Background: Poly(ADP-ribose) polymerase 1 (PARP1) exerts important functions in transcriptional regulation of transcription factors.
Results: PAPR1 interacts with estrogen receptor α (ERα), and poly(ADP-ribosyl)ation promotes ERα transactivation.
Conclusion: PARP1 is a crucial regulator of ERα-dependent gene transcription.
Significance: This might be the first study examining the interpaly between PARP1 and ERα, and identifies that PARP1 controls ERα transactivation.
Keywords: Gene Regulation, Nuclear Receptors, Protein-Protein Interactions, Signal Transduction, Transcription, ERα, PARP1
Abstract
Activation of nuclear receptor estrogen receptor α (ERα) exerts cardiovascular protective effects by modulating the expression of ERα target genes. However, the underlying mechanism remains unclear. PARP1 is a ubiquitous multifunctional nuclear enzyme. In this study, we examined the interplay between PARP1 and ERα, and identified PARP1 as an important regulator of ERα-dependent transcription. We showed that PARP1 could directly bind to ERα, and ERα could be poly(ADP-ribosyl)ated by PARP1. Poly(ADP-ribosyl)ation increased ERα binding to estrogen response element (ERE) present in the promoter of target genes and promoted ERα-mediated gene transcription. Estradiol, the ligand of ERα, increased PARP enzymatic activity and enhanced poly(ADP-ribosyl)ation of ERα. Upon treatment with estradiol, ERα binding to ERE- and ERα-dependent gene expression was dramatically increased in cultured vascular smooth muscle cells (VSMCs). Inhibition of PARP1 by PARP inhibitor or PARP1 siRNA decreased ERα binding to ERE and prevented ERα-dependent gene transcription in VSMCs. Further studies revealed that PARP1 served as an indispensible component for the formation of the ERα-ERE complex by directly interacting with ERα. Thus, our results identify PARP1 as a key regulator of ERα in controlling ERα transactivation.
Introduction
Nuclear receptors (NRs)4 are ligand-dependent transcription factors that have evolved from an ancestral orphan receptor into a highly diverse family present throughout the entire animal kingdom. These receptors play crucial roles in multiple aspects of animal physiology, ranging from development and differentiation to metabolic homeostasis (1). Previous studies demonstrate that the regulation of gene transcription by nuclear receptors is mainly due to the realization that not only the interaction of the receptors with DNA, but also that many transcription co-regulators (including both co-activators and co-repressors), which were crucial in transmitting the signals to the transcriptional machinery of the receptors (2). Estrogen receptor α (ERα, UniProt ID: P06211), known as NR3A1 (nuclear receptor subfamily 3, group A, member 1), is a member of the steroid nuclear receptor family. In the unliganded state, ERα recruits the co-repressor complexes in association with histone deacetylases to silence target genes (3). In the presence of the natural ligand of ERs, sex hormone estradiol, ERα undergoes a conformational change that dislodges the co-repressor complexes, exposing a “docking” site for co-activators, then modulates the expression of ERα target genes via directly binding to the specific response elements known as estrogen response element (ERE) (3, 4). Mediators as the general nuclear receptor-interacting co-activators play an important role in NR-dependent transcription. They act as a “bridge” between the upstream DNA-bound activator and the basal transcriptional machinery via protein-protein interactions (5–8).
Poly(ADP-ribose) polymerase 1 (PARP1, UniProt ID: P27008), an abundant and ubiquitous nuclear enzyme present in eukaryotes (9), is the most abundant isoform of the PARP enzyme family, accounting for about 90% of total cellular PARP activity (9, 10). In the nucleus, activated PARP1 catalyzes the transfer of poly(ADP-ribose) (PAR) from donor nicotinamide adenine dinucleotide (NAD+) molecules onto nuclear acceptor proteins to affect the activity of transcription factors (10–12). This modification known as poly(ADP-ribosyl)ation is transient but very extensive in vivo, as polymer chains can reach more than 200 units on protein acceptors, playing important roles in many cellular processes (9). PARP1 also poly(ADP-ribosyl)ates itself, and the auto-poly(ADP-ribosyl)ation represents a major regulatory mechanism for PARP1 resulting in the down-regulation of the enzyme activity (10, 13). PARP1 and PAR have received considerable attention in the recent literature. Previous studies have shown that PARP1 participates in the regulation of ERα-dependent transcription (14). However, the underlying mechanism remains elusive. As PARP1 has been shown to regulate the expression of various nuclear proteins at the transcriptional level (9, 15–18), we then wish to determine whether ERα could represent a target for PARP1.
In the present study, we found that PARP1 could bind to and poly(ADP-ribosyl)ate ERα. In cultured rat VSMCs, inhibition of PARP1 inhibited ERα binding to ERE and decreased ERα-dependent target gene expression. Treatment with estradiol increased poly(ADP-ribosyl)ation of ERα, and thereafter promoted ERα transactivation. Thus we identified PARP1 as a key regulator of ERα-mediated transcription.
EXPERIMENTAL PROCEDURES
Cell Culture
Rat aortic VSMCs were isolated from thoracic aorta in male Wistar rats (150–180 g, obtained from Tongji Medical College, Huazhong University of Science and Technology, China) by a tissue adherent method described previously. Freshly isolated thoracic aorta was clipped into small slices and then planted in the culture flask. After about 1 week, the primary VSMCs were digested by 0.5% trypsin, and maintained in Dulbecco's modified Eagle's medium (DMEM) without phenol red and with 20% fetal bovine serum (FBS). The purity of the VSMCs was evaluated by staining the cells with monoclonal antibodies to smooth muscle cell α-actin. More than 96% of cells expressed smooth muscle cell α-actin. Experiments were performed on cells at 3 to 10 passages after primary culture. Human embryonic kidney 293 cells were routinely maintained in DMEM supplemented with 10% FBS in humidified 5% CO2 atmosphere at 37 °C.
Plasmid Construction
The full-length cDNA of human PARP1 was cloned by RT-PCR from 293 cells. The fragments of PARP1 A (aa1–214), B (aa215–372), C (aa373–476), D (aa477–524), E (aa525–656), and F (aa657–1014) were constructed in the mammalian expression vector p3flag-CMV (Sigma-Aldrich). Human cDNAs encoding ERα was cloned by RT-PCR from 293 cells. The expression plasmids for GFP-tagged ERα A (aa1–184), B (aa185–310), C (aa311–595) were constructed by inserting PCR fragments from ERα cDNA into the pcDNA3.1-GFP vector. A catalytically inactive mutant of PARP1 (mut-PARP1), in which lysine 893 was substituted by isoleucine (K893I), was generated as previously by using the QuickChange site-directed mutagenesis kit (Stratagene) (19). Sequences of the primers are available upon request.
RNA Interference and Transfection
Small interfering RNA (siRNAs) was synthesized by RiBoBio Co. Ltd (China). The sequence of rat PARP1 siRNA was: sense 5′-GGAUGAUCUUCGACGUGGA-3′, and antisense 5′-UCCACGUCGAAGAUCAUCC-3′. The sequence of rat ERα siRNA was: sense 5′-GGAGAAUGUUGAAGCACAA-3′, and antisense 5′-CCUCUUACAACUUCGUGUU-3′. The sequence of unrelated siRNA was: sense 5′-UUCUCCGAACGUGUCACGU-3′ and antisense 5′-AAGAGGCUUGCACA GUGCA-3′. The cultured VSMCs were transfected with 50 nm siRNA using Lipofectamine 2000 (Invitrogen).
