Abstract
The Drosophila wings-up A gene encodes Troponin I. Two regions, located upstream of the transcription initiation site (upstream regulatory element) and in the first intron (intron regulatory element), regulate gene expression in specific developmental and muscle type domains. Based on LacZ reporter expression in transgenic lines, upstream regulatory element and intron regulatory element yield identical expression patterns. Both elements are required for full expression levels in vivo as indicated by quantitative reverse transcription-polymerase chain reaction assays. Three myocyte enhancer factor-2 binding sites have been functionally characterized in each regulatory element. Using exon specific probes, we show that transvection is based on transcriptional changes in the homologous chromosome and that Zeste and Suppressor of Zeste 3 gene products act as repressors for wings-up A. Critical regions for transvection and for Zeste effects are defined near the transcription initiation site. After in silico analysis in insects (Anopheles and Drosophila pseudoobscura) and vertebrates (Ratus and Coturnix), the regulatory organization of Drosophila seems to be conserved. Troponin I (TnI) is expressed before muscle progenitors begin to fuse, and sarcomere morphogenesis is affected by TnI depletion as Z discs fail to form, revealing a novel developmental role for the protein or its transcripts. Also, abnormal stoichiometry among TnI isoforms, rather than their absolute levels, seems to cause the functional muscle defects.
INTRODUCTION
Contractile protein systems are widely represented in most cell types as a force generator device (Davison et al., 2000). In muscles, troponin I (TnI) is a key element in the protein complex that regulates sliding of thin over thick filaments (Farah and Reinach, 1995; Geeves and Lehrer, 1998; Squire and Morris, 1998; Maytum et al., 2003). Several TnI protein isoforms are generated, either from transcription of independent genes (e.g., vertebrates) or from differential splicing of a single gene primary transcript (e.g., Drosophila). Amino acid substitutions in constitutive or alternatively spliced exons in TnI can lead to pathological conditions such as familial hypertrophic cardiomyopathy (Carrier et al., 1993; Coonar and McKenna, 1997) and distal arthrogryposis (Sung et al., 2003), due to abnormal interactions with other sarcomere components. Also, TnI is a relevant indicator of heart failure (Lewinter and Vanburen, 2002) and a potent angiogenesis inhibitor through its interaction with polycystin-2 (Li et al., 2003). The potential applications that this knowledge could provide, however, are handicapped by the scant information on the regulatory mechanisms of TnI gene expression. This issue is particularly relevant in the context of future gene therapy strategies and justifies this in vivo study of the regulatory mechanism of the Drosophila homologue. In addition, this study takes advantage of the fact that TnI in Drosophila is encoded by a single gene, wings up A (wupA), and that isoform replacement during normal development as in the mouse heart (Siedner et al., 2003) does not take place. These features render the task amenable.
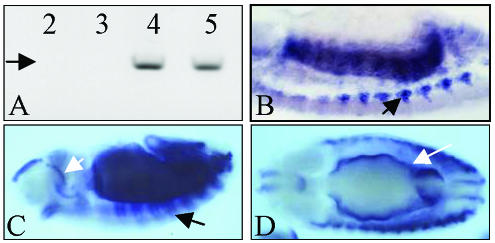
Transcription regulation is an elaborated process that requires specific interactions between genomic sequences and components of the transcriptional machinery resulting in local structural changes of the chromatin (Davidson et al., 2002). In addition to these cis-effects, transcriptional changes can be elicited in trans- on the homologous gene copy through a largely unknown mechanism thought to depend on correct chromosomal pairing at the gene locus (but see Goldsborough and Kornberg, 1996). In vertebrates, three different genes (fast, slow, and cardiac), encode specialized forms of TnI, whereas in Drosophila, the single gene wings-up A encodes 10 TnI protein isoforms (Barbas et al., 1991). The gene is expressed shortly after the monolayer of mesodermal cells is determined by the sequential expression of twist, Dmef2, tinman, and other transcription factor-encoding genes (stages 6–7 or 3–4 h of development) (Thisse et al., 1987; Azpiazu and Frasch, 1993; Bour et al., 1995; Lilly et al., 1995; Taylor, 2000). Muscle progenitor cells are determined immediately afterwards, and their descendants, the muscle founder cells, become recognizable by the end of stage 11 (7–8 h) (Carmena et al., 1995; Ruiz and Bate, 1997). Cell fusions between founder and adjacent fusion-competent cells begin at stage 13 (9–10 h) leading to muscle syncitia (Baylies et al., 1998; Paululat et al., 1999; Taylor, 2002). The earliest TnI transcripts are detected at 4 h by reverse transcription-polymerase chain reaction (RT-PCR) and are maintained thereafter as indicated by in situ hybridization (Prado et al., 1999) (Figure 1A). By stage 13, high levels of TnI transcripts can be observed in segmental arrays of muscle primordia of fused cells when they initiate differentiation and stretch toward anchoring apodemes (Figure 1B). By stage 14, TnI transcripts are prominent in visceral and somatic developing muscles (Figure 1, C and D). Thus, although TnI is usually considered a muscle differentiation marker involved in the regulation of sarcomere contraction, it is clearly expressed well before muscle founder cells begin to fuse.
Figure 1.
wings-up A expression in the embryo. (A) RT-PCR of 2- to 5-h embryo extracts primed for TnI. Expression of a 1.2-kb cDNA (arrow) can be seen from stage 7 (4 h) onwards. Numbers indicate hours of development from 30 min. Egg-laying periods of the CS strain. (B) Lateral view of a 10-μm section of a stage 12 embryo revealing TnI transcripts in somatic muscle primordia (arrow) by in situ hybridization. (C) Lateral view of a whole mount stage 12 embryo showing TnI transcripts in segment arranged somatic developing muscles (black arrow), and in the foregut (white arrow). (D) Dorsal view of a whole mount stage 14 embryo showing TnI expression in developing visceral muscles (white arrow).
After this introductory description of the early TnI gene expression, we address the regulatory mechanisms for wings-up A transcription, characterize some of the transcription factor binding sites, and measure in vivo the transcriptional cis- and trans-effects produced by rearrangements of the regulatory regions. The regulatory design described here seems conserved in other Drosophila muscle genes and, most relevant, in TnI encoding vertebrate genes. Furthermore, we demonstrate that TnI is required for muscle morphogenesis, as opposed to muscle contraction only, and that the unbalance of TnI isoforms results in severe muscle dysfunction.
MATERIALS AND METHODS
Fly Strains and Crosses
Df(1)23437 and the point mutations hdp2 and hdp3 were described previously (Beall and Fyrberg, 1991; Barbas et al., 1993; Prado et al., 1995, 1999). Regulatory mutants PL87 and PG31 were provided by H.M. Bourbon (Bourbon et al., 2002). Transcription factors null alleles mef222–21, tin346, bin22, and mincA388 have been described previously (Azpiazu and Frasch 1993; Bour et al., 1995; Zaffran et al., 2001; Ruiz-Gomez et al., 2002). In(1)zae(bx), z58g, and P{w+LacZ}Trls2325 fly stocks were obtained from the Drosophila stock center (University of Indiana, Bloomington, IN). Additional information can be obtained from FlyBase (http://flybase.bio.indiana.edu). Embryos were staged according to Campos-Ortega and Hartenstein (1997).
Transvection Index
It is defined by the algorithm 1+ (E - C/E + C), where E is percentage of adults with normal wing position of the experimental genotype and C is the equivalent for the control genotype. Both genotypes are siblings. Index values range from 1 (maximal transvection) to 0 (minimal transvection). Wing position was determined in 4-d aged adults.
Histochemistry and Immunostaining
β-Galactosidase activity was assayed in larvae and adults of transgenic lines as described in Ashburner (1989) with minor modifications. Third instars were dissected and fixed in 1% glutaraldheyde in phosphate-buffered saline during 30 min. Adult thorax and abdomen muscles were fixed in 4% paraformaldheyde in phosphate-buffered saline for the same time. To detect variations on expression levels among different constructs, we monitored the blue reaction products for 1 h 30 min at 37°C in a humidified chamber. The highest expression level is attained at this incubation time. The numbers 0–3 indicate the relative intensity that each genomic fragment yields under these incubating conditions. Fragments showing no expression were confirmed by additional 24-h incubation. Inmunohistochemical staining was performed as described previously (Marques et al., 2002). Embryos were collected, dechorionized, and stained with anti-β-galactosidase (Cappel Laboratories, Durham, NC) at 1:1000 and biotinylated horse anti-mouse IgG (Sigma-Aldrich, St. Louis, MO) at 1:200. Gene expression was monitored in whole mount in situ hybridizations with a digoxigenin-labeled RNA probe as described previously (Ruiz-Gomez et al., 1997). The RNA probe was generated from the L9-TnI cDNA template (Barbas et al., 1991). Homozygous mutant embryos were identified from balancer LacZ or GFP marked chromosomes.
