Abstract
Eukaryotic cells respond to stress caused by the accumulation of unfolded/misfolded proteins in the endoplasmic reticulum by activating the intracellular signaling pathways referred to as the unfolded protein response (UPR). In metazoans, UPR consists of three parallel branches, each characterized by its stress sensor protein, IRE1, ATF6, and PERK, respectively. In Drosophila, IRE1/XBP1 pathway is considered to function as a major branch of UPR; however, its physiological roles during the normal development and homeostasis remain poorly understood. To visualize IRE1/XBP1 activity in fly tissues under normal physiological conditions, we modified previously reported XBP1 stress sensing systems (Souid et al., Dev Genes Evol 217: 159–167, 2007; Ryoo et al., EMBO J 26: 242-252, 2007), based on the recent reports regarding the unconventional splicing of XBP1/HAC1 mRNA (Aragon et al., Nature 457: 736–740, 2009; Yanagitani et al., Mol Cell 34: 191–200, 2009; Science 331: 586–589, 2011). The improved XBP1 stress sensing system allowed us to detect new IRE1/XBP1 activities in the brain, gut, Malpighian tubules, and trachea of third instar larvae and in the adult male reproductive organ. Specifically, in the larval brain, IRE1/XBP1 activity was detected exclusively in glia, although previous reports have largely focused on IRE1/XBP1 activity in neurons. Unexpected glial IRE1/XBP1 activity may provide us with novel insights into the brain homeostasis regulated by the UPR.
Keywords: UPR, ER stress, XBP1, Drosophila, Glia
Introduction
Most of the secretory and membrane proteins in eukaryotic cells are folded and assembled in the endoplasmic reticulum (ER) with the assistance of ER chaperones and folding catalysts. Only properly folded proteins are transported to their own final destinations inside or outside the cell and function there properly. Therefore, the accumulation of the unfolded/misfolded proteins in the ER (ER stress) could impact the overall integrity of the cell. Unfolded protein response (UPR) is the transcriptional/translational regulatory pathway that mitigates such impaired cellular integrity upon the detection of ER stress by the sensor proteins. In mammalian cells, UPR consists of three parallel branches of the intracellular signaling pathway, each of which is characterized by its sensor protein, IRE1, ATF6, and PERK, respectively. Each sensor protein senses the ER stress in its own fashion and induces the expression of its target genes which facilitate the protein-folding capacity in the ER (Walter and Ron 2011; Hetz 2012; Parmar and Schröder 2012). Since the identification of IRE1 as the ER stress response-related gene in yeast (Cox et al. 1993; Mori et al. 1993), detailed molecular mechanisms of UPR, including ATF6 and PERK pathways, have been widely elucidated using yeast and mammals (Walter and Ron 2011; Hetz 2012; Parmar and Schröder 2012). On the other hand, the physiological role of UPR during normal development remains poorly understood. To reveal the role of UPR under physiological conditions, we use Drosophila melanogaster as a model organism. In D. melanogaster, three sensor molecules (IRE1, ATF6, and PERK) and XBP1 are highly conserved with their human homologues (IRE1, ∼38 % at luminal sensor domain; ∼56 % at cytoplasmic domains; XBP1, ∼43 %; ATF6, ∼46 %; PERK, ∼34 %) (Ryoo and Steller 2007).
The IRE1/XBP1 pathway is widely conserved in eukaryotic cells. IRE1, the ER-resident type 1 membrane protein, senses the accumulation of unfolded proteins in the ER with the sensor domain on its N-terminus and is activated through its oligomerization and autophosphorylation (Lee et al. 2008a; Korennykh et al. 2009; Wiseman et al. 2010; Ali et al. 2011; Chawla et al. 2011; Rubico et al. 2011; Korennykh et al. 2011a, b). Activated IRE1 splices the mRNA of XBP1 (HAC1 in yeast), using the RNase domain near its C-terminus oriented to the cytoplasm. This unconventional splicing causes a frameshift in the XBP1 coding sequence, thereby generating the active transcription factor, XBP1(s) that enhances the expression of UPR target genes. XBP1(u), derived from unspliced XBP1 mRNA, does not function as the active transcription factor, but instead, antagonizes UPR by stimulating the degradation of XBP1(s) and ATF6 (Yoshida et al. 2006). The activated XBP1(s) enhances the expression of UPR target genes encoding endoplasmic reticulum-associated protein degradation-related factors or some lipid synthetic enzymes (Yamamoto et al. 2007; Lee et al. 2008b). Thus, the production of XBP1(s) reflects the activation of IRE1/XBP1 pathway. This unconventional splicing of XBP1 mRNA is also conserved in Drosophila (Souid et al. 2007; Ryoo et al. 2007).
Taking advantage of the frameshift on the XBP1 coding sequence during the unconventional splicing, several groups have independently attempted to monitor IRE1/XBP1 activation. In those in vivo XBP1 stress sensing systems, XBP1-enhanced green fluorescent protein (EGFP) was designed to express utilizing the same mechanism as when XBP1 mRNA was spliced by IRE1 (Iwawaki et al. 2004; Shim et al. 2004; Souid et al. 2007; Ryoo et al. 2007). Although the loss of function analysis suggests a developmental role of IRE1/XBP1 pathway, those systems showed only limited IRE1/XBP1 activity in developing tissues. In Drosophila, IRE1/XBP1 activity was detected only in the larval salivary gland (Souid et al. 2007). In mice, while IRE1/XBP1 activity was not detectable in embryos, it was detected in the muscle, pancreas, brain, and heart only weeks after birth (Iwawaki et al. 2004). In consideration of the spliced form of xbp1 mRNA in the testis of the adult fly detected by RT-PCR (Souid et al. 2007), we expected that there was still room for the improvement of the XBP1 stress sensing system in Drosophila. To improve the sensitivity, we constructed a new xbp1-EGFP gene, based on the recent reports regarding the unconventional splicing of XBP1/HAC1 mRNA (Aragon et al. 2009; Yanagitani et al. 2009; Yanagitani et al. 2011). The resulting highly sensitive XBP1 stress sensing system allowed us to identify IRE1/XBP1 activation in the (a) brain, (b) gut, (c) Malpighian tubules, and (d) trachea of third instar larvae, and in (e) a specific portion of the adult male reproductive organ, under normal physiological conditions.
Materials and methods
Antibodies
Anti-green fluorescent protein (GFP) rabbit polyclonal serum (A6455) was obtained from Molecular Probes (Eugene, OR). Rat monoclonal antibody 7E8A10 against Elav and mouse monoclonal antibody 8D12 against Repo were obtained from Developmental Studies Hybrydoma Bank (University of Iowa, Iowa City, IA). Alexa Fluor 488-conjugated goat anti-rabbit IgG was from Molecular Probes. Rhodamine Red-conjugated donkey anti-rat IgG, Cy5-conjugated donkey anti-mouse IgG, and HRP-conjugated goat anti-rabbit IgG were from Jackson ImmunoResearch (West Grove, CA).
cDNA constructs
HG indicator construction
Drosophila xbp1 cDNA cloned into the EcoRI-XhoI site of pOT2 was obtained from Drosophila Genomics Resource Center (Indiana University, Bloomington, IN). pMS508 is a derivative of pBluescriptII-SK(−) carrying the xbp1 gene. For its construction, 1.8-kilobasepair BamH1-Asp718I fragment (representing the XBP1(s) coding region and 3′ UTR of xbp1 gene), amplified using primers 5′-GTCTGGATCCAATGGCACCCACAGCAAAC-3′ and 5′-GGGGTACCGTTGTTTGGTTTGGTTTA-3′, was cloned into the corresponding site of pBluescriptII-SK(−). pMS522 is a derivative of pBluescriptII-SK(−) carrying the xbp1 gene including the additional 0.5-kilobase pair immediately downstream of its 3′ UTR. In terms of pMS522, 0.5-kilobase pair HindIII-Asp718I fragment derived from pMS508 was replaced by the 1.0-kilobase pair HindIII-Asp718I fragment amplified by 5′-ACGAGGAAAGCTTCGATCCGATC-3′ and 5′-GCTCGTTGGTACCGTCATTTCTG-3′. The stop codon of the xbp1 gene on pMS522 was eliminated and an EcoRI site was generated at the same position by site-directed mutagenesis, using 5′-CTGTTTCCCAGTTTGAATTCTGAGTTTTTCAAGC-3′ and 5′-GCTTGAAAAACTCAGAATTCAAACTGGGAAACAG-3′. The resulting plasmid was named pMS524. For the construction of pMS525a, EGFP gene amplified by 5′-CTGAATTCGATGGTGAGCAAGGGC-3′ and 5′-GAATTATCGAATTCCTAGTACAGCTCGTCC-3′ was digested by EcoRI and was inserted into the corresponding site on pMS524. In pMS525a, the EGFP gene is fused to the 3′ end of the XBP1(s) coding region to be in frame with the XBP1(s) coding sequence. xbp1 clone obtained from Drosophila Genomics Resource Center lacks the guanine nucleotide at the 132nd base from the adenine in start codon of xbp1 gene. A guanine nucleotide was introduced to pMS525a by site-directed mutagenesis using the Quick Change Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). The primers used for the mutagenesis were 5′-ACGCCCTCCGCCTCGCCCACGCCCTCGAGTT-3′ and its complementary oligomer DNA. The resulting plasmid carrying the correct XBP1 coding sequence was named pMS531. pMS549 is a derivative of pUAST carrying the xbp1-EGFP gene under the control of UAS promoter. For its construction, a 9.5-kilobase pair BamHI-Asp718I fragment containing the xbp1-EGFP gene was excised from pMS531 and inserted into the corresponding site of pUAST.
