Abstract
Background
Autism is a neurodevelopmental disorder with a high estimated heritability. ATP2B2, located on human chromosome 3p25.3, encodes the plasma membrane calcium-transporting ATPase 2 which extrudes Ca2+ from cytosol into extracellular space. Recent studies reported association between ATP2B2 and autism in samples from Autism Genetic Resource Exchange (AGRE) and Italy. In this study, we investigated whether ATP2B2 polymorphisms were associated with autism in Chinese Han population.
Methods
We performed a family based association study between five SNPs (rs35678 in exon, rs241509, rs3774180, rs3774179, and rs2278556 in introns) in ATP2B2 and autism in 427 autism trios of Han Chinese descent. All SNPs were genotyped using the Sequenom genotyping platform. The family-based association test (FBAT) program was used to perform association test for SNPs and haplotype analyses.
Results
This study demonstrated a preferential transmission of T allele of rs3774179 to affected offsprings under an additive model (T>C, Z = 2.482, p = 0.013). While C allele of rs3774179 showed an undertransmission from parents to affected children under an additive and a dominant model, respectively (Z = −2.482, p = 0.013; Z = −2.591, p = 0.0096). Haplotype analyses revealed that three haplotypes were significantly associated with autism. The haplotype C-C (rs3774180–rs3774179) showed a significant undertransmission from parents to affected offsprings both in specific and global haplotype FBAT (Z = −2.037, p = 0.042; Global p = 0.03). As for the haplotype constructed by rs3774179 and rs2278556, C-A might be a protective haplotype (Z = −2.206, p = 0.027; Global p = 0.04), while T-A demonstrated an excess transmission from parents to affected offsprings (Z = 2.143, p = 0.032). These results were still significant after using the permutation method to obtain empirical p values.
Conclusions
Our research suggested that ATP2B2 might play a role in the etiology of autism in Chinese Han population.
Introduction
Autism is a neurodevelopmental disorder characterized by severe impairment in reciprocal social interaction and communication skills, and the presence of stereotyped behaviors and interests [1]. Previous researches reported that autism spectrum disorders (ASD) occurred in 1 out of 150 individuals [2]. Several lines of evidence suggested that strong genetic components were involved in the susceptibility to ASD [3]–[5]. Twin studies indicated that the estimated heritability of autism was nearly 90%. However, autism is a common disorder with multiple genetic causes. Recently, genome wide association studies (GWA) identified susceptibility common genetic variants (i.e. rs4307059 in 5p14.1 [6], rs10513025 in 5p15.2 [7], rs4141463 in the intron of MACROD2 [8]) for ASD which support the “common disease, common variant” hypothesis of this complex disease [9] . However, no definitely replicated results were reported across literatures until now. Common variants dredged from simple GWAS analysis could not explain a substantial fraction of the heritability of ASD [10]. It seems that a myriad of common variations of very small effect impacts ASD liability [11] . Rare variants such as mutations and/or copy number variants (CNV) might attributable to the “missing heritability” [12] . These lines of evidence support the other hypothesis “common disease, rare variant”, which proposes that many rare variants and/or structural variants (i.e. CNV) might contribute to the risk of common disease [13]–[15] . Candidate gene approaches are also useful to detect the common and/or rare variants associated with ASD.
ATP2B2 (ATPase, calcium-transporting, plasma membrane 2) encodes plasma membrane calcium-transporting ATPase isoform 2 (PMCA2). PMCA2 belongs to the family of P-type primary ion transport ATPases which remove bivalent calcium ions from eukaryotic cells against very large concentration gradients and play a critical role in intracellular calcium homeostasis. Intracellular calcium signaling is important for the development of specific synaptic connections. Disruptions of intracellular calcium homeostasis underlie a host of emerging diseases such as seizures and autism [16]. Evidence from biochemical and genetic studies of the mitochondrial aspartate/glutamate carrier AGC1 indicated that AGC and altered Ca2+ homeostasis play a key role in the cascade of signaling events leading to autism [17]. Therefore, understanding the role of calcium in setting up brain networks may help identifying the causes of diseases such as autism.
