Abstract
Dynamic rearrangements of cell-cell adhesion underlie a diverse range of physiological processes, but their precise molecular mechanisms are still obscure. Thus, identification of novel players that are involved in cell-cell adhesion would be important. We isolated a human kelch-related protein, Kelch-like ECT2 interacting protein (KLEIP), which contains the broad-complex, tramtrack, bric-a-brac (BTB)/poxvirus, zinc finger (POZ) motif and six-tandem kelch repeats. KLEIP interacted with F-actin and was concentrated at cell-cell contact sites of Madin-Darby canine kidney cells, where it colocalized with F-actin. Interestingly, this localization took place transiently during the induction of cell-cell contact and was not seen at mature junctions. KLEIP recruitment and actin assembly were induced around E-cadherin–coated beads placed on cell surfaces. The actin depolymerizing agent cytochalasin B inhibited this KLEIP recruitment around E-cadherin–coated beads. Moreover, constitutively active Rac1 enhanced the recruitment of KLEIP as well as F-actin to the adhesion sites. These observations strongly suggest that KLEIP is localized on actin filaments at the contact sites. We also found that N-terminal half of KLEIP, which lacks the actin-binding site and contains the sufficient sequence for the localization at the cell-cell contact sites, inhibited constitutively active Rac1-induced actin assembly at the contact sites. We propose that KLEIP is involved in Rac1-induced actin organization during cell-cell contact in Madin-Darby canine kidney cells.
INTRODUCTION
Cell-cell adhesion is crucial for development and survival of multicellular organism (Adams et al., 1998). In epithelial tissues, cadherins are required for the assembly of cells into multilayer and the establishment and maintenance of the epithelial phenotype (Braga et al., 1997). Cadherins are transmembrane proteins that bind to the same type of cadherins in adjacent cells. The exodomain of cadherins interacts with each other by calcium-dependent homophilic interaction, whereas the cytoplasmic domain binds to β-catenin, and this complex is linked to the actin filaments by α-catenin (Vasioukhin and Fuchs, 2001). Functional cadherin receptors can influence the reorganization of the actin filaments and other cytoskeletal components, and the distribution of transmembrane proteins in a polarized manner, and the formation of the tight junctions and other adhesive junctions. It is therefore important to understand how cadherin-mediated cell adhesion is regulated. Although several different mechanisms have been proposed for the regulation, the regulatory mechanisms of cadherin-mediated cell adhesion are not yet fully understood (Fukata et al., 2001; Sahai and Marshall, 2002).
Recent studies revealed that Rho family small GTPases are involved in the cadherin-mediated cell adhesion (Braga, 2002b). It has been reported that Rac1 promotes E-cadherin–dependent cell adhesion in Madin-Darby canine kidney (MDCK) cells (Takaishi et al., 1997; Jou and Nelson, 1998). Rac1 is activated by the formation of E-cadherin–based cell adhesion (Nakagawa et al., 2001). It has been demonstrated that Rac1 is spatiotemporally recruited at the sites of newest cell-cell adhesion and regulates the formation of lamellipodia, which are actin-based membrane protrusion during cell-cell adhesion (Ehrlich et al., 2002). By contrast, RhoA activity was suppressed via p190RhoGAP during cell-cell contact (Noren et al., 2003). Thus, Rac1 is likely to fulfill the task of coordinating cell adhesion and actin-based cytoskeletal remodeling. IQGAP1, one of the targets of Cdc42 or Rac1, was shown to regulate the linkage between cadherins and the actin filaments (Fukata et al., 2001). The IQGAP1-based model supports a positive role for Rac1 in regulating the cadherin–mediated cell contacts (Fukata and Kaibuchi, 2001). However, some contradictions still remain. For example, dominant negative Rac1 has no effect on cell-cell contact formation or cadherin–catenins complex recruitment, although it inhibits the actin-accumulation in myogenic C2 cells (Lambert et al., 2002). Although the modulation of the cadherin-mediated cell adhesion by Rac1 might depend on the cell type, these findings suggest that there might be additional mechanisms of the cadherin-actin linkage.
The Drosophila Kelch protein is an actin filament cross-linking protein and plays an important role on the maintenance of ring canals that regulate cytoplasmic transport from nurse cells to the developing oocyte within an egg chamber (Kelso et al., 2002). Kelch has two sequence motifs (Xue and Cooley, 1993). The first motif, broad-complex, tramtrack, bric-a-brac (BTB)/poxvirus, zinc finger (POZ) domain has been proposed to function as a protein–protein interaction interface (Godt et al., 1993; Albagli et al., 1995). The second motif, the kelch repeats contain repeated sequences, each of which is composed of 40–50 amino acids (Xue and Cooley, 1993). The kelch repeats are required for its interaction with actin cytoskeleton (Kelso et al., 2002). Several mammalian proteins that contain the kelch repeats with the BTB/POZ motif are known (Adams et al., 2000), and some of them are actin-binding proteins. Like Drosophila Kelch, Kelch-related proteins might be involved in organization of actin cytoskeleton in various situations. However, proteins containing the kelch repeats have diverse cellular functions (Robinson and Cooly, 1997). Moreover, low amino acid identity of Kelch with other members of the BTB/Kelch subfamily makes it difficult to presume their functions (Bomont and Koeing, 2003).
In this study, we identified Kelch-like ECT2 interacting protein (KLEIP) as a protein that can associate with ECT2, a Rho nucleotide exchange factor involved in cytokinesis (Miki et al., 1993; Tatsumoto et al., 1999). Whereas KLEIP seems to regulate cytokinesis, we show that KLEIP is also involved in actin assembly at the sites of cell-cell adhesion.
MATERIALS AND METHODS
Isolation of KLEIP cDNA
The original KLEIP cDNA was isolated from a human testis cDNA library in a yeast two-hybrid screening by using the N-terminal half of human ECT2 as bait. A cDNA clone containing the entire open reading frame was isolated by polymerase chain reaction (PCR) by using the sequence of the kelch repeats-containing protein (GenBank accession no. AB02610). PCR was performed in a 25-μl mixture containing 10 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl, 10 mM each dNTPs, 2.5 U of high-fidelity Taq polymerase (BD Biosciences Clontech, Palo Alto, CA), 30 mM each forward primer and reverse primer, and 0.5 μg of cDNA from testis. The sequence of the forward- and reverse-primers were 5′-GGGAGATCTATGGAAGGAAAGCCAATGCGC-3′ and 5′-GGGTCGACTCACCAAATATGGGATTCACA-3′, respectively. The mixture was heated at 95°C for 5 min and then subjected to 35 cycles of amplification at 94°C for 45 s, 56°C for 45 s, and 72°C for 2 min. PCR products were separated in 2% agarose gels and purified using Ultra clean (MO BIO Laboratory, Salana Beach, CA) and then subcloned in pGEX-T vector (Promega, Madison, WI). The DNA sequences of the isolated KLEIP cDNA were the same as that of AB026190 except for position 1726, where G has been replaced by T in the KLEIP cDNA, resulting in replacement of Trp at the amino acid 593 by Gly. Because our sequence was identical to the corresponding position of another entry of this gene (GenBank accession no. AK001430), this may not represent a mutation caused by PCR.
Construction of Expression Vectors
Full-length KLEIP, KLEIP-N (aa 1–303), and -C (aa 301–609) cDNA were subcloned in the pEGFP-C1 expression vector (BD Biosciences Clontech), which contains the green fluorescent protein (GFP) or the pCEV32-F3, which contains the FLAG-epitope.
Preparation of Antibody
KLEIP-N or KLEIP-C was introduced into pCEV30G vector, which contains glutathione S-transferase (GST). GST-KLEIP fusion protein was expressed in Escherichia coli with 0.4 mM isopropyl-β-d-thiogalactopyranoside at 25°C for 2 h. GST-KLEIP-N fusion protein was purified using glutathione-Sepharose 4B (Amersham Biosciences, Piscataway, NJ) and then eluted with 20 mM reduced glutathione from the beads. Because GST-KLEIP-C was in insoluble fraction, it was separated on SDS-PAGE gels and purified by electroelution. The samples were dialyzed in phosphate-buffered saline (PBS). The purified GST-KLEIP-N or -C fusion protein was used for immunizing rabbits. We raised two antisera against each of KLEIP-N and -C. The IgG fraction was purified from the rabbit antisera by using a protein A-Sepharose column (Amersham Biosciences). For affinity purification of anti-KLEIP antibodies, KLEIP-N and KLEIP-FL was subcloned into pET-32 vector (Novagen, Darmstadt, Germany) and pMAL2c-E (New England Biolabs, Beverly, MA), respectively. His-tagged KLEIP-N and maltose binding protein (MBP)-fused KLEIP-FL were expressed in E. coli as described above, and purified using Ni-NTA resin (Novagen) and amylose resin (New England Biolabs), respectively. Each purified fusion protein was coupled with cyanogen bromide-activated beads (Amersham Biosciences) for affinity chromatography. Anti-KLEIP-N and anti-KLEIP-C were purified through His-KLEIP-N and MBP-KLEIP-FL affinity columns, respectively.