LG indicator construction
EGFP gene was fused immediately downstream of the 3′ side of the IRE1 splice site on the xbp1 gene, giving rise to the XBP1-EGFP fusion protein whose C-terminal region of XBP1 was truncated (Ryoo et al. 2007).
Fly lines
pMS549 was utilized for the germ line transformation to generate the transgenic line, w ; UAS-xbp1-EGFP / cyo. The driver line, w ; tub-Gal4 / cyo, was used for the moderate and ubiquitous expression of xbp1-EGFP gene through the Gal4/UAS system (Brand and Perrimon 1993). (w ; UAS-xbp1-EGFP, tub-Gal4 / cyo) line was generated through the mitotic recombination of the two lines above.
Cell culture and transfection
S2 cells were cultured at 25 °C in Schneider's Drosophila medium supplemented with 10 % heat-inactivated fetal bovine serum, penicillin, and streptomycin. For exogenous protein expression, a total of 0.6 mg of plasmids was transfected into 8–10 × 106 cells in 3 ml of media using EffecteneTM (Qiagen, Valencia, CA), and the transfected S2 cells were cultured for 24 h at 25 °C.
ER stress induction in S2 cells and immunoblotting
To induce ER stress, the transfected S2 cells expressing exogenous proteins were further incubated in an equal volume of fresh medium containing final 3 mM of dithiothreitol (DTT) for 4 h. After the 4-h treatment by DTT, the medium was quickly removed from the cell culture by centrifugation and the S2 cells were thoroughly washed once by resuspending in an equal volume of phosphate-buffered saline (PBS). After the removal of PBS, the cell pellets were immediately resuspended in 5 % trichloroacetic acid, followed by the storage on ice for 20 min. The protein precipitates were collected by centrifugation, washed with acetone, and then solubilized in 1 % SDS/50 mM Tris–HCl pH 7.5 solution. Protein samples obtained from 1 × 106 cells were separated by 7.5 % SDS-PAGE (Laemmli 1970) and electrophoretically blotted onto a PVDF membrane filter (Millipore, Bedford, MA). The filters were treated with anti-GFP and HRP-conjugated goat anti-rabbit IgG. The dilution of each antibody was 1:2,000 and 1:5,000, respectively. The protein bands were detected by Super Signal West Pico (Thermo Scientific, Rockford, IL).
Immunofluorescence microscopy
Dissected larval/adult tissues were fixed with 4 % formaldehyde for 20 min at room temperature. Fixed tissues were washed twice with PBS containing 0.2 % Triton X-100 and the residual formaldehyde was neutralized by incubation for 5 min in 50 mM NH4Cl in PBS, followed by two additional washes using PBS containing 0.2 % Triton X-100. Following those washes, tissues were pre-incubated for 30 min in PBS containing the appropriate concentration of Triton X-100. One, 0.4, and 0.2 % of Triton X-100 were used for the brain, other larval organs, and adult reproductive organs, respectively. After the pre-incubation, tissues were labeled with primary antibody in PBS containing the same concentration of Triton as in the pre-incubation for 6 h at 4 °C. Anti-GFP, anti-Elav, and anti-Repo were used at 1:5,000, 1:10, and 1:50 dilution, respectively, as primary antibodies. After gently washing four times with PBS containing 0.2 % Triton X-100, cells were incubated with secondary antibody in the PBS containing same concentration of Triton X-100 as in the pre-incubation and the primary antibody labeling. All of the secondary antibodies were used at 1:500 dilutions. Following three times of gentle washings with PBS containing 0.2 % Triton X-100 and a gentle washing with PBS, labeled tissues were placed on the glass slides with 70 % glycerol, and coverslips were mounted on them. Tissues were visualized using a Zeiss Observer.Z1 (Carl Zeiss, Thornwood, NY).
Results
Construction of a new XBP1 stress sensing system
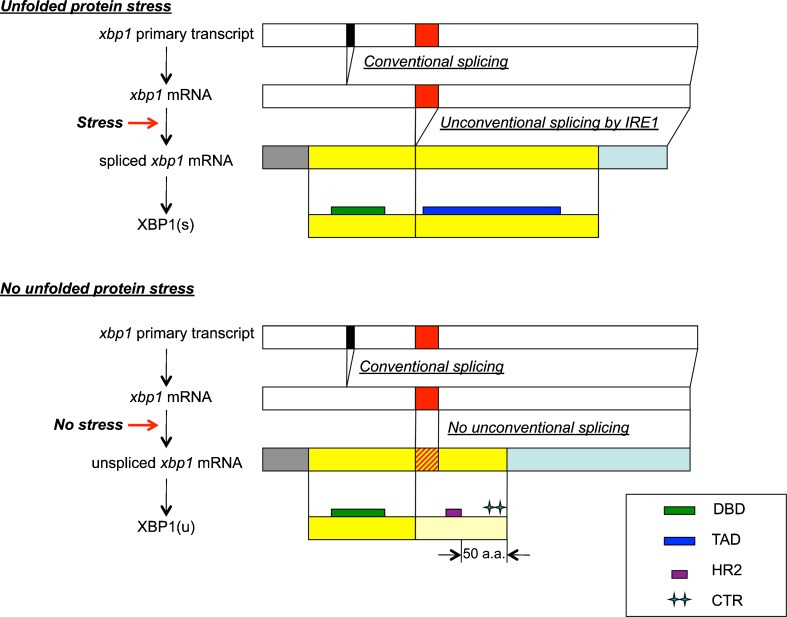
In Drosophila, XBP1 is encoded by the xbp1 gene on the 2R chromosome. Biosynthesis of XBP1 was depicted in Fig. 1. xbp1 mRNA is generated through the conventional splicing at the splice site of its primary transcript (black boxes in Fig. 1) in the nucleus and is exported to the cytoplasm. However, the open reading frame (ORF) on the resulting mRNA does not encode the active transcription factor, XBP1(s), while it encodes XBP1(u) that does not function as the transcription factor. xbp1 mRNA transported to the cytoplasm is further unconventionally spliced at the other splice site (red boxes in Fig. 1) by IRE1 on the ER membrane, only under the ER-stressed conditions. This unconventional splicing causes a frameshift on the XBP1 coding sequence and the resulting ORF encodes XBP1(s). While both XBP1(s) and XBP1(u) possess the DNA binding domain (bZIP domain) of the transcription factor, the transcription activation domain does not exist on XBP(u), but only on XBP1(s). The locations of the DNA binding domain and the transcription activation domain on XBP1 protein are depicted as green boxes and blue boxes, respectively, in Fig. 1.
Fig. 1.
