Abstract
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is often used as a stable housekeeping marker for constant gene expression. However, the transcriptional levels of GAPDH may be highly up-regulated in some cancers, including non-small cell lung cancers (NSCLC). Using a publically available microarray database, we identified a group of genes whose expression levels in some cancers are highly correlated with GAPDH up-regulation. The majority of the identified genes are cell cycle-dependent (GAPDH Associated Cell Cycle, or GACC). The up-regulation pattern of GAPDH positively associated genes in NSCLC is similar to that observed in cultured fibroblasts grown under conditions that induce anti-senescence. Data analysis demonstrated that up-regulated GAPDH levels are correlated with aberrant gene expression related to both glycolysis and gluconeogenesis pathways. Down-regulation of fructose-1,6-bisphosphatase (FBP1) in gluconeogenesis in conjunction with up-regulation of most glycolytic genes is closely related to high expression of GAPDH in the tumors. The data presented demonstrate that up-regulation of GAPDH positively associated genes is proportional to the malignant stage of various tumors and is associated with an unfavourable prognosis. Thus, this work suggests that GACC genes represent a potential new signature for cancer stage identification and disease prognosis.
Introduction
Although the gene encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is frequently used as a stable marker for constitutive gene expression, its expression is not always constant, especially in cancer. For example, in one study of non-small cell lung cancer (NSCLC), GAPDH was the least stable of the 6 “reference” genes examined [1]. This housekeeping glycolytic enzyme has been implicated in multiple functions and has been found to be over-expressed in certain cancers [2].
More than 50 years ago, Warburg hypothesized that cancer growth is facilitated by tumors generating their energy through aerobic glycolysis [3]. Recent studies aimed at evaluating this hypothesis have demonstrated that cancer cells have adapted their metabolism to facilitate the uptake and incorporation of nutrients into the biomass required to produce new cells [4]. Tumor development and progression are indeed correlated with enhanced glucose uptake and/or aberrant glucose metabolism [5]–[8]. The hypoxic environment in which tumor cells reside leads to an increase in glycolytic metabolism. As a key intermediate component of glycolysis, GAPDH could serve an important role in cancer cell development and tumor progression. While it is known that most glycolytic enzymes, including GAPDH, are activated and highly expressed to respond to oxygen deprivation in the tumor [9], the role of up-regulated GAPDH in NSCLC remains unclear. In one possibly cancer-related scenario, GAPDH was found to be a pro-survival regulator of caspase-independent cell death (CICD) [10].
In the current study, a publically available microarray database was employed to identify cell cycle-dependent genes that correlate with GAPDH up-regulation and anti-apoptotic activity. This analysis has identified a set of cell cycle-based signature genes, designated GAPDH Associated Cell Cycle (GACC) genes, whose up-regulation is correlated with the aggressiveness of several tumor types and their unfavourable prognosis. Identification of GACC genes may be useful in efforts aimed at elucidating pathways that connect carbohydrate metabolism with cell cycle-based cancer cell development, which might lead to the novel cancer targets based on GACC gene expression patterns in cancers.
Results
Up-regulation of GAPDH Associated Cell Cycle (GACC) genes in non-small cell lung cancer (NSCLC)
An integrated NSCLC gene expression dataset, based on the Affymetrix GeneChip Human Genome U133 Plus 2.0 Array, was created from three independent cohorts that were directly downloaded from the publically available European Bioinformatics Institute ArrayExpress database: E-GEOD-18842, E-GEOD-19188 and E-GEOD-19804. The combined dataset, designated the full cohort, consisted of 174 NSCLCs and 156 control tissues. Preliminary analysis suggested the transcription of some cell cycle genes in the NSCLC dataset might correlate with the transcription of GAPDH. Given the role of GAPDH in glycolysis, this finding encouraged further gene expression profile analysis of the tumors for the relationship between carbohydrate metabolism and cell cycle regulation.
Gene expression correlation analysis identified many genes whose up-regulation in the tumors is closely correlated with GAPDH expression. Specifically, within the cancer cohort, 341 up-regulated genes were identified with a GAPDH expression correlation coefficient greater than or equal to 0.6. Of these, 117 genes (34%) are described as cell cycle-related based on the Gene Ontology biological process terminology. In the original GeneChip Human Genome U133 Plus 2.0 Array with 54,613 probes, only 2,044 related genes (3.7%) are associated with Gene Ontology biological process term “cell cycle,” which implies more than a 9-fold enrichment for cell cycle-related genes in the tumors. An even higher percentage of cell cycle-related genes (18/26, 69%; a 17-fold enrichment for cell cycle-related genes) are found when the correlation coefficient is set more stringently, at greater than or equal to 0.72 (Table 1). These genes are designated here as GAPDH Associated Cell Cycle (GACC) genes. The genes in the list encode proteins related to G1/S and/or G2/M phase transitions (FOXM1, CDCA3 (Tome-1), CCNB2 (cyclin B2), BIRC5, CCNB1 (cyclin B1), CDC45, PRC1 and CCNA2 (cyclin A2)), and proteins involved in mitotic processes (NCAPD2, TPX2, AURKB, PSMD2, KIF4A, KIF2C, UBE2S, and FAM83D). KPNA2 (importin alpha 1) and CDKN3 (cyclin-dependent kinase inhibitor 3) are also cell cycle-related. Top ranked GAPDH positively associated genes in this class include triosephosphate isomerase 1 (TPI1) and glucose-6-phosphate isomerase (GPI), each of which encodes a key glycolytic enzyme. Thus, only 6 of the 26 genes in the list (CENPA, RAD51AP1, SLC2A1, KIF4A, RFC4, and RRM2) may not be designated as related to the cell cycle or glycolysis. At least a sub-set of these six genes may not actually be exceptions, as CENPA is an essential centromeric protein subject to cell cycle-dependent changes [11]. There is also some evidence that suggests RAD51AP1 may contribute to neoplastic growth [12], whereas SLC2A1 is involved in glycolysis related glucose transport. Finally, KIF4A is implicated in chromosome condensation and segregation during mitosis [13], RFC4 is essential for proliferating cell nuclear antigen (PCNA) dependent DNA synthesis [14], and RRM2 encodes ribonucleotide reductase M2, which catalyzes the formation of deoxyribonucleotides from ribonucleotides in a cell-cycle dependent fashion [15]. In sum, two gene classes have been identified in the list. Class I consists of glucose metabolic pathway related genes while Class II covers cell cycle related genes.
Table 1. Top ranked up-regulated GAPDH positively associated genes in NSCLC (correlation coefficient greater than or equal to 0.72).
| Gene Title | Symbol | Probe | r value | GO Definition | T/C | p value (t-test) |
| triosephosphate isomerase 1 | TPI1 | 213011_s_at | 0.79 | glycolysis | 1.09 | 5.06E-45 |
| non-SMC condensinI complex, subunit D2 | NCAPD2 | 201774_s_at | 0.79 | cell cycle | 1.19 | 8.33E-35 |
| cell division cycle associated 3 | CDCA3 | 221436_s_at | 0.78 | cell cycle | 1.27 | 3.05E-50 |
| forkhead box M1 | FOXM1 | 202580_x_at | 0.77 | cell cycle | 1.54 | 5.89E-49 |
| TPX2, microtubule-associated, homolog (Xenopus laevis) | TPX2 | 210052_s_at | 0.76 | cell cycle | 1.62 | 9.96E-57 |
| centromere protein A | CENPA | 204962_s_at | 0.75 | centromere protein | 1.59 | 1.29E-47 |
| karyopherin alpha 2 (RbAG cohort 1, importin alpha 1) | KPNA2 | 211762_s_at | 0.75 | cell cycle | 1.16 | 4.99E-43 |
| glucose-6-phosphate isomerase | GPI | 208308_s_at | 0.75 | glycolysis | 1.11 | 5.81E-44 |
| cyclin B2 | CCNB2 | 202705_at | 0.75 | cell cycle | 1.54 | 9.85E-56 |
| cell division cycle 20 homolog (S. cerevisiae) | CDC20 | 202870_s_at | 0.75 | cell cycle | 1.66 | 1.09E-57 |
| cyclin B1 | CCNB1 | 214710_s_at | 0.74 | cell cycle | 1.61 | 2.68E-57 |
| RAD51 associated protein 1 | RAD51AP1 | 204146_at | 0.74 | DNA repair | 1.32 | 9.97E-43 |
| solute carrier family 2 (facilitated glucose transporter), member 1 | SLC2A1 | 201250_s_at | 0.74 | glucose transport | 1.45 | 1.02E-46 |
| ribonucleotide reductase M2 | RRM2 | 209773_s_at | 0.74 | DNA replication | 1.56 | 2.01E-54 |
| aurora kinase B | AURKB | 209464_at | 0.73 | cell cycle | 1.43 | 1.81E-44 |
| replication factor C (activator 1) 4, 37kDa | RFC4 | 204023_at | 0.73 | DNA replication | 1.24 | 1.91E-38 |
| proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 | PSMD2 | 200830_at | 0.73 | cell cycle | 1.06 | 2.69E-26 |
| kinesin family member 4A | KIF4A | 218355_at | 0.73 | microtubule-based movement | 1.63 | 1.52E-56 |
| baculoviral IAP repeat-containing 5 | BIRC5 | 202095_s_at | 0.73 | cell cycle | 1.62 | 5.74E-56 |
| kinesin family member 2C | KIF2C | 209408_at | 0.72 | cell cycle | 1.42 | 6.65E-54 |
| cyclin-dependent kinase inhibitor 3 | CDKN3 | 209714_s_at | 0.72 | cell cycle | 1.53 | 2.16E-54 |
| ubiquitin-conjugating enzyme E2S | UBE2S | 202779_s_at | 0.72 | cell cycle | 1.17 | 3.38E-29 |
| family with sequence similarity 83, member D | FAM83D | 225687_at | 0.72 | cell cycle | 1.40 | 5.67E-35 |
| cell division cycle 45 homolog (S. cerevisiae) | CDC45 | 204126_s_at | 0.72 | cell cycle | 1.34 | 5.28E-43 |
| cyclin A2 | CCNA2 | 203418_at | 0.72 | cell cycle | 1.62 | 8.53E-49 |
| protein regulator of cytokinesis 1 | PRC1 | 218009_s_at | 0.72 | cell cycle | 1.47 | 7.11E-51 |
r = correlation coefficient between GAPDH and current gene expression in NSCLC.
T/C = expression level ratio between cancer and control.
The highest ranked GACC genes are highly associated to the pathway regulated by the transcriptional factor Forkhead Box M1 (FoxM1, r = 0.77) in NSCLC. Table 2 lists the genes in the FoxM1 pathway in which their expression is related to G2/M progression, chromosomal segregation, and cytokinesis. The r values between the expression of a majority of these genes (19/29, 68%) and the expression of GAPDH in the list are above 0.6.
Table 2. Correlation of gene expression between FoxM1 related genes and GAPDH in NSCLC.
| Gene in FoxM1 pathway | Symbol | Function | r value | T/C | p value (t-test) | Probe Set ID |
| NDC80 homolog, kinetochore complex component (S. cerevisiae) | NDC80 (KNTC2) | Spindle check point protein | 0.66 | 1.55 | 1.52E-44 | 204162_at |
| budding uninhibited by benzimidazoles 1 homolog beta (yeast) | BUB1B (BUBR1) | Spindle check point protein | 0.70 | 1.58 | 1.27E-55 | 203755_at |
| polo-like kinase 1 | PLK1 | Spindle check point protein | 0.71 | 1.25 | 3.58E-43 | 202240_at |
| MAD2 mitotic arrest deficient-like 1 (yeast) | MAD2L1 | Spindle check point protein | 0.71 | 1.57 | 2.29E-55 | 203362_s_at |
| budding uninhibited by benzimidazoles 1 homolog (yeast) | BUB1 | Spindle check point protein | 0.70 | 1.64 | 3.45E-50 | 209642_at |
| aurora kinase A | AURKA | Spindle check point protein | 0.65 | 1.42 | 2.15E-54 | 204092_s_at |
| polo-like kinase 4 | PLK4 | Spindle check point protein | 0.56 | 1.18 | 9.30E-37 | 204887_s_at |
| budding uninhibited by benzimidazoles 3 homolog (yeast) | BUB3 | Spindle check point protein | 0.47 | 1.05 | 3.59E-14 | 201457_x_at |
| MAD1 mitotic arrest deficient-like 1 (yeast) | MAD1L1 | Spindle check point protein | 0.08 | 1.03 | 1.88E-06 | 204857_at |
| centromere protein A | CENPA | FOXM target protein | 0.75 | 1.59 | 1.29E-47 | 204962_s_at |
| cell division cycle 20 homolog (S. cerevisiae) | CDC20 | FOXM target protein | 0.75 | 1.66 | 1.09E-57 | 202870_s_at |
| aurora kinase B | AURKB | FOXM target protein | 0.73 | 1.43 | 1.81E-44 | 209464_at |
| baculoviral IAP repeat-containing 5 | BIRC5 (survivin) | FOXM target protein | 0.73 | 1.62 | 5.74E-56 | 202095_s_at |
| NIMA (never in mitosis gene a)- related kinase 2 | NEK2 | FOXM target protein | 0.69 | 1.74 | 1.32E-57 | 204641_at |
| centromere protein F, 350/400kDa (mitosin) | CENPF | FOXM target protein | 0.67 | 1.47 | 1.01E-55 | 207828_s_at |
| cell division cycle 25 homolog B (S. pombe) | CDC25B | FOXM target protein | 0.11 | 1.02 | 0.001216 | 201853_s_at |
| mitochondrial ribosomal protein 63 | MRP63 | FOXM target protein | 0.04 | 0.99 | 0.400757 | 204387_x_at |
| antigen identified by monoclonalantibody Ki-67 | MKI67 | cell cycle marker proteins (KI-67) | 0.67 | 1.40 | 8.04E-49 | 212021_s_at |
| cell division cycle 25 homolog C (S. pombe) | CDC25C | cell cycle marker proteins (CDC25) | 0.53 | 1.27 | 3.49E-41 | 205167_s_at |
| cyclin-dependent kinase 1 | CDK1 | cell cycle marker proteins (CDC2) | 0.70 | 1.39 | 2.03E-49 | 203214_x_at |
| cyclin B2 | CCNB2 | cell cycle marker proteins | 0.75 | 1.54 | 9.85E-56 | 202705_at |
| cyclin B1 | CCNB1 | cell cycle marker proteins | 0.74 | 1.61 | 2.68E-57 | 214710_s_at |
| cyclin A2 | CCNA2 | cell cycle marker proteins | 0.72 | 1.62 | 8.53E-49 | 203418_at |
| Ubiquitin specific peptidase 22 | USP22 | cell cycle marker proteins | 0.19 | 1.02 | 7.11E-06 | 216964_at |
| cyclin D1 | CCND1 | cell cycle marker proteins | 0.19 | 0.99 | 0.436576 | 208711_s_at |
| CDC28 protein kinase regulatory subunit 1B | CKS1B | CDKI proteins degradation | 0.64 | 1.17 | 1.09E-46 | 201897_s_at |
| S-phase kinase-associated protein 2 (p45) | SKP2 | CDKI proteins degradation | 0.61 | 1.16 | 1.29E-25 | 203625_x_at |
| cyclin-dependent kinase inhibitor 1B (p27, Kip1) | CDKN1B | CDK inhibitor | 0.05 | 0.98 | 0.021125 | 209112_at |
r = correlation coefficient between GAPDH and current gene expression in lung cancers.
T/C = expression level ratio between cancer and control.
One hundred twenty-three down-regulated genes, having a correlation coefficient less than or equal to −0.6, were identified in NSCLC in the course analyzing the data (data not shown). None of the genes in this group are described by the Gene Ontology biological process as cell cycle related.
In contrast to the cancer tissues, analysis of the cancer-free control tissues in the cohort identified only 5/70 (7%) of the genes that correlated positively with GAPDH expression (r greater than or equal to 0.6). The Gene Ontology biological process describes these genes as cell cycle. The correlated cell cycle genes include CDK4 (r = 0.62) and CHK1 (r = 0.62). These two genes were also observed to be highly expressed within the cancer tissues.
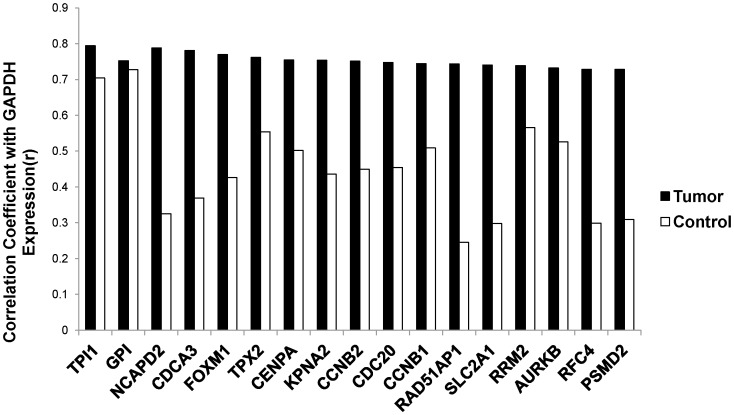
As implied by the above findings, for most of the highly expressed GACC genes in the tumors, there was only a weak correlation for GAPDH and GACC gene expression observed in the control tissue samples (correlation coefficients were mostly in 0.2–0.5 ranges). By contrast, the correlation coefficients for the 15 highest ranked GACC genes from the cancer tissues were greater than 0.7 (Figure 1). Expression of genes within the glycolytic pathway (GAPDH, TPI1 and GPI) remained highly correlated in both in NSCLCs and non-cancer control tissue samples.
Figure 1. Correlation coefficients for top ranked GAPDH associated gene and GAPDH expression in NSCLCs and controls.
Correlation coefficients of GAPDH expression levels and GAPDH associated gene expression levels were calculated in NSCLCs (black, selected genes with r>0.7) and controls (white) are plotted. TPI1 and GPI are genes encoding two enzymes in glycolysis. The remainder of genes encodes cell cycle related proteins.
Up-regulation of GAPDH in NSCLC and glycolysis/gluconeogenesis pathways
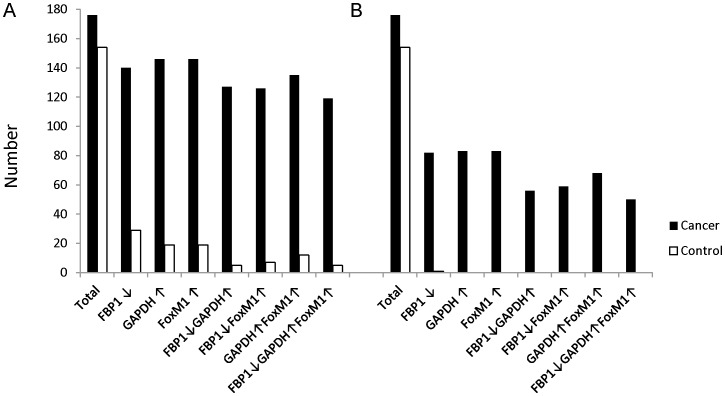
The forgoing results imply that GAPDH expression may be associated with cell cycle related activities. To explore the relationship between the up-regulated GACC genes in lung cancer and the possibly aberrant expression of glucose metabolic pathways, we analyzed the expression values of genes implicated in glycolysis and gluconeogenesis. The expression of each gene in these pathways is up-regulated or unchanged, in the tumor cells, with the exception of fructose-1,6-bisphosphatase (FBP1) (Table 3, Figure 2). Up-regulation of GAPDH is most statistically significant among all 10 glycolysis (G1-10 in Table 3) steps and 4 bypass (BP1-3 in Table 3) gluconeogenesis steps (1.07 fold increase and one-tailed t-test p = 1.44E-57). Approximately 75% or 35% of NSCLC displays both increased GAPDH expression and decreased FBP1 expression in the dataset using either the median or 25% quartile value as cut-off point. Furthermore, the expression of GAPDH and FBP1 are co-regulated with the up-regulation of the GACC gene FoxM1 (r = 0.77) in tumors (Figure 3).
Table 3. Gene expression of regulatory enzymes of glycolysis and gluconeogenesis.
| Pathway | Gene | Symbol | T/C | p value (t-test) | Probe Set ID |
| G1 | hexokinase 2 | HK2 | 1.05 | 0.0007 | 202934_at |
| G2 | glucose-6-phosphate isomerase | GPI | 1.11 | 5.81E-44 | 208308_s_at |
| G3 | phosphofructokinase, liver | PFKP | 1.14 | 8.71E-35 | 201037_at |
| G4 | aldolase A, fructose-bisphosphate | ALDOA | 1.07 | 3.26E-46 | 200966_x_at |
| G5 | triosephosphate isomerase 1 | TPI1 | 1.09 | 1.26E-45 | 200822_x_at |
| G6 | glyceraldehyde-3-phosphate dehydrogenase | GAPDH | 1.07 | 1.44E-57 | 212581_x_at |
| G7 | phosphoglycerate kinase 1 | PGK1 | 1.05 | 1.75E-28 | 200738_s_at |
| G7 | phosphoglycerate kinase 2 | PGK2 | 1.05 | 1.65E-12 | 217009_at |
| G8 | phosphoglycerate mutase family member 5 | PGAM5 | 1.1 | 3.27E-24 | 1555943_at |
| G8 | phosphoglycerate mutase 1 (brain) | PGAM1 | 1.04 | 1.18E-23 | 200886_s_at |
| G8 | phosphoglycerate mutase 2 (muscle) | PGAM2 | 1.05 | 2.92E-07 | 205736_at |
| G9 | enolase 1, (alpha) | ENO1 | 1.09 | 6.09E-45 | 201231_s_at |
| G9 | enolase 2 (gamma, neuronal) | ENO2 | 1.11 | 3.71E-16 | 201313_at |
| G9 | enolase 3 (beta, muscle) | ENO3 | 1.06 | 5.41E-10 | 204483_at |
| G10 | pyruvate kinase, muscle | PKM2 | 1.09 | 1.50E-25 | 201251_at |
| BP-1.1 | pyruvate carboxylase | PC | 1.15 | 1.90E-32 | 204476_s_at |
| BP-1.2 | phosphoenolpyruvate carboxykinase 1(soluble) | PCK1 | 1.01 | 0.0048 | 208383_s_at |
| BP-2 | fructose-1,6-bisphosphatase 1 | FBP1 | 0.84 | 8.45E-38 | 209696_at |
| BP-3 | glucose-6-phosphatase, catalytic subunit | G6PC | 1 | 0.1092 | 206952_at |
G1-G10: Glycolysis step 1 to step 10. BP1-3 = gluconeogenesis bypass step 1 to step 3.
T/C = expression level ratio between cancer and control.
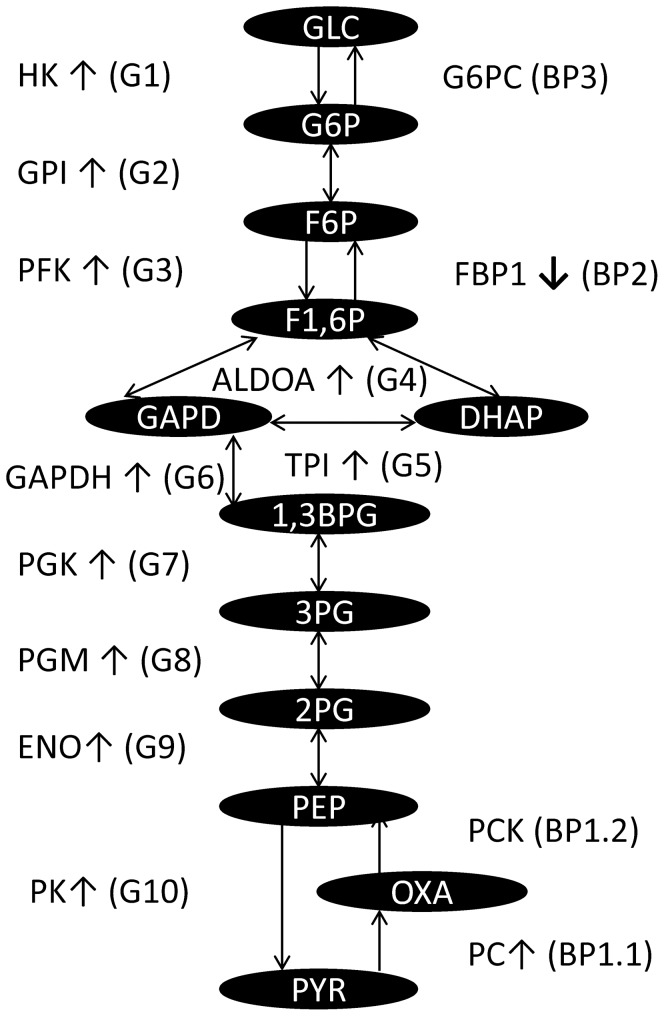
Figure 2. Gene expression regulation analysis of glycolysis and gluconeogenesis in NSCLC.
The Figure shows the pathways of cellular glycolysis and gluconeogenesis. Oval shape text represents the metabolites of the pathways. GLC = Glucose, G6P = Glucose-6-phosphate, F6P = Fructose 6-phosphate, F1,6P = Fructose 1,6-bisphosphate, GAPD = Glyceraldehyde 3-phosphate, DHAP = Dihydroxyacetone phosphate, 1,3-BPG = 1,3-Bisphosphoglyceric acid, 3PG = Glycerate 3-phosphate, 2PG = Glycerate 2-phosphate, PEP = Phosphoenolpyruvic acid, PYR = Pyruvic acid, OXA = Oxaloacetate. Gene symbols, regulation state (↑ or ↓ if detected) and pathway (in the parentheses, G1-10: glycolysis step 1–10, BP1-3: gluconeogenesis bypass step 1–3) are indicated.