Furthermore, ATP2B2 is expressed mainly in cerebellum, and also expressed in cerebral cortex, olfactory bulb, and hippocampal formation [18]–[22]. Previous researches have demonstrated that absence of ATP2B2 led to defects of both the vestibular and auditory systems [23]–[27]. Moreover, several studies suggested that ATP2B2 plays an important role in synaptic signaling and functional cerebellar output [28]–[30]. And ATP2B2 had a strong influence on development and maintenance of the cerebellar function [31].
ATP2B2 is located in human chromosome region 3p25.3. Several studies identified region of suggestive linkage with autism on chromosome 3p25 [32]–[34]. There might reside some susceptibility genes for autism. Previous GWAS did not detect the association between ATP2B2 and autism [6]–[8] . Recently, Carayol J, et al. performed a family-based association study between ATP2B2 and autism. They firstly provided the evidence for a susceptibility gene ATP2B2 for autism [35]. Then this result was replicated in another research in Italy [36]. These researches indicated that ATP2B2 might be a predisposing gene for autism.
The biological functions of ATP2B2 and the positive results of previous association made ATP2B2 become an attractive candidate gene for autism. Considering the genetic heterogeneity in different ethnicities, we performed a family-based association study to investigate the association between ATP2B2 and autism in Chinese Han population.
Materials and Methods
Ethics Statement
This research was approved by the Ethics Committee of Institute of Mental Health, Peking University. All subjects provided written informed consent, and informed written consent for children was obtained from their biological parents (the children’s legal guardians).
Subjects
We performed a family based association study in 427 autism trios of Han Chinese descent (1281 individuals, including probands and their biological parents). All subjects were recruited at the Institute of Mental Health, Peking University, China. Of the 427 autistic children, 403 were male and 24 were female. The mean age of the children at the time of testing was 6.3 years old (ranging from 2 to 16.7 years). These children were diagnosed with autism by two senior psychiatrists, and all children met the DSM-IV criteria for autism. Besides, these autistic children were assessed using Autism Behavior Checklist (ABC) [37] and Childhood Autism Rating Scale (CARS) [38]. Children who had scores ≥53 on ABC and ≥35 on CARS were included in the present study. The demographic data were listed in Table S1 . Children with Asperger’s syndrome, Rett syndrome, phenylketonuria, fragile X syndrome, tuberous sclerosis and a previously identified chromosomal abnormality were excluded. Children with pervasive developmental disorder not otherwise specified (PDD-NOS) which also called atypical autism were not included in this study. As for PDD-NOS, the criteria for autistic disorder were not met, for instance because of late age of onset, atypical symptomatology, or subthreshold symptomatology, or all of these.
SNP Selection and Genotyping
Genomic DNA was extracted from blood samples using the QIAamp DNA Blood Mini Kit. The SNPs information was obtained from dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/) and HapMap phaseII genotype Chinese Han in Beijing (CHB) dataset (http://www.hapmap.org). Previous studies had reported that four SNPs (rs35678 in exon, rs241509, rs3774180, and rs2278556 in introns) were associated with autism [35], [36]. To explore whether these results could be replicated in Chinese Han population, we selected the above-mentioned four SNPs (rs35678 (NC_000003.11:g.10379923C>T) in exon, rs241509 (NC_000003.11:g.10387059C>A), rs3774180 (NC_000003.11:g.10396988T>C), and rs2278556 (NC_000003.11:g.10402103G>A) in introns) and another SNP rs3774179 (NC_000003.11:g.10397069T>C) which is close to rs3774180 in the present association study. The minor allele frequencies (MAF) of these five SNPs in Chinese Han population were more than 0.05.
All SNPs were genotyped using the iPLEX Assay on the Sequenom MassARRAY platform (Sequenom Inc, San Diego, California), which uses the matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry primer extension assay [39], [40]. Primers were designed by Sequenom online service (http://www.sequenom.com). Information of primers was listed in Table 1. We used iPLEX genotyping assay, which has increased plexing efficiency and flexibility for the MassARRAY system through single base primer extension with mass-modified terminators [41]. Polymerase chain reaction (PCR) was performed according to the standard protocol. Spectra were analyzed using MassARRAY Typer, version 4.0 software (Sequenom). Quality control was performed by excluding individual SNPs or samples with genotype call rates less than 95% and SNP assays with poor-quality spectra/cluster plots.
