Background: The interaction of PafA, the prokaryotic ubiquitin-like ligase, with its protein substrates is poorly understood.
Results: Measurements of PafA kinetics reveal cooperative substrate binding and experiments with engineered substrates suggest that PafA forms dimers.
Conclusion: The PafA enzymatic mechanism involves allosteric transitions.
Significance: PafA interaction with its target substrates is regulated at the enzyme level.
Keywords: Allosteric Regulation, Proteasome, Protein Degradation, Tuberculosis, Ubiquitin, Dop, Mycobacterium smegmatis, PafA, Pup, Pupylation
Abstract
Protein degradation via prokaryotic ubiquitin-like protein (Pup) tagging is conserved in bacteria belonging to the phyla Actinobacteria and Nitrospira. The physiological role of this novel proteolytic pathway is not yet clear, although in Mycobacterium tuberculosis, the world's most threatening bacterial pathogen, Pup tagging is important for virulence. PafA, the Pup ligase, couples ATP hydrolysis with Pup conjugation to lysine side chains of protein substrates. PafA is the sole Pup ligase in M. tuberculosis and apparently, in other bacteria. Thus, whereas PafA is a key player in the Pup tagging (i.e. pupylation) system, control of its activity and interactions with target protein substrates remain poorly understood. In this study, we examined the mechanism of protein pupylation by PafA in Mycobacterium smegmatis, a model mycobacterial organism. We report that PafA is an allosteric enzyme that binds its target substrates cooperatively and find that PafA allostery is controlled by the binding of target protein substrates, yet is unaffected by Pup binding. Analysis of PafA pupylation using engineered substrates differing in the number of pupylation sites points to PafA acting as a dimer. These findings suggest that protein pupylation can be regulated at the level of PafA allostery.
Introduction
Protein degradation via ubiquitin tagging was long thought to be a uniquely eukaryotic proteolytic pathway. In the last few years, however, it has become clear that bacterial species belonging to the phyla Nitrospira and Actinobacteria possess a tagging and degradation pathway that is highly analogous to the eukaryotic ubiquitin-proteasome pathway (1). Initially discovered in Mycobacterium tuberculosis, the bacterial ubiquitin-like tagging pathway conjugates Pup2 to lysine side chains of protein substrates, thereby targeting them for proteasomal degradation (2). M. tuberculosis, like other actinobacterial species, contains a proteasome in addition to other smaller proteases, such as ClpXP and FtsH (3–5). Because Pup is recognized by Mpa, the proteasomal regulatory subunit, tagged (i.e. pupylated) proteins are degraded (6–10). Puppylation-deficient M. tuberculosis mutants are viable, but their virulence is compromised. Specifically, such mutants are sensitized to reactive nitrogen intermediates, antimicrobial compounds that are released into phagosomes by macrophages (11, 12). Noteworthy, the vast majority of Pup/proteasome-containing bacteria are nonpathogenic, suggesting a fundamental role of this proteolytic pathway in these species (13, 14).
Despite apparent functional similarity between Pup and ubiquitin, these degradation tags are not homologous. Unlike ubiquitin, Pup is a small, natively unstructured protein of 64 amino acids (7 kDa) that presents a GGE motif at its C terminus (2, 15, 16). Conjugation of Pup to protein substrates occurs through formation of an isopeptide bond between the γ-carboxylate of the C-terminal glutamate of Pup and an ϵ-amine group of a substrate lysine (2, 17). Like ubiquitin, Pup is activated by ATP hydrolysis prior to its conjugation (18, 19). However, the enzymatic mechanism of Pup activation and conjugation is altogether different from that of ubiquitin (1, 20). The Pup ligase, an enzyme termed PafA (proteasomal accessory factor A), both activates and conjugates Pup to protein substrates in a two-step reaction that is typical of glutamine synthetases and γ-glutamyl cysteine synthetases (2, 14, 19). Indeed, PafA was identified as a distant homologue of this group of enzymes (14). The first step in the PafA-catalyzed reaction is the phosphorylation of Pup on the γ-carboxylate of its C-terminal glutamate. Next, PafA catalyzes a nucleophilic attack on the phosphorylated carboxylate by the ϵ-amine group of a substrate lysine (19). For pupylation and PafA binding, only the C-terminal region of Pup is required, and whereas the N-terminal region of the protein is required for interaction with Mpa, it is expandable for pupylation (8, 9). In M. tuberculosis and many other related species, Pup is translated with a glutamine, rather than a glutamate at its C terminus. In these bacteria, a PafA-homologous enzyme termed Dop (deamidase of Pup) converts this glutamine to a glutamate, thereby rendering Pup available for PafA conjugation (21).
Despite remarkable progress in understanding the pupylation system, it remains poorly understood how PafA interacts with its protein substrates and how such interactions are regulated. PafA is the only Pup ligase identified to date; and in M. tuberculosis, no pupylation is detected in pafA mutants (2). Understanding the enzymatic mechanism by which PafA interacts with its substrates is central to the understanding of this novel proteolytic pathway. Furthermore, PafA is important for M. tuberculosis virulence, and, as such, detailed mechanistic analysis of this enzyme may prove useful in the global combat against this pathogen.
In this study, we analyzed protein pupylation by PafA from Mycobacterium smegmatis. M. smegmatis is a commonly used mycobacterial model bacterium, with a PafA that is 94% identical to that of M. tuberculosis. We find that PafA is an allosteric enzyme that binds its target substrates cooperatively. In contrast, binding of deamidated Pup (PupE) by PafA follows simple kinetics and is, therefore, not involved in the allosteric transitions of PafA. We also find that Pup and target substrate binding by PafA are independent events. Analysis of engineered chimeric substrates that differ in the number of pupylation sites suggested that PafA is active as a dimer. In view of these findings, we propose that protein pupylation can be regulated at the enzyme level via an allosteric mechanism.
EXPERIMENTAL PROCEDURES
Protein Purification
All proteins, except a nontagged PanB, carried N-terminal polyhistidine tags and were expressed from plasmid pSH21 under the transcriptional control of the T7 promoter. Expression was at 37 °C, unless stated otherwise, in Escherichia coli ER2566 (New England Biolabs). Cells were lysed by sonication, and purification using Ni2+-NTA-agarose (Qiagen) was carried out according to a standard protocol. For PafA and PanB, purification Ni2+-NTA buffers contained 10% glycerol (v/v). As a consequent purification step, proteins were loaded onto a Superdex 75 size exclusion column (GE Healthcare) equilibrated with 25 mm Tris-HCl, pH 8.0 (hereafter, T25) for the titin-I27 variants, with pupylation buffer (50 mm Tris-HCl, pH 7.5, 100 mm KCl, 20 mm MgCl2, and 10% (v/v) glycerol) for PafA and with T25, 50 mm NaCl, and 10% glycerol for polyhistidine-tagged PanB.
For PupE purification, pupE was cloned into plasmid pSH21 in fusion with DNA encoding titin-I27 and a TEV protease recognition sequence (His6-I27-TEV-PupE). Expression was at 30 °C, and Ni2+-NTA purification was carried out as above. Following TEV cleavage, a buffer exchange step was carried out, and the His6-I27-TEV portion of the chimera was removed by loading the solution onto a Ni2+-NTA column. The flow-through was collected, and PupE was further purified on a C18 reverse phase column, lyophilized, and resuspended in T25.
For purification of Fl-PupE, a variant encoding a N-terminal cysteine was cloned (i.e. his66 i27-tev-cys-pupE). Labeling with 5-iodoacetamidofluorescein was carried out following binding of the protein to the Ni2+-NTA column. After excess 5-iodoacetamidofluorescein was washed, purification proceeded as for PupE.
To purify a PanB variant that does not carry a polyhistidine tag, panB was cloned into plasmid pET11a, and the protein was expressed at 37 °C in E. coli ER2566. The cell pellet was suspended in T25 containing 10% glycerol (v/v), and the cells were disrupted by sonication. Following centrifugation (12,000 × g, 15 min), streptomycin sulfate (1% (w/v)) was added to the clear lysate for precipitation of nucleic acids; and, following centrifugation as above, the supernatant was loaded onto an anionic exchange column (HiPrep Q FF; GE Healthcare) pre-equilibrated with T25 containing 10% glycerol (v/v). A NaCl gradient (0–1 m) was used for elution. PanB-containing fractions were pooled, concentrated, and loaded onto a size exclusion column (Superdex 200; GE Healthcare) pre-equilibrated with T25, 50 mm NaCl, 10% glycerol (v/v). As a final purification step, a Mono Q column was used (GE Healthcare), and following a buffer exchange step into T25, 50 mm NaCl, 10% glycerol (v/v) using a PD10 column (GE Healthcare), protein aliquots were stored at −80 °C until use.
Assays
Pupylation assays were carried out at 30 °C by mixing PafA, PupE, and a target substrate in pupylation buffer. Reactions were initiated with the addition of ATP to a final concentration of 2 mm. Quantitative pupylation assays relied on the use of a polyhistidine-tagged target substrate and the use of Fl-PupE instead of PupE. Initial rates were measured by collecting 10-μl aliquots at intervals during the reactions into tubes containing 50 μl of T25 and 6 m guanidine HCl (hereafter, T25G). Next, samples were diluted such that each contained equal concentrations of a polyhistidine-tagged target protein. 50 μl of each diluted sample was transferred into filter microcentrifuge tubes (Corning ZQ-VW-8169) containing 50 μl of Ni2+-NTA-agarose beads equilibrated with 500 μl of T25G. Following a 2-min incubation, the liquid was removed by centrifugation (7000 × g, 3 min), and beads were washed twice with 500 μl of T25G. Elution was carried out by resuspension of the beads in T25G, 250 mm imidazole, and centrifugation as above. The fluorescence intensity of each sample was measured with a Synergy 2 microplate reader (Biotek instruments) at 485 nm (excitation) and 528 nm (emission).
For Fl-PupE-PafA binding assays, the proteins were mixed in pupylation buffer without ATP or target substrates. Fluorescence anisotropy was measured as above.
LC/MS analysis, bioinformatics, and pupylation site identification are described in supplemental Experimental Procedures.
RESULTS
Cooperative Substrate Binding by PafA
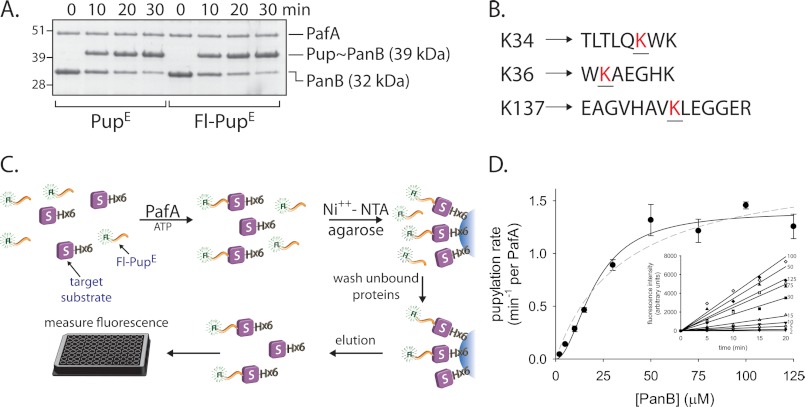
To study the interaction of M. smegmatis PafA with protein substrates, pupylation of a bona fide target substrate was examined. Earlier studies indicated that M. tuberculosis PanB (ketopantoate hydroxymethyltransferase), a homodecameric protein complex, is pupylated efficiently by the M. tuberculosis PafA both in vitro and in vivo (2, 9, 22). To test in vitro pupylation by the M. smegmatis system, the M. smegmatis panB, pafA, and pupE genes were cloned, and their encoded proteins were purified to homogeneity. In a reaction including ATP and PupE, the formation of a pupylation product with a molecular mass approximately 7 kDa heavier than PanB suggested that each PanB monomer was singly pupylated by PafA (Fig. 1A, left). MS/MS analysis indicated that PanB is pupylated primarily on either one of two lysines, namely Lys34 or Lys36. Pupylation on Lys137 was also detected, although less frequently (Fig. 1B and supplemental Fig. S1). Next, a quantitative assay able to examine M. smegmatis PafA pupylation kinetics was sought. Although the pupylation kinetics of M. tuberculosis PafA have been previously examined by monitoring the first half of the pupylation reaction cycle (i.e. ATP hydrolysis and PupE phosphorylation) (19), we wanted to analyze the full PafA reaction cycle to gain further insight into the interaction of PafA with target substrates. To this end, a quantitative pupylation assay was developed using an N-terminally fluorescein-labeled PupE variant (Fl-PupE). As PupE and Fl-PupE were conjugated equally well to protein substrates by PafA (Fig. 1A), Fl-PupE was used for quantitative pupylation analysis instead of unlabeled PupE. This novel assay, described under “Experimental Procedures,” allows quantitative measurements of end product formation by the use of Fl-PupE and a polyhistidine-tagged target substrate in a PafA-dependent reaction (Fig. 1C).
FIGURE 1.
PafA pupylation of PanB, a bona fide substrate. A, SDS-PAGE analysis of PanB (5 μm) pupylation by PafA (1 μm) and PupE or Fl-PupE (10 μm each) is shown. Following electrophoresis, Coomassie Brilliant Blue was used for staining. B, PanB pupylation sites (red) in tryptic peptides that were identified by MS/MS analysis are denoted. C, the use of Fl-PupE and a polyhistidine-tagged target substrate allows removal of unconjugated Fl-PupE via Ni2+-NTA chromatography. The fluorescence intensity of the eluate thus provides a quantitative measure of end product formation. D, steady-state rates of PafA (0.5 μm)- and Fl-PupE (15 μm)-mediated pupylation as a function of increasing polyhistidine-PanB concentration. The data were fitted to the Hill equation (rate = Vmax·[S]n/(K0.5n + [S]n); solid line) and to the Henri-Michaelis-Menten equation (rate = Vmax·[S]/(Km + [S]); dashed line). The averages ± S.D. (error bars) of three experiments are presented. Pupylation rates were derived from the measurements presented in the inset.
Titration experiments were carried out in which steady-state pupylation rates were measured at a constant Fl-PupE concentration and increasing PanB concentrations (Fig. 1D). The obtained data can be described by a sigmoidal rate equation (Fig. 1D), suggesting PanB binding by PafA to be cooperative. A Hill coefficient of 1.9 ± 0.3 (Table 1) indicates that PafA oligomers with at least two binding sites are involved in PanB pupylation. The kinetic measurements revealed a K0.5 of 21 ± 2 μm for PanB pupylation and a Vmax of 1.4 ± 0.1 Fl-PupE conjugations per min per PafA monomer (Table 1). As the assays were carried out in the presence of a saturating Fl-PupE concentration, the maximal pupylation rate is equivalent to the catalytic rate constant (kcat) of the reaction. Both the K0.5 and Vmax measured for PanB pupylation closely matched the published values for M. tuberculosis PanB pupylation by M. tuberculosis PafA (19).
TABLE 1.
Steady-state kinetic parameters for the PafA-catalyzed reactions measured in this work
| Substrate | Vmax | K0.5 | Hill coefficient |
|---|---|---|---|
| min−1/PafA | μm | ||
| PanB | 1.4 ± 0.1a | 21 ± 2 | 1.9 ± 0.3 |
| I271 | 1.4 ± 0.1a | 534 ± 87 | 1.6 ± 0.2 |
| I272 | 1.4 ± 0.1a | 348 ± 42 | 1.3 ± 0.1 |
| I273 | 1.3 ± 0.1a | 368 ± 45 | 1.2 ± 0.08 |
| Pup | 0.14 ± 0.01 | 1.3 ± 0.4 | 0.94 ± 0.2 |
a The values are equivalent to the kcat of the reactions, as measurements were carried out at saturating PupE concentrations.
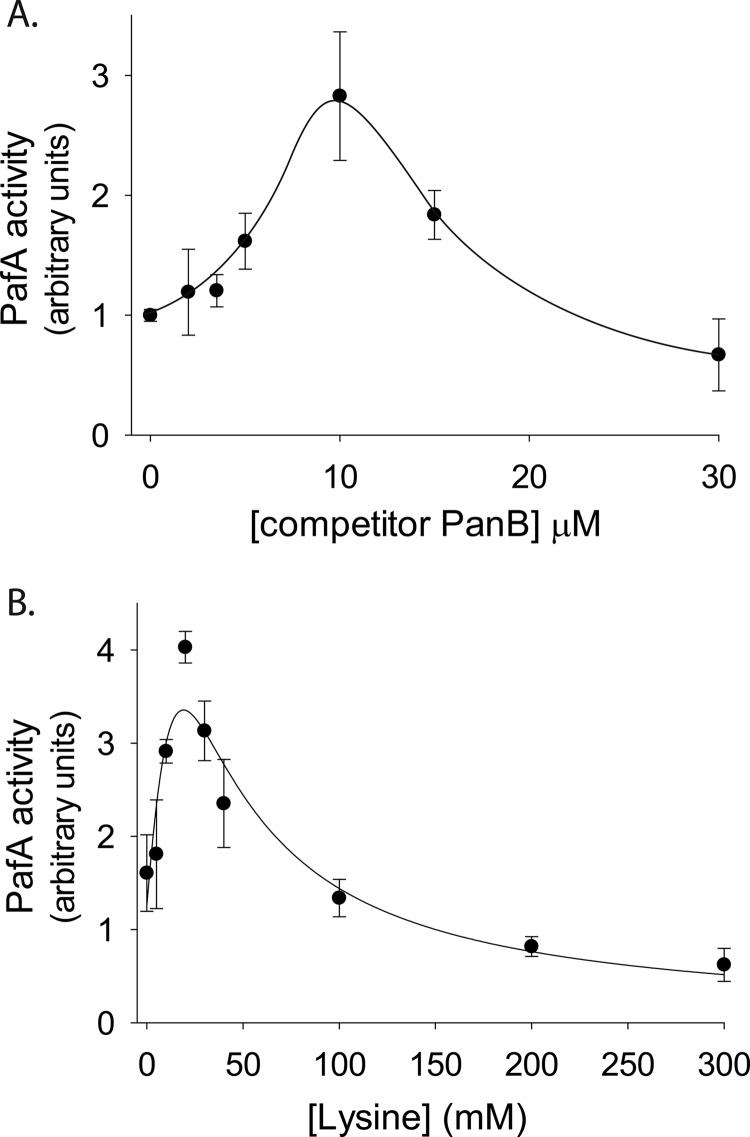
Cooperative substrate binding, as depicted in Fig. 1D, suggests that PafA is an allosteric enzyme. A hallmark of enzyme allostery is the positive effect of a competitive inhibitor on substrate processing at low inhibitor and substrate concentrations (23). An increase in enzymatic activity detected under such conditions can be attributed to stabilization of an active or high affinity enzyme conformation by the inhibitor. As a result, certain low inhibitor concentrations can lead to both inhibition and activation, such that the net effect is a facilitated substrate-processing rate (23). Similar kinetics can also be observed using a competing substrate rather than a competitive inhibitor (24). To test whether such an allosteric behavior is present in our system, pupylation reactions were carried out at a fixed low concentration of polyhistidine-PanB and at increasing concentrations of a PanB variant lacking the polyhistidine tag. As detection of pupylation in the quantitative assays employed in this study relies on the use of polyhistidine-tagged substrates (Fig. 1B), nontagged PanB thus acts as a competitor. Nevertheless, at low competitor concentrations, increased pupylation of polyhistidine-PanB was observed, providing further evidence of PafA allostery (Fig. 2A). In the presence of higher concentrations of nontagged PanB, competition dominates, resulting in a decrease in pupylation of the polyhistidine-tagged substrate (Fig. 2A). To test whether binding of a target lysine is sufficient to induce an allosteric PafA transition, a similar experiment was carried out, this time titrating free lysine instead of nontagged PanB. Again, a clear allosteric effect was observed at low lysine concentrations, whereas at higher concentrations, pupylation of polyhistidine-PanB was inhibited (Fig. 2B). Noticeably, the lysine concentrations required for induction of an allosteric PafA transition were approximately 1000-fold higher than the PanB concentrations that induced a similar effect (Fig. 2A). These differences are consistent with the ∼1000-fold lower affinity of PafA for free lysine (∼22 mm) compared with its affinity for PanB (∼21 μm) (Table 1 and Ref. 19).
FIGURE 2.
Allosteric activation by a competitor substrate. A, steady-state rates of a polyhistidine-PanB (1 μm) pupylation by PafA (0.5 μm) and Fl-PupE (15 μm) at increasing concentrations of nontagged competitor PanB. The values presented were obtained following subtraction of background fluorescence values measured in reactions lacking polyhistidine-PanB. The solid curve is a guide to the eye. B, as in A, but with free lysine (pH 8.0) instead of nontagged PanB competitor. The curve is fitted to the equation rate = C·α·(1+α+β)/(L+(1+α+β)2), where C is a scaling factor, α = [polyhistidine-PanB]/Km = 1/21, β = [Lys]/K0.5, and L is a conformational equilibrium constant (27). In both A and B, the averages ± S.D. (error bars) of three experiments are presented.
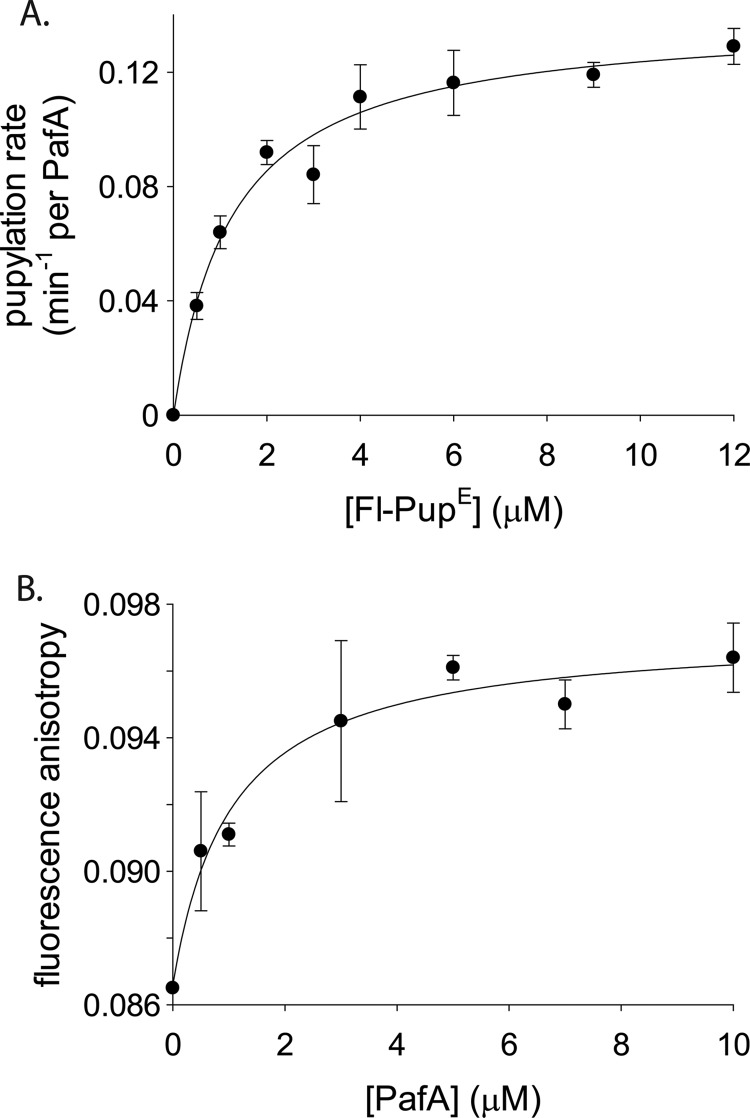
PupE Is Not Involved in Allosteric PafA Transitions
The mode of PupE binding to PafA was next considered when pupylation of low polyhistidine-PanB concentrations was measured at increasing Fl-PupE concentrations. Data fitting to the Hill equation yielded a nonsigmoidal curve with a Hill coefficient of 0.94 ± 0.2, indicating that PupE is not cooperatively bound by PafA nor does PupE affect binding of the target protein substrate to PafA (Fig. 3A and Table 1). The K0.5 value measured for Fl-PupE (1.3 ± 0.3 μm; Fig. 3A and Table 1) closely matched the KD for Fl-PupE binding to PafA, as measured in a direct binding assay in the absence of a nucleophile substrate (0.86 ± 0.3 μm; Fig. 3B). Therefore, binding of PupE to PafA is not affected by the binding of the target substrate. These results thus indicate that binding of PupE and a target substrate to PafA are independent events. This conclusion is further supported by the similarity between the obtained affinity values and the apparent binding constant measured for the interaction of M. tuberculosis PupE with M. tuberculosis PafA in the absence of a target substrate (19).
FIGURE 3.
Pup binding by PafA is noncooperative and independent of target substrate binding. A, steady-state rates of a polyhistidine-PanB (1 μm) pupylation by PafA (0.25 μm) at increasing Fl-PupE concentrations. The curve is fitted to the Hill equation. B, fluorescence anisotropy of Fl-PupE (50 nm) at increasing PafA concentrations. The curve is fitted to the binding equation, r = r0 + (rmax·[PafA])/(KD + [PafA]), where r is the fluorescence anisotropy value. In both A and B, the averages ± S.D. (error bars) of three experiments are presented.
A Dimer-based Model for PafA-Substrate Interaction
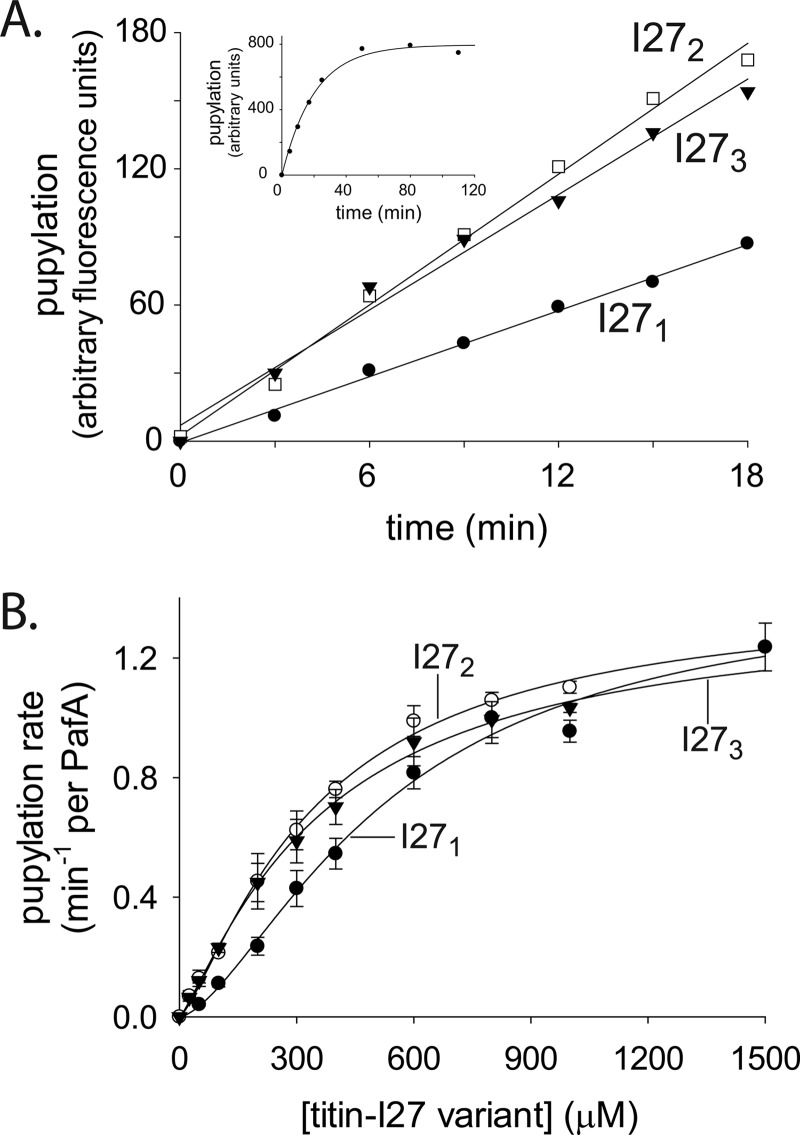
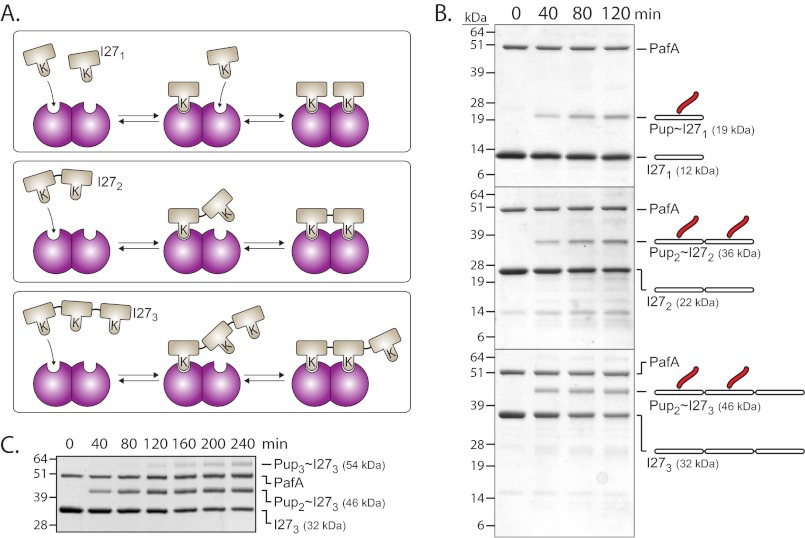
Cooperative substrate binding by PafA indicates that the active form of this enzyme is oligomeric. Indeed, Corynebacterium glutamicum PafA, an orthologue of the molecule considered here, was recently crystallized in the asymmetric unit as a dimer where swapping of an N-terminal β-strand and a concomitant α-helix was shown (25). Nonetheless, efforts to determine the stoichiometry of M. smegmatis PafA oligomers using classic methods, such as size exclusion chromatography and analytical ultracentrifugation, have been hampered by the low solubility of M. smegmatis PafA, as was also reported for M. tuberculosis PafA (19). As an alternative approach, we instead tested the pupylation of chimeric substrates presenting different numbers of pupylation sites. To this end, concatemeric variants of a nonnative substrate protein, the I27 domain of human Titin, were used. Titin-I27 is a globular domain that can be easily unfolded in vitro by chemical modification (e.g. carboxymethylation) of its two cysteines (26). We found unfolded titin-I27 to be pupylated substantially faster than was the folded form of the protein (supplemental Fig. S2A). It was also noted that titin-I27 is pupylated primarily on a single lysine, as determined by SDS-PAGE and mass spectrometry (Fig. S2, A and B). To examine the impact on pupylation kinetics upon encountering multiple substrate pupylation sites, linear fusions of two and three titin-I27 domains were constructed. Single titin-I27 and the resulting concatemeric proteins (denoted as I271, I272, and I273) were purified to homogeneity, unfolded by carboxymethylation, and the rates of their in vitro pupylation were compared (Fig. 4A). I272 was pupylated approximately twice as fast as was I271, as expected, given that I272 contains two pupylation sites, as opposed to the single such site in I271. However, no additional increase in rate was observed for pupylation of I273. Rather, this substrate was pupylated as fast as was I272 and again about twice as fast as was I271 (Fig. 4A). To better assess the pupylation kinetics of each titin-I27 variant, titration assays were carried out by measuring pupylation rates at different substrate concentrations (Fig. 4B). No significant differences in the value of Vmax (∼1.4 min−1 per PafA) were detected (Fig. 4B and Table 1). By contrast, the K0.5 value for pupylation of I271 (534 ± 87 μm; Table 1) was found to be nearly 2-fold higher than that of I272 (348 ± 42 μm) or of I273 (368 ± 45 μm). These results suggest that the rate differences observed in Fig. 4A arise mainly from affinity effects. A Hill coefficient of 1.6 ± 0.2 for I271 pupylation suggested cooperative binding of this titin-I27 variant by PafA, as in the case of PanB pupylation (Table 1). Notably, PafA binding of the I272 and I273 concatemers occurred with a lower degree of cooperativity, as determined by Hill coefficients of 1.3 ± 0.1 and 1.2 ± 0.08, respectively (Table 1). We thus conclude that the pupylation kinetics of the titin-I27 variants are consistent with PafA being active as a dimer. As illustrated in Fig. 5A, binding of the first target lysine of I272 can lead to an increase in the local concentration of the second target lysine near a free PafA binding site. As a result, binding of the second I272 target lysine is highly favorable, with the flexibility of the unfolded titin-I27 domains enabling binding without steric constrains that might otherwise limit the binding of folded proteins. A dimer-based model also explains the higher degree of cooperativity observed for I271 pupylation. Indeed, binding of I272 or I273 to a PafA dimer cannot lead to stabilization of a free substrate binding site, thus preventing cooperative binding of an additional substrate. Finally, the existence of only two binding sites explains why elongation of I272 by an additional unit to generate I273 does not improve binding.
FIGURE 4.
PafA pupylation of titin-I27 variants. A, pupylation of titin-I27 variants (15 μm each) by PafA (0.5 μm) and Fl-PupE (1 μm). The inset demonstrates an I271 pupylation reaction with PafA (0.5 μm) and Fl-PupE (7 μm) that continued to completion. B, steady-state pupylation rates by PafA (0.5 μm) and Fl-PupE (15 μm) at increasing concentrations of titin-I27 variants. The curves are fitted to the Hill equation. The averages ± S.D. (error bars) of three experiments are presented.
FIGURE 5.
A two-binding site model for PafA interaction with titin-I27 concatemers. A, two adjacent PafA subunits (purple) can bind two I271 monomers, one I272 monomer, or two target lysines in I273. B, SDS-PAGE analysis of pupylation by PafA (1.0 μm) and PupE (25 μm) of I271 (12 μm), I272 (6 μm), and I273 (4 μm). Following electrophoresis, Coomassie Brilliant Blue was used for staining. C, pupylation of I273 as in C is shown, but for a longer reaction time.
A dimer-based model, as illustrated in Fig. 5A, predicts I271 to be singly pupylated and I272 to be doubly pupylated. Importantly, according to this model, I273 should be only doubly pupylated despite presenting three pupylation sites. These predictions were indeed verified when pupylation of the titin-I27 variants was examined by SDS-PAGE (Fig. 5B). Noticeably, dual pupylation of I272 and of I273 proceeded without the formation of gel bands corresponding to single I272 and I273 pupylation intermediates. This suggests that following the initial pupylation of either I272 or I273, the second pupylation event occurs very fast and probably without substrate release. This further suggests that a third I273 pupylation event can occur, yet requires substrate release (following double pupylation) and rebinding. In agreement with this rationale, a third pupylation event was indeed observed for I273 when pupylation was allowed to proceed over an extended period of time (Fig. 5C).
DISCUSSION
By establishing a quantitative pupylation assay, we were able to measure the kinetics of M. smegmatis PafA pupylation in this study. Thus, using a bona fide PafA substrate, PanB, cooperative binding by PafA could be observed for the first time, suggesting that PafA is an allosteric enzyme. This conclusion was further supported by competition assays in which substrate pupylation was facilitated at low competitor concentrations. These experiments further indicated that binding of the target lysine to PafA is sufficient to induce the allosteric effect.
There are a number of ways in which binding of a target substrate to PafA can facilitate the pupylation rate. For example, binding of a target lysine by PafA can enhance its affinity to PupE, thereby facilitating pupylation at undersaturating PupE concentrations. Such a model, however, is highly unlikely, as our data clearly indicate that binding of PupE and the binding of target substrates by PafA are independent events. Therefore, PupE does not play a role in the allosteric transitions that are reflected in the experiments presented here. Rather, the data can be explained by the existence of alternative PafA conformations that differ either in their affinity for target substrates or in their catalytic activity. This scenario requires that the binding of target substrates to PafA would stabilize the high affinity or the more active conformation.
A Hill coefficient of 1.9 ± 0.3 for PanB pupylation (Table 1) indicates that the minimal oligomeric form of PafA is dimeric. Cooperative substrate binding was also evident when pupylation of titin-I27 was examined. In addition, these assays indicated that elongation of I271 by an additional titin-I27 unit resulted in an approximately 2-fold increase in affinity, as reflected in the K0.5 values measured for the titin-I27 variants. However, elongation by yet another titin-I27 unit to generate I273 did not improve affinity to PafA. These results are in perfect correlation with the number of Pup molecules that were conjugated to each titin-I27 variant. In other words, the 2-fold increase in the affinity of I272 and I273, compared with I271, was accompanied by an additional Pup molecule attached to these chimeric substrates. These observations can be explained by the existence of PafA oligomers containing two binding sites, as illustrated in Fig. 5A. A dimer-based model is also consistent with the reduced cooperativity observed for I272 and I273, compared with I271. Our results suggest that following binding of the first target lysine of I272 or I273, binding of the second lysine occurs almost simultaneously. This would explain the absence of detectable singly pupylated I272 and I273 in our steady-state kinetics assays. An almost simultaneous accommodation of both binding sites of a PafA dimer by I272 or I273 would result in a 2-fold increase in the pupylation rate. This interaction would, however, prevent facilitated binding of the next substrate molecule. Therefore, a less cooperative binding of I272 and I273 is observed. Unlike I272 and I273, PanB is cooperatively bound by PafA. Thus, despite being a decamer, PanB does not simultaneously accommodate the two binding sites of a PafA dimer. This is likely because for decameric PanB to bind two PafA binding sites, the PanB target lysines must be positioned in a conformation that may not exist in the folded, rigid conformation of the decamer. I272 and I273, on the other hand, are unfolded and flexible, and can thus easily assume the conformation required for the simultaneous occupation of the two PafA binding sites.
A dimer-based model for PafA allostery is consistent with the recently published C. glutamicum PafA structure (25). Altogether, our kinetic data strongly suggest that the dimeric form is the correct biological assembly and the allosterically active conformation of the protein. We cannot exclude the possibility of dimeric PafA existing as part of a higher oligomeric form, such as a dimer of dimers or a trimer of dimers. Hence, additional studies are required to decipher the oligomeric state of biologically active PafA.
As the only Pup ligase currently known, PafA plays a most central role in deciding which proteins will be pupylated. It is, therefore, reasonable that the expression level and activity of PafA be tightly regulated in vivo. This study suggests that substrate pupylation in vivo can be regulated at the level of PafA allostery. For example, pupylation can be regulated by altering the equilibrium between an active oligomeric form and an inactive conformation. Further study will be required to understand the mechanistic details of PafA allostery and the role it plays in bacterial physiology.
Supplementary Material
Acknowledgments
We thank Bob Sauer (Massachusetts Institute of Technology), Boaz Shaanan, Amir Aharoni, and Ofer Yifrach (Ben-Gurion University) for helpful advice and fruitful discussions.
This work was supported in part by the German-Israeli Foundation Grant I-2230-2055.13/2009.

This article contains supplemental Figs. S1 and S2 and supplemental Experimental Procedures.
- Pup
- prokaryotic ubiquitin-like protein
- Ni2+-NTA
- nickel-nitrilotriacetic acid
- PupE
- deamidated Pup
- TEV
- tobacco etch virus.
REFERENCES
- 1. Burns K. E., Darwin K. H. (2010) Pupylation versus ubiquitylation: tagging for proteasome-dependent degradation. Cell Microbiol. 12, 424–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pearce M. J., Mintseris J., Ferreyra J., Gygi S. P., Darwin K. H. (2008) Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science 322, 1104–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nagy I., Tamura T., Vanderleyden J., Baumeister W., De Mot R. (1998) The 20S proteasome of Streptomyces coelicolor. J. Bacteriol. 180, 5448–5453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Knipfer N., Seth A., Roudiak S. G., Shrader T. E. (1999) Species variation in ATP-dependent protein degradation: protease profiles differ between mycobacteria and protease functions differ between Mycobacterium smegmatis and Escherichia coli. Gene 231, 95–104 [DOI] [PubMed] [Google Scholar]
- 5. Darwin K. H., Lin G., Chen Z., Li H., Nathan C. F. (2005) Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Mol. Microbiol. 55, 561–571 [DOI] [PubMed] [Google Scholar]
- 6. Sutter M., Striebel F., Damberger F. F., Allain F. H., Weber-Ban E. (2009) A distinct structural region of the prokaryotic ubiquitin-like protein (Pup) is recognized by the N-terminal domain of the proteasomal ATPase Mpa. FEBS Lett. 583, 3151–3157 [DOI] [PubMed] [Google Scholar]
- 7. Burns K. E., Liu W. T., Boshoff H. I., Dorrestein P. C., Barry C. E., 3rd (2009) Proteasomal protein degradation in Mycobacteria is dependent upon a prokaryotic ubiquitin-like protein. J. Biol. Chem. 284, 3069–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burns K. E., Pearce M. J., Darwin K. H. (2010) Prokaryotic ubiquitin-like protein provides a two-part degron to Mycobacterium proteasome substrates. J. Bacteriol. 192, 2933–2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Striebel F., Hunkeler M., Summer H., Weber-Ban E. (2010) The mycobacterial Mpa-proteasome unfolds and degrades pupylated substrates by engaging Pup's N terminus. EMBO J. 29, 1262–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang T., Darwin K. H., Li H. (2010) Binding-induced folding of prokaryotic ubiquitin-like protein on the Mycobacterium proteasomal ATPase targets substrates for degradation. Nat. Struct. Mol. Biol. 17, 1352–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Darwin K. H., Ehrt S., Gutierrez-Ramos J. C., Weich N., Nathan C. F. (2003) The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 302, 1963–1966 [DOI] [PubMed] [Google Scholar]
- 12. Cerda-Maira F. A., Pearce M. J., Fuortes M., Bishai W. R., Hubbard S. R., Darwin K. H. (2010) Molecular analysis of the prokaryotic ubiquitin-like protein (Pup) conjugation pathway in Mycobacterium tuberculosis. Mol. Microbiol. 77, 1123–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lupas A., Zwickl P., Baumeister W. (1994) Proteasome sequences in eubacteria. Trends Biochem. Sci. 19, 533–534 [DOI] [PubMed] [Google Scholar]
- 14. Iyer L. M., Burroughs A. M., Aravind L. (2008) Unraveling the biochemistry and provenance of pupylation: a prokaryotic analog of ubiquitination. Biol. Direct 3, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liao S., Shang Q., Zhang X., Zhang J., Xu C., Tu X. (2009) Pup, a prokaryotic ubiquitin-like protein, is an intrinsically disordered protein. Biochem. J. 422, 207–215 [DOI] [PubMed] [Google Scholar]
- 16. Chen X., Solomon W. C., Kang Y., Cerda-Maira F., Darwin K. H., Walters K. J. (2009) Prokaryotic ubiquitin-like protein pup is intrinsically disordered. J. Mol. Biol. 392, 208–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sutter M., Damberger F. F., Imkamp F., Allain F. H., Weber-Ban E. (2010) Prokaryotic ubiquitin-like protein (Pup) is coupled to substrates via the side chain of its C-terminal glutamate. J. Am. Chem. Soc. 132, 5610–5612 [DOI] [PubMed] [Google Scholar]
- 18. Imkamp F., Rosenberger T., Striebel F., Keller P. M., Amstutz B., Sander P., Weber-Ban E. (2010) Deletion of dop in Mycobacterium smegmatis abolishes pupylation of protein substrates in vivo. Mol. Microbiol. 75, 744–754 [DOI] [PubMed] [Google Scholar]
- 19. Guth E., Thommen M., Weber-Ban E. (2011) Mycobacterial ubiquitin-like protein ligase PafA follows a two-step reaction pathway with a phosphorylated pup intermediate. J. Biol. Chem. 286, 4412–4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maupin-Furlow J. (2012) Proteasomes and protein conjugation across domains of life. Nat. Rev. Microbiol. 10, 100–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Striebel F., Imkamp F., Sutter M., Steiner M., Mamedov A., Weber-Ban E. (2009) Bacterial ubiquitin-like modifier Pup is deamidated and conjugated to substrates by distinct but homologous enzymes. Nat. Struct. Mol. Biol. 16, 647–651 [DOI] [PubMed] [Google Scholar]
- 22. Chaudhuri B. N., Sawaya M. R., Kim C. Y., Waldo G. S., Park M. S., Terwilliger T. C., Yeates T. O. (2003) The crystal structure of the first enzyme in the pantothenate biosynthetic pathway, ketopantoate hydroxymethyltransferase, from M. tuberculosis. Structure 11, 753–764 [DOI] [PubMed] [Google Scholar]
- 23. Monod J., Wyman J., Changeux J. P. (1965) On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 12, 88–118 [DOI] [PubMed] [Google Scholar]
- 24. Gur E., Sauer R. T. (2009) Degrons in protein substrates program the speed and operating efficiency of the AAA+ Lon proteolytic machine. Proc. Natl. Acad. Sci. U.S.A. 106, 18503–18508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Özcelik D., Barandun J., Schmitz N., Sutter M., Guth E., Damberger F. F., Allain F. H., Ban N., Weber-Ban E. (2012) Structures of Pup ligase PafA and depupylase Dop from the prokaryotic ubiquitin-like modification pathway. Nat. Commun. 3, 1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kenniston J. A., Baker T. A., Fernandez J. M., Sauer R. T. (2003) Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of an AAA+ degradation machine. Cell 114, 511–520 [DOI] [PubMed] [Google Scholar]
- 27. Segel IH. (1975) Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems, pp. 421–461, John Wiley & Sons, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.