Abstract
The microtubule cytoskeleton is involved in regulation of cell morphology, differentiation, and cell cycle progression. Precisely controlled dynamic properties are required for these microtubule functions. To better understand how tubulin's dynamics are embedded in its primary sequence, we investigated in vivo the consequences of altering a single, highly conserved residue in β-tubulin that lies at the interface between two structural domains. The residue differs between the cold-adapted Antarctic fish and temperate animals in a manner that suggests a role in microtubule stability. Fungi, like the Antarctic fish, have a phenylalanine in this position, whereas essentially all other animals have tyrosine. We mutated the corresponding residue in fission yeast to tyrosine. Temperature effects were subtle, but time-lapse microscopy of microtubule dynamics revealed reduced depolymerization rates and increased stability. Mitotic exit signaled by breakdown of the mitotic spindle was delayed. In meiosis, microtubules displayed prolonged contact to the cell cortex during horsetail movement, followed by completion of meiosis I but frequent asymmetric failure of meiosis II spindle formation. Our results indicate that depolymerization dynamics modulated through interdomain motion may be important for regulating a subset of plus-end microtubule complexes in Schizosaccharomyces pombe.
INTRODUCTION
The microtubule cytoskeleton is essential for normal eukaryotic cell function, cellular differentiation, and multicellular development. Dramatic changes in microtubule organization and dynamics occur at cellular transitions that include progression from interphase through mitosis (Mitchison and Salmon, 2001; Rusan et al., 2001), spindle rotation in early embryonic divisions in animal cells (Ahringer, 2003), and the formation of specialized microtubule arrays upon mitotic exit or during the meiotic horsetail movement in fission yeast (Hagan and Hyams, 1988; Yamamoto et al., 2001).
Cyclic polymerization and depolymerization of microtubules, known as dynamic instability, is required for many cytoskeletal functions (Desai and Mitchison, 1997). Tubulin primary sequence directly affects its conformational stability and presumably interactions between tubulin dimers that can modulate microtubule dynamic parameters. A factor contributing to differences in dynamics is the presence of several tubulin isotypes in cells with up to 15% sequence variations (Murphy et al., 1987) and distinctive dynamic properties (Panda et al., 1994; Bode et al., 2003). Cellular microtubule dynamics are also sensitive to single mutations in tubulin (Oakley, 1985; Gupta et al., 2001, 2002) and are affected by drugs that target tubulins (Downing, 2000). Understanding how microtubule dynamics are embedded in tubulin sequence is still in its infancy. This knowledge will be essential for a full appreciation of cytoskeletal mechanisms and for advancement of drug development for cancer treatment and for anthelminths, fungicides, and herbicides in agriculture.
α- and β-Tubulins form heterodimeric building blocks of microtubules, and each display ∼75% conservation in sequence within each family (Burns and Surridge, 1993). This similarity is presumed to derive in part from sequence constraints imposed by various microtubule structural requirements and from the interaction of tubulin with a large number of nontubulin proteins. Tubulin-associated proteins include those necessary for tubulin folding, dimer formation, and microtubule nucleation (Schiebel, 2000; Lopez-Fanarraga et al., 2001; Nogales, 2001) as well as proteins that influence assembled microtubule dynamics or organization (Hunter and Wordeman, 2000; Cassimeris and Spittle, 2001; Schroer, 2001; Cassimeris, 2002). Regions of tubulin that show almost complete conservation include surfaces involved in GTP binding, longitudinal interactions, and binding of kinesin-like proteins (Nogales, 2001; Löwe et al., 2001). The relative rates of evolution of sites within the core of tubulin seem to be faster than those on the surface (Blouin et al., 2003), but there is remarkable conservation of sequence from the lowest to highest eukaryotes.
Some differences among tubulins involve adaptations to environment, including the ability of microtubules to function at low temperatures (Detrich et al., 1989, 1992; Wallin and Stromberg, 1995; Nyporko et al., 2003). Purified mammalian tubulins form stable microtubules at temperatures >30°C, whereas tubulins from some psychrophiles, such as Antarctic fish that thrive at temperatures of –2–1°C, form microtubules at temperatures close to freezing (Detrich et al., 1989, 1992, 2000; Sidell, 2000). Phylogenetic analysis of β-tubulins from Antarctic fish has pinpointed the residue at position 200 as the single consistent sequence difference versus β-tubulins of homeotherms (Detrich et al., 2000). All of the tubulin isotypes sequenced from Antarctic fish have phenylalanine at this position, distinct from almost all other animals that have tyrosine. In fungi, which also have moderately cold-stable microtubules, a phenylalanine is also present, and mutation to tyrosine has been associated with increased resistance to benzimidazoles (Aspergillus nidulans, Jung et al., 1992; Botrytis cinerea, Yarden and Katan, 1993). The residue is positioned at an internal interface between tubulin domains. It has been hypothesized that movement of these domains is a principal conformational change required for depolymerization of microtubules (Amos and Löwe, 1999). A prediction of this hypothesis is that this residue could have an important role in microtubule stability. We have altered the corresponding residue in fission yeast as F200Y to investigate in vivo the importance of phenylalanine versus tyrosine on microtubule dynamic parameters, extreme temperature stability, and formation and function of distinct microtubule arrays. We discuss the impact of the mutation in regard to tubulin domain structure and cell cycle specific functions of cytoskeletal arrays.
MATERIALS AND METHODS
Sequence Comparison and Computational Analysis of Tubulin Structure
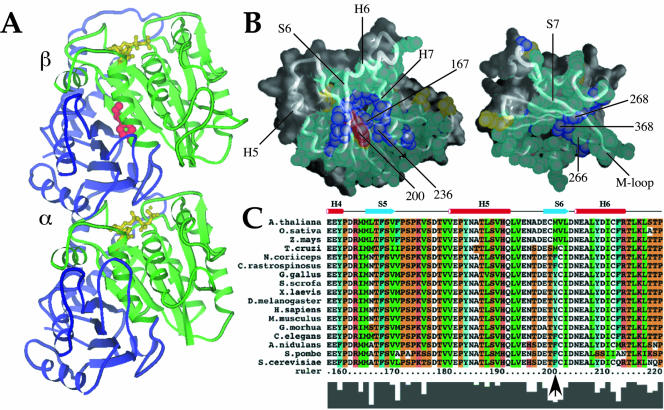
ClustalX (Thompson et al., 1997) was used to align 126 β-tubulin sequences obtained from the Swiss-Prot database. To conserve space in the alignment for Figure 1C, tubulin sequences from a smaller but representative number of species, including mainstream model organisms, are shown. Metazoan β-tubulins are sometimes described as belonging to a certain class type that is based in part on N- and C-terminal residues. Most sequenced Antarctic fish β-tubulins belong to class II, and all use phenylalanine at position 200. Among β-tubulins from warm-blooded animals, tyrosine is nearly ubiquitous at this position; however, a phenylalanine residue is present in at least one known sequence. We determined that the choice of phenylalanine versus tyrosine was not more frequent in any class. We also examined Schizosaccharomyces pombe β-tubulin in regard to class types, but it did not fit clearly within any defined class. Protein regions examined for this classification included N-terminal residues 33–57 and C-terminal residues beyond residue 430 (according to Sullivan and Cleveland, 1986). We did observe that S. pombe β-tubulin is most similar to chicken class VI β-tubulin, a divergent isotype that uses phenylalanine. The model of the tubulin dimer displayed in Figure 1A was prepared using Molscript (Kraulis, 1991). In Figure 1B, we used the program GRASP (Nicholls, 1991). The crystal coordinates of the 3.5-Å structure of tubulin (PDB ID 1JFF) are from Löwe et al. (2001).
Figure 1.
A conserved internal residue corresponding to position 200 in mammalian β-tubulin is positioned to affect interdomain motion. (A and B) Diagram of α/β tubulin dimer from the crystal structure database. Green, nucleotide binding domain comprising residues 1–240; red, residue 200; yellow, nucleotide; blue, remainder of protein. (B) Contacts made between the nucleotide-binding (left) and intermediate (right) domains of porcine β-tubulin. Atoms in residues involved in contacts are shown as spheres. Backbone worm is superimposed to indicate secondary structure with selected helices and sheets labeled for orientation. Residues colored in aqua are highly conserved among all known β-tubulins (126 sequences from Swiss Prot); purple residues show only conservative substitutions; yellow residues show nonconservative substitutions. Residue 200 falls into this last class and is colored red for distinction. Numbered atoms are those implicated in benzimidazole binding. Alpha helices H5, H6, and H7, beta strands S6 and S7, and the M-loop (Löwe et al., 2001) are indicated. (C) Primary β-tubulin sequences from representative eukaryotes showing the immediate region of interest: plants (A. thalania, Z. mays, O. Sativa), human (H. sapiens), mouse (M. musculus), chicken (G. gallus), pig (S. scrofa), frog (X. laevis), fly (D. melanogaster), psychrotolerant fish (G. morhua), worm (C. elegans), psychrophilic fish (N. coriiceps, C. rastrospinosus), and fungi (S. cerevisiae, A. nidulans, S. pombe). Numbering is according to human β-tubulin. The conserved phyla-specific residue corresponds to human Y200, Antarctic fish F200, and S. pombe F200 and is indicated by an arrow. Residues are colored as follows: orange, G, P, S, and T; red, H, K, and R; blue, W and Y; aqua, F; green, I, L, M, and V; and white, A, C, N, D, E, and Q. The histogram at the bottom of the sequences reflects conservation of amino acids at each position.
