Abstract
Nuclear factor-κB (NF-κB) is a ubiquitously expressed transcription factor that plays a central role in directing a vast range of cellular functions. Its activation is controlled by the Rac GTPase and relies on the coordinated cooperation of the E3–ligase complex SCFβTrCP, composed by Skp-1/Cullin-1, Rbx/Roc1, and the β-TrCP proteins. Recently, Cullin-1 has been reported to form a complex with the activated Rac GTPase. Here, we show that the specific activation of the Rac GTPase, besides directing its own positioning, induces the relocalization of the SCF component Cullin-1 to the ruffling membranes. This occurred only if the ruffles were stimulated by the Rac GTPase and was accompanied by the repositioning to the same intracellular compartment of the SCF protein Skp-1 and the ubiquitin-like molecule Nedd-8. The SCF substrate IkBα was also directed to the ruffling membranes in a Rac-dependent way. The novelty of these findings is in respect to the demonstration that the correct positioning at the ruffling membranes is crucial for the subsequent series of events that leads to IkBα proteasomal degradation and the resultant activation of NF-κB. Consequently, this points to the role of Rac as a docking molecule in NF-κB activation.
INTRODUCTION
The Rho GTPases are molecular switches that control complex cellular processes by using a simple biochemical strategy. This entails their ability to cycle between an inactive GDP-bound state and an active GTP-bound state under the strict control of activators (guanine nucleotide exchange factors) and inactivators (GTPase-activating proteins) (Etienne-Manneville and Hall, 2002). To date, the mammalian Rho family comprises at least 16 members, including Rho, Rac, and Cdc42. Besides the first recognized ability of regulating the actin cytoskeleton (Hall, 1998), Rho GTPases also play a pivotal role in guiding a variety of other cellular processes, such as cell proliferation, transformation, and gene transcription. In fact, they participate in the activation of nuclear factor-κB (NF-κB) (Perona et al., 1997; Montaner et al., 1998), a group of structurally related and evolutionarily conserved transcription factors involved in regulating the expression of genes required for inflammatory responses, cell growth, and suppression of apoptosis (reviewed in Ghosh et al., 1998). In mammals, NF-κB consists of five members, namely, Rel (c-Rel), RelA (p65), RelB, NF-kB1 (p50 and its precursor p105), and NF-kB2 (p52 and its precursor p100). They form both homo- and heterodimers, of which the best characterized form of NF-κB is a heterodimer composed of p50 and p65 subunits. This heterodimer, in resting cells, is sequestered in the cytoplasm by association with the inhibitory subunit IkBα (Beg et al., 1992). Signals leading to NF-κB activation trigger a transduction pathway responsible for IkBα phosphorylation at two specific serine residues. Phosphorylation allows IkBα polyubiquitination on specific lysine residues and subsequent proteolytic degradation by the 26S proteasome. After degradation of IkBα, NF-κB heterodimer is translocated into the nucleus for gene trans-activation (Baldwin, 1996).
The ubiquitination process, although involving several steps, always results in the recognition and modification of the target protein via the E3-ubiquitin-protein-ligases (Ciechanover and Schwartz, 1998). Several classes of E3s have been described and, among them, the SCF (Skp1/Cul-1/F-box protein complex) complexes that recognize phosphorylated substrates. These tetrameric complexes are composed by Skp-1/Cullin-1 and Rbx/Roc1, which are common to all SCF, and a variable F-box protein (Deshaies, 1999). Ubiquitination of IkBα, as does β-catenin, is achieved upon its association with an SCF complex that contains the β-TrCP (SCFβ-TrCP) as F-box protein (Karin and Ben-Neriah, 2000). In vitro, the capacity of SCFβ-TrCP to bind IkBα is enhanced upon Cullin-1 posttranslational neddylation, a modification consisting of the covalent addition of a Nedd-8 ubiquitin-like molecule to lysine 720 (Read et al., 2000). The Cullin-1 K720R mutant, impaired in its neddylation capacity, was localized in the nucleus, suggesting that Nedd-8 modification of Cullin-1 is required for its exit from the nucleus (Furukawa et al., 2000). Interestingly, the E3 ubiquitination ligase subunit Cullin-1 was noted to form a complex with activated Rho GTPases, and in particular with Rac (Senadheera et al., 2001), raising the hypothesis that Rac GTPase-Cullin-1 interaction could serve to regulate E3 subcellular localization.
To verify such a hypothesis, we undertook a study using an Escherichia coli protein toxin that specifically activates the Rho GTPases, named cytotoxic necrotizing factor 1 (CNF1). It acts by deamidating a pivotal glutamine residue of RhoA, Rac, and Cdc42 (glutamine 63 of Rho or glutamine 61 of Rac and Cdc42) into glutamic acid (Flatau et al., 1997; Schmidt et al., 1997; Lerm et al., 1999). Recent findings have established that CNF1-induced permanent activation of RhoA, Rac, and Cdc42 (Doye et al., 2002; Lerm et al., 2002) sensitizes them to ubiquitination and proteasome degradation. Interestingly, some carcinoma cell lines, including HEp-2 cells, respond to CNF1 with a lower efficiency of Rac ubiquitination and degradation that results in a sustained Rac activation (Doye et al., 2002). This identifies CNF1 as a preferential tool for studying cellular activities linked to Rac activation in HEp-2 epithelial cells.
In this article, we provide evidence that protein complexes involved in the activation of the transcription factor NF-κB are spatially relocalized at the level of membrane ruffles, after Rac activation.
MATERIALS AND METHODS
Cell Cultures and Antibodies
HEp-2 cells were grown in minimal Eagle's medium supplemented with 10% fetal calf serum (Flow Laboratories, Irvine, United Kingdom), 5 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells were seeded at a concentration of 8 × 104 cells/ml, and experiments were performed 24 h after seeding. Antibodies were purchased from the following sources: BD Biosciences Transduction Laboratories (Lexington, KY) (anti-Rac1 [clone 102], anti-caveolin-1 [clone C060]); Chemicon International (Temecula, CA) (antiglutathione S-transferase [GST]), Babco (Richmond, CA) (anti-HA [clone11]); Sigma-Aldrich (St. Louis, MO) (anti-α-actin [clone AC-74]); Upstate Biotechnology (Lake Placid, NY) (anti-IkBα); and Santa Cruz Biotechnology (San Diego, CA) (anti-Nedd-8 and anti-p65). If indicated, cells were treated with 10-9 M CNF1, either wild type or catalytic inactive mutant C866S purified as described previously (Flatau et al., 1997).
Inhibition Experiments
For proteasome inhibition, HEp-2 cells were preincubated for 1 h with 10 μM MG132 (Sigma-Aldrich) before being challenged with CNF1 for different time points. For experiments aimed at perturbing lipid raft organization, 2 μg/ml filipin (Sigma-Aldrich) were added after 1 h of incubation with CNF1. Toxin exposure was prolonged for an additional 3 h. To verify whether cholesterol addition would interfere with filipin activity, cells treated with CNF1 for 1 h were exposed to 60 μg/ml cholesterol together with 2 μg/ml filipin until the end of toxin treatment. Inhibition of Rho GTPases was achieved by pretreating HEp-2 cells for 2 h with 1 μg/ml of the chimeric toxin C3B (Aullo et al., 1993), LT82, or LT9048 from Clostridium sordellii (Popoff et al., 1996; El Hadj et al., 1999) before being challenged with CNF1 for 4 h.
Transfections of HEp-2 Cells
HEp-2 cells were transfected with 2 μg of pcDNA3-myc-IkBα-mutant (S32A and S36A) (Traenckner et al., 1995), pcDNA3-HA-Cullin-1 (Lisztwan et al., 1998), pcDNA3-HA-Cullin-1 K720R (Kawakami et al., 2001), pHm200-HA-Skp-1 (Winston et al., 1999), and p(6KappaB)Luc, controlled by a minimal thymidine kinase promoter and six reiterated κB sites (Bottero et al., 2001). The construction for GST-RacL61 and GST-RacN17 expressions were obtained by subcloning in the plef-vector (Rudert et al., 1996) RacL61 and RacN17 from pXJ-HA-Rac mutants (Doye et al., 2002). PEGFP-ARF6 was a gift from Philip Chavrier (Derrien et al., 2002). Exgen500 (Euromedex, France) was used to transfect cells according to the manufacturer's instructions. Eighteen hours after transfection, CNF1 was added and maintained for different incubation times.
Immunofluorescence and Confocal Microscopy
Control and CNF1-treated HEp-2 cells, either transfected or not, maintained in minimal Eagle's medium supplemented with 10% fetal calf serum were stained as follows: for p65 immunostaining, cells were fixed in acetone: methanol, 1:1 (vol/vol) for 10 min at room temperature and air dried. After 1 h of preincubation with phosphate-buffered saline (PBS) containing 10% of AB human serum, cells were incubated with the p65 antibody for 1 h at room temperature. After three washes in PBS, cells were incubated for 1 h at room temperature with fluorescein isothiocyanate-labeled anti-rabbit antibody. After three additional washings in PBS, cells were mounted on glass coverslips and analyzed with a Nikon Microphot fluorescence microscope. For immunolabeling with anti-caveolin-1 and anti-IkBα, cells were fixed in 4% paraformaldehyde and permeabilized with 1% Triton X-100. For immunolabeling with anti-β-actin, anti-hemagglutinin (HA), anti-GST, anti-Nedd-8, anti-Rac, and anti-IkBα, cells were fixed in 4% paraformaldehyde (Sigma-Aldrich) and permeabilized with 0.05% saponin (Sigma-Aldrich). Cells were processed as described previously (Doye et al., 2002) and analyzed with a TCS-SP confocal microscope (Leica, Heidelberg, Germany).
Luciferase Assay
HEp-2 cells seeded at 105 per well in a 24-well plate were transfected as described above, with both 2 μg of a luciferase reporter gene and 0.5 μg of pSV40β-galactosidase control vector (Promega, Madison, WI) per well. Eighteen hours after transfection, cells were intoxicated with 10-9 M CNF1 and lysed in 50 μl of lysis buffer (Promega) at different times. Soluble extracts were assessed for both luciferase activity and β-galactosidase activity, as recommended by the manufacturer. Results were expressed as arbitrary values corresponding to the value of the luciferase activity normalized to the β-galactosidase activity or fold activation.
Western Blots and GST-PAK Pull-Down Assay
To test the activity of Rac1 in HEp-2 cells after different times of incubation with CNF1, pull-down assay was performed by using 0.5 mg of cell lysates and 40 μg of GST-PAK70-106. Proteins were resolved on 12% SDS-PAGE followed by transfer on polyvinylidene difluoride membranes (Amersham Biosciences, Saclay, France). Rac1 binding to GST-PAK70-106, total Rac as well as IkBα protein levels were determined by Western blot analysis as described previously (Doye et al., 2002).
Statistical Analysis
The values reported in graphs are the means ± SD from three separate experiments performed in duplicate. The Student's t test was used for analysis of statistical significance. A p value <0.05 was considered significant.
RESULTS
Activated Rac Relocalizes Cullin-1 to Membrane Ruffles
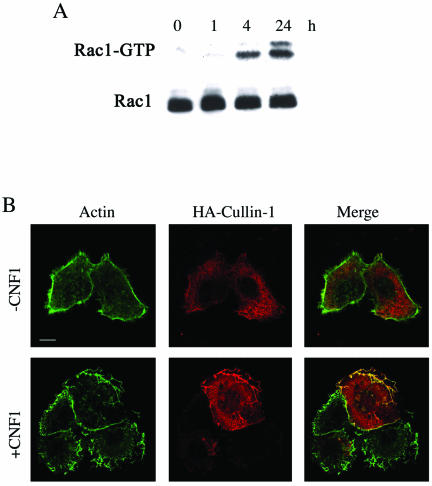
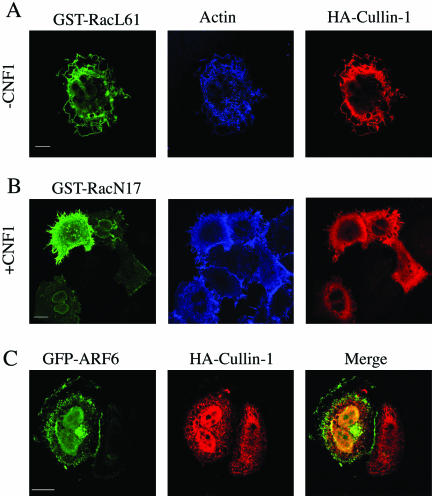
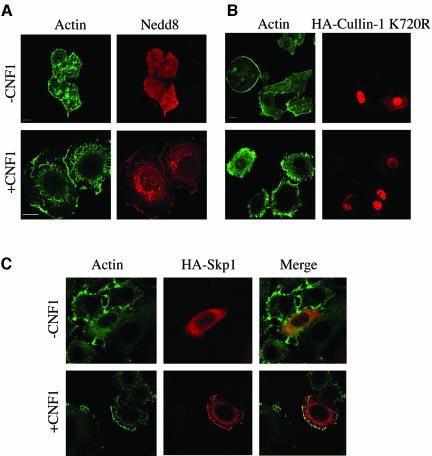
To investigate the ultimate connection linking Rac, NF-κB, and the SFC complex, we took advantage of the recent demonstration that, in HEp-2 cells, CNF1 produces a sustained activation of Rac that is due to the very low Rac ubiquitination activity in this cell type (Doye et al., 2002). To verify this finding in our experimental system, we performed a pull-down experiment by using the Rac effector PAK linked to GST protein. We found, in accordance to what was reported previously (Doye et al., 2002), that exposure to CNF1 activated Rac in a time-dependent manner, and this resulted in the activation being sustained for at least 24 h (Figure 1A). The next step was to view the localization of the components of the SCF complex after Rac activation by using confocal microscopy. We transfected HEp-2 cells with the recombinant protein HA-Cullin-1- and we found that although in control cells Cullin-1 was dispersed throughout the cytoplasm, in cells exposed to CNF1 the Cullin-1 relocalized to the actin-rich membrane ruffles (Figure 1B). Transfecting cells with the dominant positive mutant GST-RacL61 or dominant negative mutant GST-RacN17 demonstrated the specific involvement of Rac in such a phenomenon. In fact, RacL61 was capable of mimicking the displacement of Cullin-1 to ruffles promoted by CNF1 (Figure 2A), an ability that was impaired by the dominant negative mutant RacN17 (Figure 2B). As a control, we transfected cells with the ADP-ribosylation factor 6 (ARF6), a small GTPase involved in the aspects of endocytosis, exocytosis, membrane recycling, and also membrane ruffling (Hunzicker-Dunn et al., 2002). As shown in Figure 2C, ARF6-promoted ruffles were devoid of Cullin-1 signal, demonstrating that the presence of Cullin-1 at the ruffles level is specifically governed by Rac. Because it is known that to be active, Cullin-1 must be linked to the ubiquitin-like molecule Nedd-8 (Wu et al., 2000), we stained cells with an anti-Nedd-8 antibody, and we found that in control cells Nedd-8 was dispersed throughout the cytoplasm, whereas in CNF1-treated cells, it was displaced to the ruffles (Figure 3A), in the same way as that of Cullin-1. Consistent with this, the neddylation-impaired mutant Cullin-1 K720R failed in moving to the ruffling membranes after CNF1 stimulation (Figure 3B), but rather seemed localized in the nucleus, as reported previously by another group (Wu et al., 2000). To verify the presence of Skp-1, a further component of the SCF complex, at the level of membrane ruffles raised by Rac activation, HEp-2 cells were transfected with HA-Skp-1 and stained with an antibody against the tag-HA. Skp-1, a cytoplasmic/nuclear glycoprotein associated with polyubiquitination (West, 2003) was dispersed into the cell cytoplasm of control cells and relocalized to the ruffling membranes, similarly to what occurs for Cullin-1, in CNF1-treated cells (Figure 3C). Considered together, these results permit the assumption that the SCF complex is relocalized to ruffling membrane upon CNF1 exposure.
Figure 1.
Rac activation induces the relocalization of Cullin-1 to membrane ruffles. (A) Immunoblot showing the amount of activated Rac after different times of CNF1 exposure in HEp-2 cells, obtained by pulldown experiments. CNF1 induces a sustained activation of Rac that remains activated up to 24 h of toxin exposure. In parallel, total cell lysates were used to verify the equal amount of the Rac GTPase subjected to pull-down experiments. (B) Confocal microscopy analysis of control (-CNF1) and treated HEp-2 cells (+CNF1). From left to right, the first column shows the actin cytoskeleton, the second shows cells transfected by HA-Cullin-1, and the third is the merge of actin and Cullin-1. The toxin clearly promotes the recruitment of Cullin-1 to the actin-rich membrane ruffles. Bar, 10 μm.
Figure 2.
Cullin-1 is present in membrane ruffles raised by Rac but not by ARF6. (A and B) Confocal microscopy analysis showing actin localization and HA-Cullin-1 in HEp-2 cells expressing either the dominant positive mutant GST-RacL61 (A), or the dominant negative mutant GST-RacN17 (B), either untreated (-CNF1) or treated overnight with 10-9 M CNF1 (+CNF1). Rac localization was assessed by anti-GST immunolabeling. The dominant positive RacL61 mimics the CNF1-induced displacement of Cullin-1 to ruffles that is counteracted by the dominant negative RacN17, proving the specific involvement of Rac. (C) Confocal microscopy analysis showing HA-Cullin-1 in HEp-2 cells transfected with a plasmid encoding the ARF6 GTPase fused with GFP. ARF6-induced ruffles are devoid of Cullin-1 signal. Bar, 10 μm.
Figure 3.
CNF1 induces Nedd-8 relocalization to membrane ruffles. (A–C) Confocal microscopy analysis of HEp-2 cells, untreated (-CNF1) or treated overnight with 10-9 M CNF1 (+CNF1). HEp-2 cells were stained for actin together with Nedd-8 (A) or for the epitope tag-HA in the case of cells transfected with the neddylation-impaired mutant HA-Cullin-1 K720R (B) or Skp-1 (C). In cells challenged with CNF1, Nedd-8 and Skp-1 are present at the level of actin-positive ruffling membranes, whereas the mutant Cullin-1 remains in the nucleus, its ability to move to ruffles being conceivably impaired. Bar, 10 μm.
Rac Activation Produces IkBα Recruitment into Membrane Ruffles and Proteasome Degradation
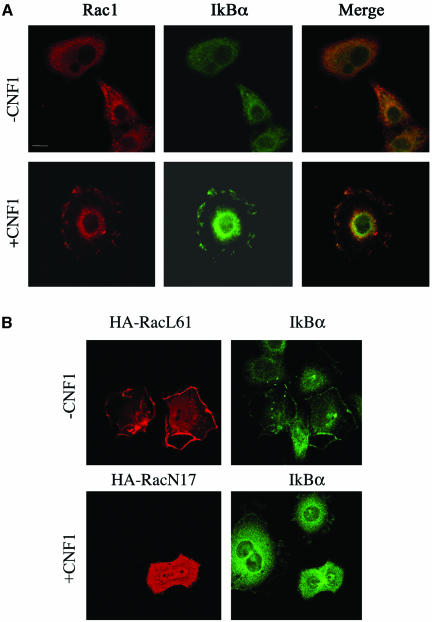
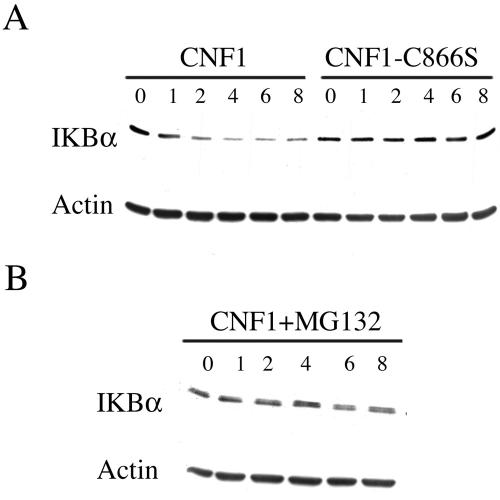
From our results on Rac-dependent SCF relocalization, we hypothesized the recruitment to the ruffles of one of the main substrates of SCF activity, that is, IkBα. As viewed by confocal microscopy, IkBα was diffusely spread throughout the cytoplasm of control cells, whereas upon exposure to CNF1, it could also be detected at the level of ruffling membranes, colocalized with Rac (Figure 4A). This phenomenon was already discernible after 2 h of toxin exposure (our unpublished data) but became particularly evident starting from 4 h of challenge with CNF1 (Figure 4A). Because these results suggested a spatial relationship between activated Rac and IkBα, we then verified whether there was an eventual dependence on Rac activation of IkBα recruitment to ruffling membranes. Hence, we transfected HEp-2 cells with the dominant positive mutant HA-RacL61 or the dominant negative mutant HA-RacN17. As shown in Figure 4B, RacL61 was capable of promoting the recruitment of IkBα to the ruffles, whereas RacN17, the dominant negative mutant, blocked the CNF1-induced relocalization of IkBα to the ruffling membranes. These data suggest the possibility that the IkBα displacement to ruffles is necessary for the subsequent SCF-dependent IkBα ubiquitination. In support of this hypothesis, we found that ubiquitin was partially redistributed to ruffling membranes after CNF1 exposure (our unpublished data). Hence, we contemplated whether the Rac-induced relocalization of IkBα, together with the SCF complex and ubiquitin, could result in its degradation. To address this question, we measured by Western blotting the amount of IkBα in CNF1-treated cells. As shown in Figure 5A, IkBα depletion had already occurred after 2 h of toxin exposure and was followed by IkBα reappearance within 24 h of challenge with CNF1 (our unpublished data). This was a proteasome-dependent phenomenon because the Rac-induced IkBα degradation was totally prevented by the addition of 10 μM proteasome inhibitor MG132 (Figure 5B). It is worth noting that the CNF1 C866S, which represents the catalytic mutant defective in the ability to activate the Rho GTPases (Schmidt et al., 1998), did not induce the proteolytic degradation of IkBα (Figure 5A), thus proving the necessity of the catalytic activity of the toxin for IkBα depletion.
Figure 4.
IkBα is recruited to membrane ruffles together with activated Rac. (A) Confocal microscopy analysis showing the localization of endogenous Rac1 and IkBα in HEp2 cells, untreated (-CNF1) or treated for 4 h with 10-9 M CNF1 (+CNF1). On exposure to the toxin, IkBα colocalized with Rac at the level of ruffles. (B) Confocal microscopy analysis of HEp-2 cells transfected with the permanently activated (HA-RacL61) or inactivated (HA-RacN17) HA-tagged Rac mutants. IkBα, analyzed in transfected cells, was relocalized together with activated HA-RacL61 to the cell periphery. Conversely, the permanently inactivated HA-RacN17 inhibited the IkBα relocalization induced by 4-h treatment with CNF1. Bar, 10 μm.
Figure 5.
IkBα is degraded by the proteasome in CNF1-treated HEp-2 cells. (A) Western blots showing the amount of IkBα in HEp-2 cells treated with wild-type CNF1 or with the catalytic inactive mutant CNF1-C866S. Starting from 2 h of CNF1 exposure, IkBα began to be depleted whereas exposure to the catalytic inactive mutant was unable to cause IkBα depletion. (B) IkBα depletion in HEp-2 cells was prevented by the addition of the inhibitor MG132 (CNF1+MG132), indicating the involvement of the proteasome in the CNF1-induced IkBα degradation. Equal amounts of total protein loaded was verified by Western blot with anti-β-actin.
Rac Activation Induces the Nuclear Translocation of the Transcription Factor NF-κB
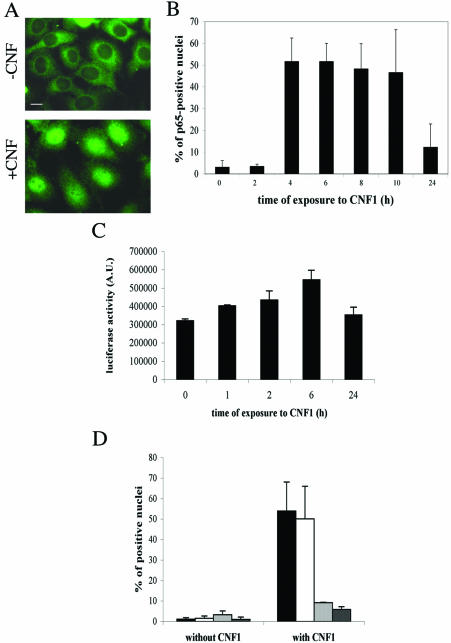
Because IkBα degradation is a crucial step for NF-κB activation, we investigated whether the CNF1-induced Rac activation could lead to its translocation and activation. First, we analyzed by fluorescence microscopy the ability of the NF-κB p65 subunit to translocate into the nucleus by using an antibody against p65 (Figure 6A). Whereas nuclei in control cells were completely devoid of signal, CNF1 promoted the nuclear translocation of p65, as evidenced by positively stained nuclei (Figure 6A). This was a time-dependent phenomenon that started 3 h after CNF1 exposure and lasted for several hours (Figure 6B). Because CNF1-induced Rac activation lasted for at least 24 h (Fig. 1A) and in contrast, p65 translocation seemed to decrease after 24 h of CNF1 treatment, we also verified in the time frame between 24 and 48 h whether CNF1 was able to induce p65 translocation. We found that, after the peak obtained between 4 and 10 h (p < 0.05), in the time frame ranging between 24 and 48 h, a minimal level of p65 translocation was observed, always lower than that observed after the initial times of CNF1 exposure. In fact, in none of the cases this was higher than 12% (our unpublished data), thus suggesting a steady state activation of the NF-κB transcription factor. NF-κB translocation correlated with its gene transactivation activity, as viewed by transfecting HEp-2 cells with a plasmid corresponding to a luciferase reporter gene under the control of six-repeated κB-binding sites. The challenge with CNF1 led to a significant increase (p < 0.05) in the luciferase activity that started 6 h after addition of the toxin (Figure 6C). The apparent discrepancy between data on p65 nuclear staining and luciferase activity (Figure 6, B and C) can be ascribed to the lack of a linear correlation between them.
Figure 6.
Rac-induced activation of NF-κB. Fluorescence micrographs of HEp-2 cells untreated (-CNF1) or treated for 4 h with 10-9 M CNF1 (+CNF1), stained with an anti-p65 antibody. The illustrations are used to note the positively stained nuclei after CNF1 exposure. (B) Graph showing the time-dependent nuclear translocation of p65 after the CNF1-induced Rac activation. (C) Graph showing the luciferase activity induced by the CNF1-induced Rac activation on HEp-2 cells transfected with a plasmid corresponding to a luciferase reporter gene under the control of six repeated κB-binding sites. It is evident that NF-κB translocation correlated with its gene transactivation activity. (D) Graph showing the effects of the Rho family inhibiting toxins on the CNF1-induced p65 nuclear translocation. Cells were preexposed to toxins for 2 h before addition of CNF1 for an additional 4 h. Note that the two Rac-inhibiting toxins (LT82, which inhibits Rac, and LT9048, which inhibits Rac and Cdc42) dramatically reduce the percentage of p65-positive nuclei, whereas C3B, the Rho-inhibiting toxin, does not. (▪, control cells; □, C3B-treated cells;  , LT82-treated cells; and
, LT82-treated cells; and  , LT9048-treated cells).
, LT9048-treated cells).
Although Rac was reported to be the principal Rho GTPase activated by CNF1 in HEp-2 cells (Doye et al., 2002), still the toxin also activates Rho and Cdc42 (Flatau et al., 1997; Schmidt et al., 1997). Furthermore, NF-κB activation has been reported to be regulated by all the three Rho GTPases (Perona et al., 1997; Cammarano and Minden, 2001). Hence, to verify that p65 translocation to the nucleus was dependent only on Rac, HEp-2 cells were for 2 h preexposed to 1 μg/ml Rho-inhibiting toxins, before being challenged with CNF1 for 4 h. We used, in particular, the chimeric toxin C3B, that specifically inhibits Rho A, B, and C (Aullo et al., 1993), Clostridium sordellii LT82 or LT9048, which inhibit Rac but not Rho and Cdc42 (Popoff et al., 1996) and Rac plus Cdc42 (El Hadj et al., 1999), respectively. C3B failed in inhibiting the CNF1-induced p65 nuclear translocation, whereas both LT toxins did have a blocking effect (Figure 6D) (LT82+CNF1 and LT9048+CNF1 versus CNF1 alone; p < 0.05). These results indicate Rac as inducer of NF-κB activation, supporting the master role of this GTPase in the positioning and function of the NF-κB–activating complex.
Is Rac-induced Protein Recruitment to Ruffling Membranes Having a Role in NF-κB Activation?
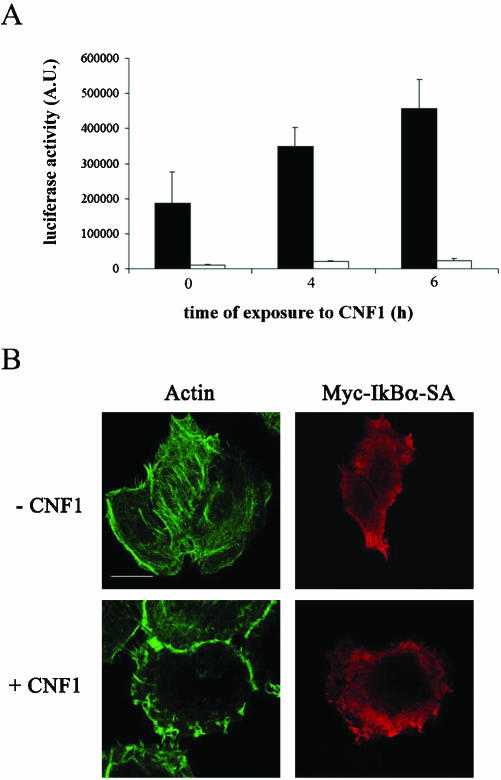
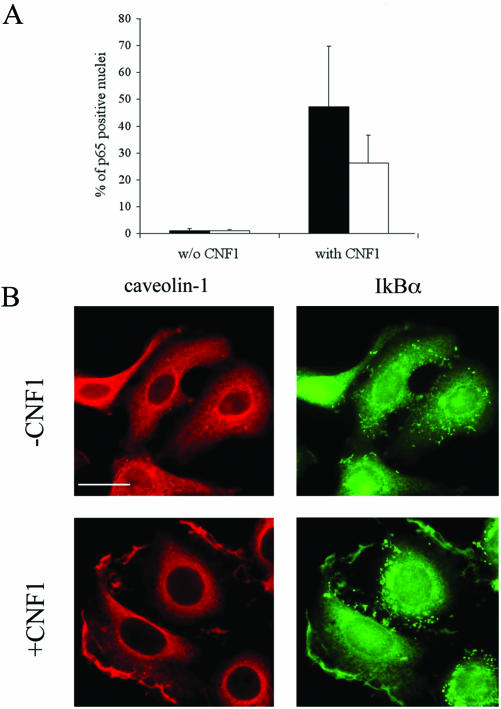
All in all, our results indicate that CNF1 induced 1) the relocalization to the ruffles of Rac together with the SCF complex and IkBα, 2) IkBα proteasomal degradation, and 3) NF-κB activation. Hence, we contemplated whether the observed protein complex relocalization to the ruffles could be an essential step for the occurrence of the subsequent events. IkBα-SA 32-36, the classical dominant negative mutant with its inability to be phosphorylated (Bottero et al., 2001), was cotransfected with the luciferase reporter gene described above. Despite the addition of CNF1, the luciferase activity resulted in being nearly eliminated by the presence of IkBα-SA 32-36 (Figure 7A). Interestingly, when CNF1 was added to cells transfected with IkBα-SA 32-36, it failed in dislocating the mutant to the ruffling membranes (Figure 7B), indicating that IkBα positioning at the level of ruffles is a prerequisite for NF-κB activation. Furthermore, because lipid rafts represents a typical domain of a membrane involved in signal transduction, we challenged HEp-2 cells with filipin, a compound known to sequester cholesterol and thus used to disrupt the organization of lipid rafts (Orlandi and Fishman, 1998). Interestingly, filipin significantly reduced the percentage of p65-positive nuclei (p < 0.05) induced by toxin treatment (Figure 8A). This was not due to alterations in the rate of CNF1-induced Rac activation, as demonstrated by a pull-down assay (our unpublished data) and in accordance to what was reported previously (Contamin et al., 2000). As a control, to verify whether the impairment of CNF1-induced p65 translocation could be due to sequestration of cholesterol by filipin, 60 μg/ml cholesterol and 2 μg/ml filipin were simultaneously added to CNF1-treated cells. What we found was that cholesterol prevented the inhibiting effect of filipin (our unpublished data), suggesting the involvement of cholesterol-rich domain in the pathway leading to NF-κB activation. Hence, we verified whether IkBα could colocalize with caveolin-1, a typical marker of lipid rafts (Harris et al., 2002), by performing a double staining for caveolin-1 and IkBα, both in control and in CNF1-treated HEp-2 cells. Whereas in control cells both proteins were dispersed throughout the cytoplasm, in HEp-2 cells challenged with the toxin caveolin-1 and IkBα colocalized at the level of ruffling membranes (Figure 8B). All in all, these findings reinforce our hypothesis indicating lipid rafts as pivotal in the CNF1-induced NF-κB activation.
Figure 7.
Protein recruitment to ruffles is necessary for NF-κB activation. (A) Graph showing the luciferase activity induced by CNF1 in HEp-2 cells transfected with a plasmid corresponding to a luciferase reporter gene under the control of six-repeated κB-binding sites (▪) and in HEp-2 cells cotransfected with this plasmid and IkBα-SA 32-36 (□). Cotransfection with IkBα-SA 32-36 results in a strong reduction of luciferase activity. (B) Confocal microscopy analysis of HEp-2 control (-CNF1) and treated cells (+CNF1) transfected with the dominant negative mutant IkBα-SA 32-36 myctagged. Left panels show the actin cytoskeleton, whereas the right panels show an anti-myc staining to visualize IkBα-SA–transfected cells. Note that IkBα-SA 32-36 failed in moving to ruffling membranes. Bar, 10 μm.
Figure 8.
Lipid rafts in CNF1-induced NF-κB activation. (A) Graph showing the inhibitory effect of filipin on the CNF1-induced p65 nuclear translocation. ▪, control cells; □, filipin-treated cells. (B) Confocal microscopy analysis of HEp-2 control (-CNF1) and cells treated for 4 h with CNF1 (+CNF1) stained with anti-caveolin-1 (left) or anti-IkBα (right) antibodies. Note that IkBα colocalizes with caveolin-1 at the level of ruffles. Bar, 10 μm.
DISCUSSION
In the current article, along with confirming the dislocation of GTP-bound Rac to the ruffles, we also provide evidence that Rac activation promoted the recruitment to the same compartment of molecules crucial for NF-κB activation, that is, the SCF complex components Cullin-1 and Skp-1, and the NF-κB inhibitor IkBα. More importantly, this occurred only if the ruffles were raised by the Rac GTPase. The novelty of these findings resides in the demonstration that the right positioning at the ruffling membranes, and more precisely in cholesterol-rich lipid rafts, is mandatory for the accomplishment of the subsequent series of events that leads to IkBα proteasomal degradation and the resultant activation of NF-κB. A scheme that summarizes our results is drawn in Figure 9.
Figure 9.
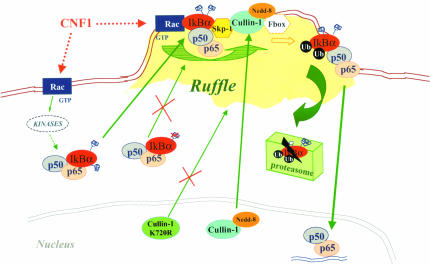
Hypothetical model by which Rac activation instructs NF-κB transactivation in HEp-2 cells. In cells challenged with CNF1, activated Rac relocalizes to membrane ruffles where it promotes the grouping of the SCF complex (Skp-1, Cullin-1, F-box) and p50/p65/IkBα, probably functioning as a docking molecule. Neddylation of Cullin-1 that occurs in the nucleus allows the displacement of Cullin-1 to the SCF complex in the ruffle, whereas lack of neddylation (as in the case of the mutant Cullin-1 K720R) forces Cullin-1 to remain inactive in the nucleus. Phosphorylation of IkBα (occurring via specific kinases) is mandatory to target the ruffles. In fact, IkBα impaired in its capacity to link Pi, is unable to localize to the plasma membrane. Hence, at the level of the ruffles, phosphorylated IkBα is ubiquitinated. Its subsequent proteosomal degradation frees p50/p65 to enter the nucleus and activate transcription. Dotted lines indicate molecules or steps that have not been investigated in the present work, and their presence in the scheme derives from literature data.
Interestingly, it has recently been reported that activated Rac family members bind to Cullin-1, and it has been suggested that this interaction may serve to regulate the E3 ligase subcellular localization (Senadheera et al., 2001). Starting from this hypothesis, we herein report the first evidence that ruffling membrane dislocation of activated Rac (Doye et al., 2002) is accompanied by the repositioning to the same intracellular compartment of Cullin-1 and Skp-1. Presumably, thus the whole SCF apparatus deputed to IkBα ubiquitination that consequently signals for IkBα degradation was at the ruffle level. Notably, Read et al. (2000) reported that phosphorylated IkBα (that is, the form targeted for ubiquitination) exclusively associates with a form of Cullin-1 that is singly modified by the ubiquitin-like protein Nedd-8. We found that not only Cullin-1 but also Nedd-8 was relocalized to the Rac-induced ruffling membranes. Because Cullins are, up to now, the only proteins reported to be modified by Nedd-8 and because Nedd-8 itself is localized primarily in the nucleus (Kamitani et al., 1997), our results suggest that the Nedd-8 protein detected at the level of the Rac-induced ruffles is part of the active Nedd-8 modified Cullin-1. The presence in the same compartment of endogenous IkBα and ubiquitin strongly supported this hypothesis. This suggests also the possibility that the activated Rac GTPase recruits to ruffles both the p65/p50/IkBα complex and the SCF complex by functioning as a docking molecule. We would like to point out that Cullin-1 was reported to be primarily localized in the nucleus and, only in a small percentage of cells, in the cytosol (Furukawa et al., 2000). Thus, this is the first report showing a Rac-dependent displacement of Cullin-1 to the ruffling compartment. All this is in accordance with the general assumption that certain proteins, after a proper activatory stimulus, need to be precisely relocalized to exert their function. Proteins of the Rho family are targeted to specific subcellular compartments, thus playing an essential role once the right positioning is achieved (Kraynov et al., 2000). In particular, the Rac GTPase targeted to the membrane has been reported to recruit the cytosolic p67phox protein for the assembly of the NADPH-oxidase complex (Gorzalczany et al., 2000). On the other hand, activation of Rac leads to a simultaneous membrane recruitment and activation of kinases involved in the production of phosphoinositides, thus regulating the membrane ruffling as well as the endocytosis and exocytosis (Wei et al., 2002). In our case, the Rac-dependent grouping of all SCF, IkBα, and ubiquitin to ruffles is a crucial newly described prerequisite for the activation of the transcription factor NF-κB.
It is worth noting that ruffles serve in numerous cellular processes and must consequently recruit all the components required for their functions. On the other hand, it is well-established that proteins involved in cell signaling can partition into subdomains of the plasma membrane, namely, lipid rafts that contain high concentrations of cholesterol and glycosphingolipids and where the cell signaling is controlled through different mechanisms (Pike, 2003). The demonstration that CNF1-treated cells require the cholesterol availability for an efficient NF-κB activation and the finding that IkBα colocalizes with caveolin-1 at the level of CNF1-induced ruffling membranes define the lipidic subdomains of recruitment that are present in the ruffling membranes. Therefore, in our case, lipid rafts can be viewed as signaling platforms where the requested components for NF-κB activation convey and colocalize. This conceivably serves to facilitate their interaction that would occur rapidly and efficiently because of the spatial proximity of the interacting components.
NF-κB plays a central role in regulating the expression of several genes involved in immune, inflammatory, and apoptotic processes (Gosh et al., 1998). Intriguingly, NF-κB also provides a link between toxins hijacking the Rho GTPases and apoptosis in the bacterial world (Fiorentini et al., 2003). Hence, it is not surprising that such a pivotal transcription factor can somehow be governed by the central switching molecule Rac. From the point of view of bacteria, challenging Rac with a protein toxin can represent, together with the others, a strategy to handle this transcription factor and to avoid their own elimination. On the other hand, the immune system has evolved a variety of receptors to recognize invading bacteria, including the so-called Toll-like receptors that are able to convey signals to transcription factors that orchestrate the inflammatory response (Zhang and Ghosh, 2001). It is worth noting that NF-κB activation can be mediated by Toll-like receptors and is controlled by Rac (Arbibe et al., 2000) and, although it involves different molecules, is dependent on the assembly of a multiprotein complex in the same manner as the one herein described. Of interest, epithelial cells have a family of cytosolic proteins, one of which, Nod1 (nucleotide-binding oligomerization domain) also mediates NF-κB activation in response to invasive bacteria (Girardin et al., 2001). Finally, the Rac-induced activation of the transcription factor NF-κB by the bacterial toxin CNF1 can furnish a molecular framework to our previous observations showing a protective effect of the toxin to apoptotic stimuli (Fiorentini et al., 1998).
Acknowledgments
We thank J.F. Peyron for pcDNA3-myc-IkBmutant and p(6KappaB)Luc, W. Krek for pcDNA3-HA-Cullin-1, K. Tanaka for pcDNA3-HA-Cullin-1 K720R, J.W. Harper for pHm200-HA-Skp-1 expression construct, and A. Doye and M. Falchi for technical assistance. We are grateful to A. Spinedi for useful suggestions and to M. Di Santolo for reviewing the manuscript. This work is supported by a Ligue National contre le Cancer fellowship to L.B. and a Grant from the Association pour la Recherche sur le Cancer (ARC no. 4463 to E.L.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–05–0301. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-05-0301.
References
- Arbibe, L., Mira, J.P., Teusch, N., Kline, L., Guha, M., Mackman, N., Godowski, P.J., Ulevitch, R.J., and Knaus, U.G. (2000). Toll-like receptor 2-mediated NF-kappa B activation requires a Rac1-dependent pathway. Nat. Immunol. 1, 533-540. [DOI] [PubMed] [Google Scholar]
- Aullo, P., Giry, M., Olsnes, S., Popoff, M.R., Kocks, C., and Boquet, P. (1993). A chimeric toxin to study the role of the 21 kDa GTP binding protein rho in the control of actin microfilament assembly. EMBO J. 12, 921-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin, A.S. (1996). The NF-kB and IkB proteins: new discoveries and insights. Annu. Rev. Immunol. 14, 649-681. [DOI] [PubMed] [Google Scholar]
- Beg, A.A., Ruben, S.M., Scheinman, R.I., Haskill, S., Rosen, C.A., and Baldwin, A.S., Jr. (1992). I kappa B interacts with the nuclear localization sequences of the subunits of NF-kappa B: a mechanism for cytoplasmic retention. Genes Dev. 6, 1899-1913. [DOI] [PubMed] [Google Scholar]
- Bottero, V., Busuttil, V., Loubat, A., Magne, N., Fischel, J.L., Milano, G., and Peyron, J.F. (2001). Activation of nuclear factor kappaB through the IKK complex by the topoisomerase poisons SN38 and doxorubicin: a brake to apoptosis in HeLa human carcinoma cells. Cancer Res. 61, 7785-7791. [PubMed] [Google Scholar]
- Cammarano, M.S., and Minden, A. (2001). Dbl and the Rho GTPases activate NF kappa B by I kappa B kinase (IKK)-dependent and IKK-independent pathways. J. Biol. Chem. 276, 25876-25882. [DOI] [PubMed] [Google Scholar]
- Ciechanover, A., and Schwartz, A.L. (1998). The ubiquitin-proteasome pathway: the complexity and myriad functions of proteins death. Proc. Natl. Acad. Sci. USA 95, 2727-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contamin, S., Galmiche, A., Doye, A., Flatau, G., Benmerah, A., and Boquet, P. (2000). The p21 Rho-activating toxin cytotoxic necrotizing factor 1 is endocytosed by a clathrin-independent mechanism and enters the cytosol by an acidic-dependent membrane translocation step. Mol. Biol. Cell 11, 1775-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien, V., Couillault, C., Franco, M., Martineau, S., Montcourrier, P., Houl-gatte, R., and Chavrier, P. (2002). A conserved C-terminal domain of EFA6-family ARF6-guanine nucleotide exchange factors induces lengthening of microvilli-like membrane protrusions. J. Cell Sci. 115, 2867-2879. [DOI] [PubMed] [Google Scholar]
- Deshaies, R.J. (1999). SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Dev. Biol. 15, 435-467. [DOI] [PubMed] [Google Scholar]
- Doye, A., Mettouchi, A., Bossis, G., Clement, R., Buisson-Touati, C., Flatau, G., Gagnoux, L., Piechaczyk, M., Boquet, P., and Lemichez, E. (2002). CNF1 exploits the ubiquitin-proteasome machinery to restrict Rho GTPase activation for bacterial host cell invasion. Cell 111, 553-564. [DOI] [PubMed] [Google Scholar]
- El Hadj, N.B., Popoff, M.R., Marvaud, J.C., Payrastre, B., Boquet, P., and Geny, B. (1999). G-protein-stimulated phospholipase D activity is inhibited by lethal toxin from Clostridium sordellii in HL-60 cells. J. Biol. Chem. 274, 14021-14031. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville, S., and Hall, A. (2002). Rho GTPases in cell biology. Nature 420, 629-635. [DOI] [PubMed] [Google Scholar]
- Fiorentini, C., Matarrese, P., Straface, E., Falzano, L., Fabbri, A., Donelli, G., Cossarizza, A., Bloom, P., and Malorni, W. (1998). Toxin-induced activation of Rho GTP-binding protein increases Bcl-2 expression and influences mitochondrial homeostasis. Exp. Cell Res. 242, 341-350. [DOI] [PubMed] [Google Scholar]
- Fiorentini, C., Falzano, L., Travaglione, S., and Fabbri, A. (2003). Hijacking Rho GTPases by protein toxins and apoptosis: molecular strategies of pathogenic bacteria. Cell Death Differ. 10, 147-52. [DOI] [PubMed] [Google Scholar]
- Flatau, G., Lemichez, E., Gauthier, M., Chardin, P., Paris, S., Fiorentini, C., and Boquet, P. (1997). Toxin-induced activation of the G protein p21 Rho by deamidation of glutamine. Nature 387, 729-733. [DOI] [PubMed] [Google Scholar]
- Furukawa, M., Zhang, Y., McCarville, J., Ohta, T., and Xiong, Y. (2000). The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification, and ubiquitin ligase activity of CUL1. Mol. Cell Biol. 20, 8185-8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosh, S., May, M., and Kopp, E.B. (1998). NF-kB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16, 225-260. [DOI] [PubMed] [Google Scholar]
- Girardin, S.E., et al. (2001). CARD4/Nod1 mediates NF-kB and JNK activation by invasive Shigella flexneri. EMBO Rep. 2, 736-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorzalczany, Y., Sigal, N., Itan, M., Lotan, O., and Pick, E. (2000). Targeting of Rac1 to the phagocyte membrane is sufficient for the induction of NADPH oxidase assembly. J. Biol. Chem. 275, 40073-40081. [DOI] [PubMed] [Google Scholar]
- Hall, A. (1998). Small GTPases and the actin cytoskeleton. Science 279, 509-514. [DOI] [PubMed] [Google Scholar]
- Harris, J., Werling, D., Hope, J.C., Taylor, G., and Howard, C.J. (2002). Caveolae and caveolin in immune cells: distribution and functions. Trends Immunol. 23, 158-164. [DOI] [PubMed] [Google Scholar]
- Hunzicker-Dunn, M., Gurevich, V.V., Casanova, J.E., and Mukherjee, S. (2002). ARF 6, a newly appreciated player in G protein-coupled receptor desensitization. FEBS Lett. 521, 3-8. [DOI] [PubMed] [Google Scholar]
- Kamitani, T., Kito, K., Nguyen, H.P., and Yeh, E.T. (1997). Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J. Biol. Chem. 272, 28557-28562. [DOI] [PubMed] [Google Scholar]
- Karin, M., and Ben-Neriah, Y. (2000). Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu. Rev. Immunol. 18, 621-663. [DOI] [PubMed] [Google Scholar]
- Kawakami, et al. (2001). NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 20, 4003-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraynov, V.S., Chamberlain, C., Bokoch, G.M., Schwartz, M.A., Slabaugh, S., and Hahn, K.M. (2000). Localized Rac activation dynamics visualized in living cells. Science 290, 333-337. [DOI] [PubMed] [Google Scholar]
- Lerm, M., Selzer, J., Hoffmeyer, A., Rapp, U.R., Aktories, K., and Schmidt, G. (1999). Deamidation of Cdc42 and Rac by Escherichia coli cytotoxic necrotizing factor 1, activation of c-Jun N-terminal kinase in HeLa cells. Infect. Immun. 67, 496-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerm, M., Pop, M., Fritz, G., Aktories, K., and Schmidt, G. (2002). Proteasomal degradation of cytotoxic necrotizing factor 1-activated Rac. Infect. Immun. 70, 4053-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisztwan, J., Marti, A., Sutterluty, H., Gstaiger, M., Wirbelauer, C., and Krek, W. (1998). Association of human CUL-1 and ubiquitin-conjugating enzyme CDC34 with the F-box protein p45(SKP2): evidence for evolutionary conservation in the subunit composition of the CDC34-SCF pathway. EMBO J. 17, 368-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montaner, S., Perona, R., Saniger, L., and Lacal, J.C. (1998). Multiple signalling pathways lead to the activation of the nuclear factor kappaB by the Rho family of GTPases. J. Biol. Chem. 273, 12779-12785. [DOI] [PubMed] [Google Scholar]
- Orlandi, P.A., and Fishman, P.H. (1998). Filipin-dependent inhibition of cholera toxin: evidence for toxin internalization and activation through caveolae-like domains. J. Cell Biol. 141, 905-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perona, R., Montaner, S., Saniger, L., Sanchez-Perez, I., Bravo, R., and Lacal, J.C. (1997). Activation of the nuclear factor-kappaB by Rho, CDC42, and Rac-1 proteins. Genes Dev. 11, 463-475. [DOI] [PubMed] [Google Scholar]
- Pike, L.J. (2003). Lipid rafts: bringing order to chaos. J. Lipid Res. 44, 655-667. [DOI] [PubMed] [Google Scholar]
- Popoff, M. R., et al. (1996). Ras, Rap, and Rac small GTP-binding proteins are targets for Clostridium sordellii lethal toxin glucosylation. J. Biol. Chem. 271, 10217-10224. [DOI] [PubMed] [Google Scholar]
- Read, M. A., et al. (2000). Nedd8 modification of cul-1 activates SCF(beta(TrCP)-dependent ubiquitination of IkappaBalpha. Mol. Cell Biol. 20, 2326-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudert, F., Visser, E., Gradl, G., Grandison, P., Shemshedini, L., Wang, Y., Grierson, A., and Watson, J. (1996). pLEF, a novel vector for expression of glutathione S-transferase fusion proteins in mammalian cells. Gene 169, 281-282. [DOI] [PubMed] [Google Scholar]
- Schmidt, G., Sehr, P., Wilm, M., Selzer, J., Mann, M., and Aktories, K. (1997). Gln 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature 387, 725-729. [DOI] [PubMed] [Google Scholar]
- Schmidt, G., Selzer, J., Lerm, M., and Aktories, K. (1998). The Rho-deamidating cytotoxic necrotizing factor 1 from Escherichia coli possesses transglutaminase activity. Cysteine 866 and histidine 881 are essential for enzyme activity. J. Biol. Chem. 273, 13669-13674. [DOI] [PubMed] [Google Scholar]
- Senadheera, D., Haataja, L., Groffen, J., and Heisterkamp, N. (2001). The small GTPase Rac interacts with ubiquitination complex proteins Cullin-1 and CDC23. Int. J. Mol. Med. 8, 127-133. [DOI] [PubMed] [Google Scholar]
- Traenckner, E.B., Pahl, H.L., Henkel, T., Schmidt, K.N., Wilk, S., and Baeuerle, P.A. (1995). Phosphorylation of human I kappa B-alpha on serines 32 and 36 controls I kappa B-alpha proteolysis and NF-kappa B activation in response to diverse stimuli. EMBO J. 14, 2876-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Y.J., Sun, H.Q., Yamamoto, M., Wlodarski, P., Kunii, K., Martinez, M., Barylko, B., Albanesi, J.P., and Yin, H.L. (2002). Type II phosphatidylinositol 4-kinase beta is a cytosolic and peripheral membrane protein that is recruited to the plasma membrane and activated by Rac-GTP. J. Biol. Chem. 277, 46586-46593. [DOI] [PubMed] [Google Scholar]
- West, C.M. (2003). Evolutionary and functional implications of the complex glycosylation of Skp-1, a cytoplasmic/nuclear glycoprotein associated with polyubiquitination. Cell Mol. Life Sci. 60, 229-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston, J.T., Strack, P., Beer-Romero, P., Chu, C.Y., Elledge, S.J., and Harper, J.W. (1999). The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and betacatenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev. 13, 270-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, K., Chen, A., and Pan, Z.Q. (2000). Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1-CUL1 complex to promote ubiquitin polymerization. J. Biol. Chem. 275, 32317-32324. [DOI] [PubMed] [Google Scholar]
- Zhang, G., and Ghosh, S. (2001). Toll-like receptor-mediated NF-kappaB activation: a phylogenetically conserved paradigm in innate immunity. J. Clin. Investig. 107, 13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]