The following antibodies were from commercial sources: anti-β-tubulin (Sigma-Aldrich, St. Louis, MO), anti-FLAG antibody (M2 FLAG antibody; Sigma-Aldrich), anti-ZO1 (Zymed Laboratories, South San Francisco, CA), and anti-E-cadherin and β-catenin (BD Transduction Laboratories, San Diego, CA).
Cell Culture
MDCK, HeLa, HEK293T, COS, Swiss 3T3, and NIH 3T3 cells were grown in 35- or 100-mm dish with DMEM (Invitrogen, Carlsbad, CA) containing 10% FBS. U2OS cells were cultured with McCoy's 5A medium (Invitrogen) containing 10% fetal bovine serum (FBS). MDCK, HeLa, and HEK293 cells are of epithelial origin, whereas Swiss 3T3 and NIH3T3 cells are mouse fibroblasts. COS cells and U2OS cells are derived from kidney of monkey and human osteosarcoma, respectively. HEK293T is a human embryonic kidney cells.
For calcium-switch assays, MDCK cells were grown in low calcium-containing media (calcium-free S-MEN containing 2% dialyzed FBS) overnight and then incubated normal media for several hours.
For transfection, LipofectAMINE 2000 (Invitrogen) was used for the transient transfection according to the manufacturer's protocols.
Immunoblotting
Cells were lysed in lysis buffer (50 mM Tris-HCl, pH 7.5, 1% NP-40, 150 mM NaCl, 2 mM EDTA, 2 mM phenylmethylsulfonyl fluoride, 0.1 mg/ml aprotinin, 0.1 mg/ml leupeptin) at 4°C for 30 min. Total cell lysates were clarified by centrifugation at 14,000 × g for 20 min at 4°C and then the protein concentration of each sample was determined using BCA assay (Pierce Chemical, Rockford, IL). Samples were separated and analyzed on 8–16% gradient SDS-PAGE gels (Invitrogen) and then transferred to polyvinylidene difluoride membranes (Invitrogen). The membranes were blocked with Tris-buffered saline containing 3% milk and 0.1% Tween 20 and then probed with primary antibodies. The membranes were washed with Tris-buffered saline containing 0.1% Tween 20 for 7 min three times and probed with horseradish peroxidase-conjugated secondary antibody. Immunoreactive bands were visualized using ECL (Amersham Biosciences).
Immunoprecipitation
An identical amount (390 μg of protein) of protein from each sample was precleared by incubation with protein G-Sepharose (Amersham Biosciences) for 1 h at 4°C. Immunoprecipitation of antigen–antibody complex was accomplished by incubation for 4 h at 4°C. The antigen–antibody complex was incubated with protein G-Sepharose for 1 h at 4°C. Bound proteins were solubilized in Laemmli buffer and analyzed by immunoblotting.
mRNA Expression Analysis
MTC panel single-strand cDNA (BD Biosciences Clontech) was used for mRNA expression analysis. PCR was performed in a 25-μl mixture containing 10 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl, 10 mM each dNTPs, 2.5 U of Taq polymerase (Promega), 30 mM each forward primer and reverse primer, and 2.5 μl of MTC panel single-strand cDNA. PCR primer sets were as follows: for KLEIP, the forward primer was 5′-GGGAGATCTCTAATGCAAGGACCAAGGACG-3′ and the reverse primer was 5′-GCATAAAGAAAGCCTCCAAGTACT-3′; and for glyceraldehyde-3-phosphate dehydrogenase, a forward primer (5′-TGAAGGTCGGAGTCAACGGATTTGGT-3′) and a reverse primer (5′-CATGTGGGC CATGAGGTCCACCAC-3′) were used. The mixtures were heated at 95°C for 5 min and then subjected to 26-cycle amplification at 94°C for 45 s, 60°C for 45 s, and 72°C for 1 min. PCR products were separated on 2% agarose gel and visualized using ethidium bromide. The sequences of the PCR products were confirmed using automated DNA sequencer.
Cross-Linking of MBP-KLEIP-N Dimers
The MBP-KLEIP-N fusion protein and MBP were expressed and purified as described above. The protein was incubated in a buffer containing 20 mM HEPES, 150 mM NaCl, and 10 mM dithiobis-(propionic acid n-hydroxysuccinimide ester) (DTSP), a bifunctional protein cross-linking reagent, freshly prepared as a 450 mM stock in dimethyl sulfoxide (Sigma-Aldrich). Cross-linking was carried out at room temperature for 30 min and terminated by adding 2× Laemmli buffer either with or without 50 mM dithiothreitol (DTT). Proteins were boiled for 5 min, resolved on an 8–16% gradient gel, and visualized by Coomassie Blue staining.
Actin Cosedimentation Assays
Actin cosedimentation assays were performed using the actin binding protein biochem kit (Cytoskeleton, Denver, CO). Briefly, rabbit muscle actin was polymerized at 24°C for 1 h under 5 mM Tris-HCl, pH 8.0, 0.2 mM CaCl2, 50 mM KCl, 2 mM MgCl2, and 1 mM ATP. MBP-fusion proteins were expressed and purified as described above. Purified proteins (2 μM) were added and incubated at 24°C for 30 min with or without the prepared polymerized actin. The samples were centrifuged at 24°C, 150,000 × g for 90 min. The precipitates were suspended in Laemmli buffer. The supernatants and precipitates were analyzed using SDA-PAGE and visualized by Coomassie Blue staining.
Immunofluorescence Microscopy
Cells were grown on coverslips and fixed for 20 min with 4% paraformaldehyde (PFA) in PBS, and then cells were permeabilized for 7 min with 0.1% Triton X-100 in PBS and washed with PBS twice. For staining against E-cadherin, cells were fixed for 7 min with 100% methanol at –20°C. Samples were incubated with PBS containing 2% horse serum for 1 h and then with primary antibody for 1 h. The cells were washed in PBS three times and incubated with Cy3-(Jackson ImmunoResearch Laboratories, West Grove, PA) or Alexa 488-conjugated second antibody (Molecular Probes, Eugene, OR). To detect actin filaments, cells were incubated with Alexa 350 or 488-conjugated phalloidin (Molecular Probes) for 20 min. Cells were observed with fluorescence microscopy. For deconvolution microscopy (Figures 4A and 5, A and D), the image of each focal plane was taken, and each image was cleaned by removing blurs from other focal planes using Open Lab software (Improvision, Lexington, MA).
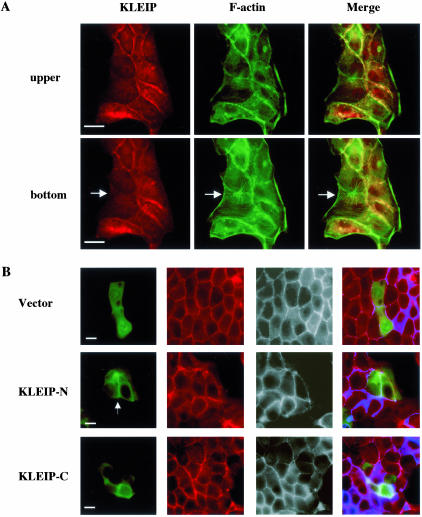
Figure 4.
KLEIP is localized at the sites of cell-cell contact in MDCK cells. MDCK cells were stained 18 h after replating by affinity-purified anti-KLEIP-N antibody. KLEIP (red) and F-actin (green) were visualized by Cy3-conjugated anti-rabbit antibody and Alexa 488-conjugated phalloidin, respectively. The images of focal planes were taken and cleaned by deconvolution. The top and bottom panels show images of the same cells at relatively top and bottom levels, respectively. Yellow indicates the overlapping region of KLEIP and F-actin (Merge). Arrows in the bottom panels show a location of stress fibers. Note that KLEIP was not localized at stress fibers. Bars, 10 μm. (B) Fluorescence images of MDCK cells transfected with GFP or GFP-tagged KLEIP derivatives. Transfected MDCK cells were fixed with 4% PFA 18 h after transfection. GFP or GFP-tagged KLEIP was visualized by fluorescence microscopy. Arrows indicate the accumulation of expressed GFP-tagged KLEIP at cell-cell contact sites. β-Catenin (red) and F-actin (grayscale) were also stained and observed. Blue expresses F-actin staining in merged images. Bars, 10 μm.
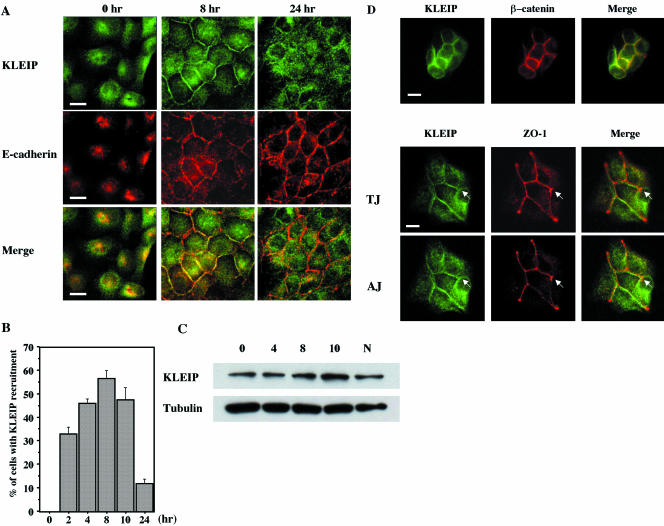
Figure 5.
KLEIP was temporally recruited at the sites of cell adhesion during cell-cell contact. (A) MDCK cells were fixed with methanol before (0 h) and after (8 and 24 h) a calcium switch. Cells were stained for KLEIP (green) and E-cadherin (red). The images were cleaned by deconvolution. Merged images are shown on the bottom. Bars, 10 μm. (B) KLEIP recruitment at the sites of cell adhesion after the calcium switch. The number of cells that exhibited KLEIP recruitment at the cell-cell adhesion sites was counted. The average of five independent experiments is shown. At least, 200 cells were observed in each experiment. Error bars indicate SD. (C) Immunoblotting by anti-KLEIP-N antibody. MDCK cells were harvested at indicated the time points (hour) after a calcium switch (0, 4, 8, and 10 h). MDCK cells were also grown in normal culture media for 24 h and then harvested (N). Total lysates (20 μg of protein) were analyzed by immunoblotting as indicated. The typical results of immunoblotting using anti-KLEIP antibody or anti-β-tubulin are shown. (D) Colocalization of KLEIP with adherens junction makers. MDCK cells were fixed 18 h after replating. KLEIP was visualized by anti-KLEIP-N antibody and then Alexa 488-conjugated anti-rabbit antibody (green), and images were observed by deconvolution microscopy. The samples were also stained for β-catenin (top) and ZO-1 (middle and bottom) were also stained (red). Merged images are shown at the right. TJ and AJ exhibit the presumed focal planes of tight junctions and adherens junctions, respectively. Bars, 10 μm.
To evaluate KLEIP recruitment at the sites of cell-cell contact, the cells that exhibited at least one clear line-like staining pattern of KLEIP along the cell-cell border were scored.
When the effects of constitutive active Rac or KLEIP-N were examined, cells with increased staining patterns of KLEIP or F-actin at the cell-cell contact sites compared with those of surrounding cells were scored positive.
E-Cadherin/Fc Beads Assays
Concanavalin A (ConA; Sigma-Aldrich) or human E-cadherin/Fc chimera (Sigma-Aldrich) was immobilized with latex beads as described previously (Burbelo et al., 1995; Lambert et al., 2000). Briefly, 200 μl of Polystyrene latex beads (6.4-μm average diameter; Sigma-Aldrich) were coated with either 100 μg/ml ConA or 100 μg/ml E-Cadherin/Fc in 0.1 M borate buffer (pH 8.0) at room temperature for 18 h. The beads were washed with PBS twice and then incubated with 2 mg/ml bovine serum albumin at room temperature for 2 h. The beads were washed using PBS three times again and suspended in 200 μl of PBS containing 1 mM CaCl2. An aliquot of beads (5 μl) was used for a 35-mm culture dish. MDCK cells were incubated in the presence of beads for 2 h at 37°C and analyzed by immunofluorescence. The beads placed on cell surfaces were identified by phase contrast image, and examined for rim-like staining of β-catenin, F-actin and KLEIP around the beads. To analyze the effect of F-actin on KLEIP recruitment, MDCK cells were incubated with E-cadherin/Fc chimera-coated beads in the presence of vehicle alone (dimethyl sulfoxide) or 10 μg/ml cytochalasin B (Sigma-Aldrich) for 2 h at 37°C.
RESULTS
KLEIP Encodes a Kelch-like Protein
We identified a human kelch repeat-containing protein, designated KLEIP, in a yeast two-hybrid screening by using ECT2 as bait. A database search revealed that KLEIP is nearly identical to a Kelch-related protein of unknown function (GenBank accession no. AB026190). A full-length cDNA was isolated by PCR cloning from a human cDNA library as a template. The predicted KLEIP protein contains a BTB/POZ domain and six-tandem kelch repeats of 40–50-amino acid sequence that contained highly conserved sequences among the repeats (Figure 1). It has been reported that the kelch repeats forms a β-sheet propeller structure (Adams et al., 2000).
Figure 1.
KLEIP belongs to the superfamily of Kelch-like proteins. Predicted structure of KLEIP and its derivatives. (A) Schematic representation of the structure of KLEIP and its derivatives. KLEIP contains the N-terminal BTB/POZ motif (aa 53–165) and C-terminal kelch repeat domains. (aa 316–609). The regions carried by KLEIP-N (aa 1–303) and -C (aa 301–609) are shown. (B) Alignment of the kelch repeats of human KLEIP. Conserved amino acids are shown in bold. The bottom line shows the consensus sequence of the kelch repeats. The C-terminal 16 amino acid residues (aa 598–613) is a part of the first kelch repeat (Adams et al., 2000). (C) Amino acid comparison of human KLEIP, Drosophila Diablo, the Anopheles Kelch-like protein, and Drosophila Kelch. Consensus sequences of the BTB motif and the kelch repeats are shown at the top.
Database search also revealed two proteins closely related to KLEIP. Drosophila Diablo (GenBank accession no. AF237711) exhibited amino acid identities of 80.8% to KLEIP (Figure 1C). In contrast, the sequence identity between human KLEIP and Drosophila Kelch was 43.0%. Therefore, Diablo seems to be the Drosophila orthologue of human KLEIP. Although the biological function of Drosophila Kelch is well documented, the function of Diablo is still unknown. Another protein in Anopheles (GenBank accession no. EAA05692) also exhibited a high amino acid identity to KLEIP (79.8%), and could be the Anopheles orthologue of human KLEIP. The biological function of the Anopheles Kelch-like protein is also not known.
Identification and Expression of Endogenous KLEIP
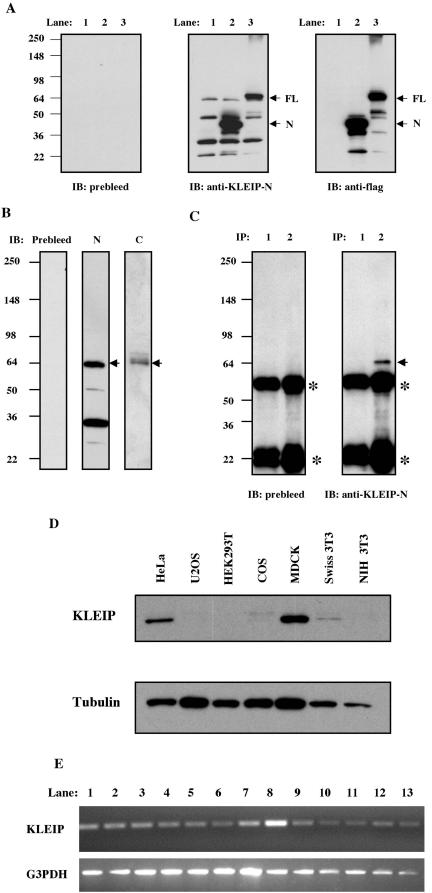
To analyze endogenous KLEIP, we raised antisera against the N-terminal (KLEIP-N) and C-terminal (KLEIP-C) half of KLEIP (Figure 1A) and affinity purified the antibodies. To test these antibodies, we expressed FLAG-tagged KLEIP-N and full-length KLEIP (KLEIP-FL) in COS cells (Figure 2A). Anti-FLAG antibody detected major bands of ∼40- and 64-kDa in FLAG-KLEIP-N and FLAG-KLEIP-FL transfectants, respectively (Figure 2A, right). The sizes of these proteins corresponded to the predicted sizes of FLAG-KLEIP-N and -FL. Anti-FLAG antibody also detected several minor bands, three of which were common in the lysates of KLEIP-N and KLEIP-FL transfectants, suggesting that these bands represented degradation products. Anti-KLEIP-N antibody detected similar proteins in addition to the band whose mobility was slightly lower than that of FLAG-KLEIP-FL (Figure 2A, middle). This band seemed to be endogenous KLEIP, because it was commonly observed in KLEIP-N, KLEIP-FL, and vector alone transfectants. Anti-KLEIP-N also detected degradation products of exogenous proteins in KLEIP-N and KLEIP-FL transfectants. However, anti-KLEIP-N also detected bands of 34 kDa in all three lanes (Figure 2, A and B). Preimmune sera did not detect any protein (Figure 2A, left). In contrast, anti-KLEIP-C recognized only a protein of 64 kDa (Figure 2B), supporting that the 64-kDa protein is endogenous KLEIP. It is possible that the 34-kDa protein might be an isoform of KLEIP that lacks the C-terminal domain, because anti-KLEIP-C did not recognize the protein. Alternatively, it might represent an unrelated protein recognized by a cross-reactivity of anti-KLEIP-N. When anti-KLEIP-N was used for immunoprecipitation, the 64-kDa protein was detected in the immunoprecipitate, whereas the 34-kDa protein was not (Figure 2C). The 64-kDa protein, but not the 34-kDa protein, was also detected by anti-KLEIP-C after immunoprecipitation by anti-KLEIP-N (our unpublished data). These results suggest that the 64-kDa protein represents endogenous KLEIP, and the 34-kDa protein is recognized by anti-LEIP-N only when it is denatured. We also performed immunoblotting and immunoprecipitation against anti-KLEIP-N by using lysates of MDCK cells and obtained the similar results to those of HeLa cells (see supplemental data, Figure S1).
Figure 2.
KLEIP is a 64-kDa protein and expressed ubiquitously in human adult tissues. (A) Immunoblotting analysis using an affinity-purified anti-KLEIP-N antibody. FLAG-vector (lane 1), FLAG-tagged KLEIP-N (lane 2) or -FL (lane 3) was transfected in COS cells. Cell lysates (30 μg of protein) were separated on 8–16% gradient SDS-PAGE gels and analyzed by the immunoglobulin fraction of preimmune sera (prebleed), purified anti-KLEIP-N antibody or anti-FLAG antibody. Predicted molecular sizes of KLEIP-N and -FL are 64- and 98-kDa, and the locations of these proteins are shown by arrows. Locations of molecular size markers are shown on the left of the panel. (B) Identification of endogenous KLEIP in cultured cells. Lysates of HeLa cells were analyzed by immunoblotting. Prebleed, N and C show immunoblots with the preimmune sera (for anti-KLEIP-N), anti-KLEIP-N or -C, respectively. The location of the predicted KLEIP is shown by an arrow. (C) Immunoprecipitation of KLEIP. HeLa cell lysates (390 μg of protein) were immunoprecipitated by 0.25 μg of the IgG fraction of preimmune sera (lane 1) or anti-KLEIP-N antibody (lane 2), followed by immunoblotting with IgG of preimmune sera or affinity-purified anti-KLEIP-N antibody. An arrow denotes the 64-kDa protein. Asterisks show bands of heavy and light chains of IgG. (D) Identification of endogenous KLEIP in cultured cells. Lysates (10 μg of protein in each) were separated by SDS-PAGE gels and analyzed by immunoblotting with anti-KLEIP-N. Tubulin was detected as a loading control (bottom). (E) mRNA expression of KLEIP in human tissues. PCR was performed to detect expression of KLEIP (top) or glyceraldehyde-3-phosphate dehydrogenase G3PDH (bottom). Lane 1, heart; 2, brain; 3, liver; 4, placenta; 5, lung; 6, kidney; 7, skeletal muscle; 8, pancreas; 9, spleen; 10, thymus; 11, small intestine; 12, testis; 13, ovary; and 14, colon.
Next, we examined the expression of KLEIP in several cultured cells (Figure 2D). The highest expression of KLEIP was observed in MDCK cells among the cultured cells examined. KLEIP expression was also detected in HeLa and Swiss 3T3 cells but not significantly in HEK293T and NIH3T3 cells. We also examined mRNA expression levels of KLEIP in several human tissues by a PCR-based analysis. KLEIP mRNA was detected among a number of adult tissues examined (Figure 2E), suggesting that KLEIP is ubiquitously expressed in various cell types.
KLEIP Can Form Dimers and Associate with F-Actin
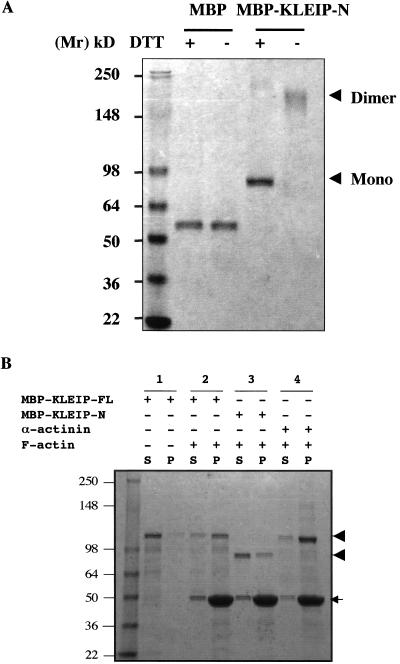
It has been reported that some Kelch-related proteins with the BTB/POZ motif can form homodimers through their BTB/POZ domains (Hernandez et al., 1997; Soltysik-Espanola et al., 1999). To determine whether KLEIP can form dimers, affinity-purified MBP-fused KLEIP-N or MBP alone was cross-linked by the bifunctional protein cross-linker DTSP and analyzed by SDS-PAGE. DTSP forms stable linkages between free amine groups of interacting proteins, and these linkages are cleavable with a S-S reducing reagent DTT (Soltysik-Espanola et al., 1999). Whereas MBP itself could not form dimers, the majority of MBP-KLEIP-N protein was detected at the position corresponding to the dimers (Figure 3A). When DTT was added to cleave the linkages, most of MBP-KLEIP-N was detected at the position corresponding to the monomeric form. These observations suggest that MBP-KLEIP-N, but not MBP itself, can form dimers under these conditions and are consistent with the finding that KLEIP-N contains the BTB/POZ motif, which is known to play a role in dimer formation.
Figure 3.
KLEIP forms dimers and interacts with F-actin in vitro. (A) Homodimer-formation through the BTB/POZ domain of KLEIP. MBP and MBP-KLEIP-N were cross-linked by DTSP in PBS. The cross-linkage was cleavable with DTT. Both reduced and non-reduced samples were separated on 8–16% gradient SDS-PAGE gels and then visualized by Coomassie staining. Locations of molecular makers are shown at the left. Note that MBP-dimers were not detected. Arrows indicate the predicted locations of monomeric (Mono) and dimeric KLEIP (Dimer). (B) Actin cosedimentation assays. The supernatants (S) and precipitates (P) after centrifugation at 150,000 × g were separated on 8–16% gradient SDS-PAGE gels and then visualized by Coomassie staining. MBP-KLEIP-FL (∼115 kDa) was analyzed after incubation without (lane 1) or with F-actin (lane 2). α-Actinin was used as a positive control (lane 4). Arrowheads indicate the locations of MBP-KLEIP-N and -FL. The position of actin is shown by an arrow. Locations of molecular size markers are indicated on the left.
Some Kelch-related proteins are known to colocalize with filamentous (F)-actin (Soltysik-Espanola et al., 1999, Sasagawa et al., 2002). To test whether purified KLEIP can interact with F-actin, we performed actin cosedimentation assays. MBP-KLEIP-FL was incubated with or without F-actin, and resulting complexes were sedimented by centrifugation (Figure 3B). When F-actin was not present in the reaction, the majority of MBP-KLEIP-FL was detected in the supernatants (Figure 3B, lane 1). However, the majority of MBP-KLEIP-FL was detected in the precipitates, when F-actin was mixed (Figure 3B, lane 2), suggesting that KLEIP interacts with F-actin in vitro. The association of KLEIP with F-actin was as efficient as the known actin-binding protein α-actinin (Figure 3B, lane 4). In contrast, MBP-KLEIP-N, which lacks the kelch repeats (Figure 1A), was not cosedimented with F-actin efficiently, although a weak association was detected (Figure 3B, lane 3). MBP itself did not associate with F-actin (our unpublished data). Because MBP-KLEIP-C and GST-KLEIP-C were insoluble, we could not test whether KLEIP-C can associate with F-actin in vitro. Nonetheless, these data suggest that the kelch repeats of KLEIP play a critical role in F-actin association. We also examined the possible interaction of KLEIP with globular (G)-actin by using pull-down assays. However, MBP, MBP-KLEIP-N or MBP-KLEIP–FL did not efficiently interact with G-actin (our unpublished data).
KLEIP Is Colocalized with Actin Bundles in Epithelial Cell Junctions
Next, we investigated subcellular localization of KLEIP in MDCK cells by immunocytochemistry using anti-KLEIP antibodies (Figure 4A). Whereas anti-KLEIP-C did not yield any significant signal in immunocytochemistry (our unpublished data), affinity-purified anti-KLEIP-N staining revealed two patterns dependent on a different compaction of colonies of MDCK cells that were formed 18 h after replating. KLEIP was detected at cell-cell adhesion sites of some cells (Figure 4A, top). In these cells, circumference actin bundles had been well established, and KLEIP was partially colocalized with F-actin, although the intensity of the staining for KLEIP was not always correlated with that of F-actin. The accumulation of KLEIP was limited to cell-cell junctions and was not observed at cell-free edges or stress fibers on the bottom of cells (Figure 4A, bottom). In the other cells, KLEIP was mainly localized in the intracellular pool (our unpublished data), and colocalization of KLEIP and F-actin was not observed. A similar staining pattern for KLEIP was also observed in pig proximal renal epithelial cell lines, LLC-PK-1 (our unpublished data). The immunoglobulin fraction of preimmune sera did not yield any significant signal (our unpublished data). These results suggest that KLEIP is transiently recruited to the cell-cell junctions of adherent cells.
To exclude the possibility that the detection of KLEIP at the cell-cell junctions was due to a cross-reactivity of the antibody, we also analyzed the localization of GFP-fused KLEIP derivatives in MDCK cells. As shown in Figure 4B, GFP-KLEIP-N, but not GFP-KLEIP-C, exhibited the localization to cell-cell junctions, suggesting that the N-terminal half of KLEIP is responsible for the localization to the cell-cell junctions. The expression level of GFP-KLEIP-FL was too low to evaluate its localization in MDCK cells (our unpublished data). We also examined the localization of Myc- or FLAG-tagged KLEIP-FL, but they also showed similar results, suggesting that KLEIP-FL overexpression is detrimental to MDCK cells.
KLEIP Is Recruited to Adherens Junctions upon Cell-Cell Contact
To verify the idea that KLEIP is temporally recruited at the sites of cell-cell junctions, we examined the KLEIP localization in MDCK cells by using the calcium-switch procedure (Figure 5A, top). E-cadherin–mediated cell adhesion is disrupted in low calcium media, because homophilic E-cadherin binding requires extracellular calcium (Braga, 2002a). After calcium addition, E-cadherin accumulates at the sites of cell-cell contact and the cell adhesion structure assembles. Like E-cadherin, KLEIP was mainly localized in the intracellular pool in low calcium media (Figure 5A, 0 h). However, KLEIP became concentrated at the sites of cell-cell contact after a calcium switch (Figure 5A). This recruitment of KLEIP took place at least within 2 h after the replacement of media and reached the peak 8 h after the calcium switch (Figure 5B). Consistent with published reports (Nakagawa et al., 2001; Rothen-Rutishauser et al., 2002), E-cadherin–mediated cell adhesion was well established 8 h after the calcium switch (Figure 5A, middle). Thus, the recruitment of KLEIP at the sites of the cell-cell contact coincided with E-cadherin–mediated cell adhesion. Interestingly, KLEIP returned to the intracellular pool in many cells 24 h after the calcium switch (Figure 5A, top), whereas E-cadherin was well maintained at cell-cell junctions 24 h after the calcium switch (Figure 5A, middle). The calcium switch itself did not affect either the protein amount or the mobility of KLEIP as determined by immunoblotting (Figure 5C). We also tested the detergent-solubility of KLEIP from MDCK cells by a procedure (Jou et al., 1998; Gimond et al., 1999) with some modifications. KLEIP was detected in the 0.5% Triton X100 insoluble fraction at three time points after the calcium switch (0, 8, and 24 h), whereas the 34-kDa protein was in the soluble fraction (see supplemental data, Figure S1).
To analyze in which site of cell-cell contact KLEIP is concentrated, we performed double staining for KLEIP and several junctional proteins that are known to localize at the sites of cell-cell contact (Figure 5A, 8 h; and D). When KLEIP became concentrated at the sites of the cell-cell contact, it was colocalized with two components of adherens junctions, E-cadherin (Figure 5A, 8 h) and β-catenin (Figure 5D, top). However, KLEIP was not exactly colocalized with the tight junction marker ZO-1 (Figure 5D, middle and bottom, see arrows). Thus, the localization of KLEIP during cell-cell contact was consistent with that at adherens junctions rather than tight junctions.
KLEIP Recruitment Is Regulated by E-Cadherin
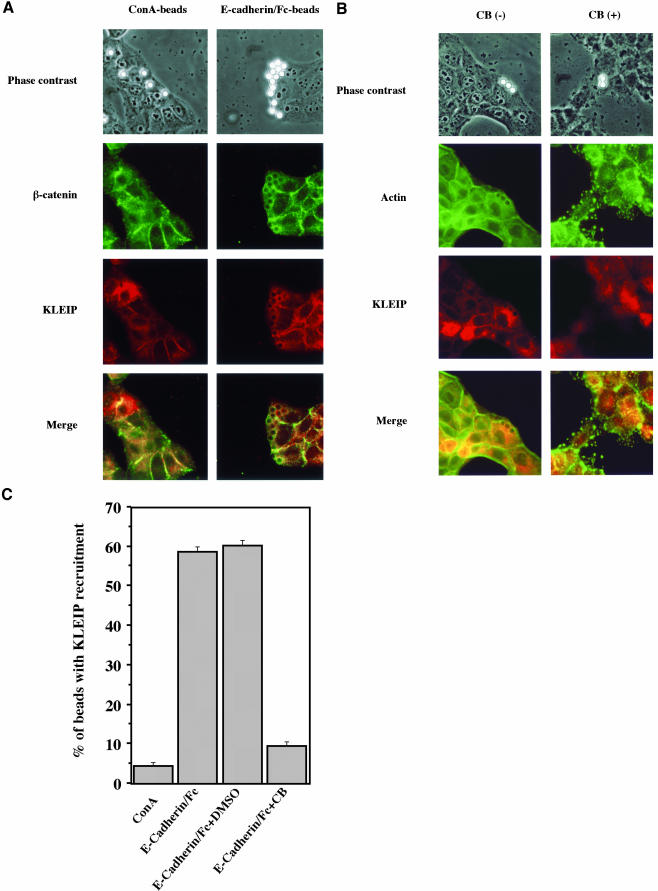
E-cadherin, the prototypical epithelial cadherin, mediates cell-cell adhesion in mature epithelia (Braga, 2002a). E-cadherin/Fc chimera (E-cadherin ectodomain fused to the Fc fragment of IgG) induces β-catenin recruitment in the stable human E-cadherin expressing Chinese hamster ovary cells (Kovacs et al., 2002). It has been reported that latex beads coated with N-cadherin/Fc chimera induce the recruitment of cadherin–catenin complexes and F-actin around the beads in myogenic cells (Lambert et al., 2002). To test whether E-cadherin ligation regulates KLEIP recruitment to the cell-cell junctions, we used the cadherin-coated beads assay (Lambert et al., 2000, 2002). We placed latex beads coated with E-cadherin/Fc chimera on MDCK cells and examined for KLEIP recruitment around the beads. Because ConA is a nonspecific adhesive ligand and binds to cell membrane glycoproteins (Trickett and Kwan, 2003), ConA-coated beads were used as negative control (Levenberg et al., 1998). The E-cadherin/Fc-coated beads, but not ConA-coated beads, efficiently induced β-catenin recruitment (Figure 6A) and F-actin recruitment (Figure 6B) around the beads that reached on the surfaces of MDCK cells, indicating that our E-cadherin/Fc-coated beads were functional.
Figure 6.
E-cadherin/Fc-beads assays. Beads assays were performed as described in MATERIALS AND METHODS. (A) ConA-coated beads (left) or E-cadherin/Fc chimera-coated beads (right) were used. Bright round beads (mean size 6.4 μm) were detected by phase contrast image (top). KLEIP (red) and β-catenin (green) were visualized (middle). Merged images of KLEIP and β-catenin are also shown (bottom). KLEIP and β-catenin were recruited around the E-cadherin/Fc chimera-coated beads on the cell surfaces, but not ConA-coated beads. (B) MDCK cells were incubated with E-cadherin/Fc beads in the presence of vehicle alone (0.1% DMSO) or cytochalasin B (CB) and stained for KLEIP (red) and actin (green). Note that CB disrupted actin structures at the cell-cell contact sites. (C) Quantitative analysis of the effects of CB on KLEIP recruitment to ConA- and E-cadherin/Fc-coated beads. Only the beads placed on the cell surfaces were analyzed for KLEIP recruitment. The averages of three independent experiments were shown. At least, 100 beads were analyzed in each experiment. Error bars show SD.
When KLEIP localization was analyzed in these assays, the recruitment of KLEIP to the beads was detected (Figure 6, A and C). The recruitment of KLEIP to the beads was detected in 58.8% of beads that were on the cell surfaces (n = 908). By contrast, most of ConA-coated beads did not induce the recruitment of KLEIP (Figure 6, A and C, n = 367). Moreover, KLEIP recruitment around the E-cadherin/Fc beads was not observed either in the presence of 2 mM EDTA or under low calcium media (our unpublished data). Therefore, E-cadherin/Fc-ligand seemed to induce the recruitment of KLEIP around the beads. Such the recruitment of KLEIP around E-cadherin/Fc-coated beads was also observed in LLC-PK1 cells (our unpublished data). These results suggest that KLEIP recruitment is induced by E-cadherin, possibly by their ligation, and spatially limited to the vicinity of E-cadherin molecules.
F-Actin Is Required for KLEIP Recruitment to the Cell-Cell Junctions
Because KLEIP bound to F-actin (Figure 3B), we tested whether F-actin are required for this KLEIP recruitment around the E-cadherin/Fc-coated beads. To inhibit the formation of actin filaments, we used cytochalasin B (CB), an inhibitor of actin polymerization (Johnson et al., 2002). MDCK cells were incubated with E-cadherin/Fc beads in the presence or absence of CB (Figure 6, B and C). In the absence of CB, 60.3% of E-cadherin/Fc beads placed on the cell surface exhibited KLEIP recruitment as well as F-actin recruitment around the beads (n = 431). In contrast, only 9.6% of E-cadherin/Fc-coated beads placed on the cells induced F-actin and KLEIP recruitment around the beads in the presence of CB (n = 611). CB also disrupted de novo actin filaments of MDCK cells. These results suggest that F-actin is required for the KLEIP recruitments around E-cadherin/Fc beads.
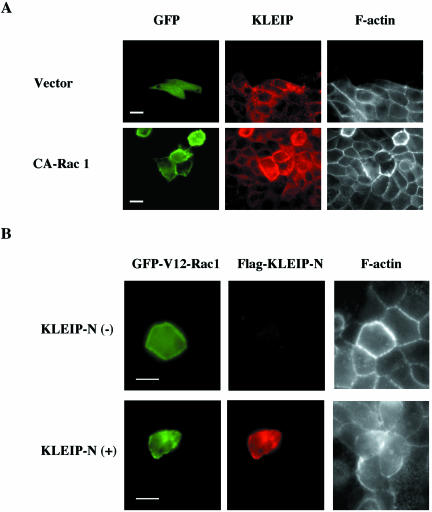
Rac1, a small GTPase of the Rho family, is activated by E-cadherin-mediated cell adhesion in MDCK cells (Nakagawa et al., 2001). It has been reported that a constitutive active (CA) form of Rac1 increases F-actin staining at the cell-cell junction (Takaishi et al., 1997). To test whether CA-Rac1 also stimulates KLEIP recruitment to the cell junction, GFP-fused CA-Rac1 (V12) was transfected into MDCK cells in low calcium media. Cells were subjected to a calcium switch and examined for KLEIP and F-actin recruitment (Figure 7A). We observed KLEIP recruitment at the cell-cell contact sites 8 h after calcium switch, which was consistent with maximum KLEIP recruitment at this time point (Figure 5B). Seventy percent of GFP-CA-Rac1–expressing cells exhibited increased staining for KLEIP as well as F-actin at the cell-cell junctions, compared with surrounding nontransfected cells (Figure 7A, bottom, n = 458). Such an increased recruitment of KLEIP and F-actin was not observed in vector-transfected cells (Figure 7A, top) or CA-Cdc42 (our unpublished data). These observations suggest that KLEIP is localized on actin filaments at the cell-cell junction during the initiation of cell-cell contact.
Figure 7.
Effects of Rac1 on KLEIP recruitment to cell-cell contact sites. (A) GFP-vector and GFP-CA-Rac1 (constitutive active) were transiently transfected in MDCK cells. Cells were subjected to a calcium switch 18 h after transfection and then fixed with 4% PFA 8 h after the calcium switch. Samples were stained for KLEIP (red) and F-actin (grayscale) by anti-KLEIP-N antibody and Alexa 350-conjugated phalloidin, respectively. Bars, 10 μm. (B) Effects of KLEIP-N on CA-Rac1 induced actin recruitment. GFP-V12-Rac1–dependent F-actin recruitment to the cell-cell contact sites was inhibited by FLAG-KLEIP-N. Allows show GFP-V12-Rac1 expressing cells. GFP-V12-Rac1 and FLAG-KLEIP-N were transiently cotransfected in MDCK cells in low calcium media. The media were changed to normal culture media containing calcium 18 h after the cotransfection. Cells were fixed with 4% PFA 10 h after the replacement of culture media. FLAG-KLEIP-N and F-actin was stained by anti-FLAG antibody and Alexa 350-conjugated phalloidin, respectively. GFP-V12-Rac1 (green), FLAG-KLEIP-N (red) and F-actin (grayscale) were observed by the fluorescence microscopy. Bars, 10 μm.
KLEIP-N Inhibits Actin Assembly to Cell-Cell Adhesion Sites
Because CA-Rac1 strongly induced the recruitment of KLEIP as well as F-actin to cell-cell junctions, we reasoned that KLEIP is an F-actin cross-linking protein like Drosophila Kelch and is involved in actin assembly through the Rac1-mediated signaling during cell adhesion. In this case, the N-terminal half of KLEIP (KLEIP-N) might exhibit a dominant negative phenotype with regard to F-actin accumulation at cell-cell adhesion sites, because it lacks the kelch repeats, which can bind to F-actin, but can still accumulate at the sites of cell-cell adhesion (Figure 4B). In Drosophila Kelch, such a dominant negative effect of N-terminal half of the protein on ring canal formation has been reported (Robinson and Cooly, 1997). To test this possibility, we transfected CA-Rac1 with or without KLEIP-N into MDCK cells and examined F-actin localization at the junctions of transfected cells (Figure 7B). Compared with surrounding cells, a strong actin recruitment induced by CA-Rac1 was observed in 80% of cells expressing GFP-CA-Rac1 alone (n = 208), whereas only 33% of cells coexpressing both GFP-CA-Rac1 and FLAG-KLEIP-N (n = 176) exhibited the strong actin recruitment to the cell-cell junctions. Therefore, KLEIP-N inhibited Rac1-induced actin recruitment to the cell junctions. In contrast, expression of KLEIP-N alone did not significantly alter the pattern of actin localization in MDCK cells (our unpublished data; but see Figure 4B). These observations suggest that KLEIP is specifically involved in Rac1-regulat actin accumulation at cell-cell adhesion sites.
DISCUSSION
In this study, we identified a novel human Kelch-related protein, KLEIP, which consisted of the BTB motif and six-tandem kelch repeats. KLEIP is mostly related to Drosophila Diablo, whose function is still unknown. KLEIP shared some characteristics with other Kelch-related proteins. First, the BTB/POZ motif of KLEIP was involved in dimer formation. Second, KLEIP interacted with F-actin in vitro. However, the low amino acid identity of KLEIP with other members of the BTB/Kelch subfamily made it difficult to presume its function (Bomont and Koeing, 2003).
We found accumulated signals at the sites of cell-cell adhesion by affinity-purified anti-KLEIP-N antibody in immunocytochemistry (Figure 4A). Anti-KLEIP-N and KLEIP-C antibodies recognized a protein of 64 kDa in immunoblotting, which corresponded to the predicted molecular size of KLEIP (Figure 2B). These results suggest that the 64-kDa protein represents endogenous KLEIP and that the isolated KLEIP cDNA contains the entire open reading frame. Although anti-KLEIP-C recognized the major single band of 64 kDa, anti-KLEIP-N also detected a 34-kDa protein. Because the 34-kDa protein was not immunoprecipitated by anti-KLEIP-N, anti-KLEIP-N seemed to recognize only denatured forms of the protein (Figure 2C). Because we used the methanol method for fixation, which should retain the native structure of proteins (Allan, 2000), the 34-kDa protein might not be detected in our immunocytochemical analysis. Additionally, as we detected the 34-kDa protein in the Triton X-soluble fraction in MDCK cells (see supplementary data, Figure S1), it would not affect the detection of KLEIP at the cell-cell junctions, even if KLEIP-N could detect the native form of the 34-kDa protein in immunocytochemical experiments. Moreover, exogenously expressed GFP-KLEIP-N also localized at the sites of cell-cell contact (Figure 4B). These results strongly suggest that KLEIP localizes to the sites of cell-cell adhesion in MDCK cells.
We found that KLEIP was temporally concentrated at the cell-cell contact sites (Figure 5A). KLEIP was partially colocalized with actin filaments at these sites (Figure 4A). We also showed that KLEIP interacted with F-actin in vitro (Figure 3B). This KLEIP recruitment to the cell-cell contact sites increased and reached to the peak 8 h after a calcium switch (Figure 5B). It has been reported that circumferential actin bundles are established during E-cadherin–mediated cell adhesion (Vasioukhin and Fuchs, 2001). Our E-cadherin/Fc beads assays revealed that KLEIP and F-actin were recruited in the vicinity of the beads placed on the cells (Figure 6). An inhibitor of actin polymerization, cytochalasin B, prevented KLEIP recruitment around E-cadherin/Fc beads, suggesting that KLEIP recruitment requires F-actin. Moreover, CA-Rac1 significantly stimulated KLEIP recruitment as well as F-actin to the cell-cell contact sites. Our observation of the increased F-actin staining in CA-Rac1 transfectants was consistent with a previous report (Takaishi et al., 1997). These observations may suggest that the recruitment of KLEIP to the cell-cell contact sites is due to its localization on the actin filaments.
KLEIP was localized to actin bundles at the cell-cell contact sites during cell adhesion but not other actin filaments such as stress fibers (Figure 4A). However, after the establishment of E-cadherin–mediated cell adhesion, KLEIP localized at the cell-cell junctions returned to the entire cells (Figure 5A). These observations suggest that the interaction between KLEIP and F-actin was regulated spatially and temporally. In Drosophila, phosphorylation of Kelch by Src64 negatively regulates the interaction of actin filaments (Kelso et al., 2002). We found that KLEIP was in the insoluble fraction in the presence of 0.5% Triton X-100 at 0, 8, and 24 h after a calcium switch (Figure S1). If KLEIP always interacted with actin filaments, it could be detected at other actin structures, such as stress fibers and actin filaments at the cell-free edge. It is still not clear why the calcium switch did not affect the detergent solubility of KLEIP. The interaction with cytoskeleton is not the only reason for the nonionic detergent insolubility (Solomon et al., 1998). One possibility is that KLEIP might be associated with the nonionic detergent-resistant membrane in their intracellular pool before the calcium switch. Because E-cadherin/Fc beads induced KLEIP recruitment around the beads, some downstream signaling of E-cadherin ligation might be required for the binding between KLEIP and F-actin. Further analysis would clarify how the interaction between KLEIP and F-actin is regulated.
Together with the presence of the kelch repeats, in vitro association and in vivo colocalization of KLEIP with F-actin suggest that KLEIP may function as a cross-linker like Drosophila Kelch and play a critical role on actin assembly in the junctions. We found that KLEIP-N, which lacks the actin-binding site and contains a sufficient sequence for the recruitment to the cell-cell adhesion sites, inhibited CA-Rac1-induced actin recruitment (Figure 7B). It has been reported that the N-terminal half of Drosophila Kelch has the dominant negative effects on ring canal formation (Robinson and Cooly, 1997). KLEIP-N might form dimers with endogenous KLEIP to inhibit its function, because the N-terminal half of KLEIP is involved in dimer formation like other Kelch-like proteins (Figure 3A). Therefore, KLEIP may play a role on actin remodeling during cell-cell contact and inhibit Rac signaling upstream of actin assembly. However, KLEIP-N alone did not inhibit physiological actin assembly during cell-cell contacts in MDCK cells (Figure 4B). This might indicate that there may be other compensatory mechanisms of actin remodeling during cell-cell contact. Increased accumulation of KLEIP by Rac activation seems to stimulate actin accumulation at the sites (Figure 7A). Considering the temporal accumulation of KLEIP at the junctions peaked 8 h after calcium switch (Figure 5B), KLEIP accumulation seems to form a positive feedback loop: Rac-activated F-actin accumulation at the cell-cell adhesion sites stimulates KLEIP accumulation at the sites and then KLEIP functions as an actin cross-linker to further accumulate F-actin at the sites. Because the engagements of actin filaments at the cell-cell contact sites enhance the clustering of E-cadherin (Braga, 2002b), KLEIP may be involved in such a feedback regulation of actin assembly. After the establishment of cell-cell contact, KLEIP may lose the ability of binding to F-actin and return to the intracellular pool. The interaction between KLEIP and F-actin seems to require the E-cadherin ligation signaling despite of their direct interaction in vitro. Based on these results, we propose that KLEIP functions as an actin-cross linker presumably at adherens junctions during the initiation of cell-cell adhesion. Further studies on KLEIP will clarify such unknown regulatory mechanisms to regulate the binding of KLEIP and F-action.
In an alternative approach to examine a function of KLEIP, we used RNA interference in HeLa cells (Elbashir et al., 2001). However, we could not efficiently knock down KLEIP, although six different pairs of small interfering RNAs (siRNAs) were used. It has been reported that some siRNAs were ineffective in several genes (Harborth et al., 2001). An improved strategy to design the siRNAs may help knock down KLEIP in future experiments.
In conclusion, we isolated and characterized a novel mammalian Kelch-related protein, KLEIP. KLEIP is recruited to the sites of cell-cell contact by the initiation of cell-cell contact and involved in Rac1-based actin assembly during cell-cell adhesion. Our findings will give an insight in understanding the regulatory mechanisms of cell-cell contacts as well as the functions of this Kelch-related protein.
Supplementary Material
Acknowledgments
We thank Dr. Douglas Lowy for support. T.H is supported by the Japan Society for the promotion of Science fellowship from the Ministry of Education, Science and Culture, Japan.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–07–0531. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-07-0531.
Abbreviations used: CA, constitutively active; ECT2, epithelial cell transforming gene 2; GFP, green fluorescence protein; GST, glutathione S-transferase; KLEIP, Kelch-like ECT2 interacting protein; PBS, phosphate-buffered saline; TBS, Tris-buffered saline.
References
- Adams, C.L., Chen, Y.-T., Smith, S.J., and Nelson, W.J. (1998). Mechanism of epithelial cell-cell revealed by high-resolution tracking of E-cadherin-green fluorescent protein. J. Cell Biol. 142, 1105-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, J., Kelso, R., and Cooly, L. (2000). The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 10, 17-24. [DOI] [PubMed] [Google Scholar]
- Albagli, O., Drordain, P., Dewwindt, C., Lecoceq, G., and Leprince, D. (1995). The BTB/POZ domain: a new protein-protein interaction motif common DNA- and actin-binding proteins. Cell Growth Differ. 6, 1193-1198. [PubMed] [Google Scholar]
- Allan, V. J. (2000). Basic immunofluorescence. In: Protein Localization by Fluorescence Microscopy: A Practical Approach, ed. V.J. Allan. Oxford, United Kingdom: Oxford University Press, 1-16.
- Bomont, P., and Koeing, M. (2003). Intermediate filament aggregation in fibroblasts of giant axonal neuropathy patients is aggravated in non dividing cells and by microtubule destabilization. Hum. Mol. Genet. 12, 813-822. [DOI] [PubMed] [Google Scholar]
- Braga, V. (2002a). Cadherin adhesion regulation in keratinocytes. In: Cell-Cell Interaction: A Practical Approach, ed. T.P. Fleming. Oxford, United Kingdom: Oxford University Press, 1-36.
- Braga, V.M.M. (2002b). Cell-cell adhesion and signalling. Curr. Opin. Cell. Biol. 14, 546-556. [DOI] [PubMed] [Google Scholar]
- Braga, V.M.M., Machesky, L.M., Hall, A., and Hotchin, N.A. (1997). The small GTPases Rho and Rac are required for the establishment of Cadherin-dependent cell-cell contacts. J. Cell Biol. 137, 1421-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbelo, P.D., Miyamoto, S., Utani, A., Brill, S., Yamada, K.M., Hall, A., and Yamada, Y. (1995). p190-B, a new member of the Rho GAP family, and Rho are induced to cluster after integrin cross-linking. J. Biol. Chem. 270, 30919-30926. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschi, T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- Ehrlich, J.S., Hansen, M.D.H., and Nelson, W.J. (2002). Spatio-temporal regulation of Rac1 localization and lamellipodia dynamics during epithelial cell-cell adhesion. Dev. Cell. 3, 259-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata, M., and Kaibuchi, K. (2001). Rho-family GTPases in cadherin-mediated cell-cell adhesion. Nat. Rev. 2, 887-897. [DOI] [PubMed] [Google Scholar]
- Fukata, M., Nakagawa, M., Itoh, N., Kawajiri, A., Yamaga, M., Kuroda, S., and Kaibuchi, K. (2001). Involvement of IQGAP1, an effector of Rac1 and Cdc42 GTPases, in cell-cell dissociation during cell scattering. Mol. Cell. Biol. 21, 2165-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimond, C., van der Flier, A., van Delft, Brakebusch, C., Kuikman, I., Collard, J.G., Fässler, R., and Sonnenberg, A. (1999) Induction of cell scattering by expression of β1-integrins in β1-deficient epithelial cells requires activation of members of Rho family of GTPase and downregulation of cadherin and catenin function. J. Cell Biol. 147, 1325-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godt, D., Couderc, J.L., Cramton, S.E., and Laski, F.A. (1993). Pattern formation in the limbs of Drosophila: bric a brac is expressed in a both gradient and wave-like pattern and is required for specification and proper segmentation of the tarsus. Development 119, 799-812. [DOI] [PubMed] [Google Scholar]
- Harborth, J., Elbashir, S.M., Bechert, K., Tushci, T., and Weber, K. (2001). Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 114, 4557-4565. [DOI] [PubMed] [Google Scholar]
- Hernandez, M., Andres-Barquin, P.J., Martinez, S., Buifone, A., Rubenstein, J.L.R., and Israel, M.A. (1997). ENC-1, a novel mammalian kelch-related gene specifically expressed in the nervous system encodes an actin-binding protein. J. Neurosci. 17, 3038-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R.G., Meyer, R.A., Li, X.-R., Preus, D.M., Tan, L., Grunenwald, H., Paulson, A.F., Laird, D.W., and Sheridan, J.S. (2002). Gap junctions assemble in the presence of cytoskeletal inhibitors, but enhanced assembly requires microtubules. Exp. Cell. Res. 275, 67-80. [DOI] [PubMed] [Google Scholar]
- Jou, T.-S., Schneeberger, E.E., and Nelson, W.J. (1998). Structural and functional regulation of tight junctions by RhoA and Rac1 small GTPases. J. Cell Biol. 142, 101-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jou, T.-S., and Nelson, W.J. (1998). Effects of regulated expression of mutant RhoA and Rac1 small GTPases on the development of epithelial (MDCK) cell polarity. J. Cell Biol. 142, 85-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs, E.M., Ali, R.G., McCormack, A.J., and Yap, A.S. (2002). E-Cadherin homophilic ligation directly signals through Rac and phosphatidylinositol 3-kinase to regulate adhesive contacts. J. Biol. Chem. 277, 6708-6718. [DOI] [PubMed] [Google Scholar]
- Kelso, R. J., Hudson, A. M., and Cooley. L. (2002). Drosophila Kelch regulates actin organization via Src64-dependent tyrosine phosphorylation. J. Cell Biol. 156, 703-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert, M., Choquest, D., and Mège, R.M. (2002). Dynamics of ligand-induced, Rac1-dependent anchoring of cadherins to the actin cytoskeleton. J. Cell Biol. 157, 469-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert, M., Padilla, F., and Mège, R.M. (2000). Immobilized dimers of N-cadherin-Fc chimera mimic cadherin-mediated cell contact formation: contribution of both outside and inside-out signals. J. Cell Sci. 113, 2207-2219. [DOI] [PubMed] [Google Scholar]
- Levenberg, S., Katz, B.-Z., Yamada, K.M., and Geiger, B. (1998). Long-range and selective autoregulation of cell-cell or cell-matrix adhesions by cadherin or integrin ligands. J. Cell Sci. 111, 347-357. [DOI] [PubMed] [Google Scholar]
- Miki, T., Smith, C., Long, J., Eva, A., and Flemmeng, T. (1993). Oncogene ect2 is related to regulators of small GTP-binding proteins. Nature 362, 462-465. [DOI] [PubMed] [Google Scholar]
- Nakagawa, M., Fukata, M., Yamagata, M., Itoh, N., and Kaibuchi, K. (2001). Recruitment and activation of Rac1 by the formation of E-cadherin-mediated cell-cell adhesion sites. J. Cell Sci. 114, 1829-1838. [DOI] [PubMed] [Google Scholar]
- Noren, K.N., Arthur, W.T., and Burridge, K. (2003). Cadherin Engagement inhibits RhoA via p190RhoGAP. J. Biol. Chem. 278, 13615-13618. [DOI] [PubMed] [Google Scholar]
- Robinson, D.N., and Cooly, L. (1997). Drosophila Kelch is an oligomeric ring canal actin organizer. J. Cell Biol. 138, 799-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai, E., and Marshall, C.J. (2002). ROCK and Dia have opposing effects on adherens junction downstream of Rho. Nat. Cell. Biol. 4, 408-415. [DOI] [PubMed] [Google Scholar]
- Solomon, K.R., Malloy, M.A., and Finberg, R.W. (1998). Determination of the non-ionic detergent insolubility and phosphoprotein associations of glycosylphosphatidylinositol-anchored proteins expressed on T cells. Biochem. J. 334, 325-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothen-Rutishauser, B., Riesen, F.K., Braun, A., Günthert, M., and Wunderli-Allenspach, H. (2002). Dynamics of tight and adherens under EGTA treatment. J. Membr. Biol. 188, 151-162. [DOI] [PubMed] [Google Scholar]
- Sasagawa, K., Mastudo, Y., Kang, M., Fujimura, L., Iitsuka, Y., Okada, S., Ochiai, T., Tokuhisa, T., and Hatano, M. (2002). Identification of Nd1, a novel murine kelch family protein, involved in stabilization of actin filaments. J. Biol. Chem. 277, 44140-44146. [DOI] [PubMed] [Google Scholar]
- Soltysik-Espanola, M., Rogers, R.A., Jiang, S., Kim, T., Gaedigk, R., White, R.A., and Avraham, H. (1999). Characterization of Mayven, a novel actin-binding protein predominantly expressed in brain. Mol. Biol. Cell 10, 2361-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaishi, K., Sasaki, T., Kotani, H., Nishioka, H., and Takai, Y. (1997). Regulation of cell-cell adhesion by Rac and Rho small G proteins in MDCK cells. J. Cell Biol. 139, 1047-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumoto, T., Xie, X., Blumenthal, R., Okamoto, I., and Miki, T. (1999). Human ECT2 is an exchange factor for Rho GTPase, phosphorylated in G2/M phases, and involved in cytokinesis. J. Cell Biol. 147, 921-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trickett, A., and Kwan, Y.L. (2003). T cell stimulation and expansion using anti-CD3/28 beads. J. Immunol. Methods 275, 251-255. [DOI] [PubMed] [Google Scholar]
- Vasioukhin, V., and Fuchs, E. (2001). Actin-dynamics and cell-cell adhesion in epithelia. Curr. Opin. Cell Biol. 13, 76-84. [DOI] [PubMed] [Google Scholar]
- Xue, F., and Cooley, L. (1993). Kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell 72, 681-693. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.