Strains, Media, and Temperature and Drug Tests
S. pombe strains used in this work are as follows: 972 h- wild type; YY105 [h90 ura4-D18, leu1-32, atb2::GFP] (Ding et al., 1998); AK101, AK102, and AK103 are described in a later section for generation of genomic F200Y; mad2D::ura4+ deletion strains mentioned in DISCUSSION were constructed by crossing [h+ ura4-D18, leu1-32, mad2D::ura4+] (He et al., 1997) with YY105 or strain AK101. The γ-tubulin gene fusion gtb1::GFP (Paluh et al., 2000) was integrated at the ars1 locus in YY105 and AK101 for examining γ-tubulin localization in meiosis. The h90 strains used are haploid but contain mating type information for both h+ and h- types of cells. Thus, any given population contains an isogenic mixture of h+ and h- cells. Cells grown at 32°C on rich media primarily undergo mitotic growth. High-frequency mating occurs when cells are moved to room temperature and minimal (MSA) media.
Genetic procedures, lithium acetate yeast transformation, rich media (YEA, yeast extract + adenine + casamino acids; or YES, YEA + supplements), and Edinburgh minimal media (EMM) or minimal sporulation media (MSA) are described in Moreno et al. (1991) and Egel et al. (1994). Adenine, 100 μg/ml, and other appropriate auxotrophic supplements were added to media as necessary (leucine, histidine, or uracil at 75 μg/ml). Both nonsynchronized cell populations and cells synchronized by 10 mM hydroxyurea as described previously (Moreno et al., 1991), were used. Temperature and drug sensitivity of cells tested on YES plates used logarithmic cultures grown to a cell density of 1 × 105 cells/ml.
Mutagenesis of β-Tubulin In Vitro
The β-tubulin gene nda3+ was amplified by polymerase chain reaction (PCR) from chromosomal DNA prepared from S. pombe strain 972 by using High Fidelity Taq polymerase (Roche Diagnostics, Indianapolis, IN). Forward and reverse oligonucleotide primers for PCR, (B5L) 5′-ATATATAACTGCAGAGGTTCGCTAATACGGGAAATTATTAAATCAATGCATGCTGGC-3′ and (B3L) 3′-ATATATAAGAGCTCAGCACAGACGATGCTTGGATCCATATGCATCGGAGT-5′ (Operon Technologies, Emeryville, CA), corresponded to flanking regions of β-tubulin initiating at positions (+538) 5′ and (-136) 3′. PstI and SacI sites incorporated into the primers allowed cloning of the amplified β-tubulin gene into the S. pombe integrable vector pRIP2/S, with coincident removal of the nmt1 promoter (pAKBF; Maundrell, 1993). The Gene-Editor in vitro site-directed mutagenesis kit (Promega, Madison, WI) and oligonucleotide 5′-GAGAACTCTGACGAAACATATTGCATTGATA-ATGAGG-3′ (Operon Technologies) were used to generate a phenylalanine to tyrosine point mutation at residue 200 in β-tubulin (designated F200Y; pAKBY). Presence of the single mutation versus wild-type nucleotide sequence was confirmed by sequencing (University of California Berkeley Sequencing Facility).
Generation of Genomic F200Y in S. pombe
Plasmid pAKBY (carrying nda3-F200Y) was linearized at an internal unique SpeI site and integrated at the nda3+ locus in YY105 via homologous recombination. Eight ura+ transformants, all of which showed similar microtubule phenotypes and cold sensitivity at 19°C, were obtained. Integration is expected to generate a single tandem repeat of nda3-F200Y and native nda3+ separated by plasmid sequences and the ura4+ gene (Figure 2A). One ura+ isolate was chosen for further characterization, termed AK102. It was used to generate strain AK101, which carries a single allele of mutant nda3-F200Y at the native locus, through counterselection of AK102 cells on 5-fluoroorotic acid (5-FOA) plates (Boeke et al., 1984). Ten 5-FOA–resistant colonies (5FOAR), which were expected to contain either a restored nda3+ wild-type allele or the nda3-F200Y allele, were transferred to rich plates (YEA) and replica plated onto selective plates (EMM + adenine + leucine + histidine) to confirm loss of the ura4+ gene. All 10 isolates failed to grow in the absence of supplemented uracil, as expected.
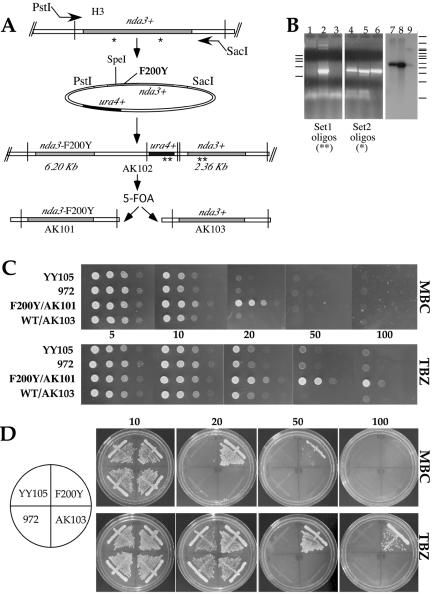
Figure 2.
Generation of S. pombe nda3-F200Y cells and resistance to antimicrotubule benzimidazole agents. (A) Flow chart of PCR cloning, generation of the mutation, and genome replacement of nda3-F200Y at the native nda3+ β-tubulin locus of YY105. Homologous recombination generates sequential β-tubulin genes (strain AK102). 5-FOA counterselection regenerates a single allele. Strains AK101 and AK103 contain either nda3-F200Y or nda3+ at the native locus, respectively. Primer pairs for PCR experiment in B are shown as asterisks in A, set 1 (*) and set 2 (**). HindIII sites used in Southern analysis are represented by vertical bars in A. See MATERIALS AND METHODS for more details to A and B. (B) PCR (lanes 1–6) and Southern analysis (lanes 7–9) of chromosomal DNA from integrants in A. F200Y/AK101, lanes 1, 5, and 7; AK102, lanes 2, 6, and 9; WT/AK103, lanes 3 and 8; and YY105 control, lane 4. DNA molecular weight standards: left bars (for PCR in kilobases): 9.4, 6.6, 4.4, 2.0, and 0.56. Right bars (for Southern in kilobases): 23.1, 9.4, 6.6, 6.1, 4.9, 4.4, 3.6, 2.3, 2.2, 1.2, and 0.49. No DNA band is generated in lanes 1 and 3 (5-FOAR, ura4-) strains, as expected. Southern confirms removal of a single copy of β-tubulin after 5-FOA treatment of AK102 that has sequential β-tubulin genes. (C and D) MBC and TBZ resistance. (C) Serial dilutions of wild-type (YY105, 972, and WT/AK103) and mutant (F200Y/AK101) cells onto YES plates containing 5, 10, 20, 50, or 100 μg/ml MBC or TBZ grown at 32°C. For the 50 μg/ml concentration, WT/AK103 is omitted. (D) Streaks of cells onto YES plates containing MBC or TBZ. Some growth is indicated at 50 μg/ml MBC with the mutant.
Chromosomal DNA from 5FOAR, ura4- (AK101) and starting ura4+ cells (AK102) was isolated into HEPES buffer by using glass beads and vortexing to disrupt cells as described previously (Moreno et al., 1991). PCR was performed using two separate sets of oligonucleotide pairs. The first set was used to detect loss of the ura4+ gene from the β-tubulin locus [URA4 oligo: 5′-GCAACTTGTCAGTCGCGGTCGATTTGACG-3′ and β-tubulin oligo: B5M, 3′-GCTTCTAAGCGCTCCTTATAGGCCTGGCATA] and the second set to confirm the presence of a single copy of β-tubulin [β-tubulin oligos: B3M 5′-CTCACTCTTTGGGTGGTGGTACTGGATCTGG and B3 3′-TACGAGTAGACGATTGGAGCTTTATCA]. Cells that had either a restored wild-type or mutant β-tubulin allele were both found in the 5FOAR population, as determined by sequencing (of 10 colonies analyzed, three were mutant). No extraneous β-tubulin mutations were detected.
Southern hybridization followed standard procedures. HindIII-digested chromosomal DNA (10–20 μg) was gel fractionated and transferred onto Hybond N+ membrane (Amersham Biosciences, Piscataway, NJ). Prehybridization and hybridization were performed at 65°C by using as probe a short internal β-tubulin fragment generated by PCR and corresponding to amino acids 136–349 in nda3+. The DNA probe was labeled by random priming.
Live Cell Analysis
Procedures for visualization of green fluorescent protein (GFP) in living cells were as described previously (Paluh et al., 2000). Coverslips were coated with 0.1% polyethyleneimine and sealed with VALAPA (1:1:1 petroleum jelly, lanolin, and paraffin). Cells were viewed primarily on a Zeiss Axiovert 200 inverted microscope equipped with a Pryor stage, by using a 63× oil differential interference contrast PlanAPOCHROMAT objective, 1.40 numerical aperture (NA), or a 100× oil PlanFLUAR lens, 1.45 NA. Images were acquired with a Hammamatsu Orca charged-coupled device by using MetaMorph software (Universal Imaging, Downingtown, PA). An Olympus IX70 inverted microscope equipped with a motorized stage, 100× oil immersion PlanApo lens, 1.35 NA, Photometrix charged-coupled device camera and Resolve 3D software (Applied Precision, Issaquah, WA) was also used. Single plane time series (2–60 s) and occasionally three-dimensional image projections (acquired at 0.2- to 0.4-μm steps, 3–20 images in stack) were examined. Images were transferred to Adobe Photoshop (Adobe Systems, Mountain View, CA) and Canvas (Deneba Systems, Miami, FL) for compilation into figures or saved as QuickTime movies in MetaMorph. No deconvolution of image projections was required.