Table 1. The information of SNPs in ATP2B2 and primers of PCR and unextended primers for Sequenom SNP genotyping.
| SNP | Location | 2nd-PCR | 1st-PCR | UEP_SEQ |
| rs35678 | Exon | ACGTTGGATGAACGAGGACGTGGAGGAGAT | ACGTTGGATGTGTCTGGATCCGATTCAGGC | GTGTAGGAGATCGACCACGC |
| rs241509 | Intron | ACGTTGGATGCGTGATTGTGGCCTTCACAG | ACGTTGGATGTCCCCCAGGCTTTGGTGTG | ACTTCATCACGCAGGTGGGTCC |
| rs3774180 | Intron | ACGTTGGATGAAACTGAGGCCATCTGTGAG | ACGTTGGATGACTCAAGTGACACAGCAGAC | GTGGCAGGGGCAGAA |
| rs3774179 | Intron | ACGTTGGATGTTCCTTCAGTCAGGGCATCC | ACGTTGGATGGTGAGACAGTTTATGACTCC | GGGGATCCTGTTGGCTCACCCCTGGG |
| rs2278556 | Intron | ACGTTGGATGAAAATCTGAGGCAAGTACGG | ACGTTGGATGGTTACGTGCCTATCATCCAG | ACGGCATAATGTTAAGATG |
SNP, single nucleotide polymorphism; PCR, polymerase chain reaction; UEP, unextended primer.
To confirm the genotype results by Sequenom, all these five SNPs were regenotyped in 10% of the samples. Direct DNA sequencing was used for analyzing rs35678, rs3774180, rs3774179 and rs2278556, which do not have natural restriction sites. The SNP rs241509 was analyzed by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis. Details for PCR-RFLP and direct sequencing were described in the Text S1.The information of primers and PCR-RFLP analysis were listed in Table S2. Raw data are available upon request. All DNA sequencing data have been deposited in GenBank. The genotyping concordance rate for Sequenom and direct sequencing was more than 99%, and that was 96% for Sequenom and PCR-RFLP.
Statistical Analysis
Prior to analysis, Mendelian inconsistencies were checked using the PEDCHECK program, version 1.1 [42]. The Hardy-Weinberg equilibrium for genotype frequency distributions was tested using the Chi-square goodness-of-fit test. We used the Haploview 4.1 (http://www.broad.mit.edu/mpg/haploview/) to calculate the pair-wise D′ values for SNPs and construct haplotypes. SNP pairs were considered to be in strong linkage disequilibrium (LD) if D′>0.8. The family-based association test (FBAT) program version 2.0.4 (beta1) (http://www.biostat.harvard.edu/~fbat/default.html) was used to perform association test for SNPs under additive model and dominant model [43]. Moreover, FBAT program provided estimated haplotype frequencies and pairwise LD between the specific markers. The individual haplotype tests were conducted under “biallelic” mode in haplotype based association test (HBAT). Meanwhile, the global haplotype tests of association were performed under “multiallelic” mode in HBAT. The significance level for all statistical tests was two-tailed (p<0.05).
The power of sample size for association tests was evaluated using Quanto software version 1.2.4 (http://hydra.usc.edu/gxe/) [44] . The sample size of 427 trios had approximately 82% power to detect risk allele frequencies ranging from 0.191 to 0.451, assuming a relative risk of 1.4, the prevalence of autism of 0.006, with additive model.
Correction for multiple testing was conducted using Bonferroni correction (α/N = 0.05/5 = 0.01). However, Bonferroni correction is too conservative to detect small or moderate effects. Considering the inter-correlation of the SNPs, SNPSpD (http://genepi.qimr.edu.au/general/daleN/SNPSpD) program was used as a simple correction for multiple testing of SNPs in LD with each other [45] . SNPSpD, on the basis of the spectral decomposition (SpD) of matrices of pairwise LD between SNPs, automatically removed redundant SNPs (i.e., SNPs in completed LD (r2 = 1) with another SNP) [46] . In our data, the effective number of independent marker loci calculated by SNPSpD was 3. The calculated significance threshold required to keep type I error rate at 5% was 0.01695.