Evaluation of Transfection Efficiency
50 nmol/liter Cy3-labeled unrelated siRNA were transfected into the cultured VSMCs in 24-well plates as described above. Transfections were performed in triplicate for each treatment. 6 h later, after several washes in PBS, one part was prepared for flow cytometric analyses. The others were fixed in 4% paraformaldehyde for 30 min in the dark, and washed again with PBS. The cover slips were mounted with PBS/glycerin. Cells were photographed under a light or fluorescence microscope (for Cy3, wavelength 555 nm, Olympus Microscope BX-51, Japan).
Preparation of Whole Extracts and Nuclear Extracts
Methods for preparation of whole cell extracts and nuclear extracts were described previously (22, 23). These whole cell and nuclear extracts were stored at −80 °C until use. Protein concentrations of these extracts were determined by the Bradford assay.
PARP Activity Assay
PARP activity was assayed using the universal colorimetric PARP assay kit (Trevigen), based on the incorporation of biotinylated ADP-ribose onto histone proteins. Cell lysates containing 50 μg of protein were loaded into a 96-well plate coated with histones and biotinylated poly ADP-ribose, allowed to incubate for 1 h, treated with strep-HRP, and read at 450 nm in a spectrophotometer.
Western Blot
After denaturation and SDS-PAGE electrophoresis, separated proteins were transferred to nitrocellulose membranes. Membranes were blocked with 5% nonfat milk in TBS (50 mmol/liter Tris-HCl, pH 7.6, 150 mmol/liter NaCl) for 3 h, and incubated with primary antibodies in TBS at 4 °C overnight. Antibodies used were anti-PARP1 (1:1000, R&D), anti-PAR (1:1000, Trevigen), anti-ERα (1:1000, Millipore), anti-hist1 (1:200, Santa Cruz Biotechnology), anti-β-actin (1:500, Santa Cruz Biotechnology), and anti-GAPDH (1:500, Santa Cruz Biotechnology). Membranes were incubated with peroxidase-conjugated secondary antibody in TBS at room temperature for 2 h. The specific band was detected with a chemiluminescence assay (ECL detection reagents, Pierce) and recorded on x-ray film. Quantity One software was used to quantify the intensities of bands.
Real-time RT-RCR
Total RNA was isolated using Trizol reagent (Takara Biotechnology, Japan) according to the manufacturer's instruction. 1 μg of total RNA was reverse transcribed using the RNA PCR kit (Takara Biotechnology, Japan), and the resulting cDNA was used as a PCR template. The mRNA levels were determined by real-time PCR with the ABI PRISM 7900 Sequence Detector system (Applied Biosystems) according to the manufacturer's instructions. GAPDH was used as an endogenous control. The PCR reaction mixture contained the SYBR Green I (Takara Biotechnology), cDNA and the primers. The relative gene expression level (the amount of target, normalized to endogenous control gene) was calculated using the comparative Ct method formula 2−ΔΔCt. Rat primers were as follows: GAPDH: 5′-ATGACTCTACCCACGGCAAG-3′ and 5′-TACTCAGCACCAGCATCACC-3′; TGF-α 5′-CCTGGCTGTCCTCATTATCACCT-3′ and 5′-AGCAGGCAGTCCTTCCTTTCAG-3′; cyclin D1: 5′-GAGGAGCAGAAGTGCGAAGAGG-3′ and 5′-GGCGGATAGAGTTGTCAGT GTAGATG-3′; IGF-1: 5′-CTTTTACTTCAACAAGCCCACAGG-3′ and 5′-GCACAGTACATCTCCAGCCTCCT-3′.
In Vitro Protein-Protein Interaction Assay (Far-Western Blot)
Far-Western blot assays were performed by resolving protein samples by SDS-PAGE and transferring them to polyvinylidene fluoride membranes. Membranes were then incubated with Hyb-75 buffer (20 mmol/liter HEPES, pH 7.6, 75 mmol/liter KCl, 0.1 mmol/liter EDTA, 2.5 mmol/liter MgCl2, 0.005% Nonidet P-40, 1 mmol/liter DTT) supplemented with 5% nonfat milk, overnight at 4 °C. Membranes were briefly washed with Hyb-75 buffer and then incubated with 1 μg/ml recombinant protein (PARP1 (Trevigen), ERα (Millipore), or β-actin (Abnova), respectively) at room temperature for 1 h. After washing with Hyb-75 buffer, membranes were incubated with anti-PARP1 antibody or anti-ERα antibody at 4 °C overnight. After washing, membranes were incubated with HRP-conjugated secondary antibody for 2 h. Specific bands were detected using the ECL detection system (Pierce).
Co-immunoprecipitation (co-IP)
Co-IP assays were performed as described previously. Briefly, 500 μg of nuclear extracts were incubated with the indicated antibodies at 4 °C for 1 h, and protein-G-agarose at 4 °C for 12 h. Immunoprecipitates were pelleted by centrifugation at 5000 × g for 1 min and washed four times with lysis buffer. The pellets were suspended in SDS gel loading buffer, boiled for 10 min, and subjected to Western blot analysis. To determine the specificity of the bands, nonspecific IgG (negative control) was used.
Electrophoretic Mobility Shift Assay (EMSA) and Supershift Assay
DNA-protein interaction was detected using LightShiftTM Chemiluminescent EMSA kit (Pierce) according to the manufacturer's protocol. The sequence of ERE consensus oligonucleotides was: 5′-CTTCGAGGAGGTCACAGTGACCTG GAGCGG-3′. Biotin was labeled at the 5′-end of the oligonucleotides. In the supershift assay, after incubation of nuclear extracts with anti-PAR, anti-PARP1 antibody, anti-ERα antibody, or nonspecific IgG at 4 °C for 30 min, biotin-labeled oligonucleotides were added to the reaction and incubated for another 20 min.
Chromatin Immunoprecipitation (ChIP)
ChIP experiments were performed as previously described (20). Cells were sonicated, and lysates were immunoprecipitated using the anti-ERα antibody (Santa Cruz Biotechnology). In re-ChIP assays, chromatin was first immunoprecipitated with anti-ERα antibody and then eluted with 100 μl of elution buffer with 10 mmol/liter DTT at 37 °C for 30 min and then, diluted (25-fold) with dilution buffer (20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 2 mm EDTA, 1% Triton X-100), and re-immunoprecipitated with IgG or anti-PAR antibody. Real-time-PCR was performed using 1 μg of template DNA with specific primers for rat IGF-1. Sequence of the primer is available upon request. The chromosomal DNA input and ChIP DNA with nonspecific IgG were subjected to the same PCR amplification. PCR products were separated on an ethidium bromide-stained 2% agarose gel.
Southwestern Blot
A Southwestern blot was performed according to the procedure of Butler and Ordahl (18) with slight modifications. Nuclear proteins (35 μg) were resolved on a 9% SDS-PAGE and then electrotransferred to a nitrocellulose membrane. Membranes were blocked with 5% BLOTTO-0.1% bovine serum albumin-1 mg/ml poly(dI-dC) in binding buffer (30 mm HEPES (pH 7.6), 1 mm dithiothreitol), followed by incubation with 1.0 pmol biotin-labeled ERE oligonucleotides in Hyb-50 buffer (30 mmol/ liter HEPES (pH 7.6), 50 mmol/liter KCl, 10 mmol/liter MgCl2, 0.1 mmol/liter EDTA, 1 mmol/liter DTT, 5% BLOTTO, 0.1% bovine serum albumin, and 1 mg/ml poly(dI-dC)) at 4 °C overnight. After washed three times (30 mmol/liter HEPES (pH7.6), 50 mmol/liter NaCl, 1% BLOTTO), membranes were incubated with streptavidin-horseradish peroxidase conjugate in blocking buffer (Pierce) for 15 min. Specific binding was detected with ECL detection reagents (Pierce), and band intensities were quantified as described above.