Measurements of microtubule dynamic parameters in vegetative cells (Table 1) used time-lapse single plane series, 2-s intervals, 200-ms exposure, of 30–60 time points (1–2 min total time) at room temperature. Rates of microtubule growth and shrinkage were determined from linear interphase microtubules in consecutive frames by using the MetaMorph Line Tracking function. At least three measurements tracking the movement of a single microtubule end were made. In some cells, fiduciary marks from uneven incorporation of GFP-α2-tubulin allowed us to further monitor rates (Figure 4E, 210 s to end). Meiotic measurements used time series acquired at 15-, 20-, or 30-s intervals for astral microtubule dynamics in horsetail movement or 30- to 60-s intervals for meiosis I. Longer intervals in meiosis I, 200-ms exposures, along with beginning image acquisition at middle to late anaphase I avoided phototoxicity. The point of near maximum spindle extension in meiosis I time series (within 1 μm) was used as the start point for timing meiosis I spindle breakdown. This was readily observable by manually switching between consecutive images in a series by using the MetaMorph Select Plane and Region Measurement functions. The final time point was that before spindle microtubules lost continuity between poles (i.e., when a gap appeared).
Table 1.
Assembly and function of microtubule arrays in nda3-F200Y
| Interphase MT dynamics (μm/min)
|
|||
|---|---|---|---|
| F200Y | YY105 | ||
| Mitotic growth | |||
| Growth | 3.93 ± 0.84 | 3.00 ± 1.20 | |
| Shrinkage | 11.80 ± 2.25 | 18.50 ± 4.22 | |
| Spindle elongation | 0.42 ± 0.17 | 0.47 ± 0.20 | |
| Spindle breakdown (minutes) | 6.13 ± 1.83 (I) | 2.00 ± 0.42 | |
| 11.88 ± 1.26 (II) | |||
| Final spindle length as % cell length | 105.20 ± 5.16 (I) | 81.00 ± 12.5 | |
| 111.00 ± 8.60 (II) | |||
| Meiotic phenotypes | |||
| Horsetail stage | |||
| Microtubule time at cortex before depolymerization (seconds) | 70.50 ± 30.04 | 37.75 ± 8.96 | |
| Number of Astral MTs | 4–7 | 4–7 | |
| Meiosis I | |||
| Spindle breakdown (minutes) | 6.88 ± 0.63 (I) | 8.13 ± 1.60 | |
| 21.17 ± 2.84 (II) | |||
| Meiosis II | Asymmetric spindle failure in F200Y versus YY105: collapse; also thin, extended meiosis II spindles | ||
| Presporulation | Short microtubule arrays in F200Y are more prominent | ||
Information is based on (number of microtubules, cells examined) as follows: growth: F200Y (15, 9), YY105 (6, 5); shrinkage, F200Y (64, 27), YY105 (9, 4); spindle elongation (5 cells each); spindle breakdown after full extension to cell ends, F200Y (group I, 12 cells; group II, 4 cells), YY105 (9 cells). Horsetail, F200Y (20, 9), YY105 (4, 4). Meiosis I, F200Y (group I, 4 cells; group II, 3 cells) and YY105 (12 cells). See text for full details.
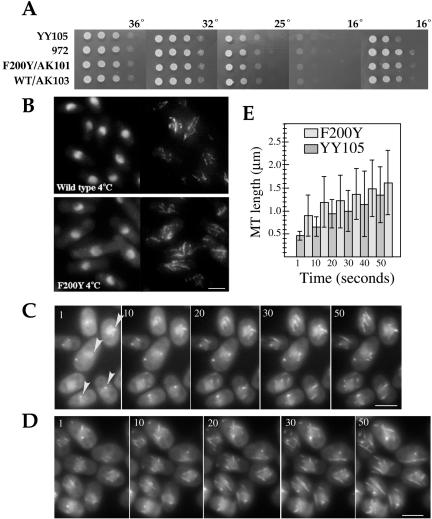
Figure 4.
Delayed mitotic exit in nda3-F200Y mutant cells. (A) Diagram of microtubule arrays during mitotic growth in S. pombe: 1) interphase; 2) anaphase; 3) telophase; 4) formation of postanaphase arrays, spindle breakdown, and mitotic exit; and 5) cytokinesis and G1/S. (B and C) Representative images of mitotic progression for YY105 (B) and nda3-F200Y (AK101) (C) cells. (D) Spindle breakdown in wild-type cells occurs within minutes after maximum spindle extension to cell ends. Time is in seconds. (E) Spindle breakdown during mitotic exit in nda3-F200Y cells is significantly delayed versus wild type. Select images from a single plane time series are shown. Images were acquired at 30-s intervals. Spindle breakdown, indicated by arrows and signified by separation of spindle ends, is seen at 540 s and more clearly at 570 s. Fiduciary marks, due to uneven incorporation of GFP-α2-tubulin, can be seen as early as 210 s, as individual spindle microtubules depolymerize. (F) Four images of contorted telophase spindles in nda3-F200Y cells. Cells with an S-shaped spindle were slightly larger in size and delayed more severely in spindle breakdown. Bar for all images, 5 μm (shown in B).
In cold-mediated depolymerization of GFP-α2-tubulin containing microtubules on slides, 2 μl of cells was placed on a slide under a coverslip, incubated on ice for 10 min, and moved quickly to the microscope for immediate time-lapse imaging. From a field of cells, repolymerizing microtubules were monitored at 5- to 10-s time points. Several time-lapse sequences were acquired for both strains and found to exhibit the same distinct trends. Measurements of microtubule length were done using the MetaMorph Region Measurement function. Ten to 29 microtubules were measured for each time point, by using those microtubules that displayed the clearest focus for accuracy. The actual numbers obtained from the images in Figure 3, C and D, for the histogram in 3E are as follows: YY105 t1 [0.463 ± 0.10], t10 [0.66 ± 0.22], t20 [0.94 ± 0.31], t30[1.00 ± 0.45], t40 [1.15 ± 0.71], and t50 [1.34 ± 0.61]; F200Y t1 [0.90 ± 0.44], t10, [1.19 ± 0.55], t20 [1.20 ± 0.57], t30, [1.37 ± 0.56], t40, [1.49 ± 0.62], t50, and [1.61 ± 0.70].
Figure 3.
S. pombe nda3-F200Y and wild-type cells display generally similar thermal characteristics. (A) Serial dilutions of cells as in Figure 2 grown at 36, 32, 25, and 16°C for 2 d. The final 16°C image was taken after 10 d. (B) Cold-mediated depolymerization of microtubules from liquid cultures. Immunofluorescence of methanol-fixed wild-type and nda3-F200Y (AK101) cells after incubation of cell cultures on ice 30 min. Left, Hoechst, right: microtubules, TAT1 monoclonal antibody. (C and D) Repolymerization of microtubule arrays in wild-type YY105 (C) or nda3-F200Y (D) cells at 24°C after cold-mediated depolymerization on slides. Images shown are in intervals of 10 s from time-lapse sequences. Arrowheads in C indicate prominent single sites for reformation of microtubule arrays. Bars, 5 μm. (E) Histogram of F200Y and YY105 microtubule lengths versus time for images in C and D, including an additional time point at 40 s. Shaded bars represent average microtubule lengths for each time point. Thin bars indicate the SD of microtubule lengths.
Immunofluorescence
Standard procedures were used to visualize microtubules (Mata and Nurse, 1997; Hagan and Ayscough, 2000). Cells were grown in appropriate media (15–20 ml) at 32°C, filtered onto Whatman glass microfiber filters, and placed in -80°C MeOH for 15 min. Primary antibody incubation of cells was done in PEMBAL (100 mM PIPES, pH 6.9, 1 mM EGTA, 0.1 mM MgSO4, 1% bovine serum albumin, 0.1% sodium azide, 100 mM lysine, and 0.5% gelatin) overnight at 4°C with 1/50 dilution of TAT1 mouse monoclonal antibody (Woods et al., 1989). Alexa Fluor 488 (Molecular Probes, Eugene, OR) secondary antibody was used at 1/200 dilution. DNA was stained with Hoechst 33342 (Sigma-Aldrich, St. Louis, MO).
RESULTS
A Conserved Internal Residue in β-Tubulin Is Positioned to Affect Interdomain Motion
A dramatic architectural change occurs in microtubules upon depolymerization to allow rapid peeling and curling of microtubule ends (Mandelkow et al., 1991; Desai and Mitchison, 1997; Tran et al., 1997). One hypothesis is that the conformational adjustment in tubulin dimers required for this change is achieved through the relative movement of the nucleotide-binding and intermediate domains (shown in Figure 1, A and B). Any inhibition or facilitation of domain motion by modulation of residues in the interface would thus impact microtubule stability. Our examination of residues at this interface in various species indicates that the bulk remain invariant or display only conservative substitutions (shown in Figure 1B as aqua and purple spheres, respectively). A few residues do show nonconservative substitutions (yellow spheres). Unique to this class is the residue corresponding to position 200 in mammalian β-tubulin (red in Figure 1, A and B), which displays a phyla-specific pattern of conservation (Figure 1C, arrow). The intraphyla conservation and interphyla diversity of this critically positioned residue in organisms living in contrasting thermal environments further supports its role in microtubule stability.
S. pombe nda3-F200Y Is Resistant to Benzimidazole Antimicrotubule Reagents Carbendazim (MBC) and Thiabendazole (TBZ)
We generated the F200Y mutation in S. pombe β-tubulin to carefully address in vivo its impact on microtubule dynamic parameters, its importance for microtubule organization during the mitotic cell cycle or upon cellular differentiation, and finally its possible role in cold lability suggested by our phylogenetic analysis. Mutations in fungal and helminth β-tubulins at residue 200 have been described in regard to benzimidazole drug resistance (Jung et al., 1992; Kwa et al., 1994). We also characterized drug sensitivity changes in S. pombe as a result of altering this residue.
The S. pombe wild-type β-tubulin gene nda3+ (Toda et al., 1983) was obtained by PCR, mutagenized in vitro, and recombined at the native locus in wild-type strain YY105 (Figure 2A; see MATERIALS AND METHODS). Treatment with 5-FOA regenerated either wild-type or mutant nda3-F200Y single alleles (Figure 2, A and B). Strain YY105 contains an additional copy of the nonessential α2-tubulin gene fused to GFP (Ding et al., 1998). It has been used extensively in S. pombe for live analysis of microtubule cytoskeletal behavior during mitotic growth (Mallavarupa et al., 1999; Brunner and Nurse, 2000a; Drummond and Cross, 2000; Paluh et al., 2000; Tran et al., 2001) and in meiosis (Ding et al., 1998; Yamamoto et al., 1999, 2001; Miki et al., 2002).
Several of the benzimidazole agents, including TBZ and MBC, induce microtubule depolymerization (Davidse and Flach, 1978; Friedman and Platzer, 1978). In Aspergillus nidulans, residues at amino acids 165 and 198–200 have been found to be mutated in drug-resistant strains and may contribute to a benzimidazole binding site (Jung and Oakley, 1990; Jung et al., 1992). We examined the F200Y mutant for resistance to TBZ and MBC. Both serial dilutions and streaks of nda3-F200Y (F200Y/AK101) and wild-type cells (control strains YY105, 972, and AK103) are shown (Figure 2, C and D). Strain F200Y/AK101 is completely resistant to TBZ (to 100 μg/ml) but only to lower levels of MBC (to 20 μg/ml). All wild-type strains showed sensitivity to both drugs at concentrations as low as 10 μg/ml, as expected.
Rapid Recovery of Ectopic Nuclear Microtubule Arrays in F200Y after Cold Treatment
Antarctic fish, like fungi, use a phenylalanine at residue 200 in contrast to tyrosine in warm-blooded animals. Conservation of this residue, exclusive of any other evident psychrophile-specific β-tubulin residues in Antarctic fish, suggests a possible role in stabilizing microtubules against cold sensitivity, although nonconserved residues may also contribute. To test for conditional growth in our mutant, we plated serial dilutions of F200Y/AK101 and wild-type cells on rich media and incubated the cells at 36, 32, 25, and 16°C (Figure 3A). No differences in growth were observed for F200Y/AK101 versus wild-type strains at any of these temperatures. We did observe that strain YY105 is slightly cold sensitive versus strain 972 and AK103 at 16°C. Additional growth tests conducted at 11°C (our unpublished data) were similar. Next, we compared microtubule stability upon sudden exposure to extreme cold temperature in the mutant versus wild-type. We incubated cells growing logarithmically in 10 ml of YES on ice for 30 min, fixed the cells, and examined microtubules by immunofluorescence (Figure 3B). Microtubules in both the mutant and wild type seemed to have depolymerized similarly under these conditions.
On examination of mitotic and meiotic phenotypes (see following sections) and determination of microtubule dynamic parameters for F200Y/AK101 (Table 1), we were prompted to reexamine recovery from cold-induced depolymerization of microtubules. This was done using small volumes on slides. Cells were preexamined for good GFP microtubule signals, spotted onto slides, incubated on ice for 5 or 10 min, quickly focused, and imaged immediately to monitor repolymerization of microtubule arrays. We hypothesized that cold-induced depolymerization of microtubules might be delayed or incomplete in the mutant versus wild type. If so, recovery of microtubule arrays would be faster in the mutant. In both the wild-type and mutant cells incubated on ice, microtubule depolymerization occurred extremely rapidly on the slides (complete within 10 min) compared with our previous experiments with cultures. Recovery of microtubule arrays was followed at 5- to 10-s intervals. In F200Y cells, microtubules can be seen extending from multiple sites on the nuclei from the earliest time points attainable. Slower recovery from 4°C occurs in YY105 cells and is evident in microtubule lengths (Figure 3E). For example at 30 s, the mean microtubule length in F200Y is 1.37 μm versus 1.00 μm in YY105. Additionally, we observed that in YY105 cells at the first time point, the earliest forming microtubules extend from a single prominent site in many cells (Figure 3C, arrowheads). This difference has not been described previously (Tran et al., 2001) and may suggest that reformation of cold-treated microtubules occurs first from the spindle pole body, preceding ectopic sites. However, because this was primarily evident at the earliest time points, we were technically limited to further clarify this observation.
Delayed Mitotic Exit
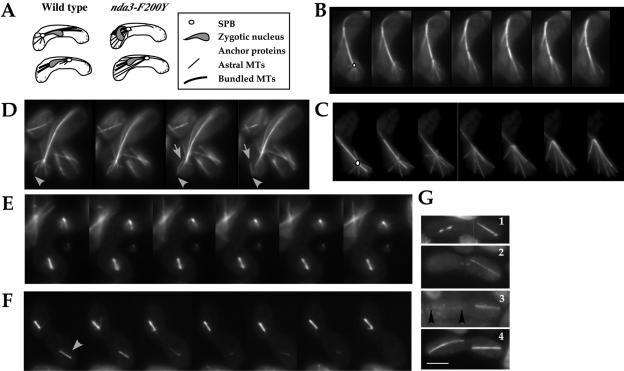
Microtubule arrays in S. pombe are well characterized in mitosis and meiosis (Hagan, 1998; Svoboda et al., 1995; Ding et al., 1998) and are summarized in Figures 4A and 5A. Changes in microtubule organization are known to affect cell morphology, spindle reorientation, chromosome segregation, and mitotic exit. We used time-lapse videomicroscopy to follow microtubule dynamics in our nda3-F200Y cells. Nonsynchronized cells or cells synchronized in early S phase by addition of and release from 10 mM hydroxyurea were examined. Progression through mitosis is shown in Figure 4, B and C, for wild-type and F200Y/AK101 cells, respectively. No differences in organization of interphase microtubule arrays were observed between mutant and wild-type cells, or during early mitosis and anaphase, in which spindle formation and elongation seemed to proceed with normal dynamics. In telophase, spindle function continued to seem normal in the mutant. However, mitotic exit, indicated by breakdown of the spindle after its extension to cell ends, was significantly delayed. In wild-type cells within 60–120 s of full extension, spindle breakage occurs (Figure 4D and Table 1). In the mutant, spindle breakdown is significantly delayed. Cells fell into two general time ranges: group I (up to 75% of cells observed) completed spindle breakdown in 5–7 min (Figure 4E and Table 1), whereas group II (up to 25% of cells observed) required 10–12 min (Table 1). The extent of delay was not predictable from initial observations of microtubule contact with cell ends. In all cases, the spindle became slightly hyperextended (Table 1) and arched or bow shaped during delay. On occasion we observed slightly larger cells with S- or scythe-shaped spindles (Figure 4F). As in wild-type cells, we observed gradual depolymerization of individual microtubules during spindle breakdown in F200Y, seen as fiduciary marks (Sagolla et al., 2003). Postanaphase arrays at the cell center formed normally. Thus, the nda3-F200Y mutation does not affect microtubule architecture during vegetative cell growth but does interfere with normal spindle breakdown and exit from mitosis. Delayed spindle breakdown was also observed for meiosis I (Table 1).
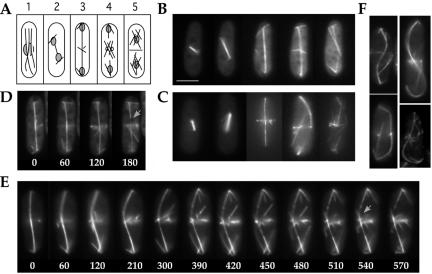
Figure 5.