Results
Five SNPs (rs35678, rs241509, rs3774180, rs3774179, and rs2278556) in ATP2B2 were genotyped in 427 autism trios (1281 individuals). The call rates of individual SNPs and samples were more than 98%. All SNP assays were with good quality cluster plots.
All of these five SNPs were polymorphic with minor allele frequencies more than 5%. Therefore, these SNPs were used as genetic markers in this study. None of the genotype distributions of these SNPs in parents significantly deviated from Hardy-Weinberg equilibrium (p>0.05). The allele frequencies, genotype frequencies, and p value for Hardy-Weinberg equilibrium of these five SNPs in children affected with autism and parents were shown in Table S3.
Univariate (single marker) test using FBAT demonstrated that T alleles of rs3774179 showed a preferential transmission from parents to children affected with autism (T>C, Z = 2.482, p = 0.013, under an additive model). While C allele of rs3774179 showed a significant undertransmission from parents to affected children under an additive model and a dominant model, respectively (Z = −2.482, p = 0.013; Z = −2.591, p = 0.0096). Allele frequencies and the association results for single SNP analyses were shown in Table 2. The results were similar in trios with male probands (data not shown).
Table 2. Results of association between 5 SNPs in ATP2B2 and autism and haplotype analyses in 427 trios of Han Chinese descent.
| Marker | Allele | Afreq | Additive model | Dominant model | ||||||||
| Families | S- E(S) | Var(S) | Z | p | Families | S- E(S) | Var(S) | Z | p | |||
| rs35678 | C | 0.429 | 287 | −9 | 94.5 | −0.926 | 0.355 | 216 | −4.8 | 48.3 | −0.683 | 0.494 |
| T | 0.571 | 287 | 9 | 94.5 | 0.926 | 162 | 4.2 | 34.8 | 0.720 | 0.471 | ||
| rs241509 | A | 0.549 | 302 | −1.5 | 102.8 | −0.148 | 0.882 | 181 | 7.2 | 38.4 | 1.169 | 0.242 |
| C | 0.451 | 302 | 1.5 | 102.8 | 0.148 | 230 | 8.7 | 50.7 | 1.229 | 0.219 | ||
| rs3774180 | C | 0.551 | 296 | −6.5 | 98.3 | −0.656 | 0.512 | 173 | −1.8 | 37.2 | −0.287 | 0.774 |
| T | 0.449 | 296 | 6.5 | 98.3 | 0.656 | 220 | 4.7 | 48.9 | 0.679 | 0.497 | ||
| rs3774179 | C | 0.191 | 211 | −19.5 | 61.8 | −2.482 | 0.013 | 202 | −18 | 48.3 | −2.591 | 0.0096 |
| T | 0.809 | 211 | 19.5 | 61.8 | 2.482 | 45 | 1.5 | 9.0 | 0.500 | 0.617 | ||
| rs2278556 | A | 0.595 | 305 | 1.5 | 95.3 | 0.154 | 0.878 | 149 | −5.5 | 32.5 | −0.965 | 0.335 |
| G | 0.405 | 305 | −1.5 | 95.3 | −0.154 | 232 | −7.0 | 53.3 | −0.959 | 0.337 | ||
| Marker | Haplotype | Afreq | Families | S- E(S) | Var(S) | Z | p | Global | p permutation a | p permutation (whole marker) | ||
| χ2 | p | |||||||||||
| rs3774180–rs3774179 | ||||||||||||
| C-C | 0.185 | 198.6 | −15.8 | 60.3 | −2.037 | 0.042 | 8.95 | 0.030 | 0.042 | 0.040 | ||
| C-T | 0.362 | 271.1 | 9.3 | 86.9 | 1.000 | 0.317 | 0.316 | |||||
| T-T | 0.447 | 286.5 | 9.7 | 96.5 | 0.985 | 0.325 | 0.320 | |||||
| rs3774179–rs2278556 | ||||||||||||
| C-A | 0.183 | 201.8 | −17.0 | 59.4 | −2.206 | 0.027 | 8.32 | 0.040 | 0.027 | 0.035 | ||
| T-A | 0.410 | 279.1 | 20.5 | 91.5 | 2.143 | 0.032 | 0.032 | |||||
| T-G | 0.400 | 294.5 | −1 | 92.8 | −0.104 | 0.917 | 0.905 | |||||
| rs3774180–rs3774179–rs2278556 | ||||||||||||
| C-C-A | 0.182 | 197.2 | −16.1 | 58.2 | −2.117 | 0.034 | 9.14 | 0.104 | 0.033 | 0.202b | ||
| C-T-A | 0.337 | 246.1 | 13.1 | 80.6 | 1.464 | 0.143 | 0.149 | |||||
| T-T-G | 0.371 | 260.4 | 3.6 | 88.3 | 0.375 | 0.708 | 0.686 | |||||
| T-T-A | 0.074 | 102.8 | 6.1 | 28.0 | 1.155 | 0.248 | 0.249 | |||||
Afreq, allele frequency; Families, number of informative families; S, test statistics for the observed number of transmitted alleles; E(S), expected value of S under the null hypothesis (i.e., no linkage and no association).