Biosynthesis of XBP1 protein is influenced by unfolded protein stress (ER stress). Biosynthesis of XBP1 proteins from xbp1 primary transcript under ER-stressed conditions (upper scheme) and no ER-stressed conditions (lower scheme) is compared. Primary transcript and mRNA generated through conventional splicing are depicted as white bars. The position of the conventional splice site and unconventional splice site are indicated as a black box and red box, respectively, on the white bars. For visual convenience, the size/width of each bar/box does not necessarily reflect the actual size of each region (e.g., 64 bp for conventional splice site and 23 bp for unconventional splice site). Below xbp1 mRNA are the spliced xbp1 mRNA generated under ER-stressed conditions and the xbp1 mRNA that is not spliced under no ER-stressed conditions. The ORFs of spliced/unspliced xbp1 mRNA are depicted as yellow bars. The unconventional splice site that is not spliced under no ER-stressed conditions is marked with red oblique lines on the yellow bar representing the ORF of unspliced xbp1 mRNA. The 5′ UTR and 3′ UTR are colored gray and light blue, respectively, on both spliced and unspliced xbp1 mRNA. At the bottom of each scheme, the final products, XBP1(s) and XBP1(u), are also depicted as yellow bars. The C-terminal region of XBP1(u) synthesized using the reading frame that is specific to unspliced xbp1 mRNA is pale yellow in color, while the region sharing the amino acid sequence with XBP1(s) is filled with yellow as well as XBP1(s). DNA binding domain (DBD:bZIP) of transcription factor, transcription activation domain (TAD), HR2, and CTR are indicated as green thin bars, a dark blue thin bar, purple box, and double green cross on the yellow bar representing XBP1 protein, respectively
To be unconventionally spliced by IRE1, xbp1 mRNA must reach the ER membrane surface where IRE1 is localized. Two different mechanisms for the recruitment of XBP1/HAC1 mRNA to the ER membrane were recently reported (Aragon et al. 2009; Yanagitani et al. 2009; Yanagitani et al. 2011). Recruitment is required for the efficient splicing of the XBP1/HAC1 mRNA. To improve the sensitivity of Drosophila XBP1 stress sensing system, we utilized the feature of those recruitment mechanisms and modified the existing stress sensing system described in Ryoo et al. (2007).
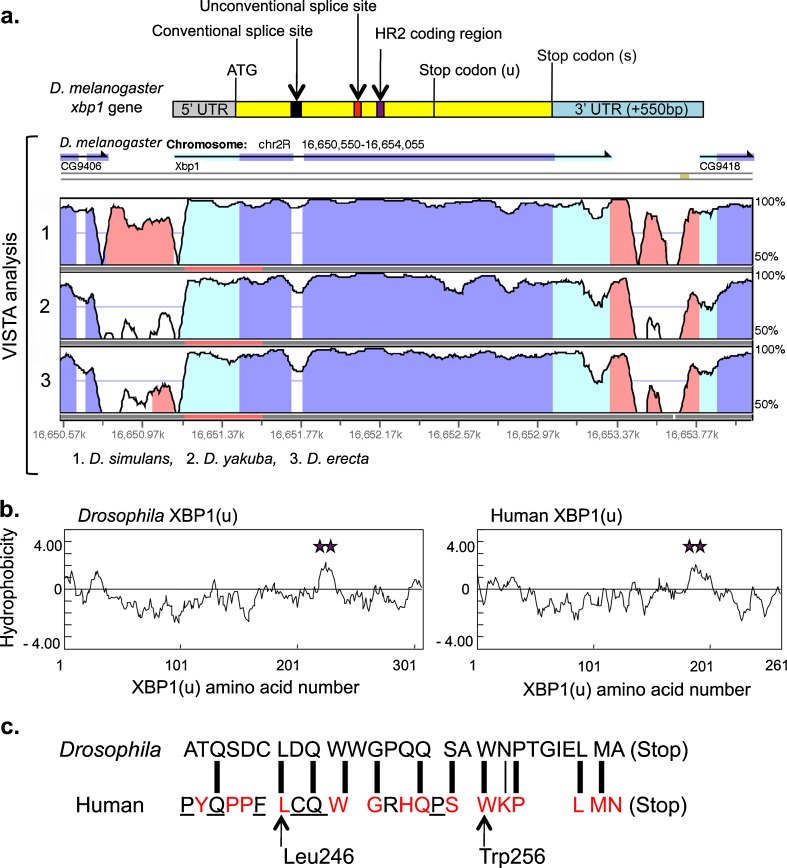
In yeast, the bipartite stem loop structure in the 3′ UTR of HAC1 mRNA (3′ BE) has affinity to the activated/oligomerized Ire1p on the ER membrane. Suggestive of its significance, two short sequence motif on 3′BE are highly conserved among Hac1 orthologues (Aragon et al. 2009). Similarly, VISTA analysis for comparative genome analysis (http://genome.lbl.gov/vista/index.shtml; Dubchak et al. 2000; Frazer et al. 2004) shows the conservation of the 3′ UTR of the xbp1 gene, the Drosophila homologue of HAC1, among four Drosophila species (D. melanogaster, Drosophila simulans, Drosophila yakuba, and Drosophila erecta). In addition, a 0.55-kilobase pair non-coding region located immediately downstream of the 3′ UTR was also conserved among these four species (Fig. 2). Thus, we predicted that the 3′ UTR on Drosophila xbp1 mRNA also has some shared function which would be the enhancement of the affinity between activated IRE1 and the 3′ UTR of xbp1 mRNA.
Fig. 2.
a VISTA analysis of xbp1 orthologues. On the top of the VISTA analysis, an outline of the Drosophila xbp1 gene that indicates the critical factors in this study is aligned. The 5′ UTR, XBP1(s) coding region, 3′ UTR including the extra 550 bp on unconventionally spliced xbp1 mRNA, conventional splice site, unconventional splice site, and HR2 coding region are depicted as gray, yellow, blue, black, red, and purple boxes/bars, respectively. The positions of the start codon and stop codons for XBP1(u) and XBP1(s) are marked with ATG, stop codon (u), and stop codon (s), respectively. VISTA analysis, 2.2 kb interval (chr2R:16,650,550-16,654,055 on the NCBI Reference Sequence) containing D. melanogaster xbp1, was analyzed by VISTA Browser through the gateway at http://genome.lbl.gov/vista/index.shtml. Each plot shows conserved sequences between D. melanogaster and one of the Drosophila counterparts. The level of conservation (vertical axis) was displayed in the coordinates of the sequence of D. melanogaster (horizontal axis). Conserved regions above the level of 70 %/100 bp were highlighted under the curve, with pink indicating a conserved non-coding region, blue, a conserved exon, and turquoise, an untranslated region. Drosophila counterparts analyzed in panels 1, 2, and 3 were D. simulans, D. yakuba, and D. erecta, respectively. CG9406 and CG9418, which are indicated under the arrows at left and right tops of VISTA figure, are the annotation ID for the genes located adjacent to the xbp1 gene on D. melanogaster chromosome 2R and covered by this analysis. The result downloaded from the VISTA browser was modified to a format suitable for this figure. b The amino acid sequence of the Drosophila and human XBP1(u)s was analyzed by the method of Kyte and Doolittle (1982). HR2 domains are marked with a purple double star. Hydrophobicity scale (vertical axis) was plotted in the coordinates of the amino acid sequence of Drosophila XBP1(u) and human XBP1(u) (horizontal axis). c Predicted Drosophila CTR is aligned with the human CTR. The amino acids colored in red are conserved among human, mouse, chicken, frog, and zebrafish. Underlined amino acids are conserved among a subset of them (Yanagitani et al. 2011). Thick lines linking the amino acids on both sequences represent identical amino acids, while the thin line represents the similarly hydrophilic amino acids, asparagine and lysine. Leu246 and Trp256 in human, which are conserved among vertebrates, are indicated by arrows
In mammals, the hydrophobic region specific to the XBP1(u) molecule (HR2) facilitates the recruitment of the cytoplasmic mRNA–ribosome–nascent polypeptide chain complex (R-RNC) to the surface of the ER membrane where IRE1 is localized (Yanagitani et al. 2009). On the other hand, the conserved amino acids (Leu246 and Trp256 in human) on the C-terminal region of XBP1(u) (CTR) support the adequate interaction between HR2 and the ER membrane through the following mechanism. The distance between the C-terminal end of XBP1(u) and HR2 is approximately 50 amino acids long (Fig. 1). Based on the rate of translation by the ribosome, which is estimated to be one to two peptide bonds per second, the synthesis of the XBP1(u) polypeptide is terminated and the nascent XBP1(u) is released from the R-RNC immediately after HR2 emerges from the exit tunnel of the ribosome in the complex. The conserved amino acids on CTR play an important role in pausing the translation of XBP1(u) to hinder the release of nascent XBP1(u) from the R-RNC, thereby increasing the efficiency of the ER membrane targeting of the R-RNC that is exposing HR2 (Ron and Ito 2011; Yanagitani et al. 2011). Through the cooperative actions taken by HR2 and CTR, an adequate amount of time is given for the interaction between IRE1 on the ER membrane and xbp1 mRNA in the R-RNC, and this promotes the production of spliced xbp1 mRNA in mammals upon the activation of IRE1. Drosophila XBP1(u) also has a conserved HR2, as predicted by the Kyte and Doolittle hydrophobicity scale (Kyte and Doolittle 1982; Fig. 2b). In addition, the C-terminal end of XBP1(u) in Drosophila and humans shows significant similarity (Fig. 2c). Therefore, production of the spliced xbp1 mRNA in Drosophila is likely to be enhanced cooperatively by its predicted HR2 and CTR in the same manner as in vertebrates.
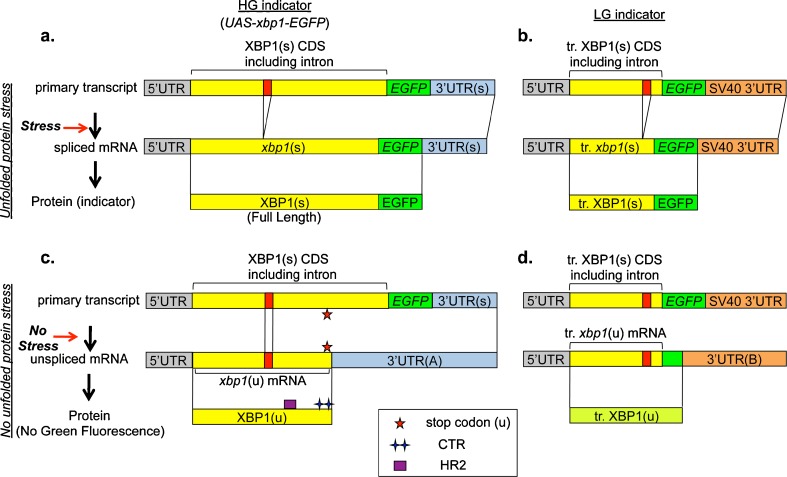
The xbp1 constructs used for the previously reported XBP1 stress sensing systems in Drosophila were lacking the endogenous 3′ UTR (Souid et al. 2007) or lacking both the endogenous 3′ UTR and the CTR coding region (Ryoo et al. 2007). Under physiological conditions, the former system detected IRE1/XBP1 activity only in the salivary gland in third instar larva, while the latter failed to detect any activity in third instar larval tissues. To visualize IRE1/XBP1 activity during Drosophila development, we improved the XBP1 stress sensing system based on our understanding described above (Fig. 3). The XBP1-EGFP molecule in this new system is the full length XBP1 fused with EGFP. For the expression of this XBP1-EGFP molecule, the 3′ UTR of xbp1 gene and the extra 0.55-kilobase pair immediately downstream of the 3′ UTR was fused to the xbp1-EGFP gene, which is under the control of UAS promoter. Henceforth, we will refer to this new XBP1-EGFP fusion protein as the high gain stress indicator (HG indicator), while the original indicator described in Ryoo et al. (2007) will be termed the low gain stress indicator (LG indicator) (Fig. 3). Under non ER-stressed conditions, xbp1-EGFP gene in the new HG indicator system is transcribed to the unspliced mRNA, and XBP1(u) carrying both HR2 and CTR is synthesized from it (Fig. 3c), while the truncated XBP1(u) like protein, which does not carry either the full size HR2 domain or the CTR, is synthesized from xbp1-EGFP gene in the LG indicator system (Fig. 3d). Based on the mechanism for the recruitment of the R-RNC to the ER membrane, the R-RNC in the HG indicator system should provide the xbp1 mRNA in the complex with the opportunity to associate with IRE1 on the ER membrane more efficiently, compared with the R-RNC in the LG indicator system. Additionally, due to the endogenous 3′ UTR and 550 bp of the extra sequence (Fig. 3a, c), the affinity between the activated IRE1 and xbp1 mRNA in the HG indicator system is expected to be much higher than that in the LG indicator system, in which SV40 3′ UTR is attached to the 3′ end of xbp1-EGFP gene (Fig. 3b, d).
Fig. 3.
The mechanism for the biosynthesis of XBP1 stress indicator. The genes we constructed for the expression of XBP1 stress indicators and the biogenesis of those indicators are depicted as follows; HG indicator under ER-stressed conditions in a, LG indicator under ER-stressed conditions in b, HG indicator under non-ER-stressed conditions in c, LG indicator under non-ER-stressed conditions in d. In each scheme, the primary transcript, processed mRNA under the stressed or not stressed condition, and synthesized protein are depicted in order, respectively. Each primary transcript is the transcription product of the each indicator gene under the control of UAS promoter. Regarding the bars representing genes (top two bars in each scheme), the 5′ UTR, XBP1(s) coding region, EGFP coding region, and unconventional splice site are filled with gray, yellow, green, and red, respectively. The 3′ UTRs containing the 3′ UTR of endogenous xbp1 gene are colored light blue, while those containing that of SV40 are colored orange. Each 3′ UTR is abbreviated as follows; “3′ UTR(s)” indicates the 3′ UTR of spliced xbp1 mRNA including the additional 550 bp immediately downstream of it 3′ UTR(s), “3′ UTR(A)” indicates the 3′ UTR of unspliced xbp1 mRNA interrupted by EGFP coding sequence, and “3′ UTR(B)” indicates the fusion of the 3′ portion of the EGFP coding sequence and the 3′ UTR of SV40. Regarding the bars representing indicator proteins (bottom bar in each scheme), both XBP1(s) and XBP1(u), and EGFP are filled with yellow and green, respectively, while the truncated XBP1(u) whose C-terminus was coded by out of frame EGPF gene is filled with yellow green. The stop codon (u), HR2, and CTR are marked with red stars, a purple box, and a green double cross, respectively. HR2 and CTR are not formed in XBP1(s) due to the frameshift caused by the unconventional splicing of xbp1 at the site colored red in the bar
New XBP1 stress sensing system (HG indicator) shows prominently increased sensitivity in vitro
The gene coding the HG indicator was cloned into pUAST for its ectopic gene expression by the Gal4/UAS system (Brand and Perrimon 1993). The gene was under the control of the UAS promoter (Fig. 3). To confirm the availability of HG indicator in vivo, ER stress response was assessed by the addition of DTT to the cultured S2 cells, in which the HG indicator was transiently expressed. The sensitivity of the HG stress indicator and that of the LG indicator reported by Ryoo et al. (2007) were compared in Fig. 4. The LG indicator is the spliced form of truncated XBP1 molecule fused with EGFP as shown in Fig. 3. Its estimated molecular weight is 55 kDa, while that of the HG indicator is 80 kDa. pMS549 and the pUAST derivative carrying the gene encoding the LG indicator are co-transfected into S2 cells with the driver gene, act-Gal4. ER stress was induced by the addition of 3 mM DTT, as indicated in “Material and methods.” The cellular accumulation of XBP1-EGFP in the treated S2 cells was assessed by immunoblotting using anti-GFP. The accumulation level of the HG indicator under the DTT-stressed conditions was significantly higher than that of the LG indicator (compare lanes 4 and 6 in Fig. 4). Additionally, the HG indicator could have detected the ER stress even in the cultured S2 cells, to which DTT was not added (Fig. 4, lane 3). Therefore, the HG indicator detected the ER stress in vitro with dramatically improved sensitivity compared with the LG indicator.
Fig. 4.
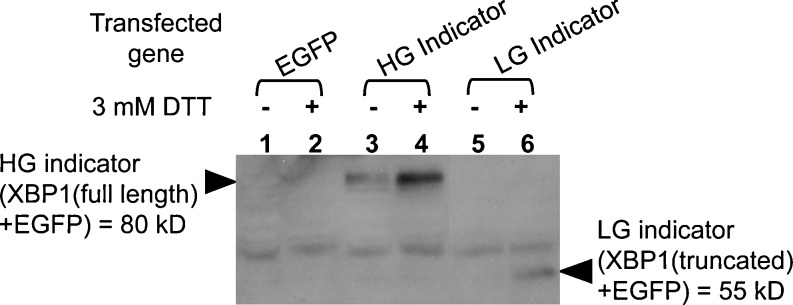
The HG stress indicator shows striking sensitivity to the ER stress in vitro. S2 cells were transfected with the same total amount of plasmids carrying EGFP, the HG indicator, and the LG indicator gene (Ryoo et al. 2007), respectively. All of these transgenes were under the control of UAS promoter. Gal4 gene under the control of actin promoter was also introduced to the cells as the driver for them. For the induction of ER stress, 3 mM of DTT were added to the cells 24 h post-transfection. Equal numbers of the cells were collected 4 h after the addition of DTT. Total cell extracts were analyzed by SDS-PAGE followed by immunoblotting with anti-GFP. Plasmids transfected with actin-gal4 promoter gene to S2 cells were: Lanes 1 and 2, coding EGFP (vector control); lanes 3 and 4, pMS549 coding the HG indicator (XBP1(full length) + EGFP) = 80kD; lanes 5 and 6, coding the LG indicator (XBP1(truncated) + EGFP) = 55kD
Visualization of IRE1/XBP1 activity in larval and adult organs through the HG indicator
Given that the second instar larval lethality of xbp1 −/− hypomorph mutant is due to the defect of IRE1/XBP1 activity, its activity is assumed to be essential for the homeostasis at the third instar larval stage. Namely, XBP1(s) is predicted to accumulate in some of the third instar larval organs whose functions are significantly affected by the malfunction of IRE1/XBP1 pathway. Therefore, we sought to detect the IRE1/XBP1 activity in third instar larval tissues using our XBP1 stress sensing system. To express the xbp1-EGFP gene moderately and ubiquitously in the larval fly body through the Gal4/UAS system, tub-Gal4 driver line was utilized. Recombinant line (w ; UAS-xbp1-EGFP, tub-GAL4 / cyo) was generated by mitotic recombination using the HG indicator transgenic line (w ; UAS-xbp1-EGFP / cyo) and the driver line (w ; tub-Gal4 / cyo). Third instar larvae of those lines were dissected and the accumulation of HG indicator (XBP1-EGFP) in each organ was monitored by immunofluorescence under the experimental conditions indicated in “Materials and methods.” Our new system detected significant IRE1/XBP1 activation in the brain, gut (especially in proventriculus), Malpighian tubules, trachea, and salivary gland (including the surrounding fat body) in third instar larvae of the recombinant line (Figs. 5, 6, and 7). Additionally, we detected significant IRE1/XBP1 activation in the adult male reproductive organs (Fig. 8).