Figure 3. Gene expression regulation analysis of GAPDH, FBP1 and FoxM1 in integrated NSCLC dataset.
Median values (A) or 25% quartile values (B) of gene expression are considered as cut-off points. The numbers of the value higher than cut-off point (GAPDH or FoxM1) or lower than cut-off point (FBP1) or various combinations are plotted for both tumors (black) and controls (white).
Relevance of up-regulated GAPDH positively correlated genes in NSCLC to regulation of cell senescence
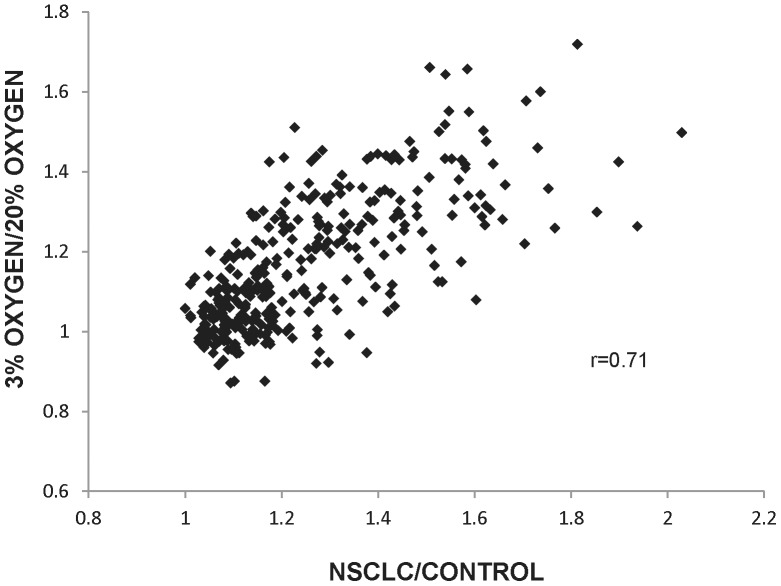
To further explore the correlation of the up-regulation of GACC genes and that of the other GAPDH positively correlated genes in NSCLC, we speculated that cancer may arise in part from a reduction in the normal regulation of senescence by genes associated with the cell cycle. We therefore examined gene expression levels in cultured cells in which anti-senescence regulation had been induced, and compared them to the genes whose up-regulated expression were found to be positively correlated with GAPDH expression in the NSCLC cohort. This comparison employed microarray dataset E-GEOD-19018, in which low passage IMR-90 human diploid fibroblasts had been cultured in 20% oxygen (normal replicative capacity) or 3% oxygen (increasing replicative capacity), after which RNA had been extracted and hybridized on microarrays. When the top 341 GAPDH positively correlated genes (r greater than or equal to 0.6) identified in the NSCLC dataset were compared with those from the cultured fibroblasts, the up-regulated GAPDH positively correlated genes (including at least 34% of the GACC genes) in both the NSCLC and the fibroblasts cultured in 3% oxygen (low oxygen tension and anti-senescence) were found to be similar (Figure 4). A good correlation (r = 0.71) was found between an expression signal ratio of cancer/control and the ratio of 3% oxygen/20% oxygen cultured cells in the expression of those genes.
Figure 4. Comparison of gene expression in NSCLC and anti-senescent cells.
Positive correlation coefficients between signal ratio of tumor/control in NSCLC dataset and signal ratio of 3% oxygen/20% oxygen cultured human diploid fibroblast dataset are plotted.
GACC genes are novel gene signatures for NSCLC and other solid tumors
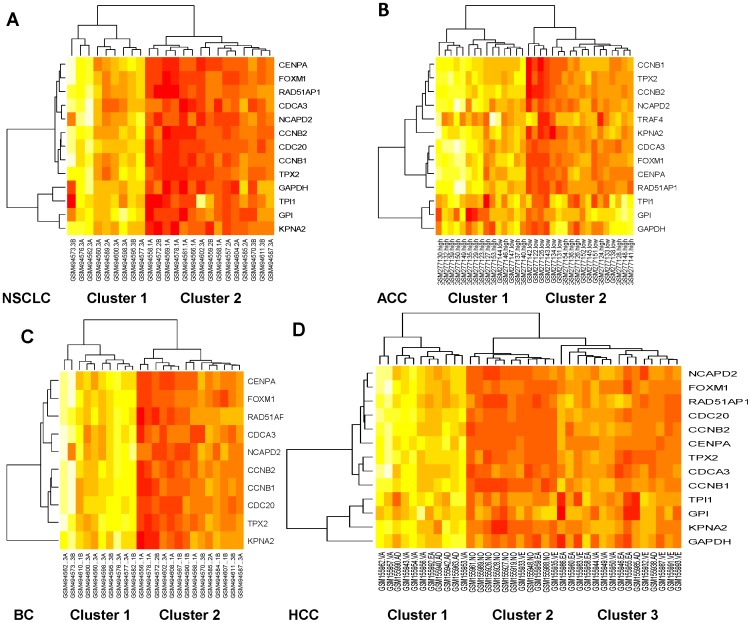
Using various datasets available to us, we evaluated whether GACC gene expression levels may be relevant to cancer stage and prognosis. As shown in the heat map plot in Figure 5, the top ranked GACC genes, as well as the glycolysis related genes, are clustered in the tumors. Furthermore, these genes can be used to distinguish different cancer stages.
Figure 5. Hierarchical clustering of GAPDH associated genes from various cancer cohorts.
The rows of a microarray heat map represent genes with each column of that row representing a different sample (source name followed by disease status). The gene expression values from four cohorts of NSCLCs with different tumor stages (I, II, III) (A), adrenocortical carcinomas (ACC) with tumor grades (high or low) (B), breast cancers (BC) with different tumor grades (I, II, III) (C), and Hepatocellular carcinomas (HCC) with different tumor stages (very early, early, advanced, very advanced) and normal liver (D) are clustered and presented by heat map.
Specifically, the cohorts from E-GEOD-19804, which include 24 NSCLCs with different tumor stages (I, II, III) that had been randomly selected from the original dataset (6 stage I, 6 stage II, and 12 stage III), are preferentially clustered according to stage. Cluster 1 is preferentially associated with more advanced stage tumors, as it has 8 stage III (89%) tumors and 1 stage II tumor. By contrast, cluster 2, which has 11 stage I/II (73%) and 4 stage III tumors, is enriched for early stage tumors (Figure 5A).
The cohorts from E-GEOD-10927, which include 33 adrenocortical cancers with tumor grades (high or low), can also be clustered. Cluster 1 is enriched for high-grade tumors, as it has 13 high grade (87%) and 2 low-grade tumors. On the other hand, cluster 2 is relatively enriched for low-grade tumors, as has 11 low-grade (61%) and 7 high-grade tumors (Figure 5B).
The cohorts from E-GEOD-29431, which contain 28 breast cancers with tumor grades (I, II, III), are clustered. Fourteen stage I/II and 14 stage III samples were randomly selected from original dataset. Cluster 1 has 8 stage III (73%) and 3 stage II tumors, while cluster 2 has 11 stage I/II (65%) and 6 stage III tumors (Figure 5C).
The cohorts from E-GEOD-6764, which contain hepatocellular carcinoma (HCC) and normal liver, are clustered. Cluster 1 is composed almost entirely of very advanced/advanced III HCC (91%), while cluster 3 has mainly very early/early tumors (67%). All 8 normal samples are in cluster 2 (73%), which also includes 2 very early and 1 early tumors (Figure 5D).
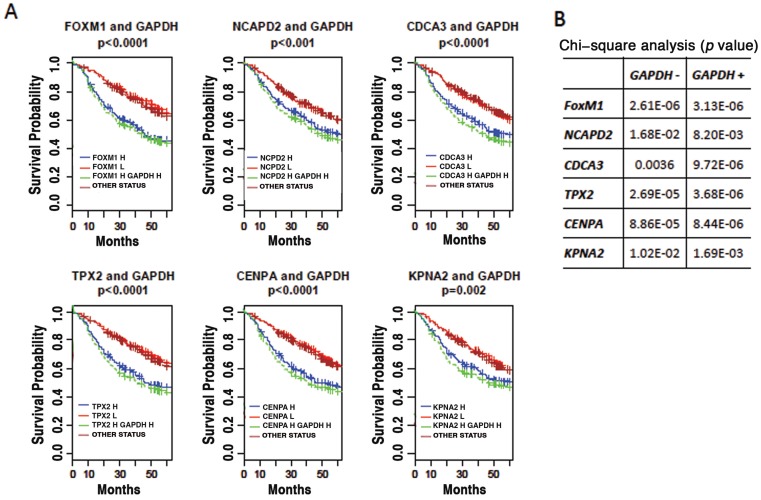
The pattern of gene clustering suggests that GACC related gene expression might be relevant as biomarkers to predict cancer prognosis. To evaluate this possibility, GACC gene expression levels were analyzed from a dataset of 442 lung adenocarcinomas [16] and correlated to 60-month survival outcomes. High expression of the top ranked GACC genes is associated with poor disease outcome (Figure 6A). Combining GACC gene expression level with GAPDH level can improve the predictive power of GACC gene level alone in most cases (Figure 6B).
Figure 6. Prognostic significance of up-regulation of top ranked GACC genes and GAPDH in lung adenocarcinomas.
(A) Director's Challenge cohort with 442 lung adenocarcinomas from caArray for Kaplan-Meier survival analysis. The up-regulation of all selected top ranked GACC gene is associated with poor prognosis. Combination of up-regulation of GAPDH with individual GACC gene improves the prediction power (high GACC gene expression and high GAPDH expression (green line) vs high GACC gene expression alone (blue line)). H = high. L = low. (B) Statistical significance of Kaplan-Meier survival analysis using individual GACC gene as marker (GAPDH−) or combination of GACC gene and GAPDH (GAPDH+) as markers. The table lists p values of chi-square analysis.
Discussion
In this study, a publically available microarray database [17] was employed to evaluate transcriptional levels of GAPDH and associated gene expression in solid tumors. The study data were analyzed using the Affymetrix GeneChip Human Genome U133 plus2 or U133A Array formats, which are commonly used for gene expression detection. Data analysis relied on a larger NSCLC cohort (total 330 samples) that combined independent data from various sources. Our identification of statistically significant changes in GAPDH expression in tumors enabled the evaluation of the gene expression profile that correlated with that of GAPDH.
Elevated levels of GAPDH have been observed in oncogene-induced transformation [18], angiogenesis [19] and anti-apoptotic function [20]–[22]. In other studies, however, GAPDH has been implicated in promoting apoptosis [23]–[25]. The reason for this paradox is poorly understood [26]. Barbini's work [27] has suggested that differential subcellular localization of GAPDH may contribute to its opposing biological activities in apoptotic and proliferating hepatocytes. The different functions of GAPDH may also be regulated by various levels of post-translational modification of the protein [28].
Our analysis shows that the profiles of GACC gene up-regulation are proportional to anti-senescence in the profiles in the tumors, rather than correlating with the promotion of senescence or apoptosis. We have identified a group of genes for which mRNA expression strongly correlates with GAPDH expression in tumor cells. In general, two such gene classes have been identified in the tumors. Class I consists of metabolic pathway related genes. Top ranked GAPDH positively associated genes in this class include triosephosphate isomerase 1 (TPI1) and glucose-6-phosphate isomerase (GPI), each of which encodes a key glycolytic enzyme. Class II covers cell cycle related genes that typically encode proteins involved in G2/M transition and M phase cell cycle regulation. Of these, the most informative protein appeared to be transcriptional factor Forkhead Box M1 (FoxM1), a crucial element for cell cycle regulation [29] and an important regulator of tumor metastasis [30]. FoxM1 associated genes CCNB2 (r = 0.75), CENPA (r = 0.75), AURKB (r = 0.73), BIRC5 (survivin r = 0.73), NEK2 (r = 0.69) are among the top GACC genes. The nuclear protein CENPF, which is a transcriptional target of FoxM1 (CENPF, r = 0.67), regulates the spindle assembly checkpoint to ensure proper chromosome stability and segregation during mitosis [31]. Up-regulation of these genes is consistently associated with high-expressions of GAPDH.
The mechanism by which GAPDH and FoxM1 may be co-regulated is unclear. In order to trigger cell cycle related events, it is possible that both GAPDH and FoxM1 translocate from the cytoplasm to the nucleus in cancer cells. Nuclear translocation of GAPDH may be regulated by acetylation [32]. FoxM1 is localized predominantly in the cytoplasm in late G1 and S phases. Nuclear translocation of FoxM1 occurs just before progression into the G2/M phase of the cell cycle and requires activity within the Raf/MEK/MAPK signaling pathway [33]. Both GAPDH and FoxM1 co-translocate to the nucleus during the G2/M transition phase through their interaction with other proteins. In this process, KPNA2 (importin alpha 1) may interact with GAPDH given that the correlation coefficient of KPNA2 expression with GAPDH in NSCLC is 0.75.
GACC genes are up-regulated in the cells with 3% oxygen incubation, which have greater replication capacity (anti-senescence) than those cultured in 20% oxygen. This result is in agreement with recent studies demonstrating that GAPDH depletion switches human tumor cells to a senescent phenotype [34]. Rescue experiments that employed metabolic and genetic models confirmed that GAPDH has important regulatory functions that link energy metabolism and cell cycle networks. In sum, our data suggest that GAPDH serves a key role in cell cycle regulation and cell senescence during cancer cell development.
In addition to NSCLC, the up-regulation of GAPDH and associated genes, including GACC genes, was observed in other tumor types. The results suggest that transcription of GAPDH in tumors is an important step in cancer development, where it may contribute to increased cell cycle-related cell proliferation. This process may also include aberrant glycolysis and gluconeogenesis, as most genes in both pathways are up-regulated. However, the gluconeogenesis gene FBP1 is down-regulated in the tumors. During normal glucose metabolism, excess GAPDH is continuously metabolized by glycolysis to pyruvic acid, which is then converted by FBP1 to fructose-1,6-bisphosphate during gluconeogenesis. In cancer, down-regulation of FBP1 can result in GAPDH accumulation within the cytoplasm and may also cause translocation of excess GAPDH to the nucleus. Down-regulation of FBP1 in cancer cells has been reported recently [35], [36]. FBP1 is considered a tumor suppressor gene in gastric cancer cells and down-regulation of FBP1 that is mediated by promoter hypermethylation is found in human hepatocellular carcinoma and colon cancer. Over-expression of GAPDH in the tumors may connect aberrant glucose metabolism with cell proliferation. However, our analysis cannot exclude the possibility that the observed up-regulation of both GAPDH and GACC genes may be a secondary effect of the cancer that is attributable to the high-energy demands required for rapid growth. While apoptosis is inhibited, cellular metabolism is increased and key metabolic pathways are activated. In addition, this study has been limited to measuring transcriptional levels based on the microarray data, and cannot establish a correlation between gene expression and cellular protein levels within the high metabolic environments. Further experimentation is required to address these issues.
Our data indicate that up-regulation of GACC genes within tumor cells is proportional to their malignant status, and thus can be a potential predictor of disease prognosis. Based on the GAPDH positively associated gene expression levels, the subclass of NSCLC, adrenocortical carcinoma, breast cancer and HCC can also be classified to various stages. The up-regulation of GAPDH positively associated genes correlates with more severe and/or later stages of cancer. Most importantly, using both GAPDH transcription and GACC gene expression levels in survival analysis greatly improve the predictive power of using GACC gene level alone in most cases.
Methods
The Gene expression microarray analyses reported in this study employed data from ArrayExpress (http://www.ebi.ac.uk/arrayexpress) of the European Institute of Bioinformatics (EBI) and caArray (https://array.nci.nih.gov/caarray/home.action) of the National Cancer Institute (NCI), both of which are publicly available. The analyses included independent cohorts from ArrayEaxpress containing NSCLC (E-GEOD-18842, E-GEOD-19188 and E-GEOD-19804), adrenocortical cancer (E-GEOD-10927), breast cancer (E-GEOD-29431), hepatocellular carcinoma (HCC) (E-GEOD-6764), cell lines in various conditions (E-GEOD-19018) and a cohort from caArray containing lung adenocarcinomas (jacob-00182) for Affymetrix GeneChip Human Genome U133 plus 2 or U133A Arrays. The CEL files containing the raw data from each experiment were directly downloaded from the EBI or NCI website with particular accession number. Data were then normalized with CEL file quality control evaluation using 3′ Expression Arrays Robust Multi-array Analysis (RMA) from the Affymetrix software Expression Console (http://www.affymetrix.com). The normalized expression values represent the probe set intensity on a log-2 scale. The integrated NSCLC dataset from ArrayExpress include three independent NSCLC datasets for analysis, in which all CEL files from these sources were combined for RMA analysis.
The student's t-test (one-tailed) and Pearson's correlation coefficient calculation were carried out using Microsoft Excel. P values of t-test less than 0.05 were considered as statistically different. Chi Square test (chisq.test), heat map drawing (heatmap) and Kaplan-Meier survival analysis were carried out using the open source statistical tool R (version 2.14.1) (Supporting Information). In the gene expression analyses, the value of a selected gene expression level was compared with the median value or 25% quartile value of the gene expression in each cohort. The numbers higher or lower than the median or 25% quartile value are plotted in the results. For survival analysis, values higher or lower than median in each gene group were placed in “high”, “low”, or different combinations for the analysis. All survival times have been adjusted to months.
Supporting Information
Data file for heat map (HCCdata.csv).
(CSV)
Data file for survival analysis (TPX2survival.csv).
(CSV)
R source code for bioinformatics analysis.
(DOCX)
Acknowledgments
We thank Drs. Jeffrey D. Kittendorf and Thomas C. Johnson of AQLS Inc., Drs. Nicholas Chi-Kwan Ling, Degang Zhong and Roger Anderson for helpful discussion of the manuscript. We thank the Affymetrix technical support team for consultations on data analysis.
Funding Statement
This research was supported in part by the Intramural Research Program, National Institutes of Health, National Cancer Institute, Center for Cancer Research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Gresner P, Gromadzinska J, Wasowicz W (2009) Reference genes for gene expression studies on non-small cell lung cancer. Acta Biochim Pol 56: 307–316. [PubMed] [Google Scholar]
- 2. Schek N, Hall BL, Finn OJ (1988) Increased glyceraldehyde-3-phosphate dehydrogenase gene expression in human pancreatic adenocarcinoma. Cancer Res 48: 6354–6359. [PubMed] [Google Scholar]
- 3. Warburg O (1956) On the origin of cancer cells. Science 123 (3191) 309–314. [DOI] [PubMed] [Google Scholar]
- 4. Vander Heiden MG, Cantley LC, Thompson CB (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324: 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dang CV (2012) Links between metabolism and cancer. Genes Dev 26: 877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bayley JP, Devilee P (2012) The Warburg effect in 2012. Curr Opin Oncol 24: 62–67. [DOI] [PubMed] [Google Scholar]
- 7. Hamanaka RB, Chandel NS (2012) Targeting glucose metabolism for cancer therapy. J Exp Med 209: 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Taubes G (2012) Cancer research. Ravenous for glucose. Science 335: 31. [DOI] [PubMed] [Google Scholar]
- 9. Denko N, Schindler C, Koong A, Laderoute K, Green C, et al. (2000) Epigenetic regulation of gene expression in cervical cancer cells by the tumor microenvironment. Clin Cancer Res 6: 480–487. [PubMed] [Google Scholar]
- 10. Colell A, Ricci JE, Tait S, Milasta S, Maurer U, et al. (2007) GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell 129: 983–997. [DOI] [PubMed] [Google Scholar]
- 11. Bui M, Dimitriadis EK, Hoischen C, An E, Quénet D, et al. (2012) Cell-cycle-dependent structural transitions in the human CENP-A nucleosome in vivo. 150: 317–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Obama K, Satoh S, Hamamoto R, Sakai Y, Nakamura Y, et al. (2008) Enhanced expression of RAD51 associating protein-1 is involved in the growth of intrahepatic cholangiocarcinoma cells. Clin Cancer Res 14: 1333–1339. [DOI] [PubMed] [Google Scholar]
- 13. Mazumdar M, Sundareshan S, Misteli T (2004) Human chromokinesin KIF4A functions in chromosome condensation and segregation. J Cell Biol 6: 613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cai J, Gibbs E, Uhlmann F, Phillips B, Yao N, et al. (1997) A complex consisting of human replication factor C p40, p37, and p36 subunits is a DNA-dependent ATPase and an intermediate in the assembly of the holoenzyme. J Biol Chem 272: 18974–18981. [DOI] [PubMed] [Google Scholar]
- 15. Pavloff N, Rivard D, Masson S, Shen SH, Mes-Masson AM (1992) Sequence analysis of the large and small subunits of human ribonucleotide reductase. DNA Seq 2: 227–234. [DOI] [PubMed] [Google Scholar]
- 16. Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, et al. (2008) Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med 14: 822–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brooksbank C, Camon E, Harris MA, Magrane M, Martin MJ, et al. (2003) The European Bioinformatics Institute's data resources. Nucleic Acids Res 31: 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Persons DA, Schek N, Hall BL, Finn OJ (1989) Increased expression of glycolysis-associated genes in oncogene-transformed and growth-accelerated states. Mol Carcinog 2: 88–94. [DOI] [PubMed] [Google Scholar]
- 19. Deindl E, Boengler K, van Royen N, Schaper W (2002) Differential expression of GAPDH and beta3-actin in growing collateral arteries. Mol Cell Biochem 236: 139–146. [DOI] [PubMed] [Google Scholar]
- 20. Lavallard VJ, Pradelli LA, Paul A, Bénéteau M, Jacquel A, et al. (2009) Modulation of caspase-independent cell death leads to resensitization of imatinib mesylate-resistant cells. Cancer Res 69: 3013–3020. [DOI] [PubMed] [Google Scholar]
- 21. Azam S, Jouvet N, Jilani A, Vongsamphanh R, Yang X, et al. (2008) Human glyceraldehyde-3-phosphate dehydrogenase plays a direct role in reactivating oxidized forms of the DNA repair enzyme APE1. J Biol Chem 283: 30632–30641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sirover MA (1999) New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochim Biophys Acta 1432: 159–184. [DOI] [PubMed] [Google Scholar]
- 23. Barbini L, Rodríguez J, Dominguez F, Vega F (2007) Glyceraldehyde-3-phosphate dehydrogenase exerts different biologic activities in apoptotic and proliferating hepatocytes according to its subcellular localization. Mol Cell Biochem 300: 19–28. [DOI] [PubMed] [Google Scholar]
- 24. Chuang DM, Ishitani R (1996) A role for GAPDH in apoptosis and neurodegeneration. Nat Med 2: 609–610. [DOI] [PubMed] [Google Scholar]
- 25. Sawa A, Khan AA, Hester LD, Snyder SH (1997) Glyceraldehyde-3-phosphate dehydrogenase: nuclear translocation participates in neuronal and nonneuronal cell death. Proc Natl Acad Sci U S A 94: 11669–11674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Colell A, Green DR, Ricci JE (2009) Novel roles for GAPDH in cell death and carcinogenesis. Cell Death Differ 16: 1573–1581. [DOI] [PubMed] [Google Scholar]
- 27. Barbini L, Rodríguez J, Dominguez F, Vega F (2007) Glyceraldehyde-3-phosphate dehydrogenase exerts different biologic activities in apoptotic and proliferating hepatocytes according to its subcellular localization. Mol Cell Biochem 300: 19–28. [DOI] [PubMed] [Google Scholar]
- 28. Lee SY, Kim JH, Jung H, Chi SW, Chung SJ, et al. (2012) Glyceraldehyde-3-phosphate, a glycolytic intermediate, prevents cells from apoptosis by lowering S-nitrosylation of glyceraldehyde-3-phosphate dehydrogenase. J Microbiol Biotechnol 22: 571–573. [DOI] [PubMed] [Google Scholar]
- 29. Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, et al. (2005) Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol 25: 10875–10894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Raychaudhuri P, Park HJ (2011) FoxM1: a master regulator of tumor metastasis. Cancer Res 71: 4329–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Costa RH (2005) Nat Cell Biol. FoxM1 dances with mitosis 7: 108–110. [DOI] [PubMed] [Google Scholar]
- 32. Ventura M, Mateo F, Serratosa J, Salaet I, Carujo S, et al. (2010) Nuclear translocation of glyceraldehyde-3-phosphate dehydrogenase is regulated by acetylation. Int J Biochem Cell Biol 42 (10) 1672–1680. [DOI] [PubMed] [Google Scholar]
- 33. Ma RY, Tong TH, Leung WY, Yao KM (2010) Raf/MEK/MAPK signaling stimulates the nuclear translocation and transactivating activity of FOXM1. Methods Mol Biol 647: 113–123. [DOI] [PubMed] [Google Scholar]
- 34. Phadke M, Krynetskaia N, Mishra A, Krynetskiy E (2011) Accelerated cellular senescence phenotype of GAPDH-depleted human lung carcinoma cells. Biochem Biophys Res Commun 411: 409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu X, Wang X, Zhang J, Lam EK, Shin VY, et al. (2010) Warburg effect revisited: an epigenetic link between glycolysis and gastric carcinogenesis. Oncogene 29: 442–450. [DOI] [PubMed] [Google Scholar]
- 36. Chen M, Zhang J, Li N, Qian Z, Zhu M, et al. (2011) Promoter hypermethylation mediated downregulation of FBP1 in human hepatocellular carcinoma and colon cancer. PLoS One 6: e25564. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data file for heat map (HCCdata.csv).
(CSV)
Data file for survival analysis (TPX2survival.csv).
(CSV)
R source code for bioinformatics analysis.
(DOCX)