The number of permutationS is 10,000;
whole marker permutation test using minimal p (when only one haplotype was significant).
The LD patterns of the SNPs were measured with D primer value using Haploview. Four SNPs (rs241509, rs3774180, rs3774179 and rs2278556) were identified in one LD block with D’ value range from 0.77 to 0.95 (Figure 1).
Figure 1. Linkage disequilibrium (LD) block constructed from 5 SNPs in ATP2B2.
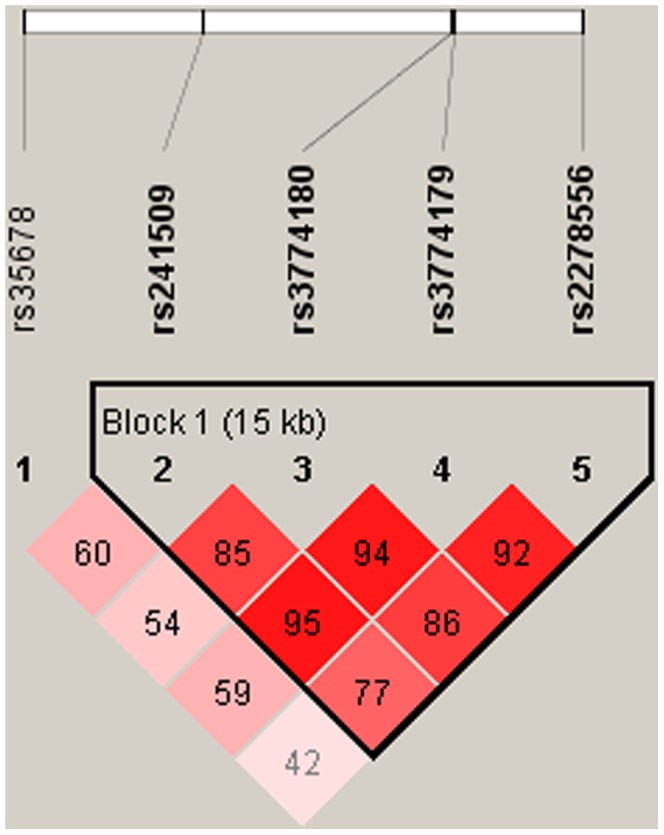
Markers with Linkage disequilibrium (LD) (D’<1 and LOD>2) are shown in red through pink (color intensity decreases with decreasing D’ value). D’ value shown within each square represents a pairwise LD relationship between the two polymorphisms. D prime values of 1.0 are never shown (the box is empty). The LD plot was generated with the Haploview program.
Bonferroni correction for multiple testing (5 tests) resulted in a significance level of p = 0.01, and only the result for rs3774179 under a dominant model remained significant. Considering Bonferroni correction is too conservative, SNPSpD approach was used as a simple correction for multiple testing of SNPs in LD with each other. The calculated significance threshold required to keep type I error rate at 5% was 0.01695. Therefore, rs3774179 was significantly associated with autism both under an additive model (p = 0.013) and a dominant model (p = 0.0096) after correction of multiple comparisons.