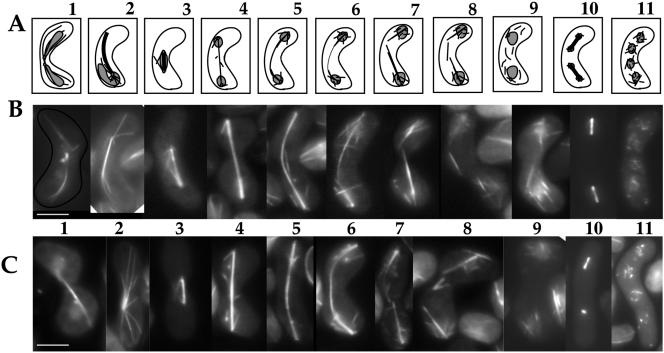
Stage-specific microtubule arrays are affected in nda3-F200Y cells during meiosis. (A) Diagram of microtubule arrays during meiotic growth in S. pombe: 1) karyogamy, 2) horsetail stage, 3–8) meiosis I early stages to breakdown, 9) premeiosis II, 10) meiosis II, and 11) presporulation. (B and C) Representative images of meiotic progression for wild-type (B) and nda3-F200Y (AK101) (C) cells. Bar, 5 μm.
Nda3-F200Y Alters Select Stages of Meiosis
The meiotic process in eukaryotes requires multiple specialized cytoskeletal events for karyogamy and segregation and reductional divisions of chromosomes. After karyogamy in fission yeast, the zygotic spindle pole body attaches to a bundle of microtubules and dynamically oscillates back and forth along the cell length. These so-called horsetail movements enhance recombination and are mediated by dynein, microtubule sliding in the bundle, and transient attachment of astral microtubules to the cell cortex (Ding et al., 1998; Yamamoto et al., 1999, 2001; Hagan and Peterson, 2000).
We examined organization and dynamics of meiotic microtubule cytoskeletal arrays in F200Y/AK101 cells versus wild-type. Representative stages of meiosis (diagrammed in Figure 5A) are shown for wild-type (Figure 5B) and mutant (Figure 5C) cells. Formation of the fused zygotic nucleus during karyogamy and the bundled microtubule array for horsetail movement were similar (Figure 5, A–C, frames 1 and 2). However, horsetail astral microtubule arrays in the mutant seemed more spidery or finger-like than those of wild-type cells. This is particularly striking in images obtained using time-lapse microscopy (Figure 6; Supplemental Movies: HorsetailF200Y, HorsetailYY105). Assembly, elongation and breakdown of meiosis I spindles seemed normal (Figure 5, frames 3–8; Supplemental Movies of spindle breakdown: MeiosisIF200Y, MeiosisIaYY105, MeiosisI-bYY105); however, as with mitosis, spindle breakdown took longer to occur in 50% of the cells examined (Table 1). After meiosis I spindle breakdown, short microtubule arrays extend from multiple sites on reformed nuclei in both wild-type and mutant cells (frame 9).
Figure 6.
nda3-F200Y cells display persistent horsetail cortical contacts and show a high frequency of spindle failure in meiosis II. (A) Diagram of cortical contacts in horsetail movement. (B and C) Time-lapse single plane images of astral microtubules during horsetail movement in wild-type (B) and nda3-F200Y (AK101) (C) cells. An open circle in the first frame of each series indicates the position of the zygotic spindle pole body. (D) Detachment of an astral microtubule from the pole region while retaining cortical contacts was observed in both wild-type and nda3-F200Y cells. Arrowhead points to cortical contact; arrow indicates point of detachment. (E and F) Time-lapse images of asymmetric meiosis II spindle failure in nda3-F200Y cells. Arrows indicate unstable spindle of pair. (G) Early and late meiosis II phenotypes in nda3-F200Y cells showing 1) single failing bipolar spindle (left), 2) a single extended meiosis II spindle, 3) one robust and one weak (flanked by arrowheads) meiosis II spindle, and 4) normal meiosis II extended spindles. Bar for all images, 5 μm (shown in G).
Meiosis II was dramatically affected in the mutant (frame 10). We observed frequent failed spindle formation in a single pair of spindles, in 35–45% of the cells, by live examination of cells carrying GFP-α2-tubulin (discussed below). This phenotype was not observed in wild-type cells. F200Y cells progressing through meiosis II showed normal formation of numerous microtubules around nuclei that preceeds prespore formation, although these microtubules were more readily observed in F200Y cells (frame 11). Spore morphology was normal; however, spore numbers per ascus varied from wild type (Figure 7, discussed below).
Figure 7.
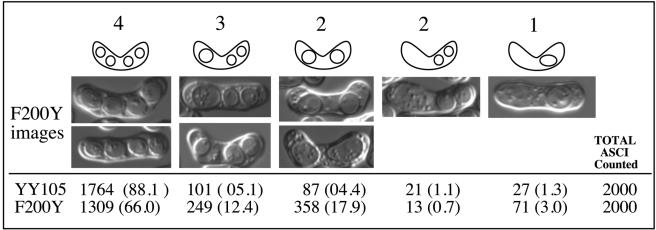
Spore analysis. Differential interference contrast images of nda3-F200Y cells and total asci counted from YY105 and F200Y/AK101.
In summary, microtubule behavior during at least three distinct stages of meiosis is clearly affected in the nda3-F200Y mutant: horsetail movement, meiosis I spindle breakdown, and meiosis II spindle formation. Microscopic analysis of these stages is consistent with sporulation analysis that indicates an increase in two and three-spored asci in the mutant and reduced spore viability (47% decrease; YY105, n = 1676; F200Y, n = 890).
Persistent Attachment of Astral Microtubules to the Cell Cortex during Horsetail Movement
During horsetail movement, astral microtubules normally extend from the fused spindle pole body of the zygotic nucleus to transiently interact with sites on the cell cortex (Yamamoto et al., 2001). These microtubules are dynamic and depolymerize quickly as the oscillating nucleus reverses direction (Figure 6A; wild type in 6B). In the nda3-F200Y mutant cells, as the nucleus reverses direction and moves away from the cell end, astral microtubule ends remain clearly attached longer at the cell cortex (Figure 6C and Table 1; Supplemental Movies: HorsetailF200Y and HorsetailYY105). The number of astral microtubules for wild-type or nda3-F200Y cells was similar (four to five microtubules), and the dynamic movement of the horsetail nucleus seemed as in wild type (our unpublished data). Interestingly, prolonged astral microtubule contacts were not observed in mitosis. The nature of cortical contacts is expected to be different in mitosis vs. meiotic horsetail movement and may account for this striking difference in behavior. Time-lapse imaging of horsetail movement in both wild-type and mutant cells revealed that an astral microtubule can remain attached at the cortex but detach from the pole region during oscillations (Figure 6D, arrowheads). We observed this to happen in at least 75% of our horsetail time series with wild-type or nda3-F200Y. No detachment of astral microtubules from the pole region has been reported previously. However, the brightness of the microtubule horsetail bundle often makes astral microtubules harder to visualize in general.
Asymmetric Spindle Collapse in Meiosis II
The transition from meiosis I to meiosis II in S. pombe requires breakdown of the meiosis I spindle, spindle pole duplication, and the reassembly of paired spindles for meiosis II equational division. In wild-type cells, meiosis II spindles form and elongate in synchrony by an unknown mechanism. In nda3-F200Y cells, we observed frequent asymmetric failure in meiosis II spindle formation. Time-lapse microscopy revealed that the failing spindles were capable of initiating assembly into short bipolar arrays but were unstable. Short 2 μm spindles formed, often with frequent shifts in length from growth to shrinkage before collapsing (Figure 6, E and F). The initial formation of both spindles was synchronized. We also observed cells in the population that had elongated a single meiosis II spindle normally, whereas the second spindle was shorter and lighter at the spindle midzone (Figure 6G, frame 1), or had a single elongated spindle without evidence of the second spindle of the pair (Figure 6G, frame 2), or had two meiosis II spindle pairs in which one of greatly reduced microtubule intensity (compare Figure 6G, frames 3 and 4). Disruption of spindle behavior in a single pair of the two meiosis II spindles has not previously been described in fungi or animal cells. The redistribution of spindle components necessary to monitor and carry out the two sequential chromosome divisions of meiosis must be required. One explanation for our observations is that γ-tubulin redistribution to spindle poles (Marschall et al., 1996) could be affected by the change in microtubule dynamics in nda3-F200Y. However, this would have to be a meiosis-specific occurrence because we saw no such defects in mitosis. We also saw no evidence of unequal distribution of γ-tubulin by use of a gtb1-GFP fusion protein in meiosis I or meiosis II (our unpublished data). Finally, initial spindle intensity, and presumably therefore the number of microtubules present, was similar between the stabilized and failing spindle pairs.
Generation of Two and Three-spored Asci and Reduced Spore Viability after Meiosis in nda3-F200Y Cells
We examined the final products of meiosis for spore viability and the presence of normal-looking, four-spored asci. As expected from our in vivo time-lapse imaging of meiotic cytoskeletal arrays, we observed an increase in two and three-spored asci in the mutant versus wild type (Figure 7). The position of the asci is also reflective of the cytoskeletal defect. For example, two-spored asci may result from failure to progress to meiosis II, with distally separated spores, or failure in meiosis II without anaphase with two more closely positioned spores in an ascus. The generation of three-spored asci is expected for failure of a single pair of meiosis II spindles. We observed no abnormal spores that could indicate a problem with prespore formation in the mutant. Formation of the forespore membrane initiates as meiosis II nears completion. It begins at the spindle poles and extends along the dividing nuclei to eventually encircle each haploid nucleus once spindle disassembly is complete (Nakamura et al., 2001, and references therein). In Figure 7, the number of three-spored asci observed is slightly less than that expected from our microscopic analysis by using GFP-α2-tubulin. This indicates either failure to undergo anaphase II chromosome segregation or rescue of the phenotype, because not all observed asymmetric meiosis II spindles were followed through to the completion of meiosis.