Plasmid Vector, Transgenic Lines and Site-directed Mutagenesis
Genomic fragments were subcloned in pBluescript KSII+. The intron regulatory element (IRE)-0.89 kb fragment was produced by PCR by using the 4.7-kb clone as template with the forward primer 5′-GCTCTAGAGACTTCGGCTATGTACACTGCG-3′ and the reverse primer 5′-GCCCTCGAGCAAGCGGAATGGAAAAACAG-3′. After amplification, the genomic fragment was digested by XbaI/XhoI, cloned into the P transformation vector, and verified by sequence analysis. Other fragments were obtained in similar ways by using primers whose sequence is available on request. For transgenic lines, we produced a new vector, YAL, derived from YES (Patton et al., 1992). YAL contains the Antennapedia promoter driving LacZ as reporter, yellow as transformation marker, and flanked by suppressor of Hairywing–binding site boundary sequences (Gerasimova et al., 1995). IRE and upstream regulatory element (URE) fragments were cloned into SalI–XbaI–XhoI targets sites of YAL polylinker, keeping the native orientation relative to the basal promoter. Germline transformation was performed in y w embryos by standard procedures (Spradling and Rubin, 1982). Site-directed mutagenesis was performed using PCR (Horton, 1995) on IRE-1.7 and IRE-0.89 templates. The sequence of oligonucleotides used to mutagenize each DMEF2 site is available on request.
Quantitative RT-PCR Assays
About 20 individuals of each genotype were used for RNA extraction by TRIzol reagent (Invitrogen, Carlsbad, CA). A total of 2 μg of RNA was reverse transcribed into cDNA by using First Strand kit (Amersham Biosciences, Piscataway, NJ) and 0.2 μg of oligo(dT) per reaction according to manufacturer's instructions. As normalizing internal control, we used the 140-kDa RNApolII subunit encoding gene (Falkenburg et al., 1987). Exon-specific oligonucleotide primers for the two genes tested, wupA and RNApolII, were designed from the databank sequences by using the Primer Select (DNA Star) software, eliminating putative dimerizing pairs of primers. Primer sequences are available on request. Fragments were verified by sequencing. Reverse transcription products were used as template for PCR reactions by using several dilutions to generate the corresponding curves. The following products were included in the reaction: SYBR Green PCR core reagents (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. The amplification was carried out in a ABI PRISM 7700 sequence detector (Applied Biosystems) by using the following conditions: 2 min 50°C, 10 min 95°C, 40 cycles (30 s 95°C, 30 s 60°C, 30 s 68°C), and 1 cycle (15 s 95°C, 1 min 60°C, 1 min 95°C). To calculate the relative index of gene expression, we used the efficiency calibrated mathematical method (Soong et al., 2000). It is based on the algorithm Ratio = [(E ref)Ct sample/(E target)Ct sample]/[(E ref)Ct calibrator/(E target)Ct calibrator, where (E ref) is the reaction efficiency for the RNApolII primers, (E target) corresponds to wupA gene, Ct sample is the average threshold cycle from the gene, the E value corresponding to the mutant genotype sample, and Ct calibrator is the average of the threshold cycles from the gene, the E value corresponding to the nonmutant genotype sample. In turn, the E values of each reaction are calculated from the standard curve slope according to E = 10-1/slope as described in Rasmussen (2001).
Western Blots
Total protein extracts were obtained from homogenized embryos. Monospecific anti-Troponin I (Barbas et al., 1993) and anti-Ariadne (Aguilera et al., 2000) antibodies were used at 1:1000 and 1:75 dilutions respectively. Signal was developed by the chemiluminiscent ECL method (Amersham Biosciences).
Electron Microscopy
Electron microscopy was performed as described previously (Therianos et al., 1995), with minor modifications. Embryos were dechorionated, devitellinized, and fixed with 6% glutaraldehyde in phosphate buffer (pH 7.2) for 5 h at room temperature. Six 10-min washes with phosphate buffer were followed by postfixation in 1% OsO4 in the same buffer for 1 h at room temperature. Dehydration was in graded ethanol series and embedded in Epon 812 resin. Samples were stained in 2% uranyl acetate (dihydrate) in aqueous solution for 60 min, followed by 15-min lead citrate incubation. Ultrathin sections of 70 nm were mounted on Formvar-coated single slot grids and viewed on a JEOL 1200 EXII electron microscope at 80 Kv.
In Silico Sequence Analysis
Sequence analyses were based on EMBL/GENEBANK/DDBJ databases and carried out using the BLAST 2.0 programs of the National Center for Biotechnology Information. For putative transcription factors binding sites, we used MatInspector version 2.2 program in the TRANSFAC databases (Wingender et al., 2000) followed by BLAST analysis with reported consensus sequences.
RESULTS
Searching for Regulatory Elements
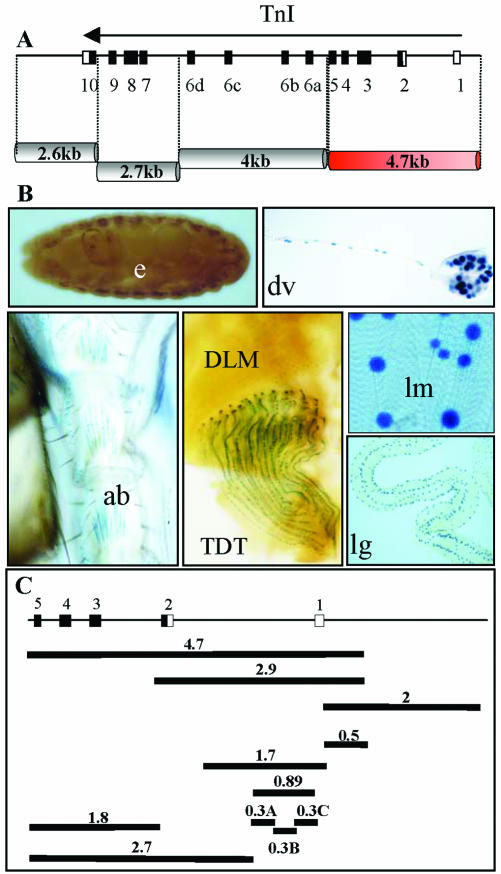
The gene wings-up A maps to an X chromosome region, 16F, whose genetic constituents are well characterized (Prado et al., 1999). We generated transgenic lines with genomic fragments that span the entire TnI encoding transcription unit. These fragments were inserted into a newly generated vector, YAL (see MATERIALS AND METHODS), whose flanking sequences are of the insulator type aiming to prevent local enhancer effects on reporter expression. Out of the four major fragments tested, only one yielded LacZ reporter signal, and it was specific for muscle tissue, including all muscle types (Figure 2, A and B). These observations indicate that downstream of the wings up A promoter, the only existing positive regulatory elements are those included within the referred 4.7-kb fragment that spans exons 1–5.
Figure 2.
Genomic fragments from the wings-up A region and its reporter expression domains. (A) Extent in kilobases of major genomic fragments tested. (B) Expression domains of the only fragment that yielded expression, the 4.7 kb. Images of the expression in the embryo (e), dorsal vessel of the larvae (dv), adult abdominal muscles (ab), adult tergal depressor of the trochanter (TDT), dorsolongitudinal muscles (DLM), somatic larval muscles (lm), and larval gut (lg). (C) Genomic subfragments tested for reporter expression. Numbers indicate size in kilobases.
Muscle Expression of TnI Is Controlled by Two Elements
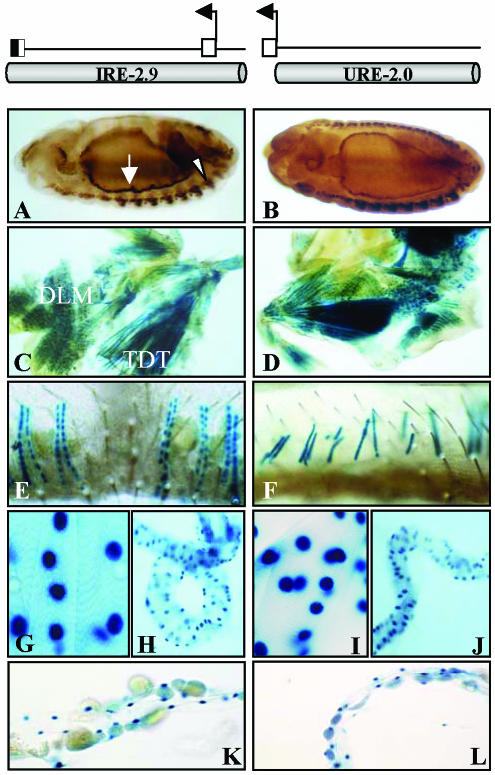
The positive regulatory elements detected in the 4.7-kb fragment were further restricted to a 2.9-kb subfragment that yielded the same expression pattern (Figure 2C). In the same way, the analysis of genomic regions upstream of the promoter region yielded an additional fragment with positive reporter activity that extended 2.5 kb and overlapped 500 base pairs with the previous element (Figure 2C). We refer to these two fragments as URE and IRE, respectively, to maintain the nomenclature as in other TnI genes (Yutzey et al., 1989). Both elements show identical muscle-specific patterns of expression at all times during development (Figure 3). The two regions, located between -2 and +2.5 kb with respect to the transcription initiation site, were further subdivided into smaller fragments and tested for reporter activity in transgenic lines (Figure 2C). The resulting expression patterns are summarized in Table 1 and reveal spatial and temporal domains of specificity. These regulatory domains, as in many other genes studied (Arnone and Davidson, 1997), may overlap with each other forming a regulatory landscape rather than a line of mutually independent stretches of regulatory DNA (see DISCUSSION). For somatic muscles, the smallest fragment that drives an expression pattern equivalent to that of the native gene is the 1.7 kb that is fully contained within the first intron of the gene.