To determine whether any specific haplotypes would confer a higher risk for autism, the specific and global haplotype association tests were performed. Haplotypes were constructed by neighboring SNPs using a sliding window approach. Three haplotypes were significantly associated with autism (Table 2). The haplotype C-C (rs3774180–rs3774179) showed a significant undertransmission from parents to affected offsprings both in specific and global haplotype FBAT (Z = −2.037, p = 0.042; Global p = 0.03). As for the haplotype constructed by rs3774179 and rs2278556, C-A might be a protective haplotype (Z = −2.206, p = 0.027; Global p = 0.04), while T-A demonstrated an excess transmission from parents to affected offsprings (Z = 2.143, p = 0.032). These results were still significant after using the permutation method to obtain empirical p values (Table 2). The haplotype C-C-A (rs3774180–rs3774179–rs2278556) revealed an undertransmission from parents to children affected with autism in the specific HBAT (Z = −2.117, p = 0.034, ppermutation = 0.033). However, the global p value of haplotypes encompassing rs3774180–rs3774179–rs2278556 did not show significant (p = 0.104). Although rs3774179 might contribute to the association of these haplotypes, haplotypes could capture the combined effects of tightly linked cis-acting causal variants [47] . Other results of haplotype analyses for haplotype constructed by the remaining SNPs were shown in Table S4. When performed association study in trios with male probands, the results had similar trend (data not shown).
Discussion
In this present study, we investigated the association between ATP2B2 polymorphisms and autism in Chinese Han population. The results of our research indicated that rs3774179 in ATP2B2 was significantly associated with autism (p = 0.013, under an additive model; p = 0.0096, under a dominant model). Haplotype analyses showed that three haplotypes were significantly associated with autism, all of which including rs3774179.
Two previous family-based association studies explored the association between ATP2B2 and ASD. One reported that SNPs (rs241509, rs3774180 and rs2278556) represented positive association with autism in their exploratory AGRE samples and in the replication samples from Italy [35]. Moreover, they did meta-analysis of the exploratory and replication results in the recessive model after correction for multiple testing, observing strongly significant association between rs35678 (p = 0.0023), rs241509 (p = 0.00026), rs3774180 (p = 0.000018), rs2278556 (p = 0.00013) and autism [35]. In the other research based on Italian ASD families, results showed that homozygotes of rs35678 (individuals with genotype CT were at lower risk compared with CC or TT individuals, p = 0.0013) were associated with ASD [36]. However, these results were not replicated in our samples in Chinese Han population. We compared the allele frequencies of these SNPs in different populations using the data downloaded from Hapmap data (Table S5). For rs35678, rs241509, rs3774180, and rs3774179, the allele frequencies in Chinese Han in Beijing, China (CHB) population were similar to those in Utah residents with Northern and Western European ancestry from the CEPH collection (CEU) population. However, the allele frequency of rs2278556 was 0.440 for G allele in CHB population while it was 0.624 in CEU population.
We detected that rs3774179 which is near to rs3774180 was significantly associated with autism. Previous research by Carayol J, et al. reported that rs3774180 was associated with autism [35]. However, they did not perform association test between rs3774179 and autism. The difference between the results in our research and previous researches might have a few reasons. One was the genetic heterogeneity in different ethnicities. For example, the allele frequency of rs2278556 is different between CHB and CEU populations. Second, the rs3774179 which was associated with autism in our research is near to rs3774180. As only a few tagSNPs were chosen to genotype according to the patterns of LD, markers tested for association must either be the causal allele or highly correlated with the causal allele [48], [49]. Another reason was that children affected with Asperger’s syndrome and PDD-NOS were excluded in our research. It was reported that the p values may decreased (i.e. the association became stronger) when the phenotype definition of autism was relaxed to a “spectrum” definition [35].