Luciferase Assay
The luciferase-reporter plasmid ERE-TK-LUC was kindly provided by Dr. Ming-Jer Tsai (Department of Molecular and Cellular Biology, Baylor College of Medicine).1 μg of ERE-TK-LUC and 10 ng of pRL-SV40 plasmids (internal control for normalization of transfection efficiency, Promega) were transfected into VSMCs using Lipofectamine 2000 (Invitrogen) in accordance with the manufacturer's instruction. After incubation for 24 h, cells were harvested, lysed, and assayed for luciferase activity with the Dual Luciferase Reporter Assay kit (Promega) according to the manufacturer's instruction.
Statistical Analysis
Values are shown as mean ± S.E. of at least three independent experiments. The significance of differences was estimated by one-way ANOVA followed by Student-Newmann-Keuls multiple comparison tests. p < 0.05 was considered significant. All statistical analyses were performed with SPSS software (version 11.0, SPSS Inc).
RESULTS
Inhibition of PARP1 Decreases ERα-mediated Gene Transcription in VSMCs
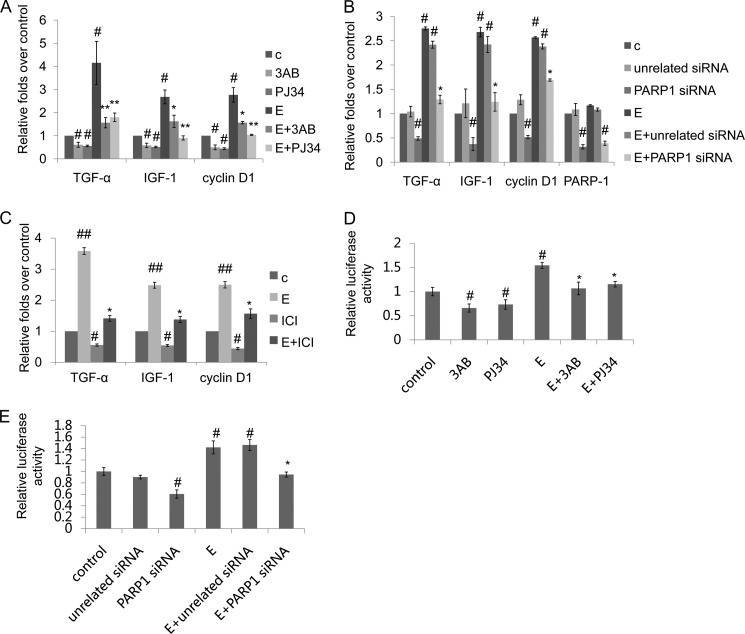
We used rat VSMCs and examined whether PARP1 is directly involved in the modulation of ERα target genes. We found that treatment of VSMCs with PARP inhibitor, 3-aminobenzamide (3AB) or N-(6-oxo-5,6-dihydrophenanthridin-2-yl)-2-(N,N-dimethylamino)acetamide (PJ34), or knockdown of PARP1 by specific siRNA significantly decreased mRNA expression of ERα target genes including cyclin D1 (UniProt ID: Q99NB4), insulin-like growth factors-1 (IGF-1, UniProt ID: P08025), and transforming growth factor-α (TGF-α, UniProt ID: P01134) (Fig. 1, A and B). Estradiol, the ligand of ERα, significantly increased mRNA expression of the above mentioned genes, which was inhibited by PARP inhibitor or PARP1 siRNA in VSMCs (Fig. 1, A and B). To confirm the estrogen regulation of these genes, we used an antiestrogen ICI 182,780 (ICI). Results showed that treatment with ICI abrogated the estradiol-induced increases in the expression of cyclin D1, IGF-1, and TGF-α (Fig. 1C). All these results suggested that PARP1 was involved in the regulation of ERα-mediated transcriptional activation.
FIGURE 1.
Inhibition of PARP1 prevents ERα transactivation in VSMCs. A, real-time RT-PCR assay of TGF-α, IGF-1, and cyclin D1 in VSMCs treated with 3AB (10 mm, 24 h) or PJ34 (10 μΜ, 24 h) in the absence or presence of estradiol (E, 10−7 m, 24 h). B, VSMCs were transfected with PARP1 siRNA (50 nm, 48 h) or unrelated siRNA (50 nm, 48 h) followed by treatment with PBS or estradiol (E, 10−7 m, 24 h). The mRNA expression of TGF-α, IGF-1, cyclin D1, and PARP1 in VSMCs was assessed by real time RT-PCR assay. C, VSMCs were treated with ICI 182,780 (ICI, 10 nm, 24 h) in the absence or presence of estradiol (E, 10−7 m, 24 h). D and E, ERE-TK-LUC and pRL-SV40 plasmids were transfected into VSMCs using Lipofectamine 2000. After 24 h, ERE-directed luciferase activity was detected with the Dual Luciferase Reporter Assay Kit (described under “Experimental Procedures”). D, VSMCs were treated with 3AB (10 mm, 24 h) or PJ34 (10 nm, 24 h) in the absence or presence of estradiol (E, 10−7 m, 24 h). E, VSMCs were transfected with PARP1 siRNA (50 nm, 48 h) or unrelated siRNA (50 nm, 48 h) followed by treatment with PBS or estradiol (E, 10−7 m, 24 h). The results are from six independent experiments. Data are expressed as the mean ± S.E. #, p < 0.05 and ##, p < 0.01 are for comparison with control group (c). *, p < 0.05 and **, p < 0.01 are for comparison with estradiol group (E).
In the nucleus, ERα directly binds to ERE in the promoter of target genes to regulate their transcription. To explore the influence of PARP1 on ERα-dependent transcriptional responses, ERE-driven luciferase reporter (ERE-TK-LUC) was constructed. Treatment with estradiol increased ERE-dependent luciferase reporter activity in VSMCs. Inhibition of PARP activity by 3AB or PJ34, or knockdown of PARP1 by siRNA decreased transcriptional output of the ERE-driven luciferase reporter under either basal or estradiol-treated condition (Fig. 1, D and E), indicating that inhibition of PARP1 prevented ERα transactivation. These results also illustrated that activation of PARP1 was critical for estradiol-induced ERα transactivation in VSMCs.
ERα Interacts Directly with PARP1
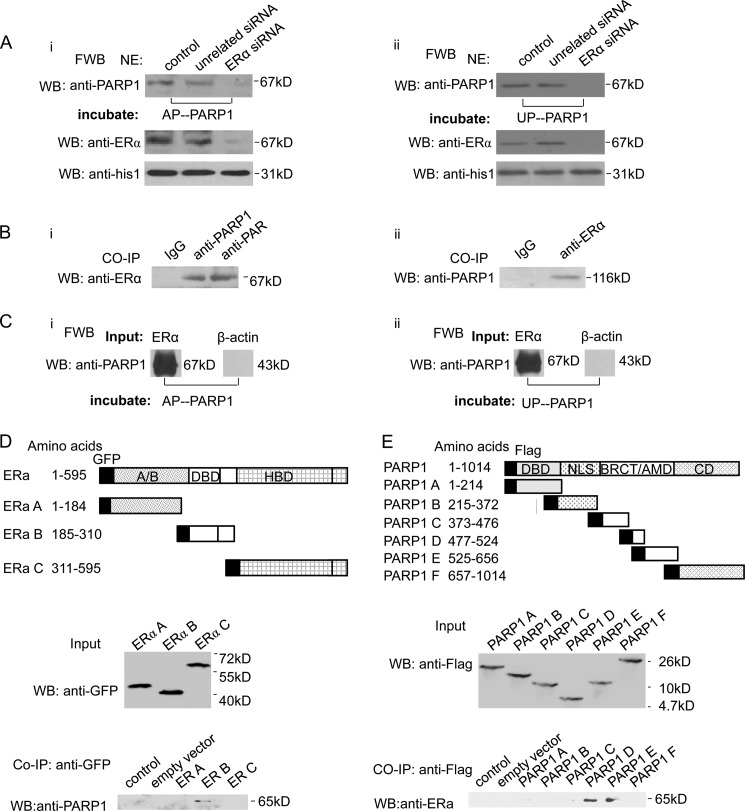
The finding that PARP1 might regulate ERα-mediated transcriptional activation led us to explore the interaction between PARP1 and ERα in detail. First, we examined whether PARP1 bound to ERα directly. Far Western blot assay showed that both un-poly(ADP-ribosyl)ated PARP1 (UP-PARP1) and auto-poly(ADP-ribosyl)ated PARP1 (AP-PARP1) could specifically bind to the 67-kDa protein in nuclear extracts from VSMCs (Fig. 2A). Since ERα was a 67-kDa nuclear protein, this protein was speculated to be ERα. In line with this speculation, both UP-PARP1 and AP-PARP1 failed to bind to the 67 kDa nuclear protein in ERα-knockdown VSMCs (Fig. 2A). To further determine if PARP1 and ERα were in the same nuclear complex in VSMCs, we performed a co-immunoprecipitation (co-IP) experiment. Endogenous PARP1 of VSMCs was immunoprecipitated by anti-PARP1 antibody followed by Western blot assay using anti-ERα antibody. Results showed that PARP1 was co-precipitated with ERα (Fig 2Bi) and vice versa (Fig 2Bii). In a cell-free system, we also found that recombinant ERα could specifically bind to recombinant UP-PARP1 and AP-PARP1, respectively (Fig. 2C). These results demonstrated that PARP1 could directly interact with ERα. To further investigate which domain of ERα-mediated protein-protein interaction with PARP1, we used the ERα deletion mutants. We then observed that endogenous PARP1 associated specifically with the DNA-binding domain (DBD) of ERα (Fig. 2D). Using PARP1 deletion mutants, we localized the ERα-binding site to the central BRCA1 C terminus (BRCT)/auto-modification domain (AMD) of PARP1 (Fig. 2E).
FIGURE 2.
ERα interacts directly with PARP1. A, VSMCs were transfected with ERα siRNA (50 nm) or unrelated siRNA (50 nm). After 48 h, nuclear extracts (NE) from VSMCs were analyzed by far-Western blot (FWB) (described under “Experimental Procedures”). After denaturation and SDS-PAGE electrophoresis, separated proteins were transferred to nitrocellulose membranes. Membranes were incubated with UP-PARP1 protein (i, 1 μg/ml) or AP-PARP1 protein (ii, 1 μg/ml), and then detected with anti-PARP1 Ab. Western blot assay with anti-ERα Ab showed the efficiency of ERα siRNA. B, co-immunoprecipitation assay of PARP1-bound proteins or poly(ADP-ribosyl)ated proteins from non-treated VSMCs, followed by Western blot assay using anti-ERα antibody (i). Co-immunoprecipitation assay of ERα-bound proteins from VSMCs, followed by Western blot assay using anti-PARP1 antibody (ii). Nonspecific IgG served as negative control. C, in a cell-free system, the binding of ERα protein to UP-PARP1 protein (i) or AP-PARP1 protein (ii) was analyzed by far-Western blot (described under “Experimental Procedures”). β-Actin protein was used as negative control. D, diagram of GFP-tagged human ERα with its domains. Fragments A–C with their amino acid coordinates are listed. Co-immunoprecipitation assays demonstrated specific binding of ERα to the DBD of ERα. E, diagram of Flag-tagged human PARP1 with its domains, DBD, nuclear localization signal (NLS), BRCA1 C terminus (BRCT)/automodification domain (AMD), and catalytic domain (CD). Fragments A–F with their amino acid coordinates are listed. Co-immunoprecipitation assays demonstrated specific binding of ERα to the BRCT/AMD of PARP1. The results are from three independent experiments.
ERα Is Poly(ADP-ribosyl)ated by PARP1
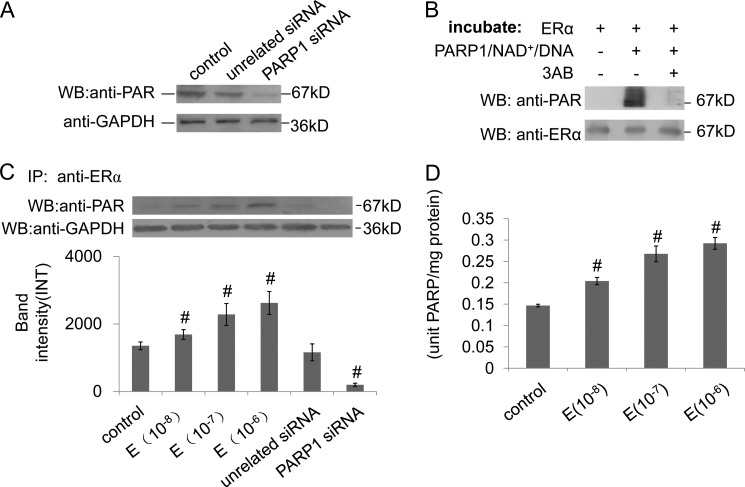
Poly(ADP-ribosyl)ation is an important post-transcriptional modification of proteins. We then detected whether or not ERα was poly(ADP-ribosyl)ated in VSMCs using IP assay with the antibody specific for poly(ADP-ribose) polymer (PAR). Western blot assay revealed that ERα was poly(ADP-ribosyl)ated (Fig. 2Bi). Knockdown of PARP1 by siRNA dramatically decreased the poly(ADP-ribosyl)ation of ERα in VSMCs, indicating that ERα was poly(ADP-ribosyl)ated by PARP1 (Fig. 3A). To investigate the role of the enzymatic activity of PARP1 in mediating ERα poly(ADP-ribosyl)ation, we used recombinant proteins in a cell-free system. Incubation of recombinant ERα protein with recombinant PARP1 protein, NAD+ and active DNA led to strong poly(ADP-ribosyl)ation of ERα; however, addition of 3AB prevented poly(ADP-ribosyl)ation of ERα (Fig. 3B). The evidence established that ERα was poly(ADP-ribosyl)ated by PARP1. IP assay with antibody specific for ERα was then carried out. Western blot assay with anti-PAR antibody revealed that estradiol treatment increased the amount of eluted poly(ADP-ribosy)lated ERα in the dose-dependent manner, while knockdown of PARP1 by siRNA abogated the eluted poly(ADP-ribosy)lated ERα (Fig. 3C), suggesting that estradiol promoted PARP1 activation and ERα poly(ADP-ribosy)lation. In line with this result, treatment with estradiol increased enzymatic activity of PARP in the dose-dependent manner in VSMCs (Fig. 3D).
FIGURE 3.