DISCUSSION
The microtubule cytoskeleton plays an active role in cell cycle progression, including roles in chromosome segregation, cellular morphology, and organelle transport. The dynamic nature of microtubules themselves contributes to these processes. Microtubules are composed of tubulins, as are in part the structures involved in nucleating microtubules. A heterodimer of α-β-tubulin forms the predominant protofilament structure of microtubules. γ-Tubulin is primarily localized to spindle poles and forms part of the microtubule-organizing center. Four additional tubulin family members, δ, ε, η, and ζ are associated with centrioles and basal bodies (for review, see Dutcher, 2001; McKean et al., 2001). In the single-celled yeasts, two isotypes of α-tubulin are present along with single alleles of β-tubulin and γ-tubulin. In vertebrates, multiple isotypes of both α- and β-tubulin exist and can be incorporated into microtubules. Tubulin isotype sequences may differ by as much as 15%, and isotype expression is often cell type specific or developmentally regulated. The ratio of mixed isotypes within microtubules can affect dynamic properties of microtubules in vitro and, in vivo, can impact cytokinesis and chromosome segregation (Matthews et al., 1993; Panda et al., 1994; Matthies et al., 1999; Bode et al., 2003).
Gross differences in tubulin sequence such as those present in isotypes are not necessary to alter microtubule dynamic properties. Numerous single residue mutations in α- and β-tubulins have been isolated in fungi that affect microtubule stability as reflected in drug resistance and cold sensitivity (Jung and Oakley, 1990; Yarden and Katan, 1993; Richards et al., 2000; Gupta et al., 2001, 2002). Recessive mutations in a testis-specific β-tubulin isotype in Drosophila are known to result in altered structure of the axoneme and the associated microtubule arrays, as well as failed cytokinesis (Fuller et al., 1988). In addition to the residues identified in tubulins by mutational studies, phylogenetic sequence analysis combined with structural information on tubulin is helping to identify phyla-specific residues of functional importance (Detrich et al., 2000; Blouin et al., 2003).
A comparison of the primary sequence of Antarctic fish tubulins with those of other animals and fungi has identified a single residue as the only consistent difference between β-tubulins of psychrophilic fish and virtually all other animals. Interestingly, the residue lies at an important domain interface in tubulin structure, and in fungi is implicated in resistance to antimicrotubule benzimidazole agents. A relative rotation at the domain interface has been hypothesized to follow GTP hydrolysis and microtubule depolymerization. Such motion may be necessary to produce the curling of protofilaments seen in rapidly depolymerizing microtubules and in GDP-tubulin rings (Erickson and Stoffler, 1996; Amos and Löwe, 1999). No information was available on the impact of this important residue, phenylalanine 200, on microtubule dynamic parameters, extreme cold sensitivity, or formation of microtubule arrays in mitosis and meiosis, leaving studies on the functional importance of this position in tubulin structure incomplete.
We chose to investigate the importance of phenylalanine versus tyrosine at position 200 in fission yeast (nda3-F200Y), the residue corresponding to Antarctic fish phenylalanine and mammalian tyrosine. We postulated that the difference in amino acid side chain character could be enough to alter the energy barrier for domain motion and thus affect the stability of microtubules in regard to depolymerization and thermal properties. Our analysis of microtubule dynamic parameters for nda3-F200Y indicates a decrease in depolymerization rate versus wild type. Altering phenylalanine to tyrosine did not affect temperature stability in vivo, but had drastic consequences on the behavior of specific microtubule arrays in both mitosis and meiosis. These affects only delayed completion of mitosis but resulted in frequent failure in meiosis.
Most striking about our in vivo observations with nda3-F200Y cells is that altered depolymerization rates dramatically affect only a subset of cellular processes that involve plus-end microtubule complexes (Howard and Hyman, 2003). This includes extended cortical contacts of meiotic horsetail astral microtubules without alteration of interphase arrays in mitosis, the delayed spindle breakdown for mitosis and meiosis I, and failed spindle formation in a single pair of meiosis II spindles. In regard to interphase and horsetail arrays, more rapid depolymerization rates may be necessary for destabilization of dynein–dynactin complexes used during the horsetail movement of meiosis (Yamamoto et al., 2001; Miki et al., 2002). Unaffected in the mutant were interphase microtubule cortical contact behavior of vegetatively growing cells (Brunner and Nurse, 2000b; Sawin, 2000; Schroer, 2001) or astral microtubule behavior that is necessary to orient the spindle before anaphase (Gachet et al., 2001).
The plus ends of microtubules that form the anaphase spindle midzone use a unique complex of proteins that includes passenger proteins and kinesin-like proteins (for review, see Severson and Bowerman, 2002). In budding yeast, Kip3p localizes to astral and central spindle microtubules (DeZwaan et al., 1997), and loss of Kip3p has been shown to delay spindle breakdown and result in elongated spindle microtubules (Straight et al., 1998). Similarly, in our F200Y cells, depolymerization of individual spindle microtubules occurred; however, the onset and completion of this event was delayed and the spindle became hyperextended. In meiosis, we observed that the failure in a single meiosis II spindle of the pair occurs at a frequency near 50%. It is interesting to note that delayed breakdown of the meiosis I spindle had a similar frequency of occurrence. In the transition between meiosis I and meiosis II, repositioning of passenger proteins has been described to occur in the mouse (Parra et al., 2002). The identification of passenger proteins and their behavior during mitosis is still at its infancy in fission yeast (Leverson et al., 2002), and no studies have been done in yeast for meiosis. If these proteins become distributed asymmetrically due to delayed meiosis I spindle breakdown in F200Y cells, meiosis II spindles could be differentially affected in their stability. Activated checkpoint proteins might be expected to monitor meiosis II behavior. If so, Mad2 is unlikely to be involved because we observed no differences in meiotic progression through meiosis II in nda3-F200Y cells carrying a mad2 deletion (our unpublished data).
Asymmetric distribution of certain regulatory components associated with spindle poles is known to occur normally in mitosis (Cerutti and Simanis, 1999). However, there is no similar evidence for such asymmetric distribution of components to spindle poles in meiosis. We cannot rule out that the meiosis II failure we see may be due to a defect at the spindle poles. Complex changes that occur in spindle pole bodies during meiosis include fusion, separation, and duplication and may require redistribution of components and be more sensitive to changes in depolymerization dynamics of microtubules than simply duplication in mitosis. However, γ-tubulin localization seems normal, as does formation of spore walls that requires intact spindle pole body complexes. A more detailed analysis of the meiosis II spindle phenotype that includes electron microscopic analysis may be necessary to unambiguously rule out the possibility of a defect at spindle poles.
Understanding the role of the conserved phenylalanine residue in drug resistance to benzimidazoles is now extended to S. pombe for thiabendazole. We also provide new information in regard to carbendazim. The F200Y mutation produces a large effect on drug resistance but only a relatively small effect on microtubule stability. This difference suggests that the mutation directly inhibits drug binding, consistent with the hypothesis that the binding site is in the vicinity of residues 6, 165, 198, and 200 (Jung et al., 1992) and residue 167 (Prichard, 2001). It seems likely that the tyrosine hydroxyl blocks part of the benzimidazole-binding region or changes the hydrophobic nature of the region. The presence of either tyrosine or phenylalanine at position 200 is strongly correlated with either asparagine or alanine, respectively, at position 165. The latter combination seems to have a hydrophobic void that may serve as part of the drug-binding site. Replacement of the phenylalanine by tyrosine would partially fill this void. We note that the most likely hydrogen-bonding partner for Y200 is N165, as judged from the crystal structure. The Antarctic fish also have N165 but clearly not the hydrogen bond with F200. The fungal A165 offers little relevant interaction with either the wild-type F200 or the mutant Y200. This region is internal to the protein, so access of the drug to the binding site apparently would involve some conformational change. It is also possible that the mutation inhibits this conformational change, and the corresponding protein stabilization results in some stabilization of the microtubules.
Only very subtle differences in cold sensitivity of microtubules, which may simply reflect the decreased microtubule depolymerization rate in nda3-F200Y, were observed in the mutant. The underlying reason for the striking phyla-specific conservation of this residue in Antarctic fish as well as the source of the low-temperature microtubule stability, thus remain unclear. It has been observed that, in microtubules formed with different tubulin isotypes, a minority population of tubulin dimers can confer properties to the bulk. For example, when Atlantic cod and bovine brain tubulins were mixed, the resulting microtubules showed temperature stability more characteristic of the minor cod component (Wallin and Billger, 1997). Thus, it may be that the stability of the Antarctic fish microtubules arises from one different isotype that has an uncommon stability. A number of differences between some of the fish tubulin isotypes and other animal tubulins were noted previously, including several at the interprotofilament interface. The results of this work point to the need to examine some of these other sites as providing possible clues to thermal differences in dynamics and stability.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant GM-46033 (to K.H.D.), by National Science Foundation grant OPP-0089451 (to H.W.D.), and by the Office of Health and Environmental Research, U.S. Department of Energy, under contract DE-AC03-76F00098. J.L.P. is supported on startup funds at Boston College.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–06–0389. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-06-0389.
Online version of this article contains video material for some figures. Online version is available at www.molbiolcell.org.
References
- Ahringer, J. (2003). Control of cell polarity and mitotic spindle positioning in animal cells. Curr. Opin. Cell Biol. 15, 73-81. [DOI] [PubMed] [Google Scholar]
- Amos, L.A., and Löwe, J. (1999). How taxol stabilizes microtubule structure. Chem. Biol. 6, R65-R69. [DOI] [PubMed] [Google Scholar]
- Blouin, C., Boucher, Y., and Roger, A.J. (2003). Inferring functional constraints and divergence in protein families using 3D mapping of phylogenetic information. Nucleic Acids Res. 31, 790-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode, C.J., Gupta, M.L., Jr., Suprenant, K.A., and Himes, R.H. (2003). The two α-tubulin isotypes in budding yeast have opposing effects on microtubule dynamics in vitro. EMBO Rep. 4, 94-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke, J.D., LaCroute, F., and Fink, G.R. (1984). A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197, 345-346. [DOI] [PubMed] [Google Scholar]
- Brunner, D., and Nurse, P. (2000a). CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell 102, 695-704. [DOI] [PubMed] [Google Scholar]
- Brunner, D., and Nurse, P. (2000b). New concepts in fission yeast morphogenesis. Phil. Trans. R. Soc. Lond. B 355, 873-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns, R.G., and Surridge, C.D. (1993). Tubulin: conservation and structure. In: Microtubules, ed. J. Hyams and C. Lloyd, New York: Wiley-Liss, 3-31.
- Cassimeris, L. (2002). The oncoprotein 18/stathmin family of microtubule destabilizers. Curr. Opin. Cell Biol. 14, 18-24. [DOI] [PubMed] [Google Scholar]
- Cassimeris, L., and Spittle, C. (2001). Regulation of microtubule-associated proteins. Int. Rev. Cytol. 210, 163-226. [DOI] [PubMed] [Google Scholar]
- Cerutti, L., and Simanis, V. (1999). Asymmetry of the spindle pole bodies and spg1p GAP segregation during mitosis in fission yeast. J. Cell Sci. 112, 2313-2321. [DOI] [PubMed] [Google Scholar]
- Davidse, L.C., and Flach, W. (1978). Interaction of thiabendazole with fungal tubulin. Biochim. Biophys. Acta 543, 82-90. [DOI] [PubMed] [Google Scholar]
- Desai, A., and Mitchison, T.J. (1997). Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13, 83-117. [DOI] [PubMed] [Google Scholar]
- Detrich, H.W., III, Johnson, K.A., and Marchese-Ragona, S.P. (1989). Polymerization of Antarctic fish tubulins at low temperatures: energetic aspects. Biochemistry 28, 10085-10093. [DOI] [PubMed] [Google Scholar]
- Detrich, H.W., III, Fitzgerald, T.J., Dinsmore, J.H., and Marchese-Ragona, S.P. (1992). Brain and egg tubulins from Antarctic fishes are functionally and structurally distinct. J. Biol. Chem. 267, 18766-18775. [PubMed] [Google Scholar]
- Detrich, H.W., III, Parker, S.K., Williams, R.C., Jr., Nogales, E., and Downing, K.H. (2000). Cold adaptation of microtubule assembly and dynamics: structural interpretation of primary sequence changes present in the α and β tubulins of Antarctic fishes J. Biol. Chem. 275, 37038-37047. [DOI] [PubMed] [Google Scholar]
- DeZwaan, T.M., Ellingson, E., Pellman, D., and Roof, D.M. (1997). Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration. J. Cell Biol. 138, 1023-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, D., Chikashige, Y., Haraguchi, T., and Hiraoka, Y. (1998). Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J. Cell Sci. 111, 701-712. [DOI] [PubMed] [Google Scholar]
- Downing, K.H. (2000). Structural basis for the interaction of tubulin with proteins and drugs that affect microtubule dynamics. Annu. Rev. Cell Dev. Biol. 16, 89-111. [DOI] [PubMed] [Google Scholar]
- Drummond, D.R., and Cross, R.A. (2000). Dynamics of interphase microtubules in Schizosaccharomyces pombe. Curr. Biol. 10, 766-775. [DOI] [PubMed] [Google Scholar]
- Dutcher, S.K. (2001). The tubulin fraternity: alpha to eta. Curr. Opin. Cell Biol. 13, 49-54. [DOI] [PubMed] [Google Scholar]
- Egel, R., Willer, M., Kjaerulff, S., and Davey, J. (1994). Assessment of pheromone production and response in fission yeast by a halo test of induced sporulation. Yeast 10, 1347-1354. [DOI] [PubMed] [Google Scholar]
- Erickson, H.P., and Stoffler, D. (1996). Protofilaments and rings, two conformations of the tubulin family conserved from bacterial FtsZ to alpha/beta and gamma tubulin. J. Cell Biol. 135, 5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, P.A., and Platzer, E.G. (1978). Interaction of anthelmintic benzimidazoles and benzimidizole derivatives with bovine brain tubulin. Biochim. Biophys. Acta 544, 605-614. [DOI] [PubMed] [Google Scholar]
- Fuller, M.T., Caulton, J.H., Hutchens, J.A., Kaufman, T.C., and Raff, E.C. (1988). Mutations that encode partially functional E2 tubulin subunits have different effects on structurally different microtubule arrays. J. Cell Biol. 107, 141-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachet, Y., Tournier, S., Millar, J.B.A., Hyams, J. S. (2001). A MAP kinase-dependent actin checkpoint ensures proper spindle orientation in fission yeast. Nature 412, 352-355. [DOI] [PubMed] [Google Scholar]
- Gupta, M.L., Jr., Bode, C.J., Dougherty, C.A., Marquez, R.T., and Himes, R.H. (2001). Mutagenesis of β-tubulin cysteine residues in Saccharomyces cerevisiae: mutation of cysteine 354 results in cold-stable microtubules. Cell Motil. Cytoskeleton 49, 67-77. [DOI] [PubMed] [Google Scholar]
- Gupta, M.L., Jr., Bode, C.J., Thrower, D.A., Pearson, C.G., Suprenant, K.A., Bloom, K.S., and Himes, R.H. (2002). β-tubulin C354 mutations that severely decrease microtubule dynamics do not prevent nuclear migration in yeast. Mol. Biol. Cell 13, 2919-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan, I.M. (1998). The fission yeast microtubule cytoskeleton. J. Cell Sci. 11, 1603-1612. [DOI] [PubMed] [Google Scholar]
- Hagan, I.M., and Ayscough, K.R. (2000). Fluorescence microscopy in yeast. In: Protein Localization by Fluorescence Microscopy, ed. V.J. Allan, Oxford: Oxford University Press, 179-206.