ATP2B2 is expressed at high level in cerebellum and abundant throughout the cerebral cortex and hippocampus, particularly clustering in the dendrites and spines of Purkinje cells in cerebellum [20], [22]. Several previous researches suggested that ATP2B2 plays an important role in modulating Ca2+ signaling at the synapse, glutamate receptor expression, and survival of Purkinje cells [30], [50]–[52]. Neural development is associated with intracellular Ca2+ levels which can activate signaling pathways to regulate neuronal survival, differentiation, migration, and synaptogenesis [53], [54]. Any defects of these developmental stages may lead to neuroanatomical abnormalities, decreases of the size and density of neurons, alterations in neural connectivity. For example, dysfunction of ATP2B2 might lead to decreased expression of metabotropic glutamate receptor and aberrant signaling in Purkinje neurons, resulting in functional deficits of cerebellum [55], which usually can be detected in autism [56]–[59]. Direct functional evidence demonstrated the influence of presynaptic ATP2B2-mediated Ca2+ extrusion for short-term plasticity at cerebellar parallel fiber to Purkinje neuron synapses. It indicated that ATP2B2 had a strong influence on the development and maintenance of cerebellar function [31]. Recent researches indicated that in Atp2b2 +/− mouse model, the dysregulation of Ca2+ signaling and the perturbations in glutamate receptor signaling complexes were paralleled by delayed Purkinje cells loss, and presumed that these mechanisms might be relevant to Purkinje cells dysfunction and loss in autistic patients [51], [60], [61].
The present study had a few limitations. First, the aim of this study was to replicate previous positive results of association between ATP2B2 and autism. Therefore, our research did not explore the association between SNPs in promoter region and 3′ untranslated region in ATP2B2 with autism. Second, rare variants were not investigated in this study. Mutation screening of ATP2B2 in children with autism could be performed in further researches. Third, the sample size of our research is needed to be expanded.
In conclusion, our present results provided evidence that ATP2B2 might be relevant to the etiology of autism. Further association study in larger samples and functional researches of ATP2B2 are needed.
Supporting Information
The demographic data of subjects affected with autism.
(DOC)
The information of primers and PCR-RFLP analysis of five SNPs in ATP2B2 .
(DOC)
Information of 5 SNPs in ATP2B2 and the genotype frequencies in 427 autism trios of Chinese Han descent.
(DOC)
Estimated haplotype frequencies and results of haplotype association analyses of ATP2B2 .
(DOC)
Comparison of the allele frequencies for the five SNPs in CHB and CEU from HapMap data.
(DOC)
Details for PCR-RFLP (polymerase chain reaction-restriction fragment length polymorphism) and direct sequencing.
(DOC)
Acknowledgments
We thank all subjects who participated in this study and our colleagues for their assistance in recruiting patients in the study.
Funding Statement
This research was supported by research grants from the National Basic Research Development Program of China (973 program 2010CB833905), and National Natural Science Foundation (grant number 81071110). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.American Psychiatric Association, Washington DC (1994) Diagnostic and statistical manual of mental disorders, Fourth Edition.
- 2. Fombonne E (2005) Epidemiology of autistic disorder and other pervasive developmental disorders. J Clin Psychiatry 66 Suppl 103–8. [PubMed] [Google Scholar]
- 3. Folstein S, Rutter M (1977) Infantile autism: a genetic study of 21 twin pairs. J Child Psychol Psychiatry 18: 297–321. [DOI] [PubMed] [Google Scholar]
- 4. Steffenburg S, Gillberg C, Hellgren L, Andersson L, Gillberg IC, et al. (1989) A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J Child Psychol Psychiatry 30: 405–416. [DOI] [PubMed] [Google Scholar]
- 5. Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, et al. (1995) Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med 25: 63–77. [DOI] [PubMed] [Google Scholar]
- 6. Wang K, Zhang H, Ma D, Bucan M, Glessner JT, et al. (2009) Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature 459: 528–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weiss LA, Arking DE, Daly MJ, Chakravarti A (2009) A genome-wide linkage and association scan reveals novel loci for autism. Nature 461: 802–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Anney R, Klei L, Pinto D, Regan R, Conroy J, et al. (2010) A genome-wide scan for common alleles affecting risk for autism. Hum Mol Genet 19: 4072–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Collins FS, Guyer MS, Charkravarti A (1997) Variations on a theme: cataloging human DNA sequence variation. Science 278: 1580–1581. [DOI] [PubMed] [Google Scholar]
- 10. Devlin B, Melhem N, Roeder K (2011) Do common variants play a role in risk for autism? