Abstract
In eukaryotic cells, packaging of DNA into highly condensed chromatin presents a significant obstacle to DNA-based processes. Cells use two major strategies including histone modifications and ATP-dependent chromatin remodeling to alter chromatin structure that allows protein factors to gain access to nucleosomal DNA. Beyond their well-established role in transcription, histone modifications and several classes of ATP-dependent chromatin-remodeling complex have been functionally linked to efficient DNA repair. Mi-2/nucleosome remodeling and histone deacetylation (NuRD) complex uniquely possess both nucleosome remodeling and histone deacetylation activities, which play a vital role in regulating transcription. However, the role of the Mi-2/NuRD complex in DNA damage response remains largely unexplored until now. Recent findings reveal that metastasis-associated protein 1 (MTA1), an integral component of the Mi-2/NuRD complex, has successfully made inroads into DNA damage response pathway, and thus, links two previously unconnected Mi-2/NuRD complex and DNA damage response research areas. In this review, we will summarize recent progress concerning the functions of histone modifications and chromatin remodeling in DNA repair, and discuss new role of Mi-2/NuRD complex in DNA damage response.
Keywords: Chromatin-remodeling complex, Histone modification, Acetylation and deacetylation, DNA repair, Mi-2/NuRD complex, MTA1
Eukaryotic genome is packaged into chromatin, which generally reduce accessibility for enzymes that mediate DNA-based cellular processes such as transcription, DNA replication, or DNA damage repair 1, 2. To overcome these regulatory barriers, eukaryotic cells use two major strategies to modify chromatin structures. The first is by post-translational modifications of histones and second by ATP-dependent nucleosome remodeling-both implicated in transcriptional regulation2–7. By analogy to transcription, DNA damage is detected and repaired in the context of chromatin. Therefore, in order for damaged DNA to be repaired efficiently, there must be restructuring of the chromatin to facilitate the accessibility of repair machinery to the site of DNA lesion. As expected, recent studies reveal that histone modifications8–10 and several classes of ATP-dependent chromatin-remodeling complexes11–14 also function in ensuring efficient DNA repair.
Role of histone acetylation/deacetylation in DNA repair
The core histone tails are susceptible to a variety of covalent modifications, including acetylation, methylation, phosphorylation, and ubiquitination15. Due to its tight association with transcriptional regulation, histone acetylation is probably one of the best studied modifications. In eukaryotes, the histones are acetylated and deacetylated on lysine residues in the N-terminal tail and on the surface of the nucleosome core, which regulate chromatin accessibility. Typically, these reactions are catalyzed by enzymes with histone acetyltransferase (HAT) or histone deacetylase (HDAC) activity16. Like its role in transcription7, histone acetylation might modulate the accessibility of damaged-DNA site to DNA-repair machinery or provide appropriate binding surfaces for repair proteins16, 17. Now it becomes increasingly clear that histone acetylation functions in specific DNA-repair pathways. For example, it was found that deletion of the N-terminal tail of or mutation of the acetylatable lysines (Lys 5, Lys 8, Lys 12, and Lys 16) of histone H4 renders cells hypersensitive to DNA double-strand break (DSB)-damaging agents 8, 18. Importantly, reintroduction of a single acetylatable lysine anywhere in the mutant histone H4 tail restores wild-type levels of resistance to DSB damage8, indicating that H4 acetylation plays an important role in DNA repair. In line with these findings, acetylation of histone H4 by the mammalian Trrap (transformation/transcription domain-associated protein)/Tip60 (Tat-interactive protein 60 kDa) HAT complex is required for recruitment/loading of repair proteins to DSBs19. In this context, Trrap depletion impairs both DNA-damage-induced histone H4 hyperacetylation and accumulation of repair molecules at DSBs. This in-turns, result in defective homologous recombination (HR) repair, albeit with the presence of a functional ataxia telangiectasia mutant (ATM)-dependent DNA-damage signaling cascade19. Importantly, the impaired loading of repair proteins and the defect in DNA repair in Trrap-deficient cells can be counteracted by chromatin relaxation, indicating that the observed DNA-repair defect in the absence of Trrap was due to impeded chromatin accessibility at the site of DNA breaks 19. Similarly, ectopic expression of mutated Tip60 lacking histone acetylase activity results in cells with defective DSB repair20.
In S. cerevisiae, acetylation of histone H4 by the catalytic subunit ESA1 (the yeast homolog of Tip60) of the NuA4 (nuclesome acetyltransferase of H4) HAT complex is also required for DSB repair 8. This was further supported by the finding that mutation in ESA1 results in increased cellular sensitivity to DSB-inducing agents and a defect in non-homologous end joining (NHEJ)8. In addition, mutation of Yng2, another component of the yeast NuA4 HAT complex, results in critical defect for genome-wide nucleosomal histone H4 acetylation, and hypersensitivity to and inefficient repair of DNA damage caused by genotoxic agents that induce replication fork stalling 21. During HR repair in yeast, the HAT Gnc5 is specifically recruited to DSB sites, where it contributes to an increased H3 and H4 histone acetylation at the site of break-flanking22. In addition, mutations in either specific lysine residues in the histone H3 tail or the yeast histone acetyltransferase Hat1p influences the HR repair of DSBs through Asf1p-dependent chromatin assembly, resulting in hypersensitivity to DSB-inducing agents 23.
Interestingly, histone deacetylation activities have also been linked to DSB repair 9. For example, it was shown that Sir2p, an NAD-dependent HDAC, and its interacting partners, Sir3p and Sir4p, influence NHEJ 24–26. Furthermore, these factors re-localize from telomeres to the site of DNA DSBs, and cells lacking these factors exhibit a defect in NHEJ24–26. A recent study further demonstrates that the Sin3p/Rpd3p HDAC complex is required for an efficient repair by NHEJ in S. cerevisiae 10. In this context, lysine 16 of histone H4 (H4K16) is deacetylated in the vicinity of chromosomal DSBs, and this deacetylation requires the HDAC Sin3-Rpd3. Furthermore, Sin3 or Rpd3 mutants are defective in NHEJ and are susceptible to DSBs 10. In line with this finding, studies from the same group also showed that acetylation of histone H3 on lysine 9 (H3K9Ac) and lysine 56 (H3K56Ac) is reduced in response to DNA damage in human cells identified by a large-scale screen for DNA-damage-responsive histone modifications, although the precise functions of H3K9Ac and H3K56Ac in the DNA damage response remains to be defined 27. Recently, H3K56Ac has been reported to have a role in DNA repair in yeast 28. Mutation of K56 in histone H3 display hypersensitivity to DNA damaging agents, specifically those that induce damage during S-phase29, 30. H3K56Ac is predicted to result in an opening of the nucleosome at the DNA entry/exit points, which could facilitate recruitment of repair factors31. Alternatively, or in addition, H3K56ac could serve as a recruitment site for bromodomain-containing protein involved in repair process31. Interestingly, bromodomains are conserved protein sequence motifs that are present in the subunits of the RSC (remodels the structure of chromatin), SWI/SNF (switching/sucrose non-fermenting), and SWR1 (SWT2-related ATPase 1) chromatin-remodeling complexes. In support of this hypothesis, a very recent study showed that acetylation of H3K56 does not directly affect the compaction of chromatin but has modest effects on remodeling by SWI/SNF and RSC complexes 32.
Interestingly, it was suggested that after ultraviolet (UV) irradiation all histones display a rapid hyperacetylation phase followed by a hypoacetylated state, although the specific histone marks that changed were unidentified 33. It is possible that acetylation is required initially to relax the chromatin, allowing access of repair proteins, and the subsequent deacetylation may serve to locally stabilize the chromatin for rejoining of the DNA ends 6, 10. Another possibility is that the HDAC is first recruited to create a hypoacetylated region to allow efficient stabilization and juxtaposition of the two broken ends. Once the repair process is completed the HAT is recruited to help re-establish the correct histone code to allow efficient DNA repair 6, 10. Thus, these two activities might even be recruited to the site of damage as parts of one complex10. Consistent with this hypothesis, it was shown that localized acetylation of histones H3 and H4 is triggered by HR through the histone acetyltransferases Gcn5 and ESA1 and subsequently removed by the histone deacetylases Rpd3, Sir2, and Hst1 22.
Role of ATP-dependent nucleosome-remodeling complexes in DNA repair
In addition to histone modifications, chromatin structure can also be altered by ATP-dependent chromatin-remodeling complexes that utilize energy from ATP hydrolysis to remove or reposition nucleosomes, thereby altering chromatin structure and influencing the accessibility of DNA to other factors 4, 12, 13, 34–37. Based on the different ATPase subunits, the remodeling complexes could be divided into three groups 15, 38. The first group uses the switch 2/sucrose non-fermenting 2 (SWI2/SNF2) or a close relative as the ATPase and includes yeast/human SWI/SNF complex, yeast RSC, and the drosophila Brahama complex15. The INO80 (inositol auxotroph 80) complex is the most recent addition to the SWI/SNF family of chromatin remodelers11. The INO80 complex includes INO80 and SWR1 in S. cerevisiae; INO80, SNF2-related CREB-binding protein (CBP) activator protein (SRCAP), and p400 in mammal; and INO80 and p400 in Drosophila melanogaster11. The second group uses imitation switch (ISWI) or a close relative as the ATPase and includes NURF (nuclesome remodeling factor), CHRAC (chromatin-accessibility complex), and ACF (ATP-utilizing chromatin assembly and remodeling factor) from Drosophila15. The third group uses Mi-2 as the ATPase and includes human nuclesome remodeling and histone deacetylase (NuRD)39–41 and its counterpart in xenopus, the Mi-2 complex42. Recently, at least four distinct ATP-dependent chromatin remodeling complexes, including SWI/SNF, INO80, SWR1, and RSC complexes, have been directly implicated in DNA repair in yeast and mammalian cells 13, 43–46.
Among them, RSC seems uniquely suited for a role in early chromatin remodeling at the DSB, because it is rapidly recruited to the DSB and functions in both NHEJ and HR 47–49. RSC participates in an early step of DSB repair by preparing the binding site for the Mre 11 and Ku complex49, as depletion of the ATPase subunit Sth1 or Rsc2 severely reduces chromatin remodeling and loading of Mre11 and Ku at the sites of DSBs. In addition, RSC also recruits Tel 1 and Mec 1 (the yeast ATR and ATM homolog, respectively) to the breaks site, and is necessary to ensure full levels of H2AX phosphorylation50. Consistent with these results, it was shown that Rsc2 is needed for efficient activation of the Rad53-dependent checkpoint, Cohesin’s association with the break site, and the DNA-damage-induced changes in nucleosome structure surrounding the DSB site50. Furthermore, the Rsc1p subunit of RSC directs nucleosome sliding immediately after DSB creation and is required for efficient induction of γ-H2A and strand resection during repair by HR 51. These findings suggest that the RSC complex participates in remodeling and in increasing the ability of MRN to bind and medicate resection of DNA ends 43.
Accessibility within chromatin is an important factor in the prompt removal of UV-induced DNA damage by nucleotide excision repair (NER). This repair pathway is used for the removal of the UV-induced cyclobutane pyrimidine dimer (CPD) and pyrimidine (6–4) pyrimidone photoproducts. Chromatin remodeling by the SWI/SNF complex has been shown to play an important modulating role in NER in vitro and in vivo 52. It was shown that Brg1, the ATPase subunit of SWI/SNF, facilitates different stages of NER by initially modulating UV-induced chromatin relaxation and stabilizing xeroderma pigmentosum group C (XPC) at the damage sites. This subsequently stimulates the recruitment of xeroderma pigmentosum group G (XPG) and proliferating cell nuclear antigen (PCNA) to successfully culminate the repair 52. In support of this observation, it was reported that the SWI/SNF-deficient human carcinoma cell line SW13 cells are sensitive to UV radiation. In contrast, SW13 cells with ectopic Brg1 expression regain active SWI/SNF and become significantly more resistance to UV radiation, suggesting that SWI/SNF protects cells against deleterious consequences of UV-induced DNA damage 53. In addition, human SWI/SNF core subunit SNF5 (hSNF5) also associates with UV damage recognition factor XPC at the damage site in response to UV irradiation and promotes NER by influencing the recruitment of ATM kinase to the damage site and activation of ATM by phosphorylation54. Consequently, SNF5 deficiency results in a defect in H2AX and breast cancer type 1 susceptibility protein (BRCA1) phosphorylation at the damage site54 and increased sensitivity to geneotoxic stress, suggesting a role of SNF5 in the DNA damage response55. A study in yeast by Gong and colleagues demonstrated enhanced interaction between the Rad4 (the yeast homologue of XPC) and two subunits of SWI/SNF complex, SNF5 and SNF6, after UV irradiation53. In S. cerevisiae, SWI/SNF complex enhances the repair of UV-induced CPDs in chromatin by photolyase 56 and stimulates the excision of chemical adducts within the core nucleosome57. In addition to its role in stimulating NER of damaged chromatin, it was found that the mammalian SWI/SNF complex is also recruited to the sites of DSBs and facilitates DSB repair by promoting γ-H2AX induction at DSB-surrounding chromatin following ionizing radiation (IR) treatment14, 47. The inactivation of SWI/SNF subunits leads to deficient H2AX phosphorylation in the presence of normal ATM, ATM and rad3-related protein (ATR), and DNA-dependent protein kinase (DNA-PK) activation and inefficient DSB repair 14.
