shRNA lentivirus or control (Ctrl) shRNA lentivirus. 72 h later (after the knockdown had occurred), cell growth was monitored in hormone-depleted medium by the MTT assay. C, inset, Western blot showing AR with GAPDH as the loading control. D, hormone-depleted C4-2 cells were infected with ELK1 shRNA 1, ELK1 shRNA 2 lentivirus, or control shRNA lentivirus. 72 h later (after the knockdown had occurred), cell growth was monitored in hormone-depleted medium by the MTT assay. D, inset, Western blot showing ELK1 with GAPDH as the loading control. E, C4-2 cells plated in hormone-depleted medium were infected with ELK1 shRNA, AR shRNA lentivirus, a combination of AR shRNA and ELK1 shRNA lentivirus, or control shRNA lentivirus. 72 h after infection, cells were seeded in 6-well plates and incubated for an additional 72 h to achieve a good combined knockdown of ELK1 and AR. Cells were harvested to measure mRNA levels for the indicated genes by quantitative real time PCR. E, inset, Western blot showing ELK1 and AR with GAPDH as the loading control. F, the RNA samples used in E were analyzed for expression of two previously known apo-AR-regulated genes by quantitative real time PCR. KD denotes knockdown. *, p < 0.001; **, p < 0.001. Error bars represent S.D.
C4-2 cells were capable of robust growth in monolayer culture in the absence of hormone (in medium containing charcoal-stripped serum). This growth was dependent on AR as depletion of AR inhibited the hormone-independent growth (Fig. 3, C and inset). Knocking down ELK1 using either one of the two ELK1 shRNA lentiviruses with different target site specificities attenuated the hormone-independent growth of C4-2 cells (Fig. 3, D and inset) as did the double knockdown of ELK1 and AR (supplemental Fig. 4). It may be noted that the ELK1-depleted cells exhibited a low basal growth rate that was absent in the AR-depleted cells, possibly reflecting the loss of repression of many growth-supporting genes by ELK1 (discussed below).
Most of the genes validated above as targets for ELK1-dependent induction by androgen, including cell growth-supporting genes, were also targets for ELK1-dependent induction by the AR apoprotein in C4-2 cells. This is evident from the observation that depletion of AR decreased gene expression in the presence of ELK1 but not when ELK1 was knocked down (Fig. 3, E and inset). As expected, the results from AR knockdown versus combined ELK1 and AR knockdown showed that ELK1 repressed the genes to a variable extent independently of AR. On the other hand the expression levels of MYC and CDC5L that we have previously shown to be supported by AR independently of hormone (9) were unaltered by depleting ELK1 (Fig. 3F), confirming that ELK1 did not regulate the level of functional AR in C4-2 cells.
Growth Effects of ELK1 Depletion Are Not Quantitatively Related to Apoptosis
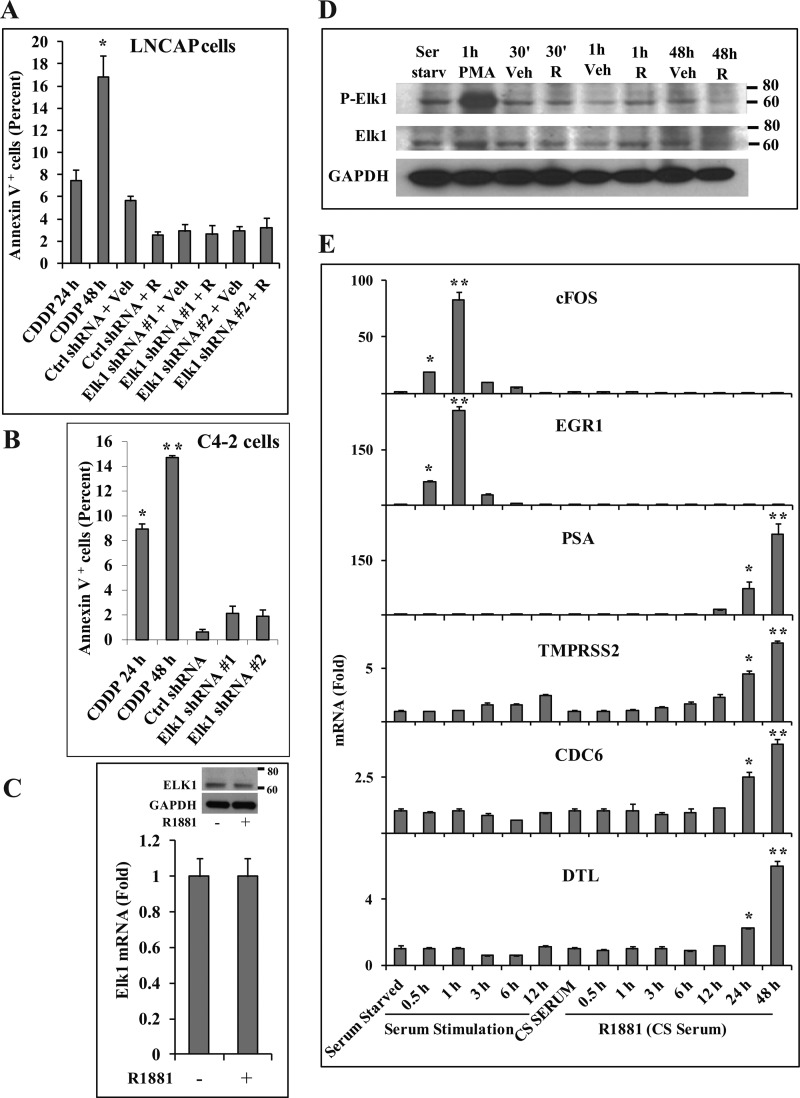
To test whether decreased cell survival contributed to the growth inhibition upon depleting ELK1, LNCaP and C4-2 cells were analyzed by the Annexin V assay at time points corresponding to the onset of the growth inhibition. As a positive control for the assay, cisplatin was used to induce apoptosis in the two cell lines. The two different ELK1 shRNA lentiviruses used in the growth studies above were used to knock down ELK1. Neither shRNA increased the number of apoptotic cells in LNCaP cells grown in the presence of androgen (Fig. 4A), and they only modestly increased apoptosis in C4-2 cells grown in hormone-depleted medium (Fig. 4B).
FIGURE 4.
Lack of influence of ELK1-dependent androgen/AR signaling on cell survival, ELK1 phosphorylation, or regulation of immediate early genes. A, hormone-depleted LNCaP cells were infected with ELK1 shRNA 1, ELK1 shRNA 2, or control (Ctrl) shRNA lentivirus. 96 h later, cells were treated with vehicle (Veh) or R1881 (1 nm) for 48 h. Apoptosis was measured by the Annexin V assay. As a positive control for the assay, apoptosis was induced by treatment with cisplatin (100 μm) for 24 and 48 h. B, hormone-depleted C4-2 cells were infected with ELK1 shRNA 1, ELK1 shRNA 2, or control shRNA lentivirus. 6 days later, apoptosis was measured by the Annexin V assay. As a positive control for the assay, apoptosis was induced by treatment with cisplatin (100 μm) for 24 and 48 h. C, LNCaP cells were treated with vehicle or R1881 (1 nm) for 48 h and harvested for RNA and protein. ELK1 expression was measured using quantitative real time PCR, and the protein lysates were analyzed by Western blot for ELK1 with GAPDH as loading control (inset). D, serum-starved (Ser starv) LNCaP cells were treated with vehicle or phorbol 12-myristate 13-acetate (PMA) (10 μm) for 1 h. LNCaP cells plated in hormone-depleted medium were treated with vehicle or R1881 (1 nm) for the indicated times. Cell lysates from all the samples were analyzed by Western blot for phospho-ELK1 (P-Elk1) or ELK1 protein with GAPDH as the loading control. E, serum-starved LNCaP cells were stimulated with 20% FBS for the indicated times. LNCaP cells plated in hormone-depleted medium in charcoal-stripped serum (CS SERUM) were treated with vehicle or R1881 (1 nm) for the indicated times. RNA was harvested, and the induction of endogenous c-FOS, EGR1, PSA, TMPRSS2, CDC6, and DTL by serum or by R1881 was measured by quantitative real time PCR. For serum stimulation, mRNA levels are plotted relative to the values for serum starvation. For R1881 treatment, the mRNA levels are plotted relative to their respective vehicle controls. R denotes R1881. * and **, p < 0.001. Error bars represent S.D.
Functional Activation of ELK1 by Androgen Is Not Associated with Altered ELK1 Expression or Phosphorylation
Androgen treatment did not affect the expression level of ELK1 mRNA or ELK1 protein (Fig. 4, C and inset). Therefore, the androgen effect that is dependent upon ELK1 is not due to regulation of ELK1 expression.
The classical mechanism of activation of ELK1 resulting in target gene induction is by phosphorylation, notably at positions Ser-383 and Ser-389. When serum-starved cells are stimulated with serum factors or steroids, including androgens, ELK1 is phosphorylated through MAPK pathways; this results in transient induction of immediate early genes such as c-FOS. In the preceding studies, ELK1 supported sustained (rather than transient) induction of growth genes by androgen or AR in medium containing charcoal-stripped serum. Therefore, it was of interest to test whether, under these conditions, the activation of ELK1 by androgen or AR involved initial ELK1 phosphorylation. In charcoal-stripped serum, androgen did not induce phosphorylation of ELK1 in LNCaP cells (Fig. 4D). As a positive control for ELK1 phosphorylation, stimulation of serum-starved LNCaP cells with phorbol 12-myristate 13-acetate caused phosphorylation of ELK1 within 1 h (Fig. 4D). Therefore, the sustained activation of ELK1 by AR observed above is independent of ELK1 phosphorylation.
In Charcoal-stripped Serum, Androgen Does Not Induce the Immediate Early Gene Response Associated with the ELK1 Ternary Complex
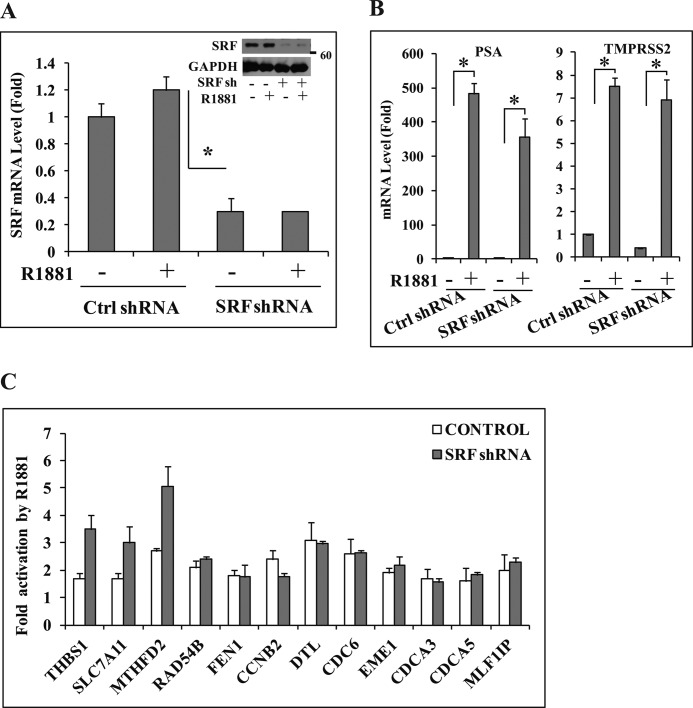
Several immediate early genes, including c-FOS and EGR1, are targets of the ternary complex of ELK1, SRF, and SRE, which is activated by MAPK-mediated phosphorylation of ELK1. In charcoal-stripped serum in which ELK1 supported sustained (long term) gene activation by androgen, the hormone treatment failed to similarly activate the TCF target genes c-FOS and EGR1 either in the short term or after prolonged exposure (Fig. 4E). In contrast, both mRNAs increased in rapid response to serum stimulation (Fig. 4E). As expected, the PSA and TMPRSS2 genes (controls; classical androgen target genes) were induced by androgen over a longer period (Fig. 4E). Examples of androgen-induced genes for which ELK1 was required, including DTL, CDC6, THBS1, and SLC7A11 (Fig. 4E and supplemental Fig. 5), followed a time course of induction similar to that for PSA and TMPRSS2. On the other hand, UBE2C, which also required ELK1 for induction by androgen (Fig. 2C), showed delayed induction by androgen (supplemental Fig. 5), suggesting that this could be one of the indirect targets of ELK1-dependent gene induction by androgen. Furthermore, depletion of SRF using lentiviral SRF shRNA (Fig. 5A) did not appreciably decrease the ability of androgen to induce either its classical target genes (PSA and TMPRSS2) (Fig. 5B) or any of the target genes tested for which ELK1 was required (Fig. 5C); as expected, depletion of SRF did attenuate the immediate early gene response (supplemental Fig. 6). Therefore, the cooperative long term transcriptional signaling of ELK1 and AR excludes the ternary complex target genes. AR elicits a sustained genotropic effect through ELK1 rather than the transient response that is typical of the classical mechanism of activation of ELK1.
FIGURE 5.
Lack of dependence on SRF for ELK1-dependent gene activation by androgen. A, hormone-depleted LNCaP cells were infected with SRF shRNA (sh) or control (Ctrl) shRNA lentivirus. 72 h later (after the knockdown had occurred), cells were treated with vehicle or R1881 (1 nm). 48 h after treatment, cells were harvested for RNA and protein. SRF mRNA expression was measured using quantitative real time PCR, and the cell lysates were analyzed by Western blot for SRF with GAPDH as loading control (inset). B, the RNA samples from A were used to measure mRNA levels of PSA and TMPRSS2 by quantitative real time PCR. C, the RNA samples used in A were analyzed for expression of the indicated genes by quantitative real time PCR. *, p < 0.001. Error bars represent S.D.
The ELK1 Binding Element and the ELK1 Protein Mediate Transactivation by AR
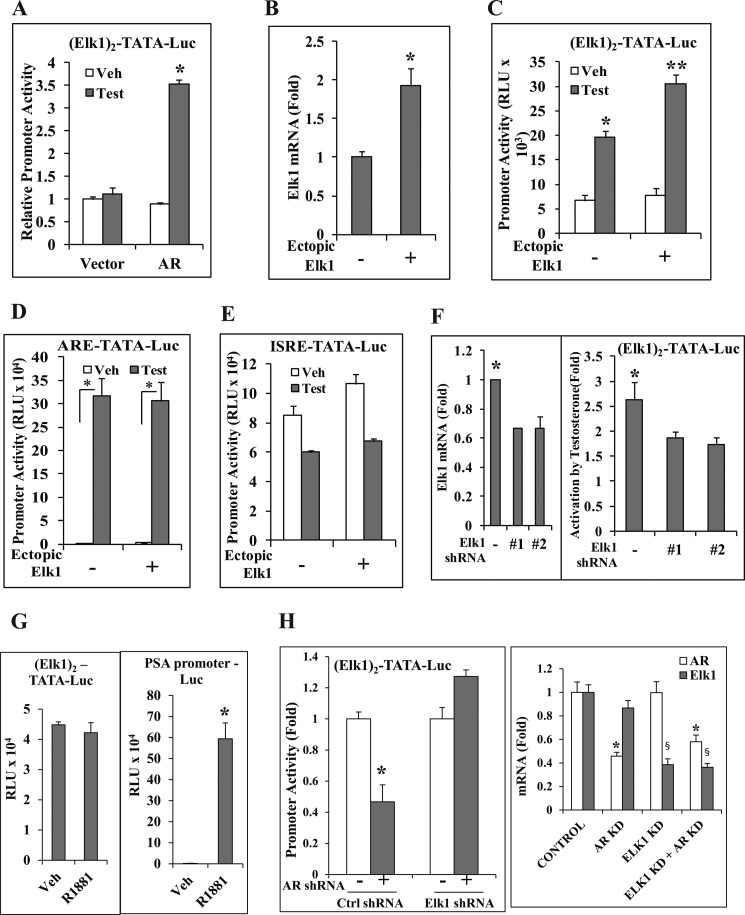
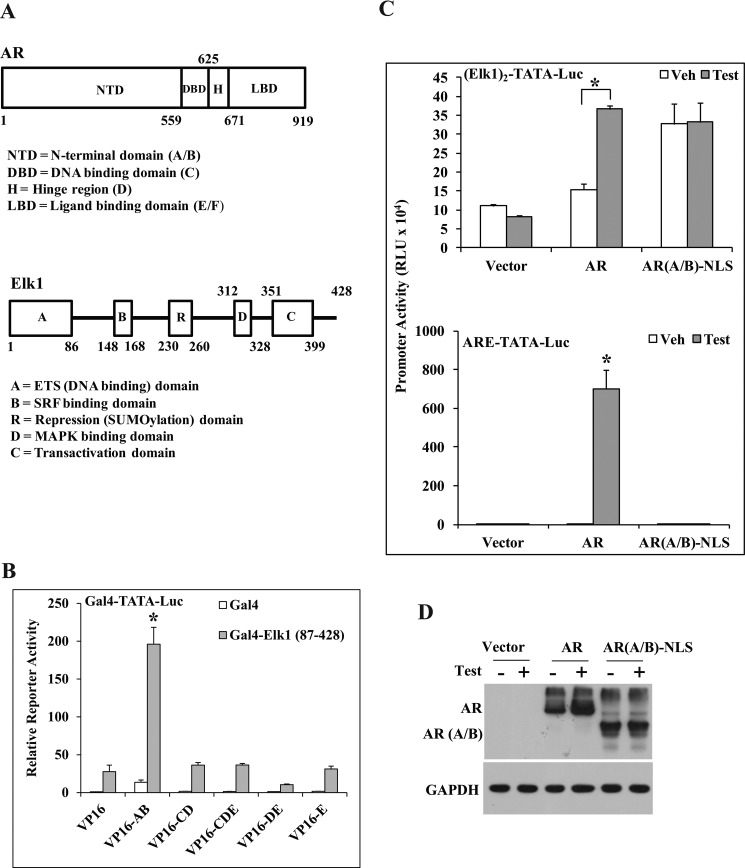
ELK1 binds to a cis-element that is shared with other ETS proteins. However, ELK1 does show sequence preference; furthermore, it is preferentially bound to certain ETS sites in the chromatin context. The cis-element preferred by ELK1 was placed as two tandem repeat elements upstream of a minimal TATA-dependent promoter-reporter luciferase construct ((ELK1)2-TATA-Luc). In the AR-negative HeLa cells in which this promoter construct was introduced by transient transfection, the promoter was activated by androgen in an AR-dependent manner (Fig. 6A). Doubling the level of ELK1 mRNA by transfecting an ELK1 expression plasmid (Fig. 6B) increased the promoter activation by androgen (Fig. 6C). In contrast, ectopic ELK1 did not affect the promoter activity when the ELK1 element was replaced by either the androgen-activated classical ARE (Fig. 6D) or an androgen-repressed ISRE (Fig. 6E). A partial knockdown of endogenous ELK1 by transient co-transfection with either of the two different ELK1 shRNA expression plasmids (Fig. 6F, left panel) significantly decreased androgen activation of (ELK1)2-TATA-Luc (Fig. 6F, right panel). Thus, both loss of function and gain of function experiments demonstrate the ability of AR to specifically activate a promoter through the ELK1 binding element and the ELK1 protein.
FIGURE 6.
Cooperative promoter activation by ELK1 and AR through the ELK1 binding element. A, HeLa cells were co-transfected with (ELK1)2-TATA-Luc and pSG5-AR or pSG5 vector control. Cells were treated with either vehicle or testosterone (10 nm), and luciferase activity was measured 48 h after transfection. B and C, HeLa cells were co-transfected with (ELK1)2-TATA-Luc and pCMV-ELK1 or pCMV vector control and pSG5-AR. Cells were treated with either vehicle (Veh) or testosterone (Test) (10 nm). 48 h after transfection, cells were harvested to measure ELK1 mRNA (B) or lysed to measure luciferase activity (C). D, HeLa cells were transfected and treated as described for C except that the promoter construct used was ARE-TATA-Luc. E, HeLa cells were transfected and treated as described for C except that the promoter construct used was ISRE-TATA-Luc. F, HeLa cells were co-transfected with (ELK1)2-TATA-Luc and ELK1 shRNA 1, ELK1 shRNA 2, or control shRNA plasmid and pSG5-AR. Cells were treated with testosterone (10 nm). 48 h after transfection, cells were harvested to measure ELK1 mRNA (left panel) or to measure luciferase activity (right panel). G, C4-2 cells were plated in hormone-depleted medium and nucleofected with either (ELK1)2-TATA-Luc (left panel) or PSA-promoter Luc (right panel) and treated with vehicle or R1881. 48 h after nucleofection, the cells were harvested for luciferase activity. H, C4-2 cells were nucleofected in hormone-depleted medium with (ELK1)2-TATA-Luc and AR shRNA, ELK1 shRNA, or both ELK1 and AR shRNAs. After 48 h of nucleofection, the cells were harvested for luciferase activity (left panel). The endogenous AR and ELK1 mRNA expression levels were measured by quantitative real time PCR (right panel). *, **, and §, p < 0.001. Error bars represent S.D. Ctrl, control; KD, knockdown; RLU, relative luciferase units.
The AR- and ELK1-dependent promoter activation studies in HeLa cells were extended to C4-2 cells by examining the effect of knocking down endogenous ELK1 specifically on the activity of transiently transfected (ELK1)2-TATA-Luc. In C4-2 cells, androgen did not activate the (ELK1)2-TATA-Luc promoter in contrast to the PSA promoter-Luc (Fig. 6G). However, the (ELK1)2-TATA-Luc promoter was activated by AR in an ELK1-dependent manner as evident from the results of the individual and combined knockdown of ELK1 and AR (Fig. 6H).
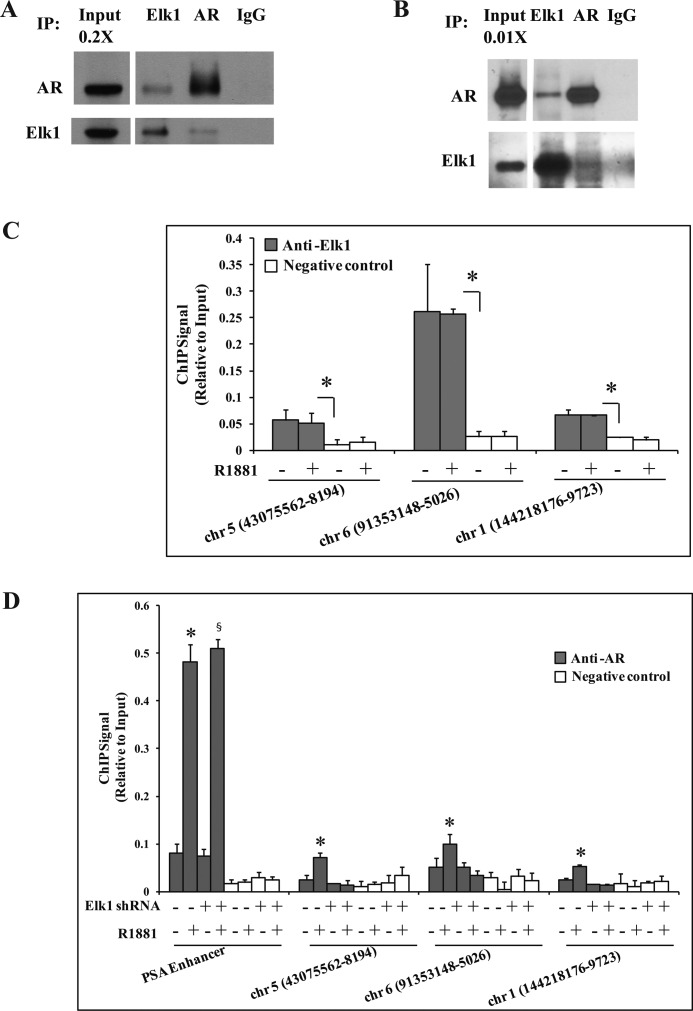
ELK1 Physically Associates with AR and Recruits It to Chromatin
Physical association between endogenous ELK1 and endogenous AR was evident from co-immunoprecipitation experiments using C4-2 (Fig. 7A) and LNCaP cells (Fig. 7B). ELK1 and AR specifically co-immunoprecipitated when antibody specific to either ELK1 or AR was used.
FIGURE 7.
Binding and chromatin recruitment of AR by ELK1. A and B, either AR or ELK1 was immunoprecipitated using the appropriate antibody from cell lysates obtained from C4-2 cells (A) and LNCaP cells (B) as described under “Experimental Procedures.” The immunoprecipitates were probed by Western blots using antibody to AR or ELK1 as indicated. Normal IgG was used as the negative control for the immunoprecipitation (IP). C, LNCaP cells plated in hormone-depleted medium were treated with either vehicle or R1881 (1 nm) for 2 h. Cells were harvested and subjected to ChIP using anti-ELK1 antibody. TaqMan probes were used to quantify the immunoprecipitated products using target sequences contained within 250 bp of the center of the chromosome (chr) regions (peak regions of ELK1 binding) indicated in the figure. D, LNCaP cells plated in hormone-depleted medium were infected with either ELK1 shRNA or control shRNA lentivirus. 72 h later, cells were treated with either vehicle or R1881 (1 nm) for 2 h. Cells were harvested and subjected to ChIP using anti-AR antibody. The TaqMan probes for the PSA enhancer and the probes used in C were used to quantify the immunoprecipitated products. * and §, p < 0.001. Error bars represent S.D.
Next, we sought evidence for the ability of ELK1 to recruit AR to chromatin. Because AR most commonly exerts its transcriptional activity by binding at relatively great distances (10–50 kb) from the target genes, we first chose seven chromatin sites selected from the published (29) HeLa cell data based on (i) the rank order of signal intensity for ELK1 binding, (ii) the presence of consensus ELK1 binding elements corresponding to the peak center and the absence of AREs, and (iii) the ability to design suitable TaqMan probes for target sequences within 250 bp of the peak center. Of the seven sites selected, three sites consistently showed ELK1 binding, and this binding was androgen-independent (Fig. 7C). Androgen induced quantifiable recruitment of AR at these sites in LNCaP cells (Fig. 7D), although as expected, the signal intensities were weaker than that at the classical enhancer site of the PSA gene. Furthermore, the androgen-induced recruitment of AR was abolished upon depleting ELK1 using lentiviral shRNA (Fig. 7D). In contrast, androgen-dependent recruitment of AR at the classical PSA enhancer was unaffected upon depletion of ELK1 (Fig. 7D). The results demonstrate the ability of ELK1 to recruit AR to ELK1 binding elements in the chromatin context.
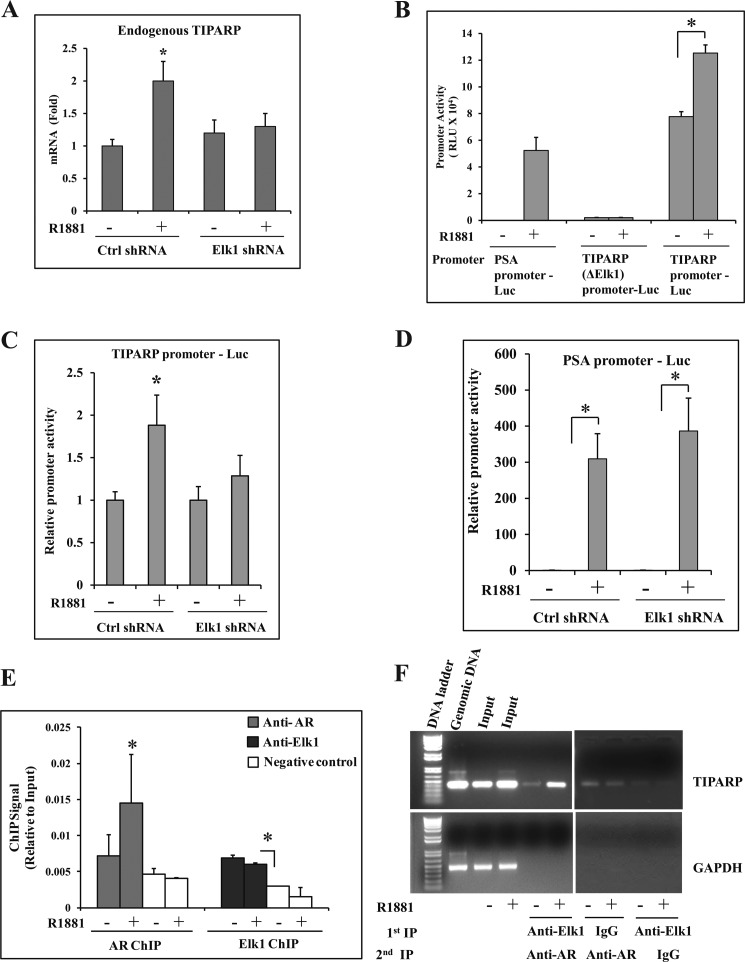
In a second approach, we sought to use both promoter activity assays and ChIP to demonstrate simultaneous occupancy of ELK1 and AR at a functional promoter-proximal site of an endogenous target gene. At such a site, we would expect the binding of AR but not ELK1 to be androgen-dependent. We used the mRNA profiling data for LNCaP cells in supplemental Table 1, the published data on chromatin sites of ELK1 binding in a limited portion of the genome in HeLa cells (29), and AR binding data in LNCaP cells (35) to identify a candidate gene. We identified a candidate ELK1 binding element at nucleotide positions −357 to −348 in the TIPARP gene, which is induced by androgen ∼2-fold in an ELK1-dependent manner. The induction of TIPARP by androgen was validated by real time quantitative RT-PCR (Fig. 8A). A TIPARP promoter-luciferase reporter construct spanning nucleotides −536 to +60 was activated by androgen in transfected LNCaP cells; androgen could not activate this construct when the ELK1 element was deleted (TIPARP (ΔELK1) promoter-Luc) (Fig. 8B) or when ELK1 was depleted (Fig. 8C) in contrast to the PSA promoter-Luc (Fig. 8D). ChIP analysis demonstrated ELK1 binding to the promoter both in the absence and in the presence of androgen, whereas the binding of AR at this chromatin site was androgen-dependent (Fig. 8E). Sequential ChIP was performed using anti-ELK1 antibody for the first ChIP followed by anti-AR antibody for the re-ChIP and vice versa. As the intensity of the re-ChIP signal was not within the reliable limit of the Ct values of the TaqMan probes, highly specific PCR primers were designed to amplify an ∼400-bp promoter fragment of the TIPARP gene; control primers that could amplify a similarly sized fragment of the GAPDH gene were also used. The results clearly demonstrated androgen-dependent binding of AR and the simultaneous binding of ELK1 (Fig. 8F and supplemental Fig. 7).
FIGURE 8.
Functional association of ELK1 and AR in the TIPARP gene promoter. A, LNCaP cells plated in hormone-depleted medium were infected with either ELK1 shRNA or control (Crtl) shRNA lentivirus. 72 h later, cells were treated with either vehicle or R1881 (1 nm) for 48 h. Total RNA from the cells was used to measure the relative mRNA levels for the endogenous TIPARP gene by real time quantitative RT-PCR. B, LNCaP cells plated in hormone-depleted medium were nucleofected with PSA promoter-Luc, TIPARP (ΔELK1) promoter-Luc, or TIPARP promoter-Luc. The cells were treated with either vehicle or R1881 (1 nm) for 48 h. The cells were then harvested, and the luciferase activities were measured. C and D, LNCaP cells plated in hormone-depleted medium were nucleofected with TIPARP promoter-Luc (C) or PSA promoter-Luc (D) and co-transfected with control shRNA or ELK1 shRNA lentivirus. The cells were treated with either vehicle or R1881 (1 nm) for 48 h. The cells were then harvested, and the luciferase activities were measured. E, LNCaP cells plated in hormone-depleted medium were treated with either vehicle or R1881 (1 nm) for 2 h. Cells were harvested and subjected to ChIP using either anti-AR antibody or anti-ELK1 antibody. TaqMan probes were used to quantify the immunoprecipitated chromatin. F, LNCaP cells plated in hormone-depleted medium were treated with either vehicle or R1881 (1 nm) for 2 h. Cells were harvested and subjected to ChIP using either anti-ELK1 antibody or normal rabbit IgG. The chromatin complexes from the first ChIP (1st IP) were subjected to re-ChIP (2nd IP) using anti-AR antibody. The immunoprecipitated chromatin was amplified by PCR using primers specific for the TIPARP gene promoter or a genomic sequence of GAPDH (non-targeting negative control). PCR products were analyzed by agarose gel electrophoresis on a 1% gel. Untreated genomic DNA was used as a control to ensure the specificity of the PCR amplification. *, p < 0.001. Error bars represent S.D. RLU, relative luciferase units.
ELK1 Binds to the N-terminal A/B Domain of AR
The interacting domains of AR and ELK1 (Fig. 9A) were mapped in situ using the mammalian two-hybrid assay. In the Gal4-ELK1(87–428) fusion construct, the N-terminal DNA binding domain of ELK1, which is highly conserved among ETS proteins, was replaced by the DNA binding domain of Gal4. VP16 fusion constructs of various domains of AR were used (Fig. 9A). The N-terminal A/B domain (residues 1–559) of AR interacted exclusively and strongly with ELK1 (Fig. 9B). The identification of a discrete segment of AR that specifically and strongly interacted with ELK1 in situ provides complementary evidence demonstrating the ability of ELK1 to physically recruit AR.
FIGURE 9.
Identification of functional domains of AR interacting with ELK1. A, schematics drawn roughly to scale indicating the structural organization of AR (top) and ELK1 (bottom), including the positions of the major functional domains. B, a mammalian two-hybrid assay was conducted as described under “Experimental Procedures.” HeLa cells plated in hormone-depleted medium were co-transfected with pG5Luc, pBind (Gal4 expression plasmid), or pBind expressing the Gal4 fusion proteins with ELK1(87–428) together with VP16 fusion proteins of different AR domains (A/B, CD, CDE, DE, and E). The hybridization signal was measured by assaying for luciferase activity. C and D, HeLa cells plated in hormone-depleted medium were transfected with either (ELK1)2-TATA-Luc (C, top panel) or ARE-TATA-Luc (C, bottom panel) and co-transfected with expression plasmids for AR, AR(A/B)-NLS, or the vector control. At the same time, the cells were treated with either vehicle (Veh) or testosterone (Test). Cells were harvested 48 h later to measure reporter luciferase activities (C) or for Western blot analysis using anti-AR antibody (D). *, p < 0.001. Error bars represent S.D. RLU, relative luciferase units.
Nuclear Targeting of the AR A/B Domain Recapitulates Promoter Activation through the ELK1 Element by Androgen and Full-length AR
The A/B domain of AR comprises about half the length of the AR polypeptide (∼550 amino acids), including sites of ligand-independent transactivation and coactivator recruitment. We attached a C-terminal nuclear localization signal to the A/B domain construct (AR(A/B)-NLS) to ensure optimal nuclear localization. When expressed in HeLa cells, AR(A/B)-NLS was able to activate (ELK1)2-TATA-Luc in the absence of hormone to the same extent that WT AR did in the presence of androgen (Fig. 9C, top panel). The expression level of the A/B domain was comparable with the androgen-stabilized level of WT AR (Fig. 9D). In contrast, ARE-driven promoter activation was only observed in the presence of WT AR plus androgen (Fig. 9C, bottom panel). The results demonstrate that the A/B domain is the functional entity required for the action of AR as a coactivator of ELK1 and that the role of ligand binding is for translocation of AR to the nucleus.
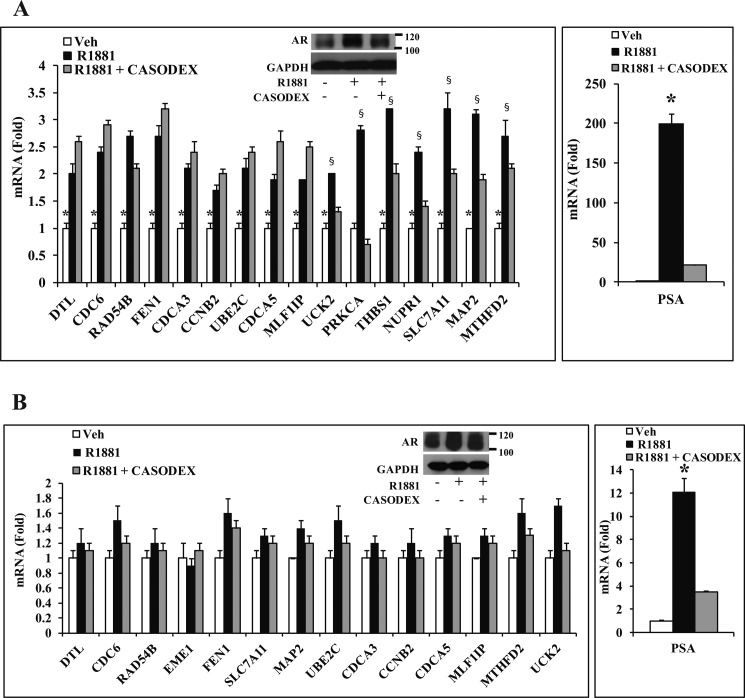
Casodex Is a Poor Antagonist of ELK1-dependent Gene Activation by Androgen or the AR Apoprotein
Casodex is an antiandrogen that competes with androgen for binding to AR and antagonizes the classical mechanism of gene activation by androgen. Although the ELK1-dependent gene activation by AR required androgen in LNCaP cells, it was only partially and variably sensitive to Casodex as demonstrated for a representative set of genes (Fig. 10A, left panel). Furthermore, in C4-2 cells, the cooperative gene activation by ELK1 and AR was largely insensitive to both androgen and Casodex (Fig. 10B, left panel). In contrast, the classical androgen target gene PSA was exquisitely sensitive to androgen as well as Casodex in both LNCaP and C4-2 cells (Fig. 10, A and B, right panels).
FIGURE 10.
Effect of Casodex on ELK1-dependent gene activation by androgen/AR. A, LNCaP cells plated in hormone-depleted medium were treated with vehicle (Veh) or R1881 (1 nm) or R1881 (1 nm) + Casodex (10 μm). 48 h after treatment, cells were harvested to measure mRNA levels of the indicated genes by quantitative real time PCR. A, inset, Western blot for AR expression following the different treatments with GAPDH as the loading control. B, C4-2 cells plated in hormone-depleted medium were treated with vehicle or R1881 (1 nm) or R1881 (1 nm) + Casodex (10 μm). 48 h after treatment, cells were harvested to measure mRNA levels of the indicated genes by quantitative real time PCR. B, inset, Western blot for AR expression following the different treatments with GAPDH as the loading control. * and §, p < 0.001. Error bars represent S.D.
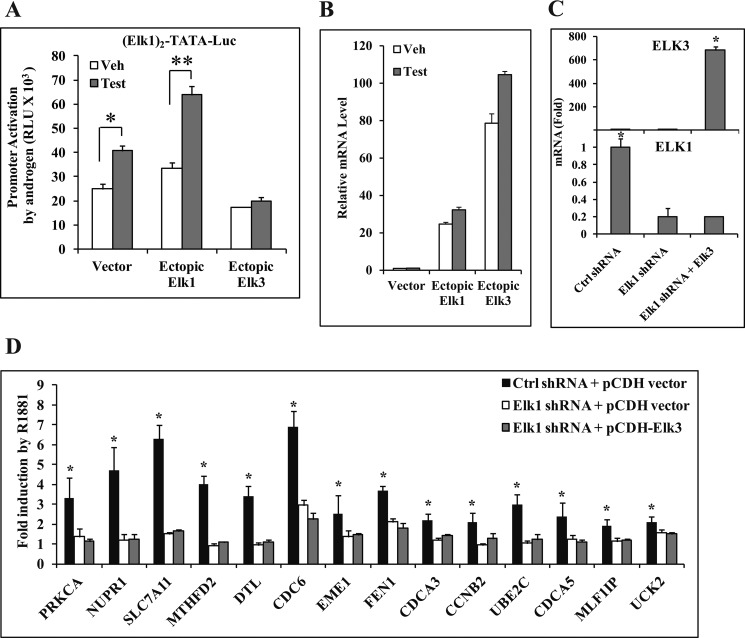
ELK3 Suppresses the Synergy between AR and ELK1
As ELK3 is the closest structural and functional analog of ELK1, the effect of ectopically overexpressing ELK3 on androgen-stimulated activation of the (ELK1)2-TATA-Luc promoter was tested. Ectopic ELK1 increased the androgen response (Fig. 11, A and B). In contrast, ectopic ELK3 (Fig. 11B) virtually completely suppressed the androgen response (Fig. 11A). Therefore, AR cannot synergize with ELK3 to activate the target promoter in contrast to ELK1.
FIGURE 11.
Inability of ELK3 to interact with AR. A and B, HeLa cells plated in hormone-depleted medium were transfected with (ELK1)2-TATA-Luc. The same amount of either the ELK1 or ELK3 expression plasmid or the pCMV vector control plasmid was co-transfected. In all cases, the AR expression plasmid was co-transfected. The cells were treated with vehicle (Veh) or testosterone (Test) (10 nm) and harvested 48 h after transfection to measure luciferase activity (A) or to measure relative mRNA levels of ELK1 and ELK3 (B). C, LNCaP cells plated in hormone-depleted medium were infected with either ELK1 shRNA or control (Ctrl) shRNA lentivirus along with pCDH empty vector lentivirus or pCDH-ELK3 lentivirus. 72 h after infection, cells were harvested. The expression levels of mRNAs for ELK3 (top panel) and ELK1 (bottom panel) were measured by quantitative real time PCR. D, LNCaP cells plated in hormone-depleted medium were infected with either ELK1 shRNA or control shRNA lentivirus along with pCDH empty vector lentivirus or pCDH-ELK3 lentivirus. 72 h later, the cells were treated with either vehicle or R1881 (1 nm) for 48 h and harvested. The expression levels of mRNAs for the indicated genes were measured by quantitative real time PCR. * and **, p < 0.001. Error bars represent S.D.
It was of further interest to confirm the inability of ELK3 to support activation of endogenous genes that are the targets of ELK1-AR. Accordingly, the ability of ectopic ELK3 to rescue gene activation by androgen in the absence of ELK1 was tested. In LNCaP cells, overexpression of ELK3 using lentivirus following knockdown of ELK1 using lentiviral shRNA (Fig. 11C) failed to rescue the activation of ELK1-dependent androgen target genes (Fig. 11D). This result further confirms that AR does not cooperate with ELK3.
DISCUSSION
Androgen is essential for all aspects of normal prostate physiology, including development, differentiation, maintenance, and function of the prostate epithelium (36). Malignant prostate epithelial cells must therefore selectively support mechanisms that direct androgen/AR signaling to strongly support growth. This study has identified one such mechanism mediated by ELK1. To our knowledge, no studies have previously reported the addiction of cancer cells of any type to ELK1. Phosphorylation of ELK1 through MAPK activates the ternary ELK1-SRF-SRE complex, resulting in transient transcriptional activation of immediate early genes such as c-FOS and EGR1. However, the sustained cooperative action of ELK1 and AR observed in this study did not involve phosphorylation of ELK1, did not require SRF, and did not activate the ternary complex target genes. Instead, ELK1 enabled sustained activation by AR of a distinct set of primarily cell growth-supporting genes. Many of those genes were targets of repression by ELK1; accordingly, under conditions of hormone depletion, hormone-dependent prostate cancer cells acquired a modest growth advantage upon depletion of ELK1. However, in the context of androgen-dependent interaction of AR with ELK1, retention of ELK1 by the tumor cells offers a profound growth advantage apparently because of the activation of many genes in addition to relief of gene repression. The same reasoning extends to the observed role of ELK1 in supporting the growth of CRPC cells whether it is androgen-dependent clonogenicity or hormone-independent growth.
The evidence indicates that the underlying mechanism for the synergy between ELK1 and AR is the recruitment of AR to the target genes as a coactivator by ELK1. The role of tethering proteins in directing the genomic actions of nuclear receptors has been well established by Safe and Kim (37, 38). In CRPC cells, although clonogenic survival was supported by androgen, the cooperative action of ELK1 and AR in supporting both cell growth and the activation of many growth-supporting genes occurred in the absence of hormone. This hormone independence was also accompanied by the lack of sensitivity of the gene activation to Casodex. In the hormone-dependent cells, the mechanistic distinction between ELK1-dependent gene activation by androgen and the classical mechanism of gene activation by androgen (e.g. activation of the PSA gene) was reflected by the absent or poor sensitivity of the former to Casodex. We have previously demonstrated that tethered association of AR with its target sites could be hormone-independent when the cells are independent of hormone for nuclear import of AR (21). The recruitment of AR by ELK1 represents the first example in which an AR-tethering protein binds to the N-terminal A/B domain of AR. This study further demonstrates that the A/B domain of AR alone is adequate to serve as a coactivator of ELK1. This is in contrast the classical mechanism of gene activation by AR that requires hormone not only for nuclear localization but also for the DNA binding and transactivation functions of AR. Although Casodex is an antagonist of the classical mechanism of gene activation by AR, the antagonist still enables localization of AR in the nucleus. Thus, the ELK1-mediated component of growth signaling by AR can also be utilized by the tumor cells after progression to CRPCs, which are refractory to traditional antiandrogens. The results are also consistent with recent reports on AR splice variants with C-terminal truncations (7, 8) that are transcriptionally active and support tumor growth in the absence of the ligand binding domain.
Most of the genes repressed by ELK1 were induced by androgen or by the AR apoprotein. Gene repression by ELK1 has been associated with SUMOylation of ELK1 that results in the recruitment of SUMO-dependent histone deacetylase. Phosphorylation of ELK1, primarily through the three MAP kinase pathways, relieves this repression and enables ELK1 to recruit coactivators with histone acetyltransferase activity (39). There is an example in the literature in which a protein, the E3 ligase PIASxα, binds to the SUMOylated form of ELK1 and relieves gene repression by blocking the recruitment of histone deacetylase 2 (40). As demonstrated in the present study, AR activates ELK1 by binding to it without an increase in ELK1 phosphorylation. However, the activation of ELK1 by AR is distinct from that induced by PIASxα in several ways. First, unlike AR, PIASxα targets immediate early genes. Second, AR not only relieved gene repression by ELK1 but also frequently further activated the genes in an ELK1-dependent manner. Finally, a large number of genes whose expression was not appreciably modulated by ELK1 alone were also activated by AR in an ELK1-dependent manner. This could be because, unlike PIASxα, AR is primarily a transcription factor that may be expected to recruit additional coactivators. Furthermore, the diverse gene/promoter contexts of the large number of genes regulated by AR through association with ELK1 must involve interplay with a variety of coregulators.
The early steps of prostate oncogenesis in approximately half of all prostate tumors have been linked to the formation of a fusion protein between the androgen-regulated TMPRSS2 and the ETS protein ERG; about 10% of prostate tumors express a fusion between TMPRSS2 and ETV1, ETV4, or ETV5 (41–43). TMPRSS2-ERG and AR modulate gene expression in an integrated manner that includes direct interaction between them (44). However, this interaction leads to suppression of many androgen target genes, possibly suppressing a differentiation program, to enable tumor growth (44). In contrast, physical association of AR with another ETS protein, ETV1, resulting in AR and ETV1 reciprocally serving each other as transcriptional coactivators of their respective target promoters has been demonstrated; the impact of this interaction on global gene expression and its physiological role in prostate cancer are still unclear, although ectopic overexpression of ETV1 did generate prostatic intraepithelial neoplasia lesions (45). ETS1 may also directly associate with AR, but its physiological significance is yet to be understood (46). As evident from the foregoing studies, interaction with ELK1 enables a distinct and key component of AR signaling in PC that is independent of TMPRSS2 gene fusions.
In the particular context of tumor growth, hormone-induced non-genomic activation of the MAPK pathway is generally believed to be of greater consequence to estrogen receptor action in breast cancer (47, 48) than to AR action in prostate cancer. Indeed, the principal effect of the cross-talk between the androgen signaling axis and MAPK signaling on prostate cancer growth appears to be via enhancement of the transcriptional activity of AR through phosphorylation of AR as noted in the Introduction.
Although ELK1 is genetically and functionally redundant, it may have acquired functional prominence in prostate tumor cells compared with normal prostate epithelial cells due to differences in the expression pattern of ELK1 and its closest structural and functional analog, ELK3 (24, 49), particularly as ELK3 did not have the ability to interact with AR. The relatively low expression of ELK3 in PC tissues compared with normal prostate (26) would suggest that changes in the TCF complement in PC cells may enable the onset of a major mechanism of growth signaling by AR that utilizes ELK1. ELK1 and ELK3 were the only TCF proteins detectable in laser-captured epithelial cells from untreated and matched CRPC tumors (31). More quantitative studies of relative ELK1 and ELK3 mRNA and protein expression levels from the epithelial cell populations derived from matched normal and malignant prostate are warranted.
Genes activated by AR play a broad spectrum of physiological roles. The pattern of expression of AR-tethering proteins during development, differentiation, and malignant transformation of the prostate could redirect AR signaling according to the physiological context. This may be exemplified by the established AR-tethering proteins, including HoxB13 (involved in development) (50) and CCAAT/enhancer-binding protein α (involved in terminal differentiation) (21, 51) as well as ELK1 (demonstrated in this study to be required for growth).
Recent reports have demonstrated enrichment of possible ELK1 binding sites in relation to chromatin sites of AR binding (52–54). The observation that ELK1, fully or in part, supported a substantial proportion (∼27%) of all gene activation (direct or indirect) by androgen in PC cells suggests that only a few AR-tethering proteins may be adequate to direct AR signaling toward supporting growth. Disrupting the interactions of AR with tethering proteins critical for growth signaling is a potential means of functionally targeting interventions selectively to prostate tumors.
Supplementary Material
Acknowledgments
We are grateful to Dr. Lirim Sheshedini, Dr. Edwin Sanchez, Dr. Kam Yeung, and Dr. Steve Patrick for generously sharing reagents, plasmids, and cells.
This work was supported by the Harold and Helen McMaster endowment (to M. R.).

This article contains supplemental Figs. 1–7 and Tables 1–3 and Supplement 8.
- AR
- androgen receptor
- PC
- prostate cancer
- TCF
- ternary complex factor
- CRPC
- castrate-recurrent PC
- ARE
- androgen response element
- SRF
- serum response element
- SRE
- serum response element
- ISRE
- interferon-stimulated response element
- SUMO
- small ubiquitin-like modifier
- Luc
- luciferase
- PSA
- prostate-specific antigen
- NLS
- nuclear localization sequence
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PIAS
- protein inhibitor of activated STATs.
REFERENCES
- 1. Ruizeveld de Winter J. A., Janssen P. J., Sleddens H. M., Verleun-Mooijman M. C., Trapman J., Brinkmann A. O., Santerse A. B., Schröder F. H., van der Kwast T. H. (1994) Androgen receptor status in localized and locally progressive hormone refractory human prostate cancer. Am. J. Pathol. 144, 735–746 [PMC free article] [PubMed] [Google Scholar]
- 2. Linja M. J., Savinainen K. J., Saramäki O. R., Tammela T. L., Vessella R. L., Visakorpi T. (2001) Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer. Cancer Res. 61, 3550–3555 [PubMed] [Google Scholar]
- 3. Zegarra-Moro O. L., Schmidt L. J., Huang H., Tindall D. J. (2002) Disruption of androgen receptor function inhibits proliferation of androgen-refractory prostate cancer cells. Cancer Res. 62, 1008–1013 [PubMed] [Google Scholar]
- 4. Li T. H., Zhao H., Peng Y., Beliakoff J., Brooks J. D., Sun Z. (2007) A promoting role of androgen receptor in androgen-sensitive and -insensitive prostate cancer cells. Nucleic Acids Res. 35, 2767–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen C. D., Welsbie D. S., Tran C., Baek S. H., Chen R., Vessella R., Rosenfeld M. G., Sawyers C. L. (2004) Molecular determinants of resistance to antiandrogen therapy. Nat. Med. 10, 33–39 [DOI] [PubMed] [Google Scholar]
- 6. Miyamoto H., Messing E. M., Chang C. (2004) Androgen deprivation therapy for prostate cancer: current status and future prospects. Prostate 61, 332–353 [DOI] [PubMed] [Google Scholar]
- 7. Guo Z., Yang X., Sun F., Jiang R., Linn D. E., Chen H., Chen H., Kong X., Melamed J., Tepper C. G., Kung H. J., Brodie A. M., Edwards J., Qiu Y. (2009) A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 69, 2305–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hu R., Dunn T. A., Wei S., Isharwal S., Veltri R. W., Humphreys E., Han M., Partin A. W., Vessella R. L., Isaacs W. B., Bova G. S., Luo J. (2009) Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 69, 16–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gonit M., Zhang J., Salazar M. d., Cui H., Shatnawi A., Trumbly R., Ratnam M. (2011) Hormone depletion-insensitivity of prostate cancer cells is supported by the AR without binding to classical response elements. Mol. Endocrinol. 25, 621–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cai C., Chen S., Ng P., Bubley G. J., Nelson P. S., Mostaghel E. A., Marck B., Matsumoto A. M., Simon N. I., Wang H., Chen S., Balk S. P. (2011) Intratumoral de novo steroid synthesis activates androgen receptor in castration-resistant prostate cancer and is upregulated by treatment with CYP17A1 inhibitors. Cancer Res. 71, 6503–6513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Larsson R., Mongan N. P., Johansson M., Shcherbina L., Abrahamsson P. A., Gudas L. J., Sterner O., Persson J. L. (2011) Clinical trial update and novel therapeutic approaches for metastatic prostate cancer. Curr. Med. Chem. 18, 4440–4453 [DOI] [PubMed] [Google Scholar]
- 12. Holzbeierlein J. M., McLaughlin M. D., Thrasher J. B. (2004) Complications of androgen deprivation therapy for prostate cancer. Curr. Opin. Urol. 14, 177–183 [DOI] [PubMed] [Google Scholar]
- 13. Pratt W. B., Toft D. O. (1997) Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr. Rev. 18, 306–360 [DOI] [PubMed] [Google Scholar]
- 14. Beato M., Herrlich P., Schütz G. (1995) Steroid hormone receptors: many actors in search of a plot. Cell 83, 851–857 [DOI] [PubMed] [Google Scholar]
- 15. Glass C. K., Rosenfeld M. G. (2000) The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14, 121–141 [PubMed] [Google Scholar]
- 16. McKenna N. J., O'Malley B. W. (2002) Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108, 465–474 [DOI] [PubMed] [Google Scholar]
- 17. Shaffer P. L., Jivan A., Dollins D. E., Claessens F., Gewirth D. T. (2004) Structural basis of androgen receptor binding to selective androgen response elements. Proc. Natl. Acad. Sci. U.S.A. 101, 4758–4763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gioeli D., Wunderlich W., Sebolt-Leopold J., Bekiranov S., Wulfkuhle J. D., Petricoin E. F., 3rd, Conaway M., Weber M. J. (2011) Compensatory pathways induced by MEK inhibition are effective drug targets for combination therapy against castration-resistant prostate cancer. Mol. Cancer Ther. 10, 1581–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen S., Xu Y., Yuan X., Bubley G. J., Balk S. P. (2006) Androgen receptor phosphorylation and stabilization in prostate cancer by cyclin-dependent kinase 1. Proc. Natl. Acad. Sci. U.S.A. 103, 15969–15974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Q., Li W., Zhang Y., Yuan X., Xu K., Yu J., Chen Z., Beroukhim R., Wang H., Lupien M., Wu T., Regan M. M., Meyer C. A., Carroll J. S., Manrai A. K., Jänne O. A., Balk S. P., Mehra R., Han B., Chinnaiyan A. M., Rubin M. A., True L., Fiorentino M., Fiore C., Loda M., Kantoff P. W., Liu X. S., Brown M. (2009) Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell 138, 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang J., Gonit M., Salazar M. D., Shatnawi A., Shemshedini L., Trumbly R., Ratnam M. (2010) C/EBPα redirects androgen receptor signaling through a unique bimodal interaction. Oncogene 29, 723–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mukhopadhyay N. K., Ferdinand A. S., Mukhopadhyay L., Cinar B., Lutchman M., Richie J. P., Freeman M. R., Liu B. C. (2006) Unraveling androgen receptor interactomes by an array-based method: discovery of proto-oncoprotein c-Rel as a negative regulator of androgen receptor. Exp. Cell Res. 312, 3782–3795 [DOI] [PubMed] [Google Scholar]
- 23. Hollenhorst P. C., Shah A. A., Hopkins C., Graves B. J. (2007) Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 21, 1882–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wei G. H., Badis G., Berger M. F., Kivioja T., Palin K., Enge M., Bonke M., Jolma A., Varjosalo M., Gehrke A. R., Yan J., Talukder S., Turunen M., Taipale M., Stunnenberg H. G., Ukkonen E., Hughes T. R., Bulyk M. L., Taipale J. (2010) Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 29, 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sharrocks A. D. (2002) Complexities in ETS-domain transcription factor function and regulation: lessons from the TCF (ternary complex factor) subfamily. The Colworth Medal Lecture. Biochem. Soc. Trans. 30, 1–9 [DOI] [PubMed] [Google Scholar]
- 26. Singh D., Febbo P. G., Ross K., Jackson D. G., Manola J., Ladd C., Tamayo P., Renshaw A. A., D'Amico A. V., Richie J. P., Lander E. S., Loda M., Kantoff P. W., Golub T. R., Sellers W. R. (2002) Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 1, 203–209 [DOI] [PubMed] [Google Scholar]
- 27. Yordy J. S., Muise-Helmericks R. C. (2000) Signal transduction and the Ets family of transcription factors. Oncogene 19, 6503–6513 [DOI] [PubMed] [Google Scholar]
- 28. Yang S. H., Jaffray E., Hay R. T., Sharrocks A. D. (2003) Dynamic interplay of the SUMO and ERK pathways in regulating Elk-1 transcriptional activity. Mol. Cell 12, 63–74 [DOI] [PubMed] [Google Scholar]
- 29. Boros J., Donaldson I. J., O'Donnell A., Odrowaz Z. A., Zeef L., Lupien M., Meyer C. A., Liu X. S., Brown M., Sharrocks A. D. (2009) Elucidation of the ELK1 target gene network reveals a role in the coordinate regulation of core components of the gene regulation machinery. Genome Res. 19, 1963–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cesari F., Rennekampff V., Vintersten K., Vuong L. G., Seibler J., Bode J., Wiebel F. F., Nordheim A. (2004) Elk-1 knock-out mice engineered by Flp recombinase-mediated cassette exchange. Genesis 38, 87–92 [DOI] [PubMed] [Google Scholar]
- 31. Best C. J., Gillespie J. W., Yi Y., Chandramouli G. V., Perlmutter M. A., Gathright Y., Erickson H. S., Georgevich L., Tangrea M. A., Duray P. H., González S., Velasco A., Linehan W. M., Matusik R. J., Price D. K., Figg W. D., Emmert-Buck M. R., Chuaqui R. F. (2005) Molecular alterations in primary prostate cancer after androgen ablation therapy. Clin. Cancer Res. 11, 6823–6834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 33. Huang da W., Sherman B. T., Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu H. C., Hsieh J. T., Gleave M. E., Brown N. M., Pathak S., Chung L. W. (1994) Derivation of androgen-independent human LNCaP prostatic cancer cell sublines: role of bone stromal cells. Int. J. Cancer 57, 406–412 [DOI] [PubMed] [Google Scholar]
- 35. Massie C. E., Lynch A., Ramos-Montoya A., Boren J., Stark R., Fazli L., Warren A., Scott H., Madhu B., Sharma N., Bon H., Zecchini V., Smith D. M., Denicola G. M., Mathews N., Osborne M., Hadfield J., Macarthur S., Adryan B., Lyons S. K., Brindle K. M., Griffiths J., Gleave M. E., Rennie P. S., Neal D. E., Mills I. G. (2011) The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J. 30, 2719–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hayward S. W., Cunha G. R. (2000) The prostate: development and physiology. Radiol. Clin. North Am. 38, 1–14 [DOI] [PubMed] [Google Scholar]
- 37. Safe S., Kim K. (2004) Nuclear receptor-mediated transactivation through interaction with Sp proteins. Prog. Nucleic Acid Res. Mol. Biol. 77, 1–36 [DOI] [PubMed] [Google Scholar]
- 38. Safe S., Kim K. (2008) Non-classical genomic estrogen receptor (ER)/specificity protein and ER/activating protein-1 signaling pathways. J. Mol. Endocrinol. 41, 263–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Besnard A., Galan-Rodriguez B., Vanhoutte P., Caboche J. (2011) Elk-1 a transcription factor with multiple facets in the brain. Front. Neurosci. 5, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang S. H., Sharrocks A. D. (2005) PIASx acts as an Elk-1 coactivator by facilitating derepression. EMBO J. 24, 2161–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tomlins S. A., Rhodes D. R., Perner S., Dhanasekaran S. M., Mehra R., Sun X. W., Varambally S., Cao X., Tchinda J., Kuefer R., Lee C., Montie J. E., Shah R. B., Pienta K. J., Rubin M. A., Chinnaiyan A. M. (2005) Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310, 644–648 [DOI] [PubMed] [Google Scholar]
- 42. Tomlins S. A., Mehra R., Rhodes D. R., Smith L. R., Roulston D., Helgeson B. E., Cao X., Wei J. T., Rubin M. A., Shah R. B., Chinnaiyan A. M. (2006) TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 66, 3396–3400 [DOI] [PubMed] [Google Scholar]
- 43. Helgeson B. E., Tomlins S. A., Shah N., Laxman B., Cao Q., Prensner J. R., Cao X., Singla N., Montie J. E., Varambally S., Mehra R., Chinnaiyan A. M. (2008) Characterization of TMPRSS2:ETV5 and SLC45A3:ETV5 gene fusions in prostate cancer. Cancer Res. 68, 73–80 [DOI] [PubMed] [Google Scholar]
- 44. Yu J., Yu J., Mani R. S., Cao Q., Brenner C. J., Cao X., Wang X., Wu L., Li J., Hu M., Gong Y., Cheng H., Laxman B., Vellaichamy A., Shankar S., Li Y., Dhanasekaran S. M., Morey R., Barrette T., Lonigro R. J., Tomlins S. A., Varambally S., Qin Z. S., Chinnaiyan A. M. (2010) An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell 17, 443–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shin S., Kim T. D., Jin F., van Deursen J. M., Dehm S. M., Tindall D. J., Grande J. P., Munz J. M., Vasmatzis G., Janknecht R. (2009) Induction of prostatic intraepithelial neoplasia and modulation of androgen receptor by ETS variant 1/ETS-related protein 81. Cancer Res. 69, 8102–8110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Massie C. E., Adryan B., Barbosa-Morais N. L., Lynch A. G., Tran M. G., Neal D. E., Mills I. G. (2007) New androgen receptor genomic targets show an interaction with the ETS1 transcription factor. EMBO Rep. 8, 871–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Duan R., Xie W., Burghardt R. C., Safe S. (2001) Estrogen receptor-mediated activation of the serum response element in MCF-7 cells through MAPK-dependent phosphorylation of Elk-1. J. Biol. Chem. 276, 11590–11598 [DOI] [PubMed] [Google Scholar]
- 48. Duan R., Xie W., Li X., McDougal A., Safe S. (2002) Estrogen regulation of c-fos gene expression through phosphatidylinositol-3-kinase-dependent activation of serum response factor in MCF-7 breast cancer cells. Biochem. Biophys. Res. Commun. 294, 384–394 [DOI] [PubMed] [Google Scholar]
- 49. Shore P., Whitmarsh A. J., Bhaskaran R., Davis R. J., Waltho J. P., Sharrocks A. D. (1996) Determinants of DNA-binding specificity of ETS-domain transcription factors. Mol. Cell. Biol. 16, 3338–3349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Norris J. D., Chang C. Y., Wittmann B. M., Kunder R. S., Cui H., Fan D., Joseph J. D., McDonnell D. P. (2009) The homeodomain protein HOXB13 regulates the cellular response to androgens. Mol. Cell 36, 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang J., Wilkinson J. E., Gonit M., Keck R., Selman S., Ratnam M. (2008) Expression and sub-cellular localization of the CCAAT/enhancer binding protein α in relation to postnatal development and malignancy of the prostate. Prostate 68, 1206–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Urbanucci A., Sahu B., Seppälä J., Larjo A., Latonen L. M., Waltering K. K., Tammela T. L., Vessella R. L., Lähdesmäki H., Jänne O. A., Visakorpi T. (2012) Overexpression of androgen receptor enhances the binding of the receptor to the chromatin in prostate cancer. Oncogene 31, 2153–2163 [DOI] [PubMed] [Google Scholar]
- 53. Chng K. R., Chang C. W., Tan S. K., Yang C., Hong S. Z., Sng N. Y., Cheung E. (2012) A transcriptional repressor co-regulatory network governing androgen response in prostate cancers. EMBO J. 31, 2810–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tan P. Y., Chang C. W., Chng K. R., Wansa K. D., Sung W. K., Cheung E. (2012) Integration of regulatory networks by NKX3–1 promotes androgen-dependent prostate cancer survival. Mol. Cell. Biol. 32, 399–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.