- Hagan, I.M., and Hyams, J.S. (1988). The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 89, 343-357. [DOI] [PubMed] [Google Scholar]
- Hagan, I.M., and Peterson, J. (2000). The microtubule organizing centers of Schizosaccharomyces pombe. Curr. Top. Dev. Biol. 49, 133-159. [DOI] [PubMed] [Google Scholar]
- He, X., Patterson, T.E., and Sazar, S. (1997). The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc. Natl. Acad. Sci. USA 94, 7965-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, J.J., and Hyman, A.A. (2003). Dynamics and mechanics of the microtubule plus end. Nature 422, 753-758. [DOI] [PubMed] [Google Scholar]
- Hunter, A.W., and Wordeman, L. (2000). How motor proteins influence microtubule polymerization dynamics. J. Cell Sci. 113, 4379-4389. [DOI] [PubMed] [Google Scholar]
- Jung, M.K., and Oakley, B.R. (1990). Identification of an amino acid substitution in the benA beta-tubulin gene of Aspergillus nidulans that confers thiabendazole resistance and benomyl supersensitivity. Cell Motil. Cytoskeleton 17, 87-94. [DOI] [PubMed] [Google Scholar]
- Jung, M.K., Wilder, I.B., and Oakley, B.R. (1992). Amino acid alterations in the benA (β-tubulin) gene of Aspergillus nidulans that confer benomyl resistance. Cell Motil. Cytoskeleton 22, 170-174. [DOI] [PubMed] [Google Scholar]
- Kraulis, P.J. (1991). MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946-950. [Google Scholar]
- Kwa, M.S.G., Veenstra, J.G., and Roos, M.H. (1994). Benzimidazole resistance in Haemonchus contortus is correlated with a conserved mutation at amino acid 200 in β-tubulin isotype 1. Mol. Biochem. Parasitol. 63, 299-303. [DOI] [PubMed] [Google Scholar]
- Leverson, J.D., Huang, H.K., Forsburg, S.L., and Hunter, T. (2002). The Schizosaccharomyces pombe aurora-related kinase Ark1 interacts with the inner centromere protein Pic1 and mediates chromosome segregation and cytokinesis. Mol. Biol. Cell 13, 1132-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Fanarraga, M., Avila, J., Guasch, A., Coll, M., and Zabala, J.C. (2001). Postchaperonin tubulin folding cofactors and their role in microtubule dynamics. J. Struct. Biol. 135, 219-229. [DOI] [PubMed] [Google Scholar]
- Löwe, J., Li, H., Downing, K.H., and Nogales, E. (2001). Refined structure of alpha beta-tubulin at 3.5 A resolution. J. Mol. Biol. 313, 1045-1057. [DOI] [PubMed] [Google Scholar]
- Mallavarupa, A., Sawin, K., and Mitchison, T. (1999). A switch in microtubule dynamics at the onset of anaphase B in the mitotic spindle of Schizosaccharomyces pombe. Curr. Biol. 9, 1423-1426. [DOI] [PubMed] [Google Scholar]
- Mandelkow, E.M., Mandelkow, E., and Milligan, R.A. (1991). Microtubule dynamics and microtubule caps: a time-resolved cryo-electron microscopy study. J. Cell Biol. 114, 977-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall, L.G., Jeng, R.L., Mulholland, J., and Stearns, T. (1996). Analysis of Tub4p, a yeast gamma-tubulin-like protein: implications for microtubule-organizing center function. J. Cell Biol. 134, 443-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata, J., and Nurse, P. (1997). Tea1 and the microtubular cytoskeleton are important for generating global spatial order with the fission yeast cell. Cell 89, 939-949. [DOI] [PubMed] [Google Scholar]
- Matthews, K.A., Rees, D., and Kaufman, T.C. (1993). A functionally specialized α-tubulin is required for oocyte meiosis and cleavage mitoses in Drosophila. Development 117, 977-991. [DOI] [PubMed] [Google Scholar]
- Matthies, H.J.G., Messina, L.G., Namba, R., Greer, K.J., Walker, M.Y. and Hawley, R.S (1999). Mutations in the α-tubulin 67C gene specifically impair achiasmate segregation in Drosophila melanogaster. J. Cell Biol. 147, 1137-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell, K. (1993). Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123, 127-130. [DOI] [PubMed] [Google Scholar]
- McKean, P.G., Vaughan, S., and Gull, K. (2001). The extended tubulin superfamily. J. Cell Sci. 114, 2723-2733. [DOI] [PubMed] [Google Scholar]
- Miki, F., Okazaki, K., Shimanoki, M., Yamamoto, A., Hiraoka, Y., and Niwa, O. (2002). The 14 kDa dynein light chain-family protein Dlc1 is required for regular oscillatory nuclear movement and efficient recombination during meiotic prophase in fission yeast. Mol. Biol. Cell 13, 939-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison, T.J., and Salmon, E.D. (2001). Mitosis: a history of division. Nat. Cell Biol. 3, E17-21. [DOI] [PubMed] [Google Scholar]
- Moreno, S., Klar, A., and Nurse, P. (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795-823. [DOI] [PubMed] [Google Scholar]
- Murphy, D. B., Wallis, K.T., Machlin, P.S., Ratarie, H., III, and Cleveland, D.W. (1987). The sequence and expression of the divergent β-tubulin in chicken erythrocytes. J. Biol. Chem. 262, 14305-14312. [PubMed] [Google Scholar]
- Nakamura, T., Nakamura-Kubo, M., Hirata, A., and Shimoda, C. (2001). The Schizosaccharomyces pombe spo3+ gene is required for assembly of the forespore membrane and genetically interacts with psy1+-encoding Syntaxin-like protein. Mol. Biol. Cell 12, 3955-3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls, A. (1991). Protein folding and association-Insights from the interfacial and thermodynamic properties of hydrocarbon. Proteins Struct. Funct. Genet. 11, 281-296. [DOI] [PubMed] [Google Scholar]
- Nogales, E. (2001). Structural insights into microtubule function. Annu. Rev. Biophys. Biomol. Struct. 30, 397-420. [DOI] [PubMed] [Google Scholar]
- Nyporko, A.Y., Demchuk, O.N., and Blume, Y.B. (2003). Cold adaptation of plant microtubules: structural interpretation of primary sequence changes in a highly conserved region of α-tubulin. Cell Biol. Int. 27, 241-243. [DOI] [PubMed] [Google Scholar]
- Oakley, B.R. (1985). Microtubule mutants. Can. J. Biochem. Cell Biol. 63, 479-488. [DOI] [PubMed] [Google Scholar]
- Paluh, J.L., Nogales, E., Oakley, B.R., McDonald, K., Pidoux, A.L., and Cande, W.Z. (2000). A mutation in γ-tubulin alters microtubule dynamics and organization and is synthetically lethal with the kinesin-like protein Pkl1p. Mol. Biol. Cell 11, 1225-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda, D., Miller, H. P., Banerjee, A., Luduena, R.F., and Wilson, L. (1994). Microtubule dynamics in vitro are regulated by the tubulin isotype composition. Proc. Natl. Acad. Sci. USA 91, 11358-11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra, M.T., Viera, A., Gomez, R., Page, J., Carmena, M., Earnshaw, W.C., Rufas, J.S., and Suja, J.A. (2002). Dynamic relocalization of the chromosomal passenger complex proteins inner centromere protein (INCENP) and aurora-B kinase during male mouse meiosis. J. Cell Sci. 116, 961-974. [DOI] [PubMed] [Google Scholar]
- Prichard, R.K. (2001). Genetic variability following selection of Haemonchus contortus with anthelmintics. Trends Parasitol. 17, 445-453. [DOI] [PubMed] [Google Scholar]
- Richards, K.L., Anders, K.R., Nogales, E., Schwartz, K., Downing, K.H., and Botstein, B. (2000). Structure-function relationships in yeast. Mol. Biol. Cell 11, 1887-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusan, N.M., Fagerstrom, C.J., Yvon, A.M., and Wadsworth, P. (2001). Cell cycle-dependent changes in microtubule dynamics in living cells expressing green fluorescent protein-alpha tubulin. Mol. Biol. Cell 12, 971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagolla, M., Uzawa, S., and Cande W.Z. (2003). Individual microtubule dynamics contribute to the function of mitotic and cytoplasmic arrays in fission yeast. J. Cell Sci. 116, 4891-4904. [DOI] [PubMed] [Google Scholar]
- Sawin, K.E. (2000). Microtubule dynamics: the view from the tip. Curr. Biol. 10, R860-R862. [DOI] [PubMed] [Google Scholar]
- Schiebel, E. (2000). Gamma-tubulin complexes: binding to the centrosome, regulation and microtubule nucleation. Curr. Opin. Cell Biol. 12, 113-118. [DOI] [PubMed] [Google Scholar]
- Schroer, T.A. (2001). Microtubules don and doff their caps: dynamic attachments at plus and minus ends. Curr. Opin. Cell Biol. 13, 92-96. [DOI] [PubMed] [Google Scholar]
- Severson, A.F., and Bowerman, B. (2002). Cytokinesis: closing in on the central spindle. Dev. Cell 2, 4-6. [DOI] [PubMed] [Google Scholar]
- Sidell, B.D. (2000). Life at body temperatures below 0 degrees C: the physiology and biochemistry of Antarctic fishes. Gravit. Space Biol. Bull. 13, 25-34. [PubMed] [Google Scholar]
- Straight, A.F., Sedat, J.W., and Murray, A.W. (1998). Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J. Cell Biol. 143, 687-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, K.F. and Cleveland, D.W. (1986). Identification of conserved isotype-defining variable region sequences for four vertebrate beta tubulin polypeptide classes. Proc. Natl. Acad. Sci. USA 83, 4327-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda, A., Bahler, J., and Kohli, J. (1995). Microtubule-driven nuclear movements and linear elements as meiosis-specific characteristics of the fission yeasts Schizosaccharomyces versatilis and Schizosaccharomyces pombe. Chromosoma 104, 203-214. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., Higgins, D.G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda, T., Umesono, K., Hirata, A., and Yanagida, M. (1983). Cold-sensitive nuclear division arrest mutants of the fission yeast Schizosaccharomyces pombe. J. Mol. Biol. 168, 251-270. [DOI] [PubMed] [Google Scholar]
- Tran, P.T., Joshi, P., and Salmon, E.D. (1997). How tubulin subunits are lost from the shortening ends of microtubules. J. Struct. Biol. 118, 107-118. [DOI] [PubMed] [Google Scholar]
- Tran, P.T., Marsh, L., Doye, V., Inoue, S., and Chang, F. (2001). A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J. Cell Biol. 153, 397-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin, M., and Stromberg, E. (1995). Cold-stable and cold-adapted microtubules. Int. Rev. Cytol. 157, 1-31. [DOI] [PubMed] [Google Scholar]
- Wallin, M., and Billger, M. (1997). Coassembly of bovine and cod microtubule proteins: the ratio of the different tubulins within hybrid microtubules determines the ability to coassemble at low temperatures, MAPs dependency and effects of Ca2+. Cell Motil. Cytoskeleton 38, 297-307. [DOI] [PubMed] [Google Scholar]
- Woods, A., Sherwin, T., Sasse, R., MacRae, T.H., Baines, A.J., and Gull, K. (1989). Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 93, 491-500. [DOI] [PubMed] [Google Scholar]
- Yamamoto, A., West, R.R., McIntosh, J.R., and Hiraoka, Y. (1999). A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J. Cell Biol. 145, 1233-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, A., Tsutsumi, C., Kojima, H., Oiwa, K., and Hiraoka, Y. (2001). Dynamic behavior of microtubules during dynein-dependent nuclear migrations of meiotic prophase in fission yeast. Mol. Biol. Cell 12, 3933-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden, O., and Katan, T. (1993). Mutations leading to substitutions at amino acid 198 to 200 of beta-tubulin that correlate with benomyl resistance phenotypes of field strains of Botrytis cinerea. Phytopathology 83, 1478-1483. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.