Similarly, the INO80 complex is recruited to a HO endonuclease-induced DSB through direct interaction of its Arp4 or Nhp10 subunit with γ-H2AX, and the loss of γ-H2AX results in reduced INO80 recruitment to the DSB 2, 3, 58. Notably, the yeast strains lacking a functional INO80 complex are hypersensitive to DSB-inducing genotoxic agents 2, 3, 5. A recent study suggests that RNA interference-mediated knockdown of INO80 increased cellular sensitivity toward UV-induced DNA damage as determined by survival assays59. Functional assays revealed that INO80 is essential for HR-based DNA repair59. It was shown that Ies4 subunit of the INO80 complex is phosphorylated by the Mec1/Tel1 (ATM/ATR in mammals) kinase in response to DNA damage, and mutation of phosphorylation sites of les4 influences DNA damage checkpoint pathways 60. The INO80 Arp4 subunit that interacts directly with γ-H2AX is also present in the histone exchange complex SWR161. Depletion of the SWR1 subunit also renders cells hypersensitive to DNA damaging agents61, 62, and this complex also catalyzes chromatin structure alternations at DSBs through exchange of γ-H2A and H2AZ 63, 64. Taken together, it is becoming increasing clear that histone acetylation/deactylation and chromatin remodeling facilitate DNA repair beyond their well-documented role in transcription, presumably by opening or loosening compact nucleosomal structure close to sites of damage.
Mi-2/NuRD complex uniquely possesses both nucleosome remodeling and HDAC activities
The Mi-2/NuRD complex is a multi-subunit complex that consists of a SNF2-related chromatin remodeling ATPase (Mi-2), a member of the MBD family of methyl CpG binding domain proteins (MBD3), histone deacetylases (HDAC1 and HDAC2), a histone binding protein (RbAp46/p48), a protein of unknown function (known as p66), an interesting subunit encoded by one of three genes (MTA1, MTA2, or MTA3) in mammals 65–68. The composition of this complex is highly conserved from xenopus to human. This complex, Mi-2/NuRD, is the only known protein entity that uniquely possesses both nucleosome remodeling and histone deacetylase activities15, 67. It has been shown to play a central role in transcriptional regulation of a number of target genes in vertebrates, invertebrates and fungi 15, 40, 66, 67, 69. Although the complex is often lined with transcriptional repression associated with the HDAC activity and the intrinsic nucleosome remodeling activity of Mi-267, the function of NuRD complex in transcriptional activation has been suggested70. For example, it was found that Mi-2α, previously studied as a subunit in the NuRD co-repressor complex, enhanced c-Myb-dependent reporter activation70. The rationale for the unexpected co-activator function seems to lie in a dual function of Mi-2α, by which this factor is able to repress transcription in a helicase-dependent and activate in a helicase-independent fashion, as revealed by Gal4-tethering experiments70. In addition, NuRD complex also plays a role in transcriptional termination71, centrosome maintenance72,73, tumorigenesis, and tumor progression74,75. Given that ATP-dependent chromatin remodeling has mechanistically similar role in transcription and DNA damage repair by disrupting chromatin to give regulatory and repair factors direct access to DNA, it is reasonably postulated that the Mi-2/NuRD complex, like other ATP-dependent chromatin-remodeling complexes11, 12, 34, 36, might be involved in DNA damage repair, beyond its well-established role in transcription 67.
MTA1, an integral component of Mi-2/NuRD complex, is a multifunctional DNA damage responsive protein
One integral subunit of the NuRD complex is the metastasis-associated protein 1 (MTA1), which was originally identified by differential cDNA library screening using the highly metastatic and nonmetastatic rat mammary adenocarcinoma cell lines76. Subsequent studies demonstrate that MTA1 is up-regulated in a wide range of human cancers and plays an important role in tumorigenesis, tumor invasion and metastasis 74, 75. As a dual-function coregulator by modulating the accessibility of DNA to transcription factors77, MTA1 functions not only as a transcriptional repressor of estrogen receptor-α78, BRCA179, Six380, and p21WAF1181 genes, but also as a transcriptional activator via interacting with RNA polymerase II on the breast cancer-amplified sequence 3 (BCAS3)82 and paired box gene 5 (Pax5) 83 promoters. The co-repressor versus co-activator activity of MTA1 might be influenced by its binding partners on the promoter region of various genes. In addition to deacetylation of histone, MTA1/2-HDAC complex has been shown to interact with and deacetylate non-histone proteins, including p53, hypoxia-inducible factor-1α, and estrogen receptor-α84. Interestingly, MTA1 also undergoes autoacetylation82. In this context, MTA1 is acetylated at lysine 626 by histone acetyltransferase p300, which might contribute to its co-activator activity on BCAS3 transcription82. However, new functions and related signaling transduction pathways of MTA1 remain to be further explored. Since it is becoming increasingly clear that chromatin structure has an impact on the DNA damage response and is modulated in response to DNA damage 85, it is therefore not surprising that our recent findings have linked the chromatin modifier MTA1 to DNA-damage response pathway, in addition to its paramount role in cancer and coregulator biology.
Initial evidence for a role of MTA1 protein in DNA damage response pathway came from experiments showing that MTA1 is stabilized in response to IR86. Mechanistically, MTA1 is targeted by the E3 ubiquitin ligase constitutive photomorphogenesis 1 (COP1) for degradation via the ubiquitin-proteasome pathway86. In response to DNA damage, ATM kinase phosphorylates COP1 on Serine 387 and promotes its auto-degradation 87, which might contribute to the increase in MTA1 stability following DNA damage by dampening the ability of COP1 to negatively regulate MTA1. One of the hallmarks of defective DNA repair is increased radiation sensitivity. We found that knockout of MTA1 (MTA1−/−) in mouse embryonic fibroblasts (MEFs) resulted in increased cellular sensitivity to IR that induces DSBs and decreased clonogenic survival, suggesting that MTA1 is important for cell survival after DNA damage 86. We further demonstrated that MTA1 is involved in DSB repair. Neutral comet assay that specifically measures DNA DSBs 14, 88 showed that MTA1−/− MEFs exhibit a decreased repair efficiency and an increased level of damaged DNA as compared with its wild-type controls after IR treatment. Given the fact that MTA1−/− MEFs still contain MTA2 and MTA3 86, 88, we concluded that MTA1 is required for efficient DSB repair.
One of the early events in mammals in response to the induction of DNA damage is the rapid phosphorylation of histone H2AX on Serine 139 by the phosphatidylinositol-3 kinase-like family of kinases at the DSB site 89–91. It is believed that ATM is the major kinase responsible for phosphorylating H2AX in response to DSBs92, where ATR is also required for UV-induced damage and DNA damage occurring at stalled replication forks93. Phosphorylated H2AX (referred to as γ-H2AX) in mammalian cells accumulates upon damage in the chromatin surrounding DSBs (known as nuclear foci), and recruits a multitude of other factors implicated in DSB repair to region of damaged chromatin in order to mend the damage14, 90, 91, 94–96. Thus, the absence of γ-H2AX foci correlates with impaired formation of repair foci at sites of damage 2, 90. Several studies in mammalian cells implicate H2AX in both NHEJ and HR repair pathways 92, 93, 97, and mouse embryonic stem cells deficient for H2AX were shown to be sensitive to IR-induced DSBs 94, 98. Interestingly, we found that knockout of MTA1 decreased the induction of γ-H2AX and compromised the γ-H2AX foci formation in response to IR86. Importantly, the observed defect in γ-H2AX induction in the MTA1-knockout cells was efficiently restored by reintroduction of MTA1 in these MTA1-deficient cells86, suggesting that MTA1 is critical for the induction of γ-H2AX and the formation of γ-H2AX foci in response to IR-induced DSBs. Therefore, it is tempting to propose that MTA1-containing Mi-2/NuRD complex, like the SWI/SNF complexes14, can function upstream of γ-H2AX, whereas the INO80 complex rather contributes to the downstream repair events3. The INO80 complex, albeit it interacts with γ-H2AX, is not required for the induction of γ-H2AX following DNA damage3, whereas mammalian SWI/SNF and Mi-2/NuRD complexes are critical for the optimal induction of γ-H2AX14, 86. Thus, different members of the ATP-dependent chromatin remodeling complex family may adopt distinct mechanisms for facilitating DSB repair. Given the crucial role for γ-H2AX in chromosomal DSB repair and cell survival after DNA damage, these results suggest that MTA1 facilitates DSB repair and hence increase the resistance to DNA damage, at least in part, by promoting γ-H2AX induction. Since γ-H2AX induction is an early event in DSB repair and has been implicated in both NHEJ and HR pathways 99, 100, MTA1 might be required for the processing of newly broken DNA ends in a nucleosomal context to facilitate HR or NHEJ. Our findings have raised a number of interesting questions to be addressed. For example, whether MTA1 directly interacts with γ-H2AX, and how MTA1 affects γ-H2AX induction in response to DNA damage would be the subject of continued studies. For example, whether MTA1 affects the expression of the kinases, ATM, ATR, DNA-PK, which are all responsible for the phosphorylation of H2AX, or directly acts on the chromatin to facilitate H2AX phosphorylation. Alternations of the structure of the Mi-2/NuRD remodeling could directly affect the accessibility of H2AX at the sites of DSBs. Thus, the Mi-2/NuRD complex might facilitate H2AX phosphorylation by influencing the higher order chromatin structure in such a way as to increase the accessibility of the H2AX-containing nucleosomes.
As mentioned above, recent studies indicate that γ-H2AX is required for the recruitment of chromatin-remodeling complex to the sites of DNA damage. In line with this notion, γ-H2AX in yeast is required for the recruitment of the NuA4 HAT complex to a region proximal to a DSB induced by HO endonuclease 58. The recruitment of this HAT complex to γ-H2AX is mediated by Arp4 and leads to acetylation of chromatin surrounding the break site, thereby facilitating efficient repair of DNA damage 58. Arp4 is also a subunit of the INO80 chromatin remodeling complex, which is also recruited to a DSB by a specific interaction with γ-H2AX 2, 3. In support of this notion, loss of γ-H2AX results in reduced INO80 recruitment to the DSBs 2, 3, which in turn is required for efficient processing of the DSB into single-stranded DNA2. Similarly, phosphorylation of H2AX results in the recruitment of the SWR1 chromatin-remodeling complex 58. Thus, whether γ-H2AX facilitates the recruitment of MTA1-containing Mi-2/NuRD complex to the damaged chromatin after DNA damage remains to be further investigated. Although MTA1 facilitating DSB repair might be through promoting the γ-H2AX induction, we cannot formally exclude the possibility that MTA1 also contributes to DSB repair by regulating the expression of yet unidentified DSB repair proteins.
The p53 protein is a critical component of the DNA damage response that plays numerous roles in a variety of DNA repair pathways, including DSB, single-strand break, base excision repair, and mismatch repair101, 102. To further investigate the potential mechanism for the role of MTA1 protein in DSB repair, we found that MTA1 interjects into the p53-dependent DNA repair88. In this context, MTA1 is required for the stabilization of p53 protein by inhibiting its ubiquitination mediated by E3 ubiquitin ligases 88. As a result, MTA1 regulates the p53-dependent transcription of p53R2, a direct p53 target gene for supplying nucleotides to repair damaged DNA 88, 103. The ability of p53R2 to supply nucleotides for repairing DNA damage requires the presence of a functional p53 protein103, and inactivation of p53R2 impairs DNA repair and sensitizes several types of cancer cells to DNA-damaging anticancer agents or to ionizing radiation (IR)103–105. We found that knockout of MTA1 impairs p53-dependent p53R2 transcription and compromises DNA repair. Interestingly, these events could be reversed by MTA1 reintroduction in the MTA1−/− cells88. Given the fact that there was no compensatory effect of MTA1 depletion in MEFs on the levels of MTA288, which as a part of the NuRD complex has been previously shown to deacetylate p53 and inhibits p53-dependent transcription of genes important in cell growth and apoptosis 106, these findings suggest that MTA1 interjects into the p53-dependent DNA repair 88 (Fig. 1).
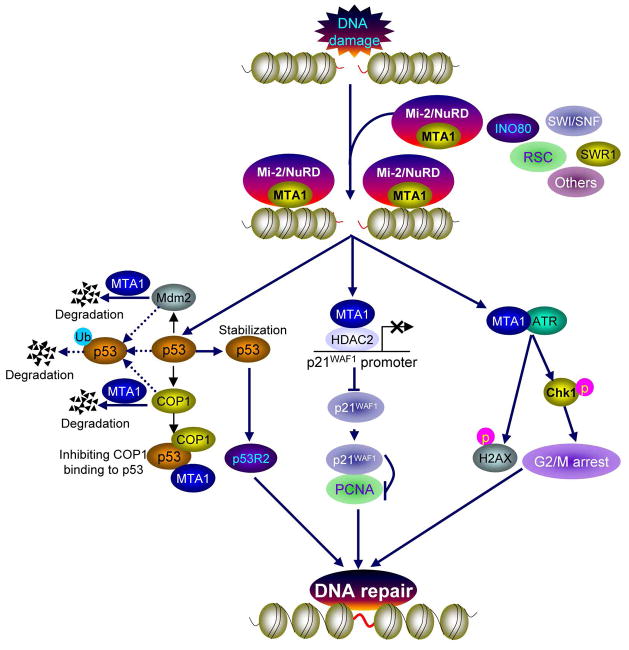
Figure 1. Summary of recently discovered functions of MTA1 in DNA damage response.
In response to DNA damage, the Mi-2/NuRD complex, like other ATP-dependent chromatin remodeling complexes (such as INO80, SWI/SNF, RSC, and SWR1), might be recruited to the site of damaged DNA, and exerts its function in DNA repair by multiple different mechanisms. It was shown that MTA1, an integral component of the Mi-2/NuRD complex, is a multiple functional DNA damage responsive protein. MTA1 regulates p53-dependent and –independent DNA repair processes following IR treatment by modulating the p53-p53R2 and p21WAF1-PCNA pathway, respectively. In this context, MTA1 controls p53 stability through inhibiting the E3 ubiquitin ligases Mdm2- and COP1-mediated ubiquitination by destabilizing of Mdm2 and COP1 and/or by competing with COP1 to bind to p53, thereby regulating the p53-dependent transcription of p53R2, a direct p53 target gene for supplying nucleotides to repair damaged DNA (left panel). In addition, MTA1 transcriptionally suppresses p21WAF1 expression by recruitment of MTA1/HDAC2 complex onto p21WAF1 promoter, and inhibits p21WAF1 binding to PCNA, thereby facilitates PCNA-dependent DNA repair. In addition, MTA1 is required for ATR-mediated DNA damage checkpoint function in response to UV irradiation. In this case, MTA1 interacts with ATR and is required for the ATR-mediated Chk1 activation and γ-H2AX induction following UV irradiation. Consequently, MTA1 deficiency results in a defective G2/M DNA damage checkpoint and increased cellular sensitivity to UV irradiation.
Following this observation, we further discovered a p53-independent role of MTA1 in DNA damage response. p21WAF1 represents one of the best characterized downstream targets of p53107, 108, and inhibits various PCNA-dependent DNA repair processes by binding to PCNA and interfering with PCNA-dependent DNA polymerase activity109–112. Our recent findings reveal that MTA1 is a p53-independent transcriptional co-repressor of p21WAF1 by recruitment of MTA1/HDAC2 complexes onto p21WAF1 promoter81. As a result, MTA1 depletion, in spite of its effect on p53 down-regulation, superinduces p21WAF1, increases p21WAF1 binding to PCNA, and decreases the nuclear accumulation of PCNA in response to IR-induced damage81, 88. Consequently, MTA1 expression in p53-null cells results in increased induction of γ-H2AX foci and DSBs repair, and decreased DNA damage sensitivity following IR treatment. These findings uncover the existence of an additional p53-independent role of MTA1 in DNA damage response, at least in part, by modulating p21WAF1-PCNA pathway81(Fig.1).
Several lines of evidence have implicated the SWI/SNF chromatin remodeling complex in efficient repair of UV-induced DNA damage 52–54, 56, 113. In addition to its role in the repair of DSBs caused by IR, recent studies reveal that MTA1 also participates in UV-induced DNA damage checkpoint pathway. It was shown in an earlier study that the components of Mi-2/NuRD complex, including MTA1, MTA2, HDAC1, HDAC2, and Mi-2, could be detected in the immunoprecipitates of ATR114, one of key regulators of the checkpoint pathways in the mammalian DNA damage response115. These results suggest that there may be a linkage between the role of ATR in mediating checkpoints induced by DNA damage and chromatin modulation via remodeling and deacetylation114. It is well accepted that ATM is primarily activated by DSB-inducing agents including IR, while ATR is activated by stalled replication forks and agents that produce bulky adducts, such as UV irradiation115, 116. Furthermore, a recent study showed that UV irradiation induces the protein expression of Mi-2, a core subunit of the Mi-2/NuRD complex39–42, by regulating protein translation and stability 117. Based on these findings, we hypothesized that Mi-2/NuRD complex may implicate in the UV-induced DNA damage response in mammalian cells and contribute to the regulation of DNA damage checkpoints. In support of this hypothesis, we found that MTA1 is required for activation of ATR following UV irradiation, as depletion of MTA1 severely impaired ATR-dependent phosphorylation of checkpoint kinase 1 (Chk1) and H2AX118. These findings further support the notion that MTA1 might act as an upstream regulator of γ-H2AX in response to IR- or UV-induced DNA damage. As a result, depletion of MTA1 results in the abrogation of G2-M checkpoint and increased cellular sensitivity to UV-induced DNA damage118. The molecular mechanism for the requirement of MTA1 in the ATR-mediated checkpoint activation is currently being investigated in our lab. One possibility is that MTA1 as a chromatin modifier could alter chromatin structure in an unknown way in response to DNA damage, resulting in increased accessibility of damaged DNA to repair factors. Taken together, these findings suggest that MTA1 also plays a role in UV-induced ATR-mediated DNA damage checkpoint pathway (Fig.1).
Conclusion and perspective
In summary, it is becoming increasingly clear that, like other chromatin-remodeling complexes, Mi-2/NuRD complex is also implicated in DNA damage repair, emphasizing the evolutionally conserved functions of this family of chromatin-remodeling complexes in DNA repair. In the context, we found that MTA1, an integral component of Mi-2/NuRD complex, is a multifunctional DNA damage responsive protein and involved in multiple DNA damage pathways (Fig. 1). Based on lessons learned from transcription, the Mi-2/NuRD remodeling complex might be used to generate nucleosome-free regions around the DNA damage sites in order to facilitate the access of large DNA repair machinery or to create specific chromatin structure suitable for DNA repair3. Studies addressing the molecular mechanism by which it does so are in their infancy and many more questions remain to be addressed. Such studies may reveal where and how Mi-2/NuRD complex induces changes in chromatin near sites of damage, and how this influences DNA repair. Importantly, DNA damage responsive proteins play key roles in tumorigenesis, and their activities, in part, determine the outcome of cancer radiotherapy and chemotherapy that function by generating DNA damage119. DNA repair provides as a common mechanism for cancer-therapy resistance. Given the fact that MTA1 is widely up-regulated in human cancers and facilitates the repair of the damaged DNA, it is speculated that drugs or inhibitors targeting MTA1 may effectively sensitize tumor cells to radiotherapy and DNA-damaging chemotherapies.
Acknowledgments
We are grateful to the members of the Kumar laboratory for their inputs throughout this work, and apologize to our many colleagues whose relevant studies could not be cited here due to space limitations. This work was supported by NIH grants CA98823 and CA98823-S1 (to RK).
Abbreviations
- ATM
ataxia telangiectasia mutant
- ATR
ATM and Rad-3 related
- DSB
double-strand break
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- HR
homologous recombination
- INO80
inositol auxotroph 80
- IR
ionizing radiation
- MEF
mouse embryonic fibroblast
- MTA1
metastasis-associated protein 1
- NER
nucleotide excision repair
- NHEJ
non-homologous end joining
- NuA4
nucleosome acetyltransferase of H4
- NuRD
nucleosome remodeling and histone deacetylase
- RSC
remodels the structure of chromatin
- SWI/SNF
switching/sucrose non-fermenting
- SWR1
SWI2-realted ATPase 1
- UV
ultraviolet
References
- 1.Shimada K, Oma Y, Schleker T, Kugou K, Ohta K, Harata M, Gasser SM. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr Biol. 2008;18:566–575. doi: 10.1016/j.cub.2008.03.049. [DOI] [PubMed] [Google Scholar]
- 2.van Attikum H, Fritsch O, Hohn B, Gasser SM. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell. 2004;119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 3.Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, Shen X. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 4.Vignali M, Hassan AH, Neely KE, Workman JL. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 6.Ehrenhofer-Murray AE. Chromatin dynamics at DNA replication, transcription and repair. Eur J Biochem. 2004;271:2335–2349. doi: 10.1111/j.1432-1033.2004.04162.x. [DOI] [PubMed] [Google Scholar]
- 7.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 8.Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez-Capetillo O, Nussenzweig A. Linking histone deacetylation with the repair of DNA breaks. Proc Natl Acad Sci U S A. 2004;101:1427–1428. doi: 10.1073/pnas.0307342101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jazayeri A, McAinsh AD, Jackson SP. Saccharomyces cerevisiae Sin3p facilitates DNA double-strand break repair. Proc Natl Acad Sci U S A. 2004;101:1644–1649. doi: 10.1073/pnas.0304797101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrison AJ, Shen X. Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat Rev Mol Cell Biol. 2009;10:373–384. doi: 10.1038/nrm2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bao Y, Shen X. Chromatin remodeling in DNA double-strand break repair. Curr Opin Genet Dev. 2007;17:126–131. doi: 10.1016/j.gde.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 13.van Attikum H, Gasser SM. ATP-dependent chromatin remodeling and DNA double-strand break repair. Cell Cycle. 2005;4:1011–1014. doi: 10.4161/cc.4.8.1887. [DOI] [PubMed] [Google Scholar]
- 14.Park JH, Park EJ, Lee HS, Kim SJ, Hur SK, Imbalzano AN, Kwon J. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. Embo J. 2006;25:3986–3997. doi: 10.1038/sj.emboj.7601291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng Q, Zhang Y. The NuRD complex: linking histone modification to nucleosome remodeling. Curr Top Microbiol Immunol. 2003;274:269–290. doi: 10.1007/978-3-642-55747-7_10. [DOI] [PubMed] [Google Scholar]
- 16.Kurdistani SK, Grunstein M. Histone acetylation and deacetylation in yeast. Nat Rev Mol Cell Biol. 2003;4:276–284. doi: 10.1038/nrm1075. [DOI] [PubMed] [Google Scholar]
- 17.van Attikum H, Gasser SM. The histone code at DNA breaks: a guide to repair? Nat Rev Mol Cell Biol. 2005;6:757–765. doi: 10.1038/nrm1737. [DOI] [PubMed] [Google Scholar]
- 18.Megee PC, Morgan BA, Smith MM. Histone H4 and the maintenance of genome integrity. Genes Dev. 1995;9:1716–1727. doi: 10.1101/gad.9.14.1716. [DOI] [PubMed] [Google Scholar]
- 19.Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- 20.Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 21.Choy JS, Kron SJ. NuA4 subunit Yng2 function in intra-S-phase DNA damage response. Mol Cell Biol. 2002;22:8215–8225. doi: 10.1128/MCB.22.23.8215-8225.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamburini BA, Tyler JK. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol. 2005;25:4903–4913. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin S, Parthun MR. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol Cell Biol. 2002;22:8353–8365. doi: 10.1128/MCB.22.23.8353-8365.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsukamoto Y, Kato J, Ikeda H. Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature. 1997;388:900–903. doi: 10.1038/42288. [DOI] [PubMed] [Google Scholar]
- 25.Martin SG, Laroche T, Suka N, Grunstein M, Gasser SM. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 1999;97:621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- 26.Mills KD, Sinclair DA, Guarente L. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell. 1999;97:609–620. doi: 10.1016/s0092-8674(00)80772-2. [DOI] [PubMed] [Google Scholar]
- 27.Tjeertes JV, Miller KM, Jackson SP. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. Embo J. 2009;28:1878–1889. doi: 10.1038/emboj.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 29.Hyland EM, Cosgrove MS, Molina H, Wang D, Pandey A, Cottee RJ, Boeke JD. Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25:10060–10070. doi: 10.1128/MCB.25.22.10060-10070.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ozdemir A, Spicuglia S, Lasonder E, Vermeulen M, Campsteijn C, Stunnenberg HG, Logie C. Characterization of lysine 56 of histone H3 as an acetylation site in Saccharomyces cerevisiae. J Biol Chem. 2005;280:25949–25952. doi: 10.1074/jbc.C500181200. [DOI] [PubMed] [Google Scholar]
- 31.Wong LY, Recht J, Laurent BC. Chromatin remodeling and repair of DNA double-strand breaks. J Mol Histol. 2006;37:261–269. doi: 10.1007/s10735-006-9047-4. [DOI] [PubMed] [Google Scholar]
- 32.Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramanathan B, Smerdon MJ. Changes in nuclear protein acetylation in u.v-damaged human cells. Carcinogenesis. 1986;7:1087–1094. doi: 10.1093/carcin/7.7.1087. [DOI] [PubMed] [Google Scholar]
- 34.Morrison AJ, Shen X. DNA repair in the context of chromatin. Cell Cycle. 2005;4:568–571. [PubMed] [Google Scholar]
- 35.Pandita TK, Richardson C. Chromatin remodeling finds its place in the DNA double-strand break response. Nucleic Acids Res. 2009;37:1363–1377. doi: 10.1093/nar/gkn1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrison AJ, Shen X. Chromatin modifications in DNA repair. Results Probl Cell Differ. 2006;41:109–125. doi: 10.1007/400_008. [DOI] [PubMed] [Google Scholar]
- 37.Karagiannis TC, El-Osta A. Chromatin modifications and DNA double-strand breaks: the current state of play. Leukemia. 2007;21:195–200. doi: 10.1038/sj.leu.2404478. [DOI] [PubMed] [Google Scholar]
- 38.Bao Y, Shen X. SnapShot: chromatin remodeling complexes. Cell. 2007;129:632. doi: 10.1016/j.cell.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 39.Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 40.Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 42.Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 43.Huertas D, Sendra R, Munoz P. Chromatin dynamics coupled to DNA repair. Epigenetics. 2009;4:31–42. doi: 10.4161/epi.4.1.7733. [DOI] [PubMed] [Google Scholar]
- 44.Escargueil AE, Soares DG, Salvador M, Larsen AK, Henriques JA. What histone code for DNA repair? Mutat Res. 2008;658:259–270. doi: 10.1016/j.mrrev.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 45.Dinant C, Houtsmuller AB, Vermeulen W. Chromatin structure and DNA damage repair. Epigenetics Chromatin. 2008;1:9. doi: 10.1186/1756-8935-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Osley MA, Shen X. Altering nucleosomes during DNA double-strand break repair in yeast. Trends Genet. 2006;22:671–677. doi: 10.1016/j.tig.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 47.Chai B, Huang J, Cairns BR, Laurent BC. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 2005;19:1656–1661. doi: 10.1101/gad.1273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shim EY, Ma JL, Oum JH, Yanez Y, Lee SE. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol Cell Biol. 2005;25:3934–3944. doi: 10.1128/MCB.25.10.3934-3944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shim EY, Hong SJ, Oum JH, Yanez Y, Zhang Y, Lee SE. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol Cell Biol. 2007;27:1602–1613. doi: 10.1128/MCB.01956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang B, Qiu J, Ratnakumar K, Laurent BC. RSC functions as an early double-strand-break sensor in the cell’s response to DNA damage. Curr Biol. 2007;17:1432–1437. doi: 10.1016/j.cub.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kent NA, Chambers AL, Downs JA. Dual chromatin remodeling roles for RSC during DNA double strand break induction and repair at the yeast MAT locus. J Biol Chem. 2007;282:27693–27701. doi: 10.1074/jbc.M704707200. [DOI] [PubMed] [Google Scholar]
- 52.Zhao Q, Wang QE, Ray A, Wani G, Han C, Milum K, Wani AA. Modulation of nucleotide excision repair by mammalian SWI/SNF chromatin-remodeling complex. J Biol Chem. 2009;284:30424–30432. doi: 10.1074/jbc.M109.044982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gong F, Fahy D, Liu H, Wang W, Smerdon MJ. Role of the mammalian SWI/SNF chromatin remodeling complex in the cellular response to UV damage. Cell Cycle. 2008;7:1067–1074. doi: 10.4161/cc.7.8.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ray A, Mir SN, Wani G, Zhao Q, Battu A, Zhu Q, Wang QE, Wani AA. Human SNF5/INI1, a component of the human SWI/SNF chromatin remodeling complex, promotes nucleotide excision repair by influencing ATM recruitment and downstream H2AX phosphorylation. Mol Cell Biol. 2009;29:6206–6219. doi: 10.1128/MCB.00503-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klochendler-Yeivin A, Picarsky E, Yaniv M. Increased DNA damage sensitivity and apoptosis in cells lacking the Snf5/Ini1 subunit of the SWI/SNF chromatin remodeling complex. Mol Cell Biol. 2006;26:2661–2674. doi: 10.1128/MCB.26.7.2661-2674.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gaillard H, Fitzgerald DJ, Smith CL, Peterson CL, Richmond TJ, Thoma F. Chromatin remodeling activities act on UV-damaged nucleosomes and modulate DNA damage accessibility to photolyase. J Biol Chem. 2003;278:17655–17663. doi: 10.1074/jbc.M300770200. [DOI] [PubMed] [Google Scholar]
- 57.Hara R, Sancar A. The SWI/SNF chromatin-remodeling factor stimulates repair by human excision nuclease in the mononucleosome core particle. Mol Cell Biol. 2002;22:6779–6787. doi: 10.1128/MCB.22.19.6779-6787.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Côté J. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 59.Wu S, Shi Y, Mulligan P, Gay F, Landry J, Liu H, Lu J, Qi HH, Wang W, Nickoloff JA, Wu C, Shi Y. A YY1-INO80 complex regulates genomic stability through homologous recombination-based repair. Nat Struct Mol Biol. 2007;14:1165–1172. doi: 10.1038/nsmb1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morrison AJ, Kim JA, Person MD, Highland J, Xiao J, Wehr TS, Hensley S, Bao Y, Shen J, Collins SR, Weissman JS, Delrow J, Krogan NJ, Haber JE, Shen X. Mec1/Tel1 phosphorylation of the INO80 chromatin remodeling complex influences DNA damage checkpoint responses. Cell. 2007;130:499–511. doi: 10.1016/j.cell.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 61.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 62.Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A. Z into euchromatin. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Papamichos-Chronakis M, Krebs JE, Peterson CL. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 2006;20:2437–2449. doi: 10.1101/gad.1440206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 65.Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 66.Bowen NJ, Fujita N, Kajita M, Wade PA. Mi-2/NuRD: multiple complexes for many purposes. Biochim Biophys Acta. 2004;1677:52–57. doi: 10.1016/j.bbaexp.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 67.Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–5438. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- 68.Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 69.Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saether T, Berge T, Ledsaak M, Matre V, Alm-Kristiansen AH, Dahle O, Aubry F, Gabrielsen OS. The chromatin remodeling factor Mi-2alpha acts as a novel co-activator for human c-Myb. J Biol Chem. 2007;282:13994–14005. doi: 10.1074/jbc.M700755200. [DOI] [PubMed] [Google Scholar]
- 71.Alen C, Kent NA, Jones HS, O’Sullivan J, Aranda A, Proudfoot NJ. A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol Cell. 2002;10:1441–1452. doi: 10.1016/s1097-2765(02)00778-5. [DOI] [PubMed] [Google Scholar]
- 72.Hakimi MA, Bochar DA, Schmiesing JA, Dong Y, Barak OG, Speicher DW, Yokomori K, Shiekhattar R. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature. 2002;418:994–998. doi: 10.1038/nature01024. [DOI] [PubMed] [Google Scholar]
- 73.Sillibourne JE, Delaval B, Redick S, Sinha M, Doxsey SJ. Chromatin remodeling proteins interact with pericentrin to regulate centrosome integrity. Mol Biol Cell. 2007;18:3667–3680. doi: 10.1091/mbc.E06-07-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kumar R, Wang RA, Bagheri-Yarmand R. Emerging roles of MTA family members in human cancers. Semin Oncol. 2003;30:30–37. doi: 10.1053/j.seminoncol.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 75.Manavathi B, Singh K, Kumar R. MTA family of coregulators in nuclear receptor biology and pathology. Nucl Recept Signal. 2007;5:e010. doi: 10.1621/nrs.05010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Toh Y, Pencil SD, Nicolson GL. A novel candidate metastasis-associated gene, mta1, differentially expressed in highly metastatic mammary adenocarcinoma cell lines. cDNA cloning, expression, and protein analyses. J Biol Chem. 1994;269:22958–22963. [PubMed] [Google Scholar]
- 77.O’Malley BW, Kumar R. Nuclear receptor coregulators in cancer biology. Cancer Res. 2009;69:8217–8222. doi: 10.1158/0008-5472.CAN-09-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mazumdar A, Wang RA, Mishra SK, Adam L, Bagheri-Yarmand R, Mandal M, Vadlamudi RK, Kumar R. Transcriptional repression of oestrogen receptor by metastasis-associated protein 1 corepressor. Nat Cell Biol. 2001;3:30–37. doi: 10.1038/35050532. [DOI] [PubMed] [Google Scholar]
- 79.Molli PR, Singh RR, Lee SW, Kumar R. MTA1-mediated transcriptional repression of BRCA1 tumor suppressor gene. Oncogene. 2008;27:1971–1980. doi: 10.1038/sj.onc.1210839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Manavathi B, Peng S, Rayala SK, Talukder AH, Wang MH, Wang RA, Balasenthil S, Agarwal N, Frishman LJ, Kumar R. Repression of Six3 by a corepressor regulates rhodopsin expression. Proc Natl Acad Sci U S A. 2007;104:13128–13133. doi: 10.1073/pnas.0705878104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li DQ, Pakala SB, Reddy SD, Ohshiro K, Peng SH, Lian Y, Fu SW, Kumar R. Revelation of p53-independent function of MTA1 in DNA damage response via modulation of p21WAF1-PCNA pathway. J Biol Chem. 2010;285 doi: 10.1074/jbc.M109.079095. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gururaj AE, Singh RR, Rayala SK, Holm C, den Hollander P, Zhang H, Balasenthil S, Talukder AH, Landberg G, Kumar R. MTA1, a transcriptional activator of breast cancer amplified sequence 3. Proc Natl Acad Sci U S A. 2006;103:6670–6675. doi: 10.1073/pnas.0601989103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Balasenthil S, Gururaj AE, Talukder AH, Bagheri-Yarmand R, Arrington T, Haas BJ, Braisted JC, Kim I, Lee NH, Kumar R. Identification of Pax5 as a target of MTA1 in B-cell lymphomas. Cancer Res. 2007;67:7132–7138. doi: 10.1158/0008-5472.CAN-07-0750. [DOI] [PubMed] [Google Scholar]
- 84.Manavathi B, Kumar R. Metastasis tumor antigens, an emerging family of multifaceted master coregulators. J Biol Chem. 2007;282:1529–1533. doi: 10.1074/jbc.R600029200. [DOI] [PubMed] [Google Scholar]
- 85.Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li DQ, Ohshiro K, Reddy SD, Pakala SB, Lee MH, Zhang Y, Rayala SK, Kumar R. E3 ubiquitin ligase COP1 regulates the stability and functions of MTA1. Proc Natl Acad Sci U S A. 2009;106:17493–17498. doi: 10.1073/pnas.0908027106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dornan D, Shimizu H, Mah A, Dudhela T, Eby M, O’rourke K, Seshagiri S, Dixit VM. ATM engages autodegradation of the E3 ubiquitin ligase COP1 after DNA damage. Science. 2006;313:1122–1126. doi: 10.1126/science.1127335. [DOI] [PubMed] [Google Scholar]
- 88.Li DQ, Divijendra Natha Reddy S, Pakala SB, Wu X, Zhang Y, Rayala SK, Kumar R. MTA1 coregulator regulates p53 stability and function. J Biol Chem. 2009;284:34545–34552. doi: 10.1074/jbc.M109.056499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 90.Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. H2AX: the histone guardian of the genome. DNA Repair (Amst) 2004;3:959–967. doi: 10.1016/j.dnarep.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 91.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 93.Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J Biol Chem. 2001;276:47759–47762. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- 94.Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, Redon C, Pilch DR, Olaru A, Eckhaus M, Camerini-Otero RD, Tessarollo L, Livak F, Manova K, Bonner WM, Nussenzweig MC, Nussenzweig A. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kuo LJ, Yang LX. Gamma-H2AX - a novel biomarker for DNA double-strand breaks. In Vivo. 2008;22:305–309. [PubMed] [Google Scholar]
- 96.Fillingham J, Keogh MC, Krogan NJ. GammaH2AX and its role in DNA double-strand break repair. Biochem Cell Biol. 2006;84:568–577. doi: 10.1139/o06-072. [DOI] [PubMed] [Google Scholar]
- 97.Stiff T, O’Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004;64:2390–2396. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- 98.Bassing CH, Chua KF, Sekiguchi J, Suh H, Whitlow SR, Fleming JC, Monroe BC, Ciccone DN, Yan C, Vlasakova K, Livingston DM, Ferguson DO, Scully R, Alt FW. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc Natl Acad Sci U S A. 2002;99:8173–8178. doi: 10.1073/pnas.122228699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, Gleason M, Bronson R, Lee C, Alt FW. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114:359–370. doi: 10.1016/s0092-8674(03)00566-x. [DOI] [PubMed] [Google Scholar]
- 100.Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA, Eckhaus M, Ried T, Bonner WM, Nussenzweig A. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114:371–383. doi: 10.1016/s0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 102.Albrechtsen N, Dornreiter I, Grosse F, Kim E, Wiesmuller L, Deppert W. Maintenance of genomic integrity by p53: complementary roles for activated and non-activated p53. Oncogene. 1999;18:7706–7717. doi: 10.1038/sj.onc.1202952. [DOI] [PubMed] [Google Scholar]
- 103.Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 104.Devlin HL, Mack PC, Burich RA, Gumerlock PH, Kung HJ, Mudryj M, deVere White RW. Impairment of the DNA repair and growth arrest pathways by p53R2 silencing enhances DNA damage-induced apoptosis in a p53-dependent manner in prostate cancer cells. Mol Cancer Res. 2008;6:808–818. doi: 10.1158/1541-7786.MCR-07-2027. [DOI] [PubMed] [Google Scholar]
- 105.Yokomakura N, Natsugoe S, Okumura H, Ikeda R, Uchikado Y, Mataki Y, Takatori H, Matsumoto M, Owaki T, Ishigami S, Aikou T. Improvement in radiosensitivity using small interfering RNA targeting p53R2 in esophageal squamous cell carcinoma. Oncol Rep. 2007;18:561–567. [PubMed] [Google Scholar]
- 106.Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000;408:377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- 107.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 108.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 109.Waga S, Hannon GJ, Beach D, Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 110.Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shivji MK, Grey SJ, Strausfeld UP, Wood RD, Blow JJ. Cip1 inhibits DNA replication but not PCNA-dependent nucleotide excision-repair. Curr Biol. 1994;4:1062–1068. doi: 10.1016/s0960-9822(00)00244-x. [DOI] [PubMed] [Google Scholar]
- 112.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 113.Lee K, Kim DR, Ahn B. Chromatin remodeling facilitates DNA incision in UV-damaged nucleosomes. Mol Cells. 2004;18:100–106. [PubMed] [Google Scholar]
- 114.Schmidt DR, Schreiber SL. Molecular association between ATR and two components of the nucleosome remodeling and deacetylating complex, HDAC2 and CHD4. Biochemistry. 1999;38:14711–14717. doi: 10.1021/bi991614n. [DOI] [PubMed] [Google Scholar]
- 115.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 117.Burd CJ, Kinyamu HK, Miller FW, Archer TK. UV radiation regulates Mi-2 through protein translation and stability. J Biol Chem. 2008;283:34976–34982. doi: 10.1074/jbc.M805383200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li DQ, Khan MN, Ohshiro K, Kumar R. Requirement of MTA1 in ATR-mediated DNA damage checkpoint function. J biol Chem. 2010:285. doi: 10.1074/jbc.M109.085258. In revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]