Figure 3.
Identical reporter expression patterns from IRE and URE regulatory regions. (A and B) Reporter expression in somatic (arrow head) and visceral (arrow) muscles in the embryo. (C and D) Adult dorsolongitudinal (DLM) and tergal depressor of the trochanter (TDT) muscles. (E and F) Adult abdominal muscles. (G and I) Larval somatic muscles. (H and J) Larval midgut visceral muscles. (K and L) Larval heart. In all cases, reporter expression is revealed by β-galactosidase reaction in the cell nucleus. Top diagrams indicate the location and extent of fragments tested as transgenes.
Table 1.
Reporter gene expression domains from IRE and URE subfragments
| Somatic muscles
|
Visceral muscles
|
Dorsal vessel
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E | LIII | TDT | DLM | DVM | DFM legs | ABD | E | LIII | A | E | LIII | A | |
| IRE-2.9 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| URE-2.0 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| IRE-1.7 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 1 | 3 | 3 | 0 | 0 | 0 |
| IRE-0.89 | 2 | 3 | 0 | 0 | 0 | 3 | 3 | 1 | 3 | 3 | 0 | 0 | 0 |
| IRE-0.3A | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| IRE-0.3B | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| IRE-0.3C | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| IRE-1.8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| IRE-2.7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| URE-0.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Numbers indicate the relative intensity (0, no detectable signal; 3, maximal signal) of reporter gene expression under standard conditions. E, embryo; LIII, third instra larvae; A, Adult; TDT, tergal depressor of the trochanter muscle; DLM, dorsolongitudinal muscle; DVM, dorsoventral muscle; DFM, direct flight muscles; Legs, tubular leg muscles; ABD, abdominal somatic muscles.
Transcription Factor Binding Sites within IRE and URE Regions
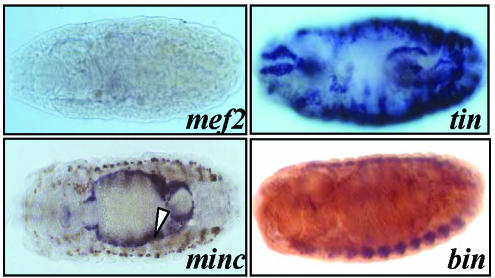
To identify transcription factors involved in wupA regulation, we first tested available null mutants for known transcription factors on their effects on TnI expression in vivo. As previously described, DMEF2 is required for transcription of muscle genes (Lin et al., 1996; Stronach et al., 1999; Arredondo et al., 2001; Kelly et al., 2002), and the corresponding mutant embryos showed absence of TnI RNA in all muscle types (Figure 4). By contrast, in mutants for TIN-MAN (Azpiazu and Frasch, 1993; Bodmer, 1993), BINIOU (Zaffran et al., 2001), and MINC (Ruiz-Gomez et al., 2002), TnI expression is prevented in selected muscle types only. The first two seem to be required for visceral expression of TnI, and the third one is required only for somatic muscles.
Figure 4.
TnI expression in mutant embryos. Dorsal view of whole mount embryos degoxygenin stained for TnI RNA expression in homozygous mutant genotypes for the indicated genes. Note the lack of TnI expression in somatic and visceral muscles in the mef2 null embryo, whereas tinman deficiency prevents expression in the heart and midgut only. Also, the biniou mutant prevents visceral expression only, whereas minc eliminates somatic expression. The dark signal observed in selected somatic muscle cells in the minc embryo (arrowhead) corresponds to an enhancer trap reporter within the dumbfounded gene that serves to mark the founder cells (Ruiz-Gomez et al., 2002).
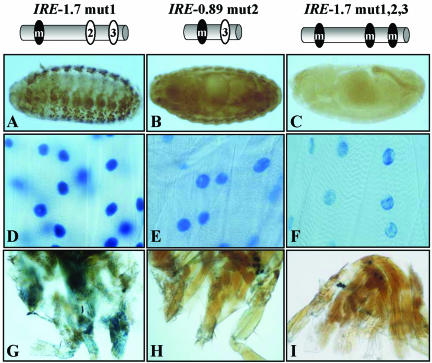
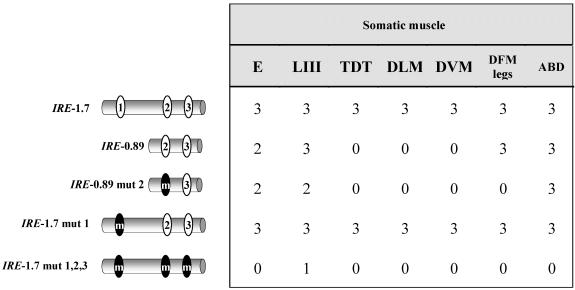
An in silico analysis identified three putative DMEF2 binding sites within the IRE whose sequence seemed similar to that considered as consensus (Black and Olson, 1998) (e.g., consensus, YTAAAAATAR; site 1, TTAAAAATAC; site 2, TTAAAAATAA; and site 3, CTAAAAATG). These putative DMEF2 binding sites fulfill the muscle, as opposed to neural, sequence criteria (Andrés et al., 1995). To assay for their actual DMEF2 binding activity, we carried out in vivo and in vitro tests. Reporter transgenes of fragments containing normal and mutated versions of these sites were analyzed in all tissues and developmental stages (Figure 5 and Table 2). The significance of site 1 is indicated by the loss of reporter expression in adult tergal depressor of the trochanter, dorsolongitudinal muscles, and dorsoventral muscles and a reduction in embryo somatic muscles when the IRE-0.89 fragment was assayed (compare Table 2, top two fragments). In addition, this site was tested for actual DMEF2 binding activity in band shift assays (see Supplementary Material). In the same way, site 2 activity is demonstrated by comparing reporter expression (see Table 2, second and third fragment) and band shift assays (see Supplementary Material). Finally, the activity of site 3 is supported by the remaining reporter activity of IRE-0.89 mut 2 fragment (Table 2). Mutations in the three sites (Table 2, lower fragment) yield a drastic reduction in all tissues, suggesting that the three sites act cooperatively. It is worth noting that although a null mutation in Dmef2 abolished TnI expression in all muscle types (Figure 4A), a fragment with the three sites mutated still leaves a detectable expression in larval somatic muscles (Table 2 and Figure 5F) as well as in visceral muscles (our unpublished data). Because no other putative DMEF2 binding site was identified within IRE upon close inspection of its sequence, this weak signal should result from another transcription factor activity that is present at the larval, but not at the embryo, stage. This additional transcription factor would be required for basal maintenance, but not initiation, of transcription of this gene (see DISCUSSION).
Figure 5.
In vivo effects of MEF2 sites inactivation. Diagrams indicate the fragments extent, and status, mutated (black) or native (white) of each MEF2 site. (A–C) Transgenic embryos. Note total absence of signal at this stage when the three MEF2 sites are mutated (C), indicating that no additional MEF2 sites, active in the embryo, exist within the IRE fragment. (D–F) Somatic larval muscles. Note the progressive reduction of expression as individual sites are mutated, albeit maintaining a residual activity when the three sites are inactivated. These observations indicate that MEF2 is necessary for initiation, but not maintenance, of wup A basal transcription. (G–I) Adult leg muscles. Note the same effect of mutated MEF2 sites as in the embryo. MEF2 mutations are the same as in band shift assays (see Supplementary Material).
Table 2.
Reporter expression from D-MEF2 mutated IRE fragments
The independent activity of URE as a regulator of TnI expression was documented in the initial screening where this region yielded an expression pattern identical to that of IRE (Figure 3). We analyzed its genomic sequence and found a number of transcription factor binding sites that occurred in a very similar arrangement compared with those in IRE (Figure 6, top diagram). The similarity includes the type of binding sites as well as their number and relative spacing. The apparent structural and functional redundancy between these two regions, however, proved misleading when the role of these regions in the expression of the native gene was studied (see below).
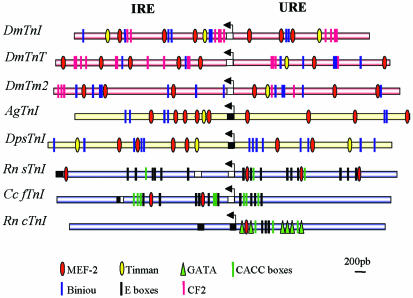
Figure 6.
Conservation of the TnI regulatory array. Diagrams of three D. melanogaster genes (TnI, TnT, and Tm2), TnI from D. pseudoobscura and An. gambiae, and three vertebrate genes encoding slow, fast, and cardiac TnI, respectively. As with the IRE and URE regions of wupA, all these genes exhibit similar clusters of transcription factors, binding sites upstream and downstream of the transcription initiation site (arrow in each diagram). For D. melanogaster TnT gene, an independent demonstration of IRE and URE regulatory regions is also available (Mas et al., 2004; this issue). Also, for D. melanogaster Tm 2, an IRE-like region has been experimentally demonstrated (Lin and Storti, 1997). Finally, for vertebrate's TnI genes, with the exception of the cardiac case, there is also experimental evidence of regulatory activities upstream and downstream of the initiation site (Banerjee-Basu and Buonanno, 1993; Nakayama et al., 1996; Murphy et al., 1997). The accepted binding site sequences for Drosophila and Anopheles have been MEF2, YTAAAAATAR (Black and Olson, 1998); TIN, TYAAGTG (Gajewski et al., 1997); BIN, RTAAAYA (Zaffran et al., 2001); and CF2, RTATATRTA (TRANSFAC databases at www.gene-regulation.com; Wingender et al., 2000). Coding (black) and noncoding (white) exons are indicated as boxes.