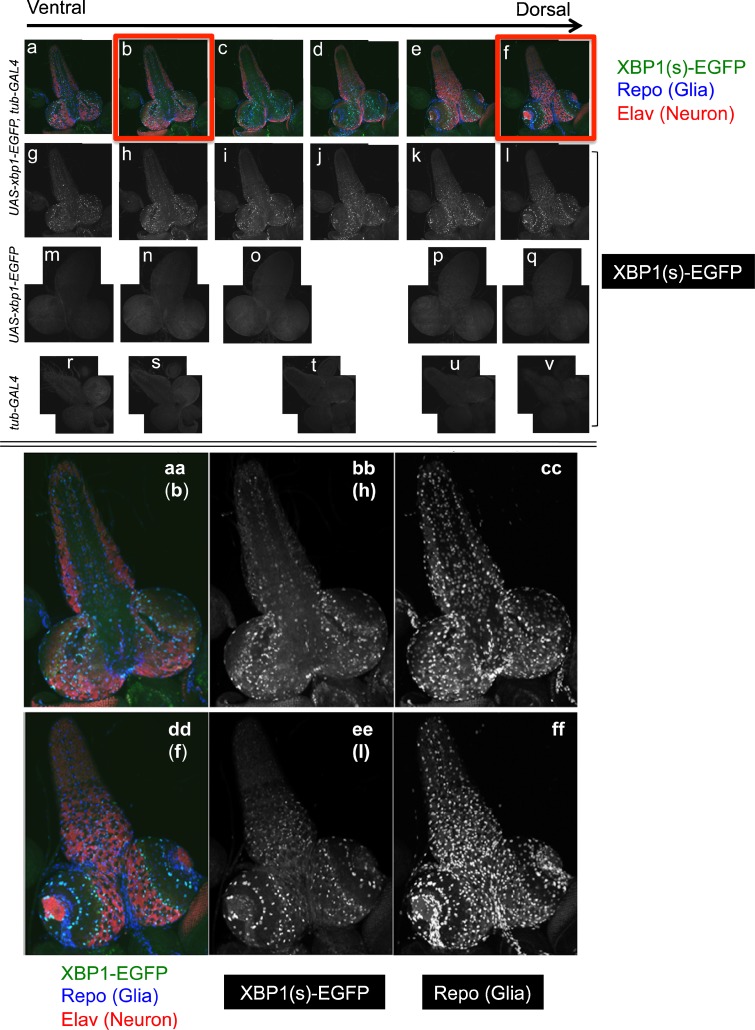
Fig. 5.
Third instar larval brain expressing the HG indicator was fixed with formaldehyde and were incubated with rabbit polyclonal anti-GFP, rat monoclonal anti-Elav, and mouse monoclonal anti-Repo, followed by labeling with Alexa Fluor 488-conjugated goat anti-rabbit IgG, Rhodamine Red-conjugated donkey anti-rat IgG, and Cy5-conjugated donkey anti-mouse IgG. a–f The brains were visualized by fluorescence microscopy with fluorescence from the HG indicator, Elav, and Repo. Sectional pictures between the ventral side and the dorsal side of a brain are lined up from left to right, as indicated by the arrow. g–l The fluorescence from Alexa Fluor 488 was extracted from a to f, respectively. m–v Brains from the lines (w ; UAS-xbp1-EGFP) (m–q) and (w ; tub-Gal4) (r–v) were treated as above and only the fluorescence from Alexa Fluor 488 was extracted. The horizontal positioning of each picture (a–v) approximately corresponds to position of the section. aa–ff The pictures shown in b, f, h, and l were magnified in aa, dd, bb, and ee, respectively. In cc and ff, the fluorescence from Cy5 was extracted from aa and dd, respectively. The merged image indicates the colocalization of HG indicator/XBP1(s)-EGFP and Repo
Fig. 6.
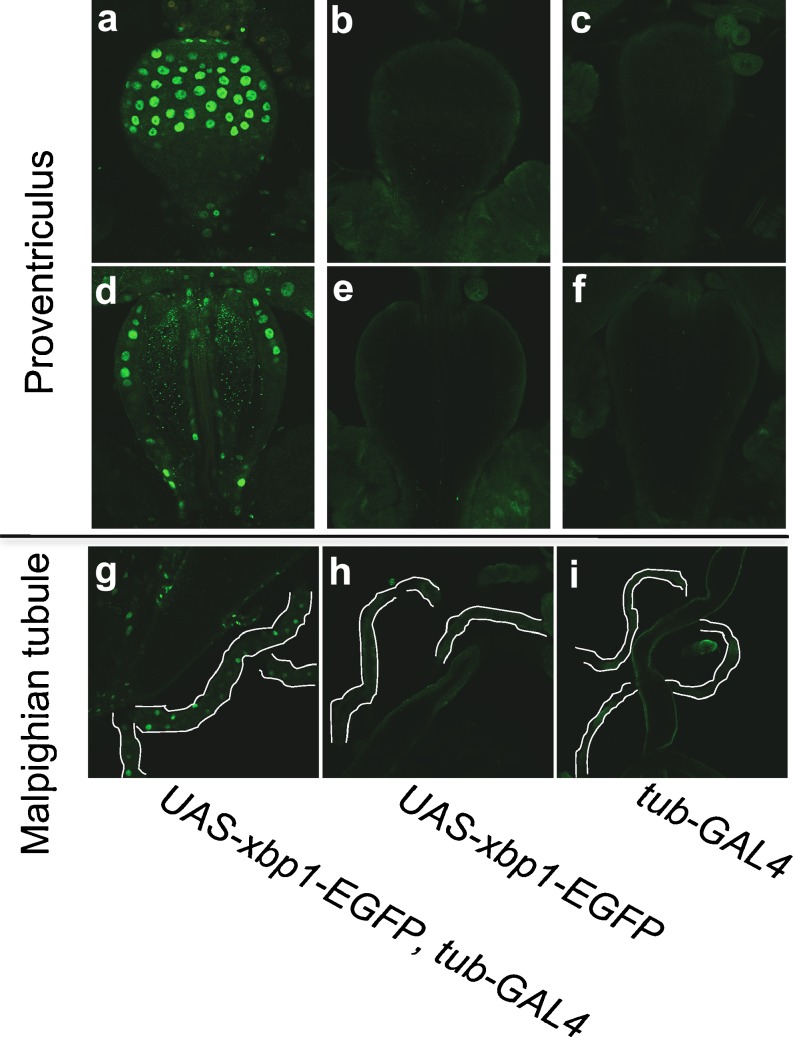
Third instar larval proventriculus (a–f) and Malpighian tubule (g–i) were fixed with formaldehyde and were incubated with rabbit polyclonal anti-GFP followed by labeling with Alexa Fluor 488-conjugated goat anti-rabbit IgG. Three lines; (w ; UAS-xbp1-EGFP, tub-GAL4) (a, d, g), (w ; UAS-xbp1-EGFP) (b, e, h), and (w ; tub-Gal4) (c, f, i), were analyzed. Regarding the proventriculus, two sectional pictures were taken for one proventriculus. One was focused on the surface (a–c), and the other was focused on the inside, approximately in the middle of the proventriculus (d–f). In panels g–i, the Malpighian tubules were outlined with white lines. The fluorescence from EGFP was detected at the surface of the upper side of the proventriculus, the tube penetrating inside the proventriculus (a, d), and throughout the Malpighian tubules derived from the line where the HG indicator was expressed
Fig. 7.
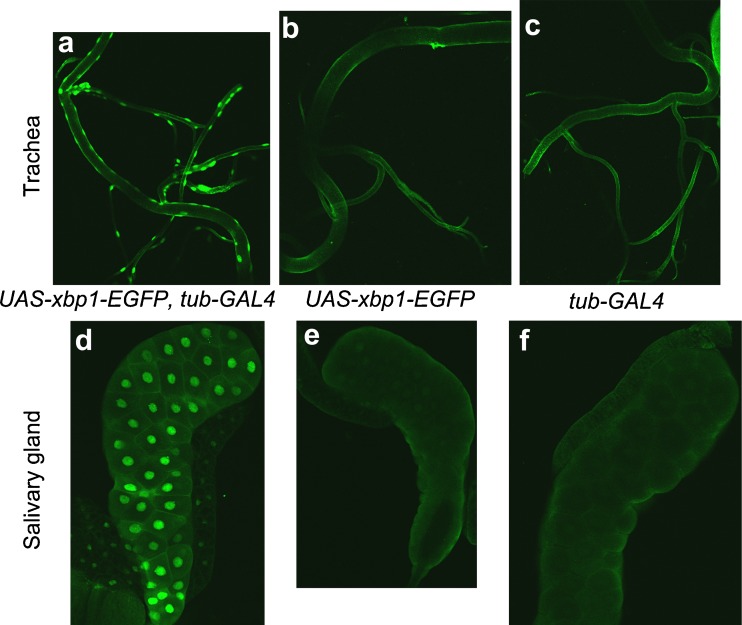
Third instar larval trachea (a–c) and salivary gland (d–f) were fixed with formaldehyde and were incubated with rabbit polyclonal anti-GFP followed by labeling with Alexa Fluor 488-conjugated goat anti-rabbit IgG. Three lines, (w ; UAS-xbp1-EGFP, tub-GAL4) (a, d), (w ; UAS-xbp1-EGFP) (b, e), and (w ; tub-Gal4) (c, f), were analyzed. In both trachea and salivary gland, the fluorescence from EGFP was detected in the line where the HG indicator was expressed (a, d). In panel d, fluorescence was also detected in the fat body attached to the salivary gland
Fig. 8.
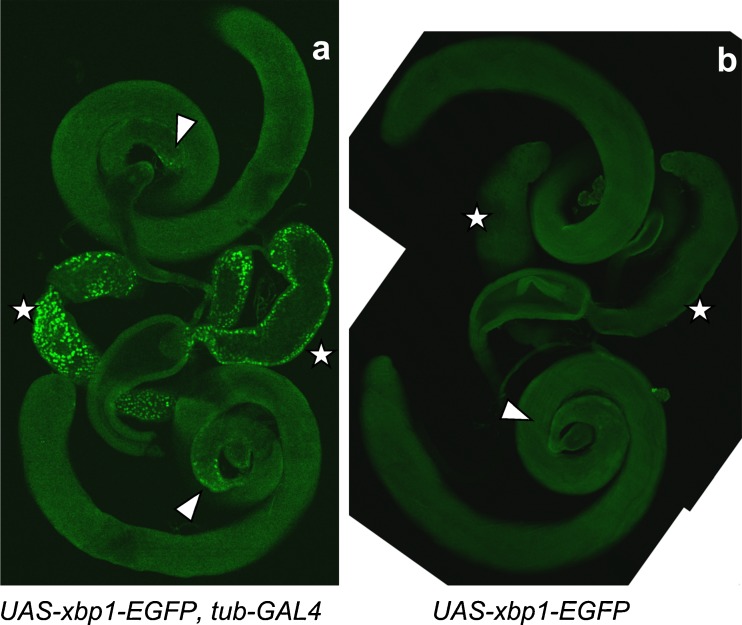
Adult male reproductive organs were fixed with formaldehyde and were incubated with rabbit polyclonal anti-GFP followed by labeling with Alexa Fluor 488-conjugated goat anti-rabbit IgG. Three lines, (w ; UAS-xbp1-EGFP, tub-GAL4) (a), (w ; UAS-xbp1-EGFP) (b), and (w ; tub-Gal4), were analyzed. The fluorescence from EGFP was detected at the accessory gland (marked with a white star), and the limited area of the testis close to the testicular duct (indicated by an arrowhead), in the line where HG indicator was expressed (a). In panel b, the corresponding areas where the fluorescence was detected in panel a are also marked by a star and an arrowhead
In the larval brain, we consistently saw the pronounced pattern of GFP staining in the recombinant line, (w ; UAS-xbp1-EGFP, tub-GAL4 / cyo) (Fig. 5(g–l)), suggestive of cell type specific IRE1/XBP1 activity. Therefore, we attempted to determine whether the activated cells were neurons or glia, by triple immunostaining using anti-Elav (neuronal marker), anti-Repo (glial marker), and anti-GFP antibodies. Various confocal sections from the dorsal to ventral side of the brain were analyzed to monitor the whole area of the larval brain (Fig. 5(a–v)). In every section, all of the HG indicators in brain were colocalized with Repo, although the colocalization of the HG indicator and Elav was not observed. Namely, IRE1/XBP1 pathway in the brain was exclusively activated in glia, rather than in neuron (Fig. 5(aa–ff)).
In the gut, the distribution of the IRE1/XBP1 activity was irregular, although IRE1/XBP1 active cells were consistently observed somewhere in the whole gut. Significant levels of IRE1/XBP1 activity was detected reproducibly only in the proventriculus, which is located at the junction between the forgut and the midgut (Fig. 6a–f). It was detected both in the forgut-derived area (center pipe) and in the midgut-derived area (outer surface). In the midgut-derived area of the proventriculus, most of the IRE1/XBP1 active cells were concentrated in the upper half (Fig. 6a, c).
In other larval organs, such as the Malpighian tubules (Fig. 6g–i), trachea (Fig. 7a–c), and salivary glands (Fig. 7d–f), the signals from the HG indicator were detected uniformly. In adult reproductive organs, significant HG activity was detected in the accessory glands and a limited area of the testis attached to the testicular duct. The fluorescence in the accessory gland was limited to its surface area (Fig. 8a).
As negative controls, we examined parental transgenic lines with only the UAS-transgene (w ; UAS-xbp1-EGFP / cyo) or the driver alone (w ; tub-Gal4 / cyo). We did not detect any significant EGFP signal in those lines. Therefore, we concluded that the EGFP signal we detected in this study reflected the expression of HG indicator/XBP1-EGFP and was not a result of epifluorescence (Figs. 5, 6, 7, and 8).
Discussion
Inadequate sensitivity of existing XBP1 stress sensing systems can be overcome by improving the efficiency of unconventional splicing of xbp1 mRNA. Recent reports regarding the cellular localization of XBP1/HAC1 mRNA during its splicing allowed us to construct a highly sensitive HG stress indicator (Fig. 4) that can visualize the activation of IRE1/XBP1 pathway at the third instar larval stage during normal Drosophila development (Figs. 5, 6, 7, and 8). Several types of cells in the organs where we detected IRE1/XBP1 activity are known for having high secretory capacity.
In the larval brain, we found significant IRE1/XBP1 activity in glial cells (Fig. 5). While glia had been originally thought to function as the structural support cells in the nervous system, it has been revealed that they play several important roles in the development and homeostasis of the nervous system. In Drosophila, glial cells are classified into three classes (surface-, cortex-, and neuropil-associated glia), each of which is subdivided further morphologically (Stork et al. 2010). Whether IRE1/XBP1 active glia is restricted to only a subtype of those glia, or more broadly, is currently under investigation.
In mammals, oligodendrocytes in the central nerve system and Schwann cells in the peripheral nerve system myelinate axons by producing a large amount of myelin membrane proteins, cholesterol, and membrane lipids through the secretory pathway. Recent reports suggested that ER stress in myelinating cells is important in the pathogenesis of various disorders of myelin (Pennuto et al. 2008; Lin and Popko 2009). Neuropil glia and peripheral glia in Drosophila are the counterparts of oligodendrocytes and Schwann cells, respectively. Therefore, these cells are the candidates that show constitutive IRE1/XBP1 activity. Although Drosophila glia do not generate myelin sheaths, they form multi-layered membrane sheaths around neurons that are morphologically similar to the myelin sheaths in mammals (Freeman and Doherty 2005). Thus, it is possible that the IRE1/XBP1 active glia protect neurons from their deterioration through this ensheathment, thereby contributing to brain homeostasis. Further studies are expected to inform us of the pathological significance of IRE1/XBP1 functions in human glia.
As shown in Fig. 5, IRE1/XBP1 pathway does not appear to be active in neuron. However, we do not exclude the possibility of neuronal IRE1/XBP1 activation in the brain. In fact, slight neuronal IRE1/XBP1 activity was occasionally observed in the ventral nerve cord during our repeated experiments. In this study, we conclude that in the third instar larval brain, the IRE1/XBP1 pathway is predominantly activated in glia while the activation is not detectable in neurons.
The importance of IRE1/XBP1 activity in the gut has already been studied in Caenorhabditis elegans and mammals (Shen et al. 2001; Kaser et al. 2008; Richardson et al. 2010). We identified intra-tissue distribution of IRE1/XBP1 activity in the proventriculus region of the gut (Fig. 6a–f). In the larval midgut and hindgut, we observed an irregular distribution of IRE1/XBP1 active cells. These were not entero-endocrine cells, as they did not colocalize with anti-Prospero antibody that marks those cells (data not shown). Secretory intestinal cells in the midgut other than entero-endocrine cells (Casali and Batlle 2009) including the intestinal stem cells are possible candidates for these IRE1/XBP1 active cells.
IRE1/XBP1 activity in the fly Malpighian tubules (analogous to the kidney in mammals) was also unexpected. The activity was detected throughout the organ, but not all of the cells were IRE1/XBP1 active (Fig. 6g–i). Although the Malpighian tubules are attached at the junction of the midgut and the hindgut, they are morphologically and functionally independent from both of them. Identification of the IRE1/XBP1 active cells in the gut and the Malpighian tubules might reflect a shared physiological function of both organs. One possible shared function may be the selective uptake of the essential molecules, including several metal ions, from the contents passing through those organs. IRE1/XBP1 pathway might regulate the function of some transporter channels in these organs. Drosophila Malpighian tubules are expected to be one of the models for the mammalian diabetic kidney diseases that are associated with UPR activation (Cunard and Sharma 2011).
In this study, we also identified IRE1/XBP1 activity in the trachea (Fig. 7a–c). Previous reports lead us to point out its relevance to glial IRE1/XBP1 activity (Pereanu et al. 2007; Tsarouhas et al. 2007). One of them showed that tracheal development in Drosophila brain was controlled by signals from glia (Pereanu et al. 2007). According to the report, the branches of cerebral trachea grow around the neuropile. If IRE1/XBP1 active glia were neuropile-associated glia, assessing IRE1/XBP1 activity at neuropile-associated glia is likely to allow us to reveal the shared physiological function of IRE1/XBP1 pathway between brain and trachea. The other report, using embryonic trachea, indicated that the proper combination of secretory activity and endocytotic activity was important for the maturation of trachea as an airway. In tracheal maturation, Sar1, one of the core COPII proteins, was required for the secretion of protein, the luminal matrix assembly, and the following expansion of tube diameter to avoid the clogging of protein, while Rab5, the small GTPase that regulates the early stage of endocytosis, was required for the clearance of deposited materials in the lumen (Tsarouhas et al. 2007). It can be predicted that, even in larval trachea, IRE1/XBP1 pathway plays a crucial role in tracheal maturation by supplying the properly folded proteins to the transport machinery. In that case, in view of second instar larval lethality of xbp1 −/− hypomorph mutant, we could also hypothesize that the tracheal maturation/maintenance is still important for larval lethality, in addition to its importance for the embryonic development.
IRE1/XBP1 activity in the salivary gland has already been reported in a previous study (Souid et al. 2007). The salivary gland is commonly used for the determination of the subcellular localization of the protein in Drosophila cells due to its morphological features. Figure 7d–f clearly indicated the nuclear localization of HG indicator, XBP1-EGFP molecule. In addition, we observed weak IRE1/XBP1 activity in the fat body that was attached to the salivary gland (Fig. 7d–f). Generally, the Drosophila fat body, which is equivalent to mammalian adipose tissue, functions as the organ for energy/lipid storage and is distributed throughout the larval body.
In addition to the larval tissues, we analyzed IRE1/XBP1 activity in the adult male reproductive organs. Though the previous RT-PCR study by Souid et al. (2007) suggested the activity in the testis, the areas we detected IRE1/XBP1 activity were the accessory glands and a limited area of the testis close to the testicular duct (Fig. 8). In the accessory gland, seminal fluid containing several hormones, which facilitate reproductive traits such as sperm transfer, sperm storage, female receptivity, ovulation, and oogenesis, are produced and secreted (Wolfner 1997; Chapman 2001). There are two morphologically distinct secretory cell types in Drosophila accessory gland. Ninety-six percent of the secretory cells are categorized as main cells and the others are secondary cells (Kalb et al. 1993). Based on the intra-tissue distribution of IRE1/XBP1 active cells in the accessory gland, the active cells are likely to be main cells. Since each of these cell types expresses a unique set of genes, the confirmation of IRE1/XBP1 active cell type is expected to allow us to narrow down the proteins related to IRE1/XBP1 activity. IRE1/XBP1 pathway is likely to function, to some extent, in maintaining proper fertility.
On the other hand, we considered a possibility that the EGFP signal we detected in each organ might not necessarily reflect the unconventional splicing of xbp1-EGFP mRNA. Higher concentrations of the spliced xbp1-EGFP mRNA and resulting XBP1-EGFP in the cells induced by the Gal4/UAS system might cause the artifactual EGFP signal. We excluded the possibility that the EGFP signal in this study was detected independently of the unconventional splicing, based on our results in this study and the following reasoning.
There are two possible molecular mechanisms that cause the artifactual EGFP signal which is not derived from the unconventional splicing of xbp1-EGFP mRNA. One is the generation of EGFP or abnormal EGFP fusion proteins, resulting from translation initiation at the start codon of the EGFP coding sequence or at ATG codons coding Met residues in XBP1(s), respectively. The other is the proteolytic digestion of XBP1-EGFP fusion protein at the junction of XBP1 and EGFP portions. Both of these are prone to happen upon overexpression of fusion proteins in cells. In particular, the proteolytic digestion is often observed in the overexpression of GST fusion protein in Escherichia coli.
In our system, there is no nuclear localization signal (NLS) on the EGFP molecule. In contrast, xbp1 gene carries a NLS coding sequence located upstream of the unconventional splice site. There are a total of 11 ATG codons that code the Met residues of XBP1(s) molecule. Eight of the ATG codons are located downstream of NLS coding sequence. Therefore, due to the lack of NLS, both EGFP and the EGFP fusion proteins using these eight ATG codons as start codons should diffuse all over the cell upon their synthesis, if they were generated. As shown in Fig. 7d–f, EGFP signal was detected exclusively in the nucleus in the salivary gland, which is often used for the analysis of the cellular localization of the proteins in Drosophila. Hence, it is not reasonable to conclude that either EGFP or the possible eight EGFP fusion proteins above were expressed in the cells. Only the EGFP fusion proteins that use the other three ATG codons that are upstream of the NLS as start codons should be synthesized upon unconventional splicing and localized in the nucleus. The estimated molecular weights of those fusion proteins are 73.7, 74.3, and 80.0 kDa, respectively. As shown in lane 4 (also lane 3) of Fig. 4, only a significant single band that represented the intact XBP1-EGFP (80.0 kDa) was detected in the S2 cell extract, in which XBP1-EGFP was overexpressed through the Gal4/UAS system. Therefore, we concluded that the EGFP fusion protein synthesized in this study was the intact XBP1-EGFP.
Additionally, there are several ATG codons that are located upstream of the unconventional splice site and are in frame with EGFP coding sequence on unspliced xbp1 mRNA. However, inside the 23 bp of unconventional-spliced fragment, there is a TGA stop codon that is also in frame with EGFP coding sequence. Even if the translation initiated from these start codons, the synthesis of these products should be terminated at this TGA stop codon before the ribosome would reach the EGFP coding region.
Regarding the proteolytic digestion of XBP1-EGFP fusion protein, the resulting EGFP should diffuse all over the cell upon its synthesis due to the lack of NLS. Therefore, the possibility of the proteolytic digestion is also excluded based on the same reasoning as above. Taken together, we concluded that the detected EGFP signal in this study exclusively reflected the occurrence of unconventional splicing of xbp1-EGFP mRNA.
In summary, we improved the sensitivity of the XBP1 stress sensing system and newly identified several organs where IRE1/XBP1 pathway is constitutively activated under normal physiological conditions. In particular, in the larval brain, significant glial specific activation was detected. Our improved system is expected to provide us with a number of clues to reveal the molecular mechanisms underlying the normal development and homeostasis controlled by IRE1/XBP1 pathway.
Acknowledgments
The authors thank Professor Toshiya Endo (Nagoya University) and Professor Shuh-ichi Nishikawa (Niigata University) for their helpful encouragement throughout this study. This work was supported by grants from the NEI (R01EY020866) and the Ellison Medical Foundation to H.D.R. and an NIDDK training grant to J.L. (5T35DK007421).
References
- Ali MM, Bagratuni T, Davenport EL, Nowak PR, Silva-Santisteban MC, Hardcastle A, McAndrews C, Rowlands MG, Morgan GJ, Aherne W, Collins I, Davies FE, Pearl LH. Structure of the Ire1 autophosphorylation complex and implications for the unfolded protein response. EMBO J. 2011;30:894–905. doi: 10.1038/emboj.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragon T, van Anken E, Pincus D, Serafimova IM, Korennykh AV, Rubio CA, Walter P. Messenger RNA targeting to endoplasmic reticulum stress signaling sites. Nature. 2009;457:736–740. doi: 10.1038/nature07641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Casali A, Batlle E. Intestinal stem cells in mammals and Drosophila. Cell Stem Cell. 2009;4:124–127. doi: 10.1016/j.stem.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Chapman T. Seminal fluid-mediated fitness traits in Drosophila. Heredity. 2001;87:511–521. doi: 10.1046/j.1365-2540.2001.00961.x. [DOI] [PubMed] [Google Scholar]
- Chawla A, Chakrabarti S, Ghosh G, Niwa M. Attenuation of yeast UPR is essential for survival and is mediated by IRE1 kinase. J Cell Biol. 2011;193:41–50. doi: 10.1083/jcb.201008071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-A. [DOI] [PubMed] [Google Scholar]
- Cunard R, Sharma K. The endoplasmic reticulum stress response and diabetic kidney disease. Am J Physiol Renal Physiol. 2011;300:1054–1061. doi: 10.1152/ajprenal.00021.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubchak I, Brudno M, Loots GG, Mayor C, Pachter L, Rubin EM, Frazer KA. Active conservation of noncoding sequences revealed by 3-way species comparisons. Genome Res. 2000;10:1304–1306. doi: 10.1101/gr.142200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, et al. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman MR, Doherty J. Glial cell biology in Drosophila and vertebrates. Trends Neurosci. 2005;29:82–90. doi: 10.1016/j.tins.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Mol Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- Iwawaki T, Akai R, Kohno K, Miura M. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat Med. 2004;10:98–102. doi: 10.1038/nm970. [DOI] [PubMed] [Google Scholar]
- Kalb JM, Dibenedetto AJ, Wolfner MF. Probing the function Drosophila melanogaster accessory glands by directed cell ablation. Proc Natl Acad Sci. 1993;90:8093–8097. doi: 10.1073/pnas.90.17.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EE, Higgins DE, Schreiber S, Glimcher LH, Blumberg RS. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korennykh AV, Egea PF, Korostelev AA, Finer-Moore J, Zhang C, Shokat KM, Stroud RM, Walter P. The unfolded protein response signals through high-order assembly of Ire1. Nature. 2009;457:687–693. doi: 10.1038/nature07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korennykh AV, Korostelev AA, Egea PF, Finer-Moore J, Stroud RM, Zhang C, Shokat KM, Walter P. Structural and functional basis for RNA cleavage by Ire1. BMC Biol. 2011;9:47. doi: 10.1186/1741-7007-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korennykh AV, Korostelev AA, Egea PF, Finer-Moore J, Stroud RM, Zhang C, Shokat KM, Walter P. Cofactor-mediated conformational control in the bifunctional kinase/RNase Ire1. BMC Biol. 2011;9:48. doi: 10.1186/1741-7007-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee KP, Dey M, Neculai D, Cao C, Dever TE, Sicheri F. Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell. 2008;132:89–100. doi: 10.1016/j.cell.2007.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Popko B. Endoplasmic reticulum stress in disorders of myelinationg cells. Nature Neurosci. 2009;12:379–385. doi: 10.1038/nn.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/CDC28- related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-Q. [DOI] [PubMed] [Google Scholar]
- Parmar VM, Schröder M. Sensing endoplasmic reticulum stress. Adv Exp Med Biol. 2012;738:153–168. doi: 10.1007/978-1-4614-1680-7_10. [DOI] [PubMed] [Google Scholar]
- Pennuto M, et al. Ablation of the UPR-mediator CHOP restores motor function and reduces demyelination in Charcot-Marie-Tooth 1B mice. Neuron. 2008;57:393–405. doi: 10.1016/j.neuron.2007.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereanu W, Spindler S, Cruz L, Hartenstein V. Tracheal development in the Drosophila brain is constrained by glial cells. Dev Biol. 2007;302:169–180. doi: 10.1016/j.ydbio.2006.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CE, Kooistra T, Kim DH. An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature. 2010;463:1092–1095. doi: 10.1038/nature08762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Ito K. A translational pause to localize. Science. 2011;331:543–544. doi: 10.1126/science.1202075. [DOI] [PubMed] [Google Scholar]
- Rubio C, Pincus D, Korennykh A, Schuck S, El-Samad H, Walter P. Homeostatic adaptation to endoplasmic reticulum stress depends on Ire1 kinase activity. J Cell Biol. 2011;193:171–184. doi: 10.1083/jcb.201007077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo HD, Steller H. Unfolded protein response in Drosophila: why another model can make it fly. Cell Cycle. 2007;6(7):830–835. doi: 10.4161/cc.6.7.4064. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Domingos PM, Kang MJ, Steller H. Unfolded protein response in a Drosophila model for retinal degeneration. EMBO J. 2007;26:242–252. doi: 10.1038/sj.emboj.7601477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Ellis RE, Lee K, Liu CY, Yang Q, Solomon A, Yoshida H, Morimoto R, Kurnit DM, Mori K, Kaufman RJ. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. doi: 10.1016/S0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- Shim J, Umemura T, Nothstein E, Rongo C. The unfolded protein response regulates glutamate receptor export from the endoplasmic reticulum. Mol Biol Cell. 2004;15:4818–4828. doi: 10.1091/mbc.E04-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souid S, Lepesant JA, Yanicostas C. The xbp-1 gene is essential for development in Drosophila. Dev Genes Evol. 2007;217:159–167. doi: 10.1007/s00427-006-0124-1. [DOI] [PubMed] [Google Scholar]
- Stork T, Bernardos R, Freeman MR (2010) Analysis of glial cell development and function in Drosophila. In: Zhang B, Freeman MR, Waddell S (eds) Drosophila neurobiology: a laboratory manual. Cold Spring Harbor Laboratory Press, New York, pp 53–74
- Tsarouhas V, Senti KA, Jayaram SA, Tiklova K, Hemphala J, Adler J, Samakovlis C. Sequential pulses of apical epithelial secretion and endocytosis drive airway maturation in Drosophila. Dev Cell. 2007;13:214–225. doi: 10.1016/j.devcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wiseman RL, Zhang Y, Lee KPK, Harding HP, Haynes CM, Price J, Sicheri F, Ron D. Flavonol activation defines an unanticipated ligand binding site in the kinase-rnase domain of IRE11. Mol Cell. 2010;38:291–304. doi: 10.1016/j.molcel.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfner MF. Tokens of love: functions and regulation of Drosophila male accessory gland products. Insect Biochem Mol Biol. 1997;27:179–192. doi: 10.1016/S0965-1748(96)00084-7. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Yanagitani K, Imagawa Y, Iwawaki T, Hosoda A, Saito M, Kimata Y, Kohno K. Cotranslational targeting of XBP1 protein to the membrane promotes cytoplasmic splicing of its own mRNA. Mol Cell. 2009;34:191–200. doi: 10.1016/j.molcel.2009.02.033. [DOI] [PubMed] [Google Scholar]
- Yanagitani K, Kimata Y, Kadokura H, Kohno K. Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science. 2011;331:586–589. doi: 10.1126/science.1197142. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Oku M, Suzuki M, Mori K. pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J Cell Biol. 2006;172:565–575. doi: 10.1083/jcb.200508145. [DOI] [PMC free article] [PubMed] [Google Scholar]