ERα is poly(ADP-ribosy)lated by PARP1. A, VSMCs were transfected with PARP1 siRNA (50 nm) or unrelated siRNA (50 nm) for 48 h. The poly(ADP-ribosyl)ation levels in VSMCs were determined by Western blotting with anti-PAR Ab. GAPDH was used for standardization. B, in a cell-free system, recombinant ERα protein was incubated with vehicles, PARP1/NAD+/active DNA, or PARP1/NAD+/active DNA/3AB, respectively. Poly(ADP-ribosyl)ation levels of recombinant ERα were determined by Western blotting with anti-PAR Ab. Western blotting with anti-ERα Ab showed the loading control. The results are from three independent experiments. C, VSMCs were treated with estradiol (E, 10−8, 10−7, 10−6 m, 24 h), or transfected with PARP1 siRNA (50 nm, 48 h) or unrelated siRNA (50 nm, 48 h). Immunoprecipitation (IP) of ERα from VSMCs treated as indicated, followed by Western blot assay using anti-PAR antibody. D, VSMCs were treated with estradiol (E, 10−8, 10−7, 10−6 m, 24 h). PARP activity in VSMCs was assayed as described under “Experimental Procedures.” The results are from six independent experiments. Data are expressed as the mean ± S.E. #, p < 0.05 is for comparison with control group.
Poly(ADP-ribosyl)ation Promotes ERα Binding to ERE in the Nucleus
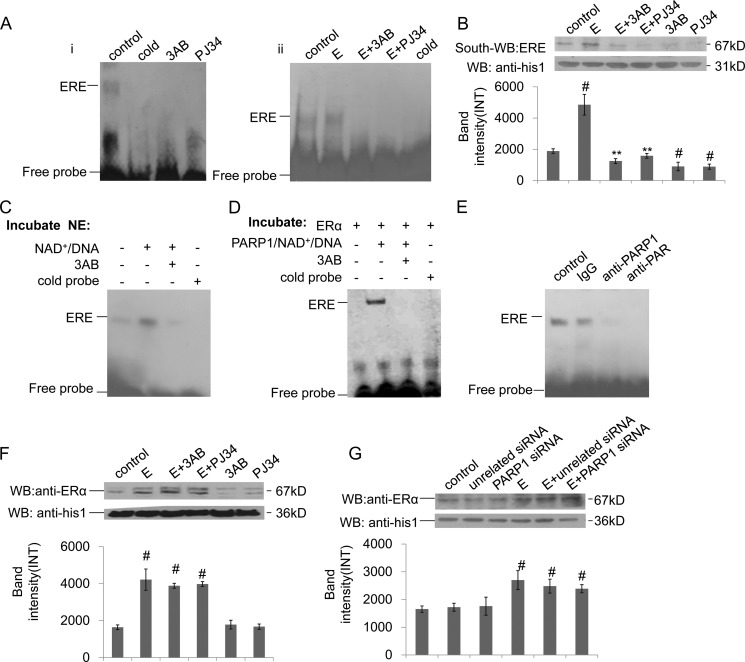
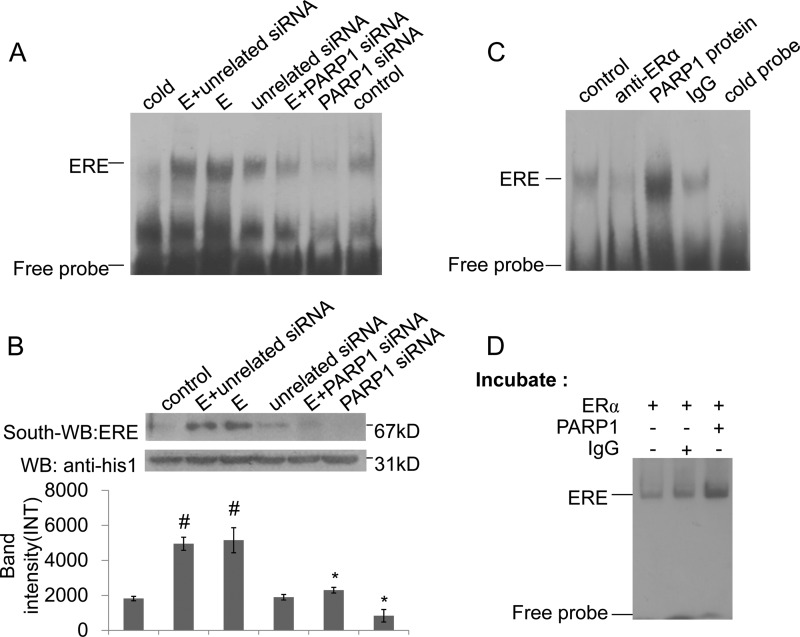
ERα can directly bind to ERE in the promoter of target gene, and the classical pathway for ERα signaling is mediated by ERα binding to ERE (21). To investigate the influence of PARP1 on ERα-ERE formation, we performed EMSA and Southwestern blot assays. Results showed that inhibition of PARP activity by 3AB or PJ34 prevented the binding of ERα to ERE in VSMCs (Fig. 4, Ai and B). Estradiol, which promoted PARP1 activation, increased ERα binding to ERE; and treatment of VSMCs with PARP inhibitor prevented this increase (Fig. 4, Aii and B). These results suggested that the activation of PARP1 promoted ERα-ERE complex formation. Thereby, the direct influence of poly(ADP-ribosyl)ation on the binding of ERα to ERE was investigated. The EMSA assay showed that incubation of nuclear extracts from non-treated VSMCs with NAD+ and active DNA increased ERα-ERE complex formation; while this increase was inhibited by co-incubation with 3AB (Fig. 4C). Similar results were obtained in a cell-free system using recombinant proteins (Fig. 4D). All the data indicated that poly(ADP-ribosyl)ation increased ERα binding to ERE. In line with this finding, a supershift assay showed that incubation of nuclear extracts with anti-PAR antibody could abrogate the band of ERα-ERE complex, while incubation with IgG failed to abrogate or shift the band of the ERα-ERE complex (Fig. 4E).
FIGURE 4.
Poly(ADP-ribosyl)ation promotes ERα binding to ERE in the nucleus. A, binding of ERα to ERE was detected by EMSA assay. (i) VSMCs were treated with 3AB (10 mm, 24 h) or PJ34 (10 mm, 24 h). (ii) VSMCs were treated with 3AB (10 mm, 24 h) or PJ34 (10 mm, 24 h) in the presence of estradiol (E, 10−7 m, 24 h). B, binding of ERα to ERE was detected by Southwestern blot assay. VSMCs were treated with 3AB (10 mm, 24 h) or PJ34 (10 mm, 24 h) in the absence or presence of estradiol (E, 10−7 m, 24 h). C, nuclear extracts from non-treated VSMCs were incubated with vehicles, PARP1/NAD+/active DNA, or PARP1/NAD+/active DNA/3AB, respectively. Binding of ERα to ERE was detected by EMSA assay. D, in a cell-free system, recombinant ERα protein was incubated with vehicles, PARP1/NAD+/active DNA, or PARP1/NAD+/active DNA/3AB, respectively. Binding of ERα to ERE was detected by EMSA assay. E, supershift assay was performed as described under “Experimental Procedures.” Nuclear extracts from non-treated VSMCs were incubated with anti-PAR Ab, anti-PARP1 Ab, or vehicles. F, nuclear expression of ERα was detected by Western blot assay. VSMCs were treated with 3AB (10 mm, 24 h) or PJ34 (10 mm, 24 h) in the absence or presence of estradiol (E, 10−7 m, 24 h). G, nuclear expression of ERα was detected by Western blot assay. VSMCs were transfected with PARP1 siRNA (50 nm, 48 h) or unrelated siRNA (50 nm, 48 h), followed by treatment with estradiol (E, 10−7 m, 24 h). Data are expressed as the mean ± S.E. #, p < 0.05 and ##, p < 0.01 are for comparison with control group. *, p < 0.05 and **, p < 0.01 are for comparison with estradiol group (E).
The nuclear accumulation of ERα also influenced the ERα-ERE complex formation in the nucleus; we thus analyzed the nuclear expression of ERα in VSMCs using Western blot assay. We found that treatment with estradiol resulted in the increased nuclear accumulation of ERα, while inhibition of PARP1 did not influence its nuclear accumulation in cells treated with or without estradiol (Fig. 4, F and G).
Poly(ADP-ribosyl)ation Enhances ERα Recruitment to the IGF-1 Promoter
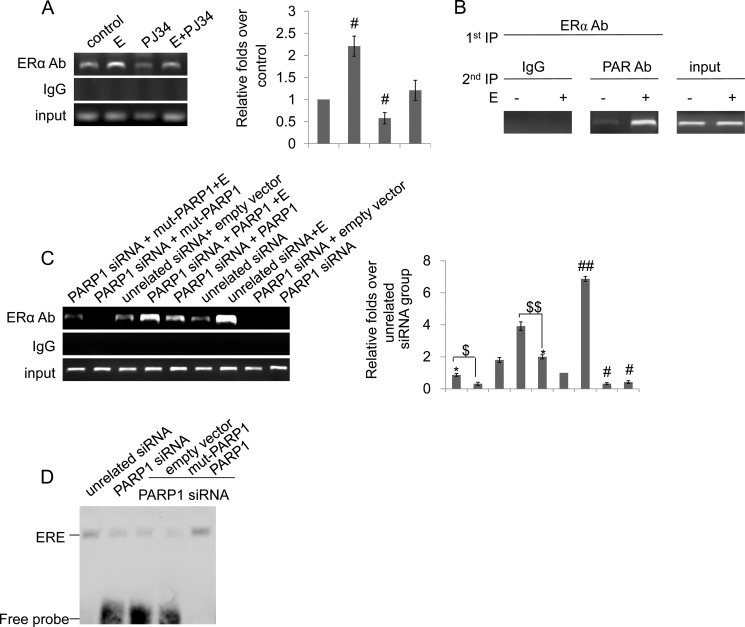
We further examined the influence of poly(ADP-ribosyl)ation on the recruitment of ERα to the promoter of IGF-1, a target gene of ERα, by ChIP assays. Enrichment of the promoter fragments in ChIP using anti-ERα antibody versus nonspecific IgG was examined by real-time PCR. Results showed that inhibition of PARP activity by PJ34 attenuated the recruitment of ERα to the IGF-1 promoter (Fig. 5A), suggesting that poly(ADP-ribosyl)ation enhanced ERα recruitment to the IGF-1 promoter. To test whether ERα with poly(ADP-ribosyl)ation is indeed associated with IGF-1 promoter, we performed re-ChIP assays using anti-ERα antibody and anti-PAR antibody. The result suggested that poly(ADP-ribosyl)ated ERα was recruited to the IGF-1 promoter (Fig. 5B).
FIGURE 5.
Poly(ADP-ribosyl)ation enhances ERα recruitment to the IGF-1 promoter. A, ChIP-PCR assay using anti-ERα antibody for amplification of IGF-1 promoters in VSMCs. Cells were treated with estradiol (E, 10−7 m, 24 h), PJ34 (10 mm, 24 h), or E+PJ34. B, in re-ChIP assays, chromatin was first immunoprecipitated with anti-ERα antibody and then, re-immunoprecipitated with anti-PAR antibody or IgG. C, ChIP-PCR assay using anti-ERα antibody for amplification of IGF-1 promoters in VSMCs. Cells were transfected with PARP1 siRNA (50 nm) or unrelated siRNA (50 nm) for 24 h, and then were treated with empty vector (p3flag-CMV), full-length (hPARP1) or the plasmid expressing an enzymatically inactive PARP1 protein (mut-PARP1) for 48 h in the absence or presence of estradiol (E, 10−7 m). D, binding of ERα to ERE was detected by EMSA assay. VSMCs were transfected with PARP1 siRNA (50 nm) or unrelated siRNA (50 nm) for 24 h, and then were treated with empty vector (p3flag-CMV), full-length (hPARP1), or the plasmid expressing an enzymatically inactive PARP1 protein (mut-PARP1) for 48 h. Data are expressed as the mean ± S.E. In Fig. 5A, #, p < 0.05 is for comparison with control group. In Fig. 5C, #, p < 0.05 and ##, p < 0.01 are for comparison with unrelated siRNA group. *, p < 0.05 is for comparison with PARP1 siRNA group. $$, p < 0.05 is for the indicated comparison.
To investigate whether ERα poly(ADP-ribosyl)ation by PARP1 are required for ERα binding to DNA and ERα transcriptional function, we constructed enzyme-defective mutants of PARP1 (mut-PARP1) in which lysine 893 was substituted with isoleucine (K893I). We first knocked down PARP1 by siRNA in VSMCs, and transfected the cells with either the mut-PARP1 or the full-length PARP1 vector. Our ChIP data suggested that transfection of mut-PARP1 in PARP1 knockdown cells led to significantly decreased recruitment of ERα to the IGF-1 promoter under either basal or estradiol-treated conditions, while transfection of full-length PARP1 in PARP1 knockdown cells induced recruitment of ERα to the target promoter (Fig. 5C). In line with this result, we found that the PARP1 knockdown cells transfected with mut-PARP1 showed a much lower level of the ERα-ERE complex than that shown in the full-length PARP1-transfected cells (Fig. 5D). These results indicated that ERα poly(ADP-ribosyl)ation by PARP1 was required for ERα binding to DNA and ERα transcriptional function.
PARP1 Is an Indispensable Component of the ERα-ERE Complex
Previous studies from our laboratory and others have shown that PARP1 modulates transactivation of several transcription factors through physical interaction. To investigate whether the physical interaction with PARP1 influenced ERα binding to ERE, we knocked down endogenous PARP1 by siRNA in VSMCs. EMSA and Southwestern blot assays showed that knockdown of PARP1 by siRNA not only prevented estradiol-induced increases in ERα binding to ERE but also inhibited binding of ERα to ERE in VSMCs under basic conditions (Fig. 6, A and B). ChIP assay also showed that depletion of the PARP1 gene by siRNA attenuated the recruitment of ERα to the target promoter (Fig. 5C). To explore the underlying mechanism, a supershift assay was carried out. Results showed that incubation of nuclear extracts from VSMCs with anti-PARP1 antibody abrogated the band of the ERα-ERE complex (Fig. 4E), indicating that PARP1 was an intrinsic component of the complex. Thereafter, the direct influences of physical interaction between PARP1 and ERα on ERα-ERE complex formation were explored. The EMSA assay showed that incubation of nuclear extracts from VSMCs with the PARP1 protein resulted in increased ERα-ERE complex formation (Fig. 6C). In line with this result, incubation of ERα with PARP1 protein promoted ERα-ERE complex formation in a cell-free system (Fig. 6D). These results suggested that the interaction with PARP1 enhanced the DNA binding activity of ERα.
FIGURE 6.
PARP1 is an indispensable component of ERα-ERE complex. A and B, binding of ERα to ERE was detected by EMSA (A) and Southwestern blot assay (B). VSMCs were transfected with PARP1 siRNA (50 nm) or unrelated siRNA (50 nm) for 48 h, followed by treatment with estradiol (E, 10−7 m, 24 h). C, binding of ERα to ERE was detected by EMSA assay. Nuclear extracts from non-treated VSMCs were incubated with PARP1 protein or IgG (served as negative control). Incubation of nuclear extracts with anti-ERα Ab showed the specificity of the ERα-ERE complex band. D, in a cell-free system, recombinant ERα protein was incubated with vehicles, PARP1 protein, or IgG (served as negative control), respectively. The binding of ERα to ERE was detected by EMSA assay. Results are from three independent experiments.
DISCUSSION
PARP1 is the dominant member of the PARP family. It has been reported to interact with a variety of nuclear-located transcription factors and thereby alter their regulatory function (9, 15–18). This ability of PARP1 is certainly important as it will ultimately change the transcriptional pattern of numerous genes (22). ERα belongs to nuclear receptors that are thought to mediate their transcriptional effects in concert with co-regulator proteins that modulate receptor interactions with components of the basal transcription machinery. In this study, we showed that ERα could bind to PARP1 in the absence of DNA or any other cofactors, implying that ERα interacted directly with PARP1. Given that PARP1 was also an intrinsic component of the ERα-ERE complex, the physical interaction between PARP1 and ERα might function importantly to the binding of ERα to ERE. In line with this notion, incubation of recombinant ERα protein or nuclear extracts with PARP1 increased the binding of ERα to ERE. Thereby, association with PARP1 increased the DNA binding capacity of ERα. PARP1 is a 116-kDa protein consisting of three main domains: the N-terminal DNA-binding domain containing two zinc fingers, the auto-modification domain (AMD), and the C-terminal catalytic domain. We further investigated the interacting domains of ERα and PARP1. By using the ERα deletion mutants, we observed that endogenous PARP1 associated specifically with the DBD of ERα. It has been reported that the BRCA1 C terminus (BRCT)/AMD of PARP1 is involved in directing protein-protein interactions (23). Consistent with such a notion, we localized the ERα-binding site to the central BRCT/AMD of PARP1 by using PARP1 deletion mutants.
Nuclear receptors serve as integrating platforms for a variety of stimuli and are the targets for post-translational modifications, such as phosphorylation, ubiquitination, acetylation, sumolyation, and poly(ADP-ribosyl)ation (24, 25). Poly(ADP-ribosyl)ation is an important post-transcriptional modification of protein catalyzed by PARP enzymes, especially PARP1. During poly(ADP-ribosyl)ation, PARP1 synthesizes polymer of ADP-ribose (PAR) from NAD+ to target proteins to change their respective structure and DNA binding activity (26). An increasing number of transcriptional co-factors of PARP1 are being reported every year, and some of them have been shown to be poly(ADP-ribosyl)ated by PARP1 (15, 16, 27, 28). In some cases, poly(ADP-ribosyl)ation is found to be required for the activation of some transcription factors, while in other cases it is not (16, 29, 30). In the present study, ERα could be poly(ADP-ribosyl)ated by PARP1 in VSMCs. Poly(ADP-ribosyl)ation promoted ERα-ERE complex formation in nuclear extracts and increased the DNA binding capacity of ERα in a cell-free system, suggesting that poly(ADP-ribosyl)ation of ERα is very important for its transactivation. In line with this notion, inhibition of PARP1 activity not only prevented the binding of ERα-ERE complex formation in nuclear extracts, but also inhibited ERα transactivation and dependent gene transcription in VSMCs. To investigate whether the ERα poly(ADP-ribosyl)ation by PARP1 was required for ERα binding to DNA and ERα transcriptional function, we constructed an enzyme-defective mutants of PARP1 (mut-PARP1) in which lysine 893 was substituted with isoleucine (K893I). We first knocked down PARP1 by siRNA in VSMCs, and transfected the cells with either the mut-PARP1 or the full-length PARP1 vector. Then we compared the ERα binding to DNA and ERα transcriptional function between the mut-PARP1 and full-length PARP1 transfected cells by ChIP and EMSA assays. Our ChIP data suggested that transfection of mut-PARP1 in PARP1 knockdown cells led to significantly decreased recruitment of ERα to the IGF-1 promoter under either basal or estradiol-treated conditions, while transfection of full-length PARP1 in PARP1 knockdown cells induced recruitment of ERα to the target promoter. By using the EMSA assay, we found that the PARP1-knockdown cells transfected with mut-PARP1 showed a much lower level of ERα-ERE complex than that shown in the full-length PARP1-transfected cells. These results indicated that ERα poly(ADP-ribosyl)ation by PARP1 was required for ERα binding to DNA and ERα transcriptional function. Moreover, we demonstrated that incubation of nuclear extracts with anti-PARP1 antibody abrogated the band of the ERα-ERE complex; and incubation of nuclear extracts from VSMCs or recombinant ERα protein with PARP1 protein resulted in the increased ERα-ERE complex formation. These data indicated that PARP1 also served as an indispensible component for the formation of the ERα-ERE complex and ERα-dependent transcription by directly interacting with ERα. Thus, both the poly(ADP-ribosyl)ation and physical interaction are required for ERα binding to DNA and ERα transcriptional activation.
Nucleocytoplasmic shuttling of transcription factors plays a critical role in the transcriptional regulation of target genes (31). ERα exerts its function through translocation from the cytoplasm to the nucleus. It has been demonstrated that the nucleocytoplasmic shuttling of ERα is mainly regulated by estradiol (32). In this study, we also analyzed the nuclear accumulation of ERα in VSMCs. Although we found that estradiol treatment resulted in the increased nuclear accumulation of ERα, inhibition of PARP1 did not influence its nuclear accumulation in cells treated with or without estradiol. These results indicated that the influence of estradiol on ERα nucleocytoplasmic shuttling was not mediated through the PARP1 pathway.
Many studies have shown that both natural and synthetic estrogens have vasoprotective effects. Estrogen has been demonstrated to directly modulate VSMC expression of genes controlling migration of adventitial fibroblasts via an ERα-dependent mechanism (33). We tested the influence of estradiol treatment on PARP activity in VSMC cells. Our results showed that the estradiol treatment increased PARP activity and enhanced the poly(ADP-ribosyl)ation of ERα in a dose-dependent manner. Further studies demonstrated that inhibition of PARP1 prevented estradiol-induced ERα transactivation and -dependent gene transcription. These findings suggested that estradiol treatment increased the binding of ERα to ERE- and ERα-dependent gene transcription through activating PARP1.
In summary, the data reveal a novel mechanism underlying ERα transactivation in VSMCs. PARP1 can bind to and poly(ADP-ribosyl)ate ERα in the nucleus. Poly(ADP-ribosyl)ation of ERα by PARP1 increases the DNA binding capacity of ERα and thereby, promotes ERα-dependent gene transcription. PARP1 also served as an indispensible component for the formation of the ERα-ERE complex and ERα-dependent transcription by directly interacting with ERα. Activation of PARP1 mediates the estradiol-induced transcriptional activation of ERα. These findings indicate that PARP1 is a crucial regulator of ERα-dependent gene transcription. Moreover, data also suggest that PARP1 might be a promising therapeutic target for proliferative vascular diseases and other diseases induced by dysregulation of ERα signaling.
Acknowledgment
We thank Dr. Ming-Jer Tsai for kindly providing plasmids.
This work was supported by the National Natural Science Foundation of China (30971245 and 81170239, to K. H. and 81000112, to D.H.).
- NR
- nuclear receptor
- 3AB
- 3-aminobenzamide
- AP-PARP1
- auto-poly(ADP-ribosy)lated PARP1
- ERα
- estrogen receptor α
- ERE
- estrogen response element
- HDACs
- histone deacetylases
- IGF-1
- insulin-like growth factors-1
- NAD+
- nicotinamide adenine dinucleotide
- PARP1
- poly(ADP-ribose) polymerase 1
- PAR
- poly(ADP-ribose) polymer
- PJ34
- N-(6-oxo-5,6-dihydro-phenanthridin-2-yl)-2-(N,N-dimethylamino)acetamide
- TGF-α
- transforming growth factor-α
- UP-PARP1
- un-poly(ADP-ribosy)lated PARP1
- VSMCs
- vascular smooth muscle cells
- AMD
- auto-modification domain
- BRCT
- BRCA1 C terminus.
REFERENCES
- 1. Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schütz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R. M. (1995) The nuclear receptor superfamily: the second decade. Cell 83, 835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Smirnov A. N. (2002) Nuclear receptors: nomenclature, ligands, mechanisms of their effects on gene expression. Biochemistry 67, 957–977 [DOI] [PubMed] [Google Scholar]
- 3. Glass C. K., Rosenfeld M. G. (2000) The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14, 121–141 [PubMed] [Google Scholar]
- 4. Benecke A., Chambon P., Gronemeyer H. (2000) Synergy between estrogen receptor alpha activation functions AF1 and AF2 mediated by transcription intermediary factor TIF2. EMBO Rep. 1, 151–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen W., Roeder R. G. (2011) Mediator-dependent nuclear receptor function. Semin. Cell Dev. Biol. 22, 749–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boyer T. G., Martin M. E., Lees E., Ricciardi R. P., Berk A. J. (1999) Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature 399, 276–279 [DOI] [PubMed] [Google Scholar]
- 7. Lewis B. A., Reinberg D. (2003) The mediator coactivator complex: functional and physical roles in transcriptional regulation. J. Cell Sci. 116, 3667–3675 [DOI] [PubMed] [Google Scholar]
- 8. Taatjes D. J., Tjian R. (2004) Structure and function of CRSP/Med2; a promoter-selective transcriptional coactivator complex. Mol. Cell 14, 675–683 [DOI] [PubMed] [Google Scholar]
- 9. D'Amours D., Desnoyers S., D'Silva I., Poirier G. G. (1999) Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 342, 249–268 [PMC free article] [PubMed] [Google Scholar]
- 10. Virág L., Szabó C. (2002) The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol. Rev. 54, 375–429 [DOI] [PubMed] [Google Scholar]
- 11. Kim M. Y., Zhang T., Kraus W. L. (2005) Poly(ADP-ribosyl)ation by PARP-1: 'PAR-laying' NAD+ into a nuclear signal. Genes Dev. 19, 1951–1967 [DOI] [PubMed] [Google Scholar]
- 12. Kraus W. L. (2008) Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr. Opin. Cell Biol. 20, 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ba X., Gupta S., Davidson M., Garg N. J. (2010) Trypanosoma cruzi induces the reactive oxygen species-PARP-1-RelA pathway for up-regulation of cytokine expression in cardiomyocytes. J. Biol. Chem. 285, 11596–11606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim M. Y., Mauro S., Gévry N., Lis J. T., Kraus W. L. (2004) NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell 119, 803–814 [DOI] [PubMed] [Google Scholar]
- 15. Huang D., Wang Y., Yang C., Liao Y., Huang K. (2009) Angiotensin II promotes poly(ADP-ribosyl)ation of c-Jun/c-Fos in cardiac fibroblasts. J. Mol. Cell Cardiol. 46, 25–32 [DOI] [PubMed] [Google Scholar]
- 16. Huang D., Yang C., Wang Y., Liao Y., Huang K. (2009) PARP-1 suppresses adiponectin expression through poly(ADP-ribosyl)ation of PPARγ in cardiac fibroblasts. Cardiovasc. Res. 81, 98–107 [DOI] [PubMed] [Google Scholar]
- 17. Cohen-Armon M., Visochek L., Rozensal D., Kalal A., Geistrikh I., Klein R., Bendetz-Nezer S., Yao Z., Seger R. (2007) DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: a link to histone acetylation. Mol. Cell 25, 297–308 [DOI] [PubMed] [Google Scholar]
- 18. Butler A. J., Ordahl C. P. (1999) Poly(ADP-ribose) polymerase binds with transcription enhancer factor 1 to MCAT1 elements to regulate muscle-specific transcription. Mol. Cell Biol. 19, 296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Simonin F., Ménissier-de Murcia J., Poch O., Muller S., Gradwohl G., Molinete M., Penning C., Keith G., de Murcia G. (1990) Expression and site-directed mutagenesis of the catalytic domain of human poly(ADP-ribose)polymerase in Escherichia coli. Lysine 893 is critical for activity. J. Biol. Chem. 265, 19249–19256 [PubMed] [Google Scholar]
- 20. Xu W., Cho H., Evans R. M. (2003) Acetylation and methylation in nuclear receptor gene activation. Methods Enzymol. 364, 205–223 [DOI] [PubMed] [Google Scholar]
- 21. Glidewell-Kenney C., Weiss J., Lee E. J., Pillai S., Ishikawa T., Ariazi E. A., Jameson J. L. (2005) ERE-independent ERα target genes differentially expressed in human breast tumors. Mol. Cell Endocrinol. 245, 53–59 [DOI] [PubMed] [Google Scholar]
- 22. Zaniolo K., Desnoyers S., Leclerc S., Guérin S. L. (2007) Regulation of poly(ADP-ribose) polymerase-1 (PARP-1) gene expression through the post-translational modification of Sp1: a nuclear target protein of PARP-1. BMC Mol. Biol. 8, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Paddock M. N., Buelow B. D., Takeda S., Scharenberg A. M. (2010) The BRCT domain of PARP-1 is required for immunoglobulin gene conversion. PLoS Biol. 8, e1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lalevée S., Ferry C., Rochette-Egly C. (2010) Phosphorylation control of nuclear receptors. Methods Mol. Biol. 647, 251–266 [DOI] [PubMed] [Google Scholar]
- 25. Miyamoto T., Kakizawa T., Hashizume K. (1999) Inhibition of nuclear receptor signalling by poly(ADP-ribose) polymerase. Mol. Cell Biol. 19, 2644–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schreiber V., Dantzer F., Ame J. C., de Murcia G. (2006) Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 7, 517–528 [DOI] [PubMed] [Google Scholar]
- 27. Huang D., Wang Y., Wang L., Zhang F., Deng S., Wang R., Zhang Y., Huang K. (2011) Poly(ADP-ribose) polymerase 1 is indispensable for transforming growth factor-β Induced Smad3 activation in vascular smooth muscle cell. PLoS One 6, e27123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Almeida K. H., Sobol R. W. (2007) A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair 6, 695–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kotova E., Jarnik M., Tulin A. V. (2009) Poly (ADP-ribose) polymerase 1 is required for protein localization to Cajal body. PLoS Genet. 5, e1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang D., Yang C. Z., Yao L., Wang Y., Liao Y. H., Huang K. (2008) Activation and overexpression of PARP-1 in circulating mononuclear cells promote TNF-α and IL-6 expression in patients with unstable angina. Arch. Med. Res. 39, 775–784 [DOI] [PubMed] [Google Scholar]
- 31. Xu L., Massagué J. (2004) Nucleocytoplasmic shuttling of signal transducers. Nat. Rev. Mol. Cell Biol. 5, 209–219 [DOI] [PubMed] [Google Scholar]
- 32. Lombardi M., Castoria G., Migliaccio A., Barone M. V., Di Stasio R., Ciociola A., Bottero D., Yamaguchi H., Appella E., Auricchio F. (2008) Hormone-dependent nuclear export of estradiol receptor and DNA synthesis in breast cancer cells. J. Cell Biol. 182, 327–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li G., Chen Y. F., Greene G. L., Oparil S., Thompson J. A. (1999) Estrogen inhibits vascular smooth muscle cell-dependent adventitial fibroblast migration in vitro. Circulation 100, 1639–1645 [DOI] [PubMed] [Google Scholar]