Conservation of the TnI Regulatory Array
For TnI genomic sequence comparisons, we chose another Drosophila species (D. pseudoobscura) and mosquito (Anopheles gambiae) because their genome sequences are available, and also those of rat (Ratus norvegicus) and quail (Coturnix coturnix) because in vitro functional data on regulatory regions are available. When comparing the genomic regions upstream and downstream to the promoter of insect TnI genes, there is a striking conservation of the type and array of putative transcription factor binding sites (Figure 6). For the rat slow, quail fast, and rat cardiac TnI genes, there are expression studies similar to those reported here, albeit in transfected cells (Yutzey et al., 1989; Banerjee-Basu and Buonanno, 1993; Nakayama et al., 1996; Murphy et al., 1997). With the exception of the rat cardiac gene, two regulatory regions upstream and downstream of the transcription initiation site have been documented. From the comparison between all TnI genes, it seems likely that the rat cardiac gene has lost the IRE region (Figure 6).
Considering other muscle genes in Drosophila, the available data on Troponin T-(Mas et al., 2004; this issue) and Tropomyosin 2 (Lin et al., 1996; Lin and Storti, 1997)-encoding genes show putative binding sites for the same transcription factors as in the TnI gene. In particular, the existence of two regions, URE and IRE-like, seems evident (Figure 6). Based on these observations, it seems that the Drosophila TnI regulatory arrangement is fairly well conserved in the orthologous counterparts and, possibly, in other genes encoding components of the muscle thin filament. The significance of this conservation is further supported by the unique nature of these regulatory regions. Three other genomic fragments tested for reporter expression and spanning ∼12 kb distal to IRE failed to drive expression in any tissue or developmental stage (Figure 2A).
Testing the Native Gene Regulatory Mechanisms
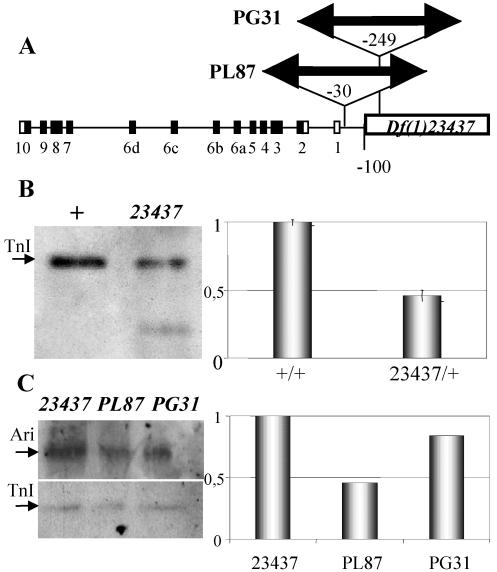
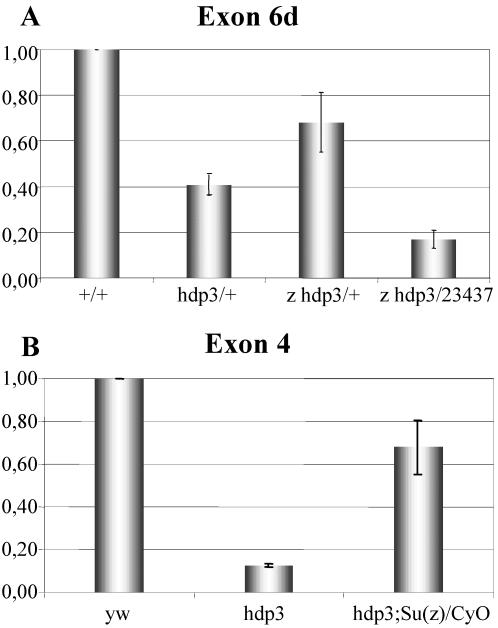
The previous data were obtained by the procedure of dissecting the corresponding genomic fragments and testing their effect on the expression of a reporter gene. Here, we analyze the regulatory activity of URE and IRE on the native gene. To that end, we used three chromosomal rearrangements that delete URE [Df(1)23437] or increase the IRE-URE spacing [P(w+LacZ)PL87 and P(w+Gal4)PG31]. The locations of the corresponding breakpoints were determined by plasmid rescue or genomic PCR (Figure 7A). The three rearrangements are lethal at the embryo stage, and their muscle phenotype is described further below.
Figure 7.
Expression effects of rearrangements. (A) Diagram indicating the location of the three rearrangements used here. PL87 and PG31 are insertions of about the same length (see MATERIALS AND METHODS) located at positions–30 and–249 base pairs with respect to the transcription initiation site. Df(1)23437 is a 2-kb deletion located at position–100. Coding (black) and noncoding (white) exons are indicated as numbered boxes. (B) Western blot (left) from normal (+) and mutant (23437) embryos revealing that TnI is still expressed, albeit reduced, when the URE region is deleted, demonstrating that IRE can drive TnI expression independently from URE. Transcription levels are demonstrably reduced by QRT-PCR in heterozygous mutant adults (right). Transcription was normalized with respect to RNA polymerase II. Error bars indicate average SE from three independent RNA extractions. (C) Western blot (left) from mutant embryos showing TnI expression with respect to that of Ariadne-1 (Ari) used here as an internal control. This blot is shown as a quantitative densitometry (right) to illustrate the effect of PL87 and PG31 where IRE and URE have been separated but remain intact. None of the rearrangements are null. Data normalized with respect to the Ari-1/TnI ratio in 23437 embryos.
The effects of these rearrangements on wup A expression were studied in Western blots and qualitative (Q)RT-PCR assays. In hemizygous Df(1)23437 embryos, where URE is virtually eliminated, TnI is still detected by Western blot (Figure 7B). These data demonstrates that IRE is able to drive transcription independently from URE. The levels of expression, however, are insufficient for viability. In heterozygous females, this deletion reduces transcription, as indicated by QRT-PCR, to 50% (Figure 7B). The other two rearrangements were analyzed by Western blots in hemizygous embryos, by using the expression of another gene, ari-1, as an internal control, and normalizing the corresponding Ari-1/TnI levels to those of Df(1)23437 (Figure 7C). Neither of the two rearrangements where URE is separated from IRE by ∼11 kb, PL87 and PG31, results in a null condition for wup A. These data demonstrate that IRE and URE are independently active in the native gene, albeit both must be within short distance to each other to synergize and yield full levels of transcription, and thus viability.
Transvection at the wupA Locus
The synergistic requirement of URE and IRE represent cis-interactions, presumably between enhancers and promoter. Chromosome pairing seems to be an additional requirement for proper transcription when two copies of the locus are present (i.e., in females, in this case) (Cook, 1997; Henikoff, 1997). In that context, we tested for transvection in heterozygotes between the three rearrangements, and between these and two point mutations in TnI. The latter correspond to wupAhdp2, a A116V change in a constitutive exon (Beall and Fyrberg, 1991; Prado et al., 1995), and to wupAhdp3, a single nucleotide change at the splice acceptor site for exon 6d (Barbas et al., 1993). In effect, allele hdp3 can be considered a regulatory mutation because it leads to the absence of a subfamily of TnI isoforms, whereas allele hdp2 is a structural mutation that does not interfere with transcription levels.
The homozygous genotypes for PL87 or PG31 result in viable and flying adults. This is the most efficient case of transvection observed at the wup A gene, and demonstrates that the deleterious cis-effects caused by the increased IRE-URE spacing can be repaired by an identical spacing in the homologous chromosome, presumably because chromosomal pairing at a critical region is now possible again. Similarly, the trans-combination between PL87 and PG31 (Table 3) yielded some viable adults with normal muscle structure, supporting flight in young as well as in 10-d aged individuals. It should be noted, however, that pairing between these rearrangements cannot be perfect along the inserted 11 kb because the transgene in each case differs, LacZ and Gal4. It seems that this mismatch is tolerated by the transcription mechanisms to the point of yielding near complete transvection. Combinations of either of these two rearrangements over 23437 remain lethal. Heterozygotes over allele hdp2 showed strong evidences of transvection because a large fraction of individuals exhibit normal wing position (transvection index values close to 1; see MATERIALS AND METHODS) (Table 3). Heterozygotes over allele hdp3 showed the same effect, although with somewhat reduced transvection index values in the case of PL87 and 23437 heterozygotes.
Table 3.
Transvection index in wings up A
| Genotype | Transvection index | |
|---|---|---|
| PG31/PL87 | 1 | |
| PG31/hdp2 | 1 | |
| PL87/hdp2 | 1 | |
| 23437/hdp2 | 1 | |
| PG31/hdp3 | 1 | |
| PL87/hdp3 | 0.7 | |
| 23437/hdp3 | 0.6 | |
| Zeste | ||
| PG31/hdp2z58g | 1 | |
| PL87/hdp2z58g | 0.9 | |
| 23437/hdp2z58g | 1 | |
| PG31/hdp2zae(bx) | 1 | |
| PL87/hdp2zae(bx) | 0.9 | |
| 23437/hdp2zae(bx) | 0.9 | |
| PG31/hdp3z58g | 1 | |
| PL87/hdp3z58g | 0.4 | |
| 23437/hdp3z58g | 0 | |
| PG31 hdp3zae(bx) | 1 | |
| PL87/hdp3zae(bx) | 0.8 | |
| 23437/hdp3zae(bx) | 0 | |
| Suppressor (z)3 | ||
| PG31/hdp2; Su(z)3/+ | 1 | |
| PL87/hdp2; Su(z)3/+ | 1 | |
| 23437/hdp2; Su(z)3/+ | 1 | |
| PG31/hdp3; Su(z)3/+ | 1 | |
| PL87/hdp3; Su(z)3/+ | 0.2 | |
| 23437/hdp3; Su(z)3/+ | 0 | |
Maximum transvection is indicated by value index 1 and minimum by 0. Index algorithm (see MATERIALS AND METHODS) was determined according to wing position in four days aged adults. All PL87/PG31 flies exhibit flight ability.
Transvection Is Dependent on zeste Activity
Transvection in other Drosophila genes is known to be influenced by DNA binding proteins involved in transcription and chromatin remodeling (Duncan, 2002; Kennison and Southworth, 2002). We tested the well characterized member of the Trithorax group of ubiquitously expressed cofactors encoded in zeste (z) (Laney and Biggin, 1996; Orlando et al., 1998; Hur et al., 2002), and the relatively less known Suppressor of zeste 3 [Su(z)3] gene. Two zeste mutant alleles were assayed in heterozygotes between hdp mutations and the three rearrangements (Table 3). The zeste background results in a reduction of transvection in virtually all genotypes tested (index values shift toward 0). The remarkable exception is the case of PG31. Heterozygotes over this rearrangement consistently maintain the highest transvection index. The observation seems to indicate that the location of this breakpoint, 249 base pairs upstream from the promoter, is beyond a critical distance for transcriptional effects in trans due to reduction of Zeste activity. Consistent with this interpretation, the shift of transvection index values is stronger in heterozygotes with PL87, which breaks at 30 base pairs upstream of the initiation site (see DISCUSSION). Heterozygotes including Df(1)23437, where URE is deleted, show the strongest shift yielding the minimum index value in most genotypes. Concerning the wupA alleles tested, hdp2 shows consistently better transvection than hdp3, and seems virtually insensitive to either of the two zeste backgrounds.
In the case of the relatively uncharacterized Su(z)3, there is a strong reduction of transvection in heterozygotes involving the hdp3, but not hdp2, allele. The effect is strong enough as to be detectable in males (genotype: hdp3, Su(z)3/+). These become progressively unpaired to move and die in a few days. These observations suggest that Su(z)3 play a role in wupA transcription similar to that of zeste.
Transvection Is Based on Transcriptional Changes
To know the molecular bases of the observed phenotypes, we measured transcription in various genotypes by QRT-PCR, discriminating among mRNA isoforms (Figure 8). The data confirm the reduction of exon 6d containing mRNAs due to the mutation hdp3 (Figure 8A, first two histograms). In addition, the data show that a zeste mutant background elicits an increase of transcription. Because these QRT-PCR assays were done with exon 6d probes, the observed increase in transcription must originate from the wild-type chromosome only. Similar tests carried out with Su(z)3 in males show that this gene acts, like zeste, as a repressor on wupA transcription (Figure 8B). Consistent with the shift of transvection index values and the enhanced severity of the motility trait in hdp3 males, the transcriptional effect of Su(z)3 seems stronger than that of zeste.
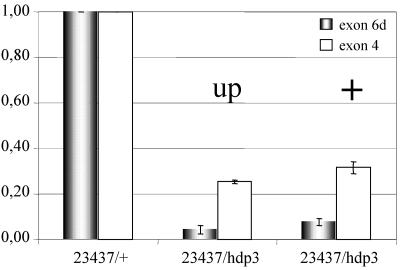
Figure 8.
Transcriptional effects of z and Su(z)3. QRT-PCR assays on wupA expression in various genotypes. Transcripts were detected with primers from the alternative exon 6d (A) or constitutive exon 4 (B) and normalized with respect to the parental strain y w by using RNApolII as an internal standard. Error bars indicate average SE from a minimum of three RT-PCR assays.
In a further attempt to correlate wing position phenotypes and transcriptional changes, we carried out QRT-PCR tests in flies sorted by wing phenotype, while identifying the chromosomal origin of TnI transcripts. The data show (Figure 9) that flies with normal wing position (evidencing transvection) yield higher levels of transcription than those with up wings position, in spite of being of the same genotype. This observation demonstrates that transvection is based on transcriptional changes. Furthermore, because the mutation hdp3 prevents the generation of exon 6d-containing transcripts (Barbas et al., 1993), those detected here must come from the Df(1)23437 chromosome. Consequently, their increased levels reflect a bona fide trans-effect on transcription.
Figure 9.
Relationship between phenotype and transcription in transvection. QRT-PCR assays in a given genotype that exhibits transvection and, in addition, allows identifying TnI transcripts produced by Df(1)23437-bearing chromosome. The mutation hdp3 prevents the generation of exon 6d containing transcripts, whereas exon 4 reveals all transcripts. Four days aged adult females were sorted according to wing position indicative of transvection (+) or not (up). Note the increased transcription in the transvecting flies (+) with respect to those not exhibiting the phenomenon (up). The levels of exon 6d-containing transcripts must come from the 23437 chromosome, thus demonstrating a bona fide trans-effect. Error bars indicate average SE from a minimum of three independent RNA extractions.
TnI Is Required for Muscle Morphogenesis
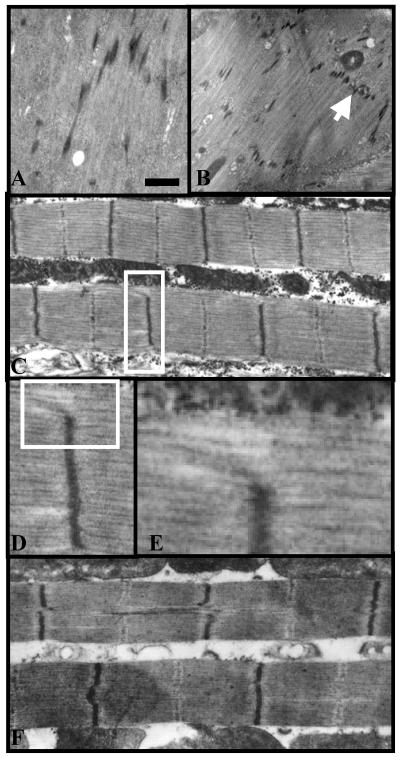
The early expression of TnI, in addition to the severe transcriptional reduction caused by the three rearrangements, prompted a search for structural defects in muscle morphogenesis. Mutant embryos can be identified by genetic markers (see MATERIALS AND METHODS) and subject to electron microscopy (Figure 10). The two mutants analyzed, Df(1)23437 and PL87, show remnants of somatic muscles with poorly oriented thin filaments that never attain a sarcomere-like aspect. One consistent feature of these filaments, however, is the presence of regularly spaced electron-dense accumulations reminiscent of Z disk fragments. In the relatively less extreme phenotype of PL87, these fragments may look aligned as to form a proto-Z disk. Spacing between these proto-Z discs is the regular 80 nm, suggesting that proteins that facilitate the antiparallel organization of thin filaments and mark sarcomere dimension (i.e., kettin) are normally incorporated (Van Straaten et al., 1999; Kulke et al., 2001). In the more extreme phenotypes, however (Figure 10A), proto-Z discs begin to form at much shorter intervals, indicating that incorporation of other protein components to thin filaments is not a prior requirement for Z discs formation.
Figure 10.
Muscle phenotypes of wup A mutants. Electron micrographs of mutant somatic muscles. (A) Df(1)23437 embryo showing thin filaments with putative Z disk electrodense material at random locations along the filaments. (B) PL87 embryo with better organized parallel thin filaments and a lined up proto Z disk (arrow). (C) Adult indirect flight muscles with mutated Troponin I and Myosin heavy chain proteins (genotype: males wup Ahdp2; MhcD41/+). Note the nearly normal sarcomere, albeit some thin filaments bypass a Z disk and continue to the adjacent sarcomere (inset). (D) Detail of inset in C. (E) Detail of inset in D. (F) Adult indirect flight muscles with double mutant TnI (genotype: males wup Ahdp2 wup AD3). Note the abnormal central part of some Z discs. Bar, 0.5 μm (A and D); 0.25 μm (B and E); 1 μm (C and F).
To further document the possible involvement of TnI in Z disk morphogenesis, we analyzed adult indirect flight muscles of genotypes in which a defined (Ala115Val) structural mutation in TnI, hdp2, is coupled to either a second structural mutation (Leu188Phe) in TnI, D3 or to an independent structural mutation (2-base pair insertion at exon 7) in the myosin heavy chain, D41. These second mutations suppress almost completely the functional defects of hdp2 and show normal levels of gene expression (Prado et al., 1995; Kronert et al., 1999). In both genotypes, Z discs can be found with subtle abnormalities, including failures in their ordered alignment or failure to anchor all thin filaments from a sarcomere (Figure 10, C–F). Together, these data suggest a developmental role for TnI in Z disk that could result either directly from a structural role of TnI in thin filament morphogenesis, or indirectly from the down-regulation of Z disk constituents.
DISCUSSION
We address here the transcriptional mechanisms of the TnI encoding gene of Drosophila. This in vivo study reveals two regulatory regions, URE and IRE, located immediately upstream and downstream to the transcription initiation site and defined by a characteristic array of binding sites for the transcription factors DMEF2, BINIOU, and TINMANN. The regions are qualitatively identical in their effects, but both are required for proper levels of transcription. Given the span of the genomic fragments tested for the reporter expression, it seems that the full set of positive regulatory elements has been identified. Putative repressor sites, however, remain to be identified. Finally, the transvection experiments suggest that, in addition to the cis-requirements, transcription is also dependent on trans-effects occurring probably at a small critical region close to the putative promoter.
Regulatory Modules in wings up A
The genomic fragments analyzed in LacZ reported transgenes allow identifying regions that contain positive regulatory elements that direct expression to specific tissues and developmental stages. These regulatory modules are revealed as overlapping stretches of DNA rather than separate and mutually exclusive units. For example, the modules for somatic and visceral muscles share ∼1 kb of sequences. None of the smaller fragments tested that subdivide this 1 kb, however, could reproduce the original somatic or visceral expression patterns. The case has precedents in other genes such as mef2 (Cripps et al., 1999; Nguyen and Xu, 1998), tubulin (Damm et al., 1998; Kremser et al., 1999), or tropomyosin 2 (Lin et al., 1996; Lin and Storti, 1997), and illustrate the intimate relationship between specific sequences (i.e., enhancers or repressors) and the topology of the surrounding chromatin in the context of proper gene expression (Yuh et al., 2001).
Mechanism of IRE + URE Activity for Transcription
The location of the two regulatory regions could sustain a particular chromatin structure, perhaps of a hair-pin type, for normal transcription. The spacing requirement and the linear order of interference effects shown by the three rearrangements support this speculation. In females, normal transcription requires correct pairing between both IRE + URE complements, and the spacing becomes less critical as long as it is the same in both chromosomes. The transvection effect that takes place when two homologous copies of the gene are present clearly implies enhanced transcription from the trans-homologue, as demonstrated by QRT-PCR assays in genotypes that allow to discriminate the chromosomal origin of some transcripts (Figure 9).
The initiation of transcription is clearly dependent on DMEF2. Maintenance, however, does not seem to rely exclusively on this transcription factor. The difference between these two types of transcription has been recently documented (Wheeler et al., 2002), and the present case may indicate that it is a general phenomenon. Because the analyzed regions contain canonical binding sites for TINMAN and BINIOU, it seems puzzling why these factors are not able to drive gene expression in a mef2 null background. One possibility is that DMEF2, in addition to its direct DNA binding activity on wupA and its role as a transcription factor, acts also as a trancriptional cofactor for TINMAN and BINIOU. Also, the role of other transcription factors such as the one encoded in Dmeso18E (Taylor, 2000) remains to be integrated into this scenario. Purification and analysis of the corresponding protein complexes would be required to test these speculations.
Concerning the Trithorax group of transcriptional cofactors assayed, the data demonstrate that zeste acts as a repressor for wupA. In addition, we show that Su(z)3 is also a repressor and it behaves similarly to zeste with respect to transvection effects. Additional data on a third gene, Trithorax-like, which encodes the GAGA factor yielded similar effects (our unpublished data). They represent cis- and trans-requirements for normal transcription. There seems to be, however, a critical domain near the promoter where the effects of these repressors become evident. Based on the immunity of PG31 heterozygotes to Trithorax group mutant backgrounds, this critical region could be defined by the–249 position as the upstream limit. In addition, the perfect transvection in PL87/PG31 heterozygotes suggest also a critical region of pairing for transcription. It is plausible that both critical regions are coincident. Presumably, pairing of these two rearrangements will be facilitated by the common sequences and size of the inserts. Their different site of insertion and the different transgenes, however, most likely will distort pairing to some extent. Thus, the critical region for transvection might be as small as 30 base pairs upstream of the initiation site. Extensive studies in the gene yellow have reached the same conclusion where the critical region for transvection seems to be the TATA box and an initiator element located in cis (Morris et al., 1999). The case of wupA, which does not contain TATA box, suggests that the critical region is the promoter per se, independently of its type. These observations should help to direct future in vitro studies with chromatin fragments.
Wing Position Phenotypes and Transcriptional Changes
It may seem counterintuitive the observation that z and Su(z)3 mutant backgrounds result in an increase of transcription at wupA, whereas the phenotypic effect shows a loss of transvection. Because the transcriptional change has been consistently observed in all genotypes assayed, including the flies sorted by wing position, it is evident that the phenotype does not correlate with the absolute levels of TnI transcripts. Furthermore, in spite of the very low levels of transcription in 23437/hdp3 females, they are viable and muscles are functional, except those involved in flight. The most plausible interpretation of these observations is that the deleterious effect on muscle structure results from changes in the stoichiometry of certain TnI isoforms rather than in their absolute levels (Gunthorpe et al., 1999). It is likely that, as in TnI isoform replacement experiments with the vertebrate homologues (Metzger et al., 2003), the unbalance of certain TnI isoforms lead to unsuitable thin filaments in vivo. If this is also the case in humans, certain muscular diseases, particularly those revealed under intense exercise, may result from mutations in regulatory regions, and thus may have escaped detection under standard sequencing procedures. In this context, the conserved gene regulatory array should be useful to guide future mutant screenings in humans.
Z discs are thought to be the anchoring points where thin filaments exert force and contract the sarcomere during muscle activity. It is somewhat unexpected that reduced levels or structural modifications of TnI can result in defective Z discs. The aspect and spacing of these Z disk-like structures suggest that a Z disk results from the lining of independent substructures deposited on the thin filaments and latter organized in register. It is worth noting, however, that a thin filament does not necessarily anchor at a Z disk. The quasi-normal sarcomeres from double mutants in troponin I, hdp2, and myosin or tropomyosin, Su(hdp2) (Figure 10), show frequent cases of thin filaments extending more than one sarcomere in length. All these abnormal features of Z discs observed in mutants that involve TnI may indicate additional developmental functions of this protein beyond the well known regulatory role in sarcomere mechanics. Alternatively, these features may result from depletion of bona fide Z disk components whose expression is downregulated because of TnI mutations. This possibility would require a coordinated regulation of gene expression among thin filament components.
A Conserved Regulatory Scenario: Biological Significance
Vertebrate TnI encoding genes are not yet amenable to the in vivo analysis that Drosophila allows. Nevertheless, previous studies on the quail fast and slow TnI genes show functional evidences of a very similar regulatory structure to that described here for wupA (Yutzey et al., 1989; Banerjee-Basu and Buonanno, 1993; Nakayama et al., 1996). Equivalent regions to IRE and URE can be revealed by sequence analysis in other TnI members with the exception of the cardiac gene. The array of regulatory regions and their characteristic features seem fairly well conserved in TnI encoding genes of insects and vertebrates. This observation will be relevant toward the design of tools that aim to mimic the native gene expression in otherwise pathological conditions. Beyond this utilitarian use, the conservation of the TnI regulatory landscape in other genes that encode thin filament components might be indicative of common trends that would ensure proper quantitative expression of these components and, eventually, could help to translate gene regulation into physiology.
Supplementary Material
Acknowledgments
We are grateful for the exchange of data and manuscripts with Dr. M. Cervera (Universidad Autónoma de Madrid, Madrid, Spain) before publication. Dr. F. Azorín (Consejo Superior de Investigaciones Cientificas, Barcelona, Spain)provided Trl mutants. Also, Dr. S. Bray (Cambridge University, Cambridge, United Kingdom) hosted M.-C.M. during a training stay. Laboratory members made critical comments to the manuscript. The technical contribution of M.C. Álvarez is most appreciated. Research was funded by grants 01/1185 and BMC-05051 from the Ministries of Health, and Science and Technology, respectively.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–09–0663. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-09-0663.
Online version of this article contains supplementary material for some figures. Online version is available at www.molbiolcell.org.
References
- Aguilera, M., Oliveros, M., Martinez-Padron, M., Barbas, J.A., and Ferrús, A. (2000). Ariadne-1, a vital Drosophila gene is required in development and defines a new conserved family of ring-finger proteins. Genetics 155, 1231-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés, V., Cervera, M., and Mahdavi, V. (1995). Determination of the consensus binding site for MEF2 expressed in muscle and brain reveals tissuespecific sequence constraints. J. Biol. Chem. 270, 23246-23249. [DOI] [PubMed] [Google Scholar]
- Arnone, M.I., and Davidson, E.H. (1997). The hardwiring of development: organization and function of genomic regulatory systems. Development 124, 1851-1864. [DOI] [PubMed] [Google Scholar]
- Arredondo, J.J., Ferreres, R.M., Maroto, M., Cripps, R.M., Marco, R., Bernstein, S.I., and Cervera, M. (2001). Control of Drosophila paramyosin/miniparamyosin gene expression. Differential regulatory mechanisms for muscle-specific transcription. J. Biol. Chem. 276, 8278-8287. [DOI] [PubMed] [Google Scholar]
- Ashburner, M. (1989). Drosophila: A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press.
- Azpiazu, N., and Frasch, M. (1993). tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 7, 1325-1340. [DOI] [PubMed] [Google Scholar]
- Banerjee-Basu, S., and Buonanno, A. (1993). cis-acting sequences of the rat troponin I slow gene confer tissue- and development-specific transcription in cultured muscle cells as well as fiber type specificity in transgenic mice. Mol. Cell. Biol. 13, 7019-7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbas, J.A., Galceran, J., Krah-Jentgens, I., de la Pompa, J.L., Canal, I., Pongs, O., and Ferrús, A. (1991). Troponin I is encoded in the haplolethal region of the Shaker gene complex of Drosophila. Genes Dev. 5, 132-140. [DOI] [PubMed] [Google Scholar]
- Barbas, J.A., Galceran, J., Torroja, L., Prado, A., and Ferrús, A. (1993). Abnormal muscle development in the heldup3 mutant of Drosophila melanogaster is caused by a splicing defect affecting selected troponin I isoforms. Mol. Cell. Biol. 13, 1433-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylies, M.K., Bate, M., and Ruiz Gomez, M. (1998). Myogenesis: a view from Drosophila. Cell 93, 921-927. [DOI] [PubMed] [Google Scholar]
- Beall, C.J., and Fyrberg, E. (1991). Muscle abnormalities in Drosophila melanogaster heldup mutants are caused by missing or aberrant troponin-I isoforms. J. Cell Biol. 114, 941-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, B.L., and Olson, E.N. (1998). Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell. Dev. Biol. 14, 167-196. [DOI] [PubMed] [Google Scholar]
- Bodmer, R. (1993). The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development 118, 719-729. [DOI] [PubMed] [Google Scholar]
- Bour, B.A., O'Brien, M.A., Lockwood, W.L., Goldstein, E.S., Bodmer, R., Taghert, P.H., Abmayr, S.M., and Nguyen, H.T. (1995). Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes Dev. 9, 730-741. [DOI] [PubMed] [Google Scholar]
- Bourbon, H.M., et al. (2002). A P-insertion screen identifying novel X-linked essential genes in Drosophila. Mech. Dev. 110, 71-83. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega, J.A., and Hartenstein, V. (1997). The Embryonic Development of Drosophila melanogaster, 2nd ed. Berlin: Springer.
- Carmena, A., Bate, M., and Jimenez, F. (1995). Lethal of scute, a proneural gene, participates in the specification of muscle progenitors during Drosophila embryogenesis. Genes Dev. 9, 2373-2383. [DOI] [PubMed] [Google Scholar]
- Carrier, L., Hengstenberg, C., Beckman, J.S., Guicheney, P., Dufour, C., Bercovici, J., Dausee, E., Berebbi, B.I., Wisnewsky, C., and Pulvenis, D. (1993). Mapping of a novel gene for a familial hypertrophic cardiomyopathy to chromosome 11. Nat. Genet. 4, 311-313. [DOI] [PubMed] [Google Scholar]
- Cook, P.R. (1997). The transcriptional basis of chromosome pairing. J. Cell Sci. 110, 1033-1040. [DOI] [PubMed] [Google Scholar]
- Coonar, A.S., and McKenna, W.J. (1997). Molecular genetics of familial cardiomyopathies. Adv. Genet. 35, 285-324. [DOI] [PubMed] [Google Scholar]
- Cripps, R.M., Zhao, B., and Olson, E.N. (1999). Transcription of the myogenic regulatory gene Mef2 in cardiac, somatic, and visceral muscle cell lineages is regulated by a Tinman-dependent core enhancer. Dev. Biol. 215, 420-430. [DOI] [PubMed] [Google Scholar]
- Damm, C., Wolk, A., Buttgereit, D., Loher, K., Wagner, E., Lilly, B., Olson, E.N., Hasenpusch-Theil, K., and Renkawitz-Pohl, R. (1998). Independent regulatory elements in the upstream region of the Drosophila beta 3 tubulin gene (beta Tub60D) guide expression in the dorsal vessel and the somatic muscles. Dev. Biol. 199, 138-149. [DOI] [PubMed] [Google Scholar]
- Davidson, E.H., et al. (2002). A genomic regulatory network for development. Science 295, 1669-1678. [DOI] [PubMed] [Google Scholar]
- Davison, F.D., D'Cruz, L.G., and McKenna, W.J. (2000). Molecular motors in the heart. Essays Biochem. 35, 145-158. [DOI] [PubMed] [Google Scholar]
- Duncan, I.W. (2002). Transvection effects in Drosophila. Annu. Rev. Genet. 36, 521-556. [DOI] [PubMed] [Google Scholar]
- Falkenburg, D., Dwornickzak, B., Faust, D.M., and Bautz, E.K.F. (1987). RNA polymerase of Drosophila. Relation of its 140, 000 Mr subunit to the β subunit of Escherichia coli RNA polymerase. J. Mol. Biol. 195, 929-937. [DOI] [PubMed] [Google Scholar]
- Farah, C.S., and Reinach, F.C. (1995). The troponin complex and regulation of muscle contraction. FASEB J. 9, 755-767. [DOI] [PubMed] [Google Scholar]
- Gajewski, K., Kim, Y., Lee, Y.M., Olson, E.N., and Schulz, R.A. (1997). d-mef2 is a target for Tinman activation during Drosophila heart development. EMBO J. 16, 515-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeves, M.A., and Lehrer, S. (1998). The muscle thin filament as a classical cooperative/allosteric regulatory system. J. Mol. Biol. 277, 1081-1089. [DOI] [PubMed] [Google Scholar]
- Gerasimova, T.I., Gdula, D.A., Gerasimov, D.V., Simonova, O., and Corces, V.G. (1995). A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell 82, 587-597. [DOI] [PubMed] [Google Scholar]
- Goldsborough, A.S., and Kornberg, T.B. (1996). Reduction of transcription by homologue asynapsis in Drosophila imaginal discs. Nature 381, 807-810. [DOI] [PubMed] [Google Scholar]
- Gunthorpe, D., Beatty, K.E., and Taylor, M.V. (1999). Different levels, but not different isoforms, of the Drosophila transcription factor DMEF-2 affect distinct aspects of muscle differentiation. Dev. Biol. 215, 130-145. [DOI] [PubMed] [Google Scholar]
- Henikoff, S. (1997). Nuclear organization and gene expression: homologous pairing and long-range interactions. Curr. Opin. Cell. Biol. 9, 388-395. [DOI] [PubMed] [Google Scholar]
- Horton, R.M. (1995). PCR-mediated recombination and mutagenesis. SOEing together tailor-made genes. Mol. Biotechnol. 3, 93-99. [DOI] [PubMed] [Google Scholar]
- Hur, M.W., Laney, J.D., Jeon, S.H., Ali, J., and Biggin, M.D. (2002). Zeste maintains repression of Ubx transgenes: support for a new model of Polycomb repression. Development 129, 1339-1343. [DOI] [PubMed] [Google Scholar]
- Kelly, K.K., Meadows, S.M., and Cripps, R.M. (2002). Drosophila MEF2 is a direct regulator of Actin57B transcription in cardiac, skeletal, and visceral muscle lineages. Mech. Dev. 110, 39-50. [DOI] [PubMed] [Google Scholar]
- Kennison, J.A., and Southworth, J.W. (2002). Transvection in Drosophila. Adv. Genet. 46, 399-420. [DOI] [PubMed] [Google Scholar]
- Kremser, T., Hasenpusch-Theil, K., Wagner, E., Buttgereit, D., and Renkawitz-Pohl, R. (1999). Expression of the beta3 tubulin gene (beta Tub60D) in the visceral mesoderm of Drosophila is dependent on a complex enhancer that binds Tinman and UBX. Mol. Gen. Genet. 262, 643-658. [DOI] [PubMed] [Google Scholar]
- Kronert, W.A., Acebes, A., Ferrús, A., and Bernstein, S.I. (1999). Specific myosin heavy chain mutations suppress troponin I defects in Drosophila muscles. J. Cell Biol. 144, 989-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulke, M., Neagoe, C., Kolmerer, B., Minajeva, A., Hinssen, H., Bullard, B., and Linke, W.A. (2001). Kettin, a major source of myofibrillar stiffness in Drosophila indirect flight muscle. J. Cell Biol. 154, 1045-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laney, J.D., and Biggin, M.D. (1996). Redundant control of Ultrabithorax by zeste involves functional levels of zeste protein binding at the Ultrabithorax promoter. Development 122, 2303-2311. [DOI] [PubMed] [Google Scholar]
- Lewinter, M.M., and Vanburen,. P. (2002). Myofilament remodeling during the progression of heart failure. J. Card. Fail. 8, 271-275. [DOI] [PubMed] [Google Scholar]
- Li, Q., Shen, P.Y., Wu, G., and Chen, X.Z. (2003). Polycystin-2 interacts with troponin I, an angiogenesis inhibitor. Biochemistry 42, 450-457. [DOI] [PubMed] [Google Scholar]
- Lin, M.H., Nguyen, H.T., Dybala, C., and Storti, R.V. (1996). Myocyte-specific enhancer factor 2 acts cooperatively with a muscle activator region to regulate Drosophila tropomyosin gene muscle expression. Proc. Natl. Acad. Sci. USA 93, 4623-4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S.C., and Storti, R.V. (1997). Developmental regulation of the Drosophila Tropomyosin 2 (Tm2) gene is controlled by a muscle activator enhancer region that contains multiple cis-elements and binding sites for multiple proteins. Dev. Genet. 20, 297-306. [DOI] [PubMed] [Google Scholar]
- Lilly, B., Zhao, B., Ranganayakulu, G., Paterson, B.M., Schulz, R.A., and Olson, E.N. (1995). Requirement of MADS domain transcription factor d-MEF2 for muscle formation in Drosophila. Science 267, 688-693. [DOI] [PubMed] [Google Scholar]
- Marques, G., Bao, H., Haerry, T.E., Shimell, M.J., Duchek, P., Zhang, B., and O'Connor, M.B. (2002). The Drosophila BMP type II receptor Wishful Thinking regulates neuromuscular synapse morphology and function. Neuron 33, 529-543. [DOI] [PubMed] [Google Scholar]
- Mas, J.A., García-Zaragoza, E., and Cervera, M. (2004). Two functionally identical molecular enhancers in Drosophila Troponin T gene establish the correct protein levels in different muscle types. Mol. Biol. Cell (in press). [DOI] [PMC free article] [PubMed]
- Maytum, R., Westerdirf, B., Jaquet, K., and Geeves, M.A. (2003). Differential regulation of the actomyosin interaction by skeletal and cardiac troponin isoforms. J. Biol. Chem. 278, 6696-6701. [DOI] [PubMed] [Google Scholar]
- Metzger, J.M., Michele, D.E., Rust, E.M., Borton, A.R., and Westfall, M.V. (2003). Sarcomere thin filament regulatory isoforms: evidence of a dominant effect of slow skeletal troponin I on cardiac contraction. J. Biol. Chem. 278, 13118-13123. [DOI] [PubMed] [Google Scholar]
- Morris, J.R., Geyer, P.K., and Wu, C. (1999). Core promoter elements can regulate transcription on a separate chromosome in trans. Genes Dev. 13, 253-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, A.M., Thompson, W.R., Peng, L.F., and Jones, L 2nd. (1997). Regulation of the rat cardiac troponin I gene by the transcription factor GATA-4. Biochem. J. 322, 393-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama, M., Stauffer, J., Cheng, J., Banerjee-Basu, S., Wawrousek, E., and Buonanno, A. (1996). Common core sequences are found in skeletal muscle slow- and fast-fiber-type-specific regulatory elements. Mol. Cell. Biol. 16, 2408-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, H.T., and Xu, X. (1998). Drosophila mef2 expression during mesoderm development is controlled by a complex array of cis-acting regulatory modules. Dev. Biol. 204, 550-566. [DOI] [PubMed] [Google Scholar]
- Orlando, V., Jane, E.P., Chinwalla, V., Harte, P.J., and Paro, R. (1998). Binding of trithorax and Polycomb proteins to the bithorax complex: dynamic changes during early Drosophila embryogenesis. EMBO J. 17, 5141-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, J.S., Gomes, X.V., and Geyer, P.K. (1992). Position-independent germline transformation in Drosophila using a cuticle pigmentation gene as a selectable marker. Nucleic Acids Res. 20, 5859-5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paululat, A., Holz, A., and Renkawitz-Pohl, R. (1999). Essential genes for myoblast fusion in Drosophila embryogenesis. Mech. Dev. 83, 17-26. [DOI] [PubMed] [Google Scholar]
- Prado, A., Canal, I., Barbas, J.A., Molloy, J., and Ferrús, A. (1995). Functional recovery of troponin I in a Drosophila heldup mutant after a second site mutation. Mol. Biol. Cell 6, 1433-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado, A., Canal, I., and Ferrús, A. (1999). The haplolethal region at the 16F gene cluster of Drosophila melanogaster: structure and function. Genetics 151, 163-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen, R. (2001). Quantification on the LightCycler. In: Cycle Real Time PCR, Methods and Applications, ed. S. Meuer, C. Wittwer, and K. Nakagawara, Heidelberg: Rapid Springer Press, 21-34.
- Ruiz, G.M., and Bate, M. (1997). Segregation of myogenic lineages in Drosophila requires numb. Development 124, 4857-4866. [DOI] [PubMed] [Google Scholar]
- Ruiz-Gomez, M., Romani, S., Hartmann, C., Jackle, H., and Bate, M. (1997). Specific muscle identities are regulated by Kruppel during Drosophila embryogenesis. Development 124, 3407-3414. [DOI] [PubMed] [Google Scholar]
- Ruiz-Gomez, M., Coutts, N., Suster, M.L., Landgraf, M., and Bate, M. (2002). myoblasts incompetent encodes a zinc finger transcription factor required to specify fusion-competent myoblasts in Drosophila. Development 129, 133-141. [DOI] [PubMed] [Google Scholar]
- Siedner, S., Kruger, M., Schroeter, M., Metzler, D., Roell, W., Fleischmann, B.K., Hescheler, J., Pfitzer, G., and Stehel, R. (2003). Developmental changes in contractility and sarcomeric proteins from the early embryonic to the adult stage in the mouse heart. J. Physiol. 548, 493-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soong, R., Ruschoff, J., and Tabiti, K. (2000). Detection of colorectal micrometastasis by quantitative RT-PCR of cytokeratin 20 mRNA. Roche Diagnostic internal publication.
- Spradling, A.C., and Rubin, G.M. (1982). Transposition of cloned P elements into Drosophila germ line chromosomes. Science 218, 341-347. [DOI] [PubMed] [Google Scholar]
- Squire, J.M., and Morris, E.P. (1998). A new look at thin filament regulation in vertebrate skeletal muscle. FASEB J. 12, 761-771. [DOI] [PubMed] [Google Scholar]
- Stronach, B.E., Renfranz, P.J., Lilly, B., and Beckerle, M.C. (1999). Muscle LIM proteins are associated with muscle sarcomeres and require dMEF2 for their expression during Drosophila myogenesis. Mol. Biol. Cell 10, 2329-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, S.S., Brassington, A.M., Grannatt, K., Rutherford, A., Whitby, F.G., Krakowiak, P.A., Jorde, L.B., Carey, J.C., and Bamshad, M. (2003). Mutations in genes encoding fast-twitch contractile proteins cause distal arthrogryposis syndromes. Am. J. Hum. Genet. 72, 681-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, M.V. (2000). A novel Drosophila, mef-2-regulated muscle gene isolated in a subtractive hybridisation-based molecular screen using small amounts of zygotic mutant RNA. Dev. Biol. 220, 37-52. [DOI] [PubMed] [Google Scholar]
- Taylor, M.V. (2002). Muscle differentiation: how two cells become one. Curr. Biol. 12, 224-228. [DOI] [PubMed] [Google Scholar]
- Therianos, S., Leuzinger, S., Hirth, F., Goodman, C.S., and Reichert, H. (1995). Embryonic development of the Drosophila brain: formation of commissural and descending pathways. Development 121, 3849-60. [DOI] [PubMed] [Google Scholar]
- Thisse, B., Messal, M., and Perrin-Schmitt, F. (1987). The twist gene: isolation of a Drosophila zygotic gene necessary for the establishment of dorsoventral pattern. Nucleic Acids Res. 15, 3439-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Straaten, M., Goulding, D., Kolmerer, B., Labeit, S., Clayton, J., Leonard, K., and Bullard, B. (1999). Association of kettin with actin in the Z-disc of insect flight muscle. J. Mol. Biol. 285, 1549-1562. [DOI] [PubMed] [Google Scholar]
- Wheeler, J.C., Vanderzwan, C., Xu, X., Swantek, D., Tracey, W.D., and Gergen, J.P. (2002). Distinct in vivo requirements for establishment versus maintenance of transcriptional repression. Nature Genet. 32, 206-210. [DOI] [PubMed] [Google Scholar]
- Wingender, E., Chen, X., Hehl, R., Karas, H., Liebich, I., Matys, V., Meinhardt, T., Pruss, M., Reuter, I., and Schacherer, F. (2000). TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 28, 316-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuh, C.H., Bolouri, H., and Davidson, E.H. (2001). Cis-regulatory logic in the endo16 gene: switching from a specification to a differentiation mode of control. Development 128, 617-629. [DOI] [PubMed] [Google Scholar]
- Yutzey, K.E., Kline, R.L., and Konieczny, S.F. (1989). An internal regulatory element controls troponin I gene expression. Mol. Cell. Biol. 9, 1397-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffran, S., Kuchler, A., Lee, H.H., and Frasch, M. (2001). Biniou (FoxF), a central component in a regulatory network controlling visceral mesoderm development and midgut morphogenesis in Drosophila. Genes Dev. 15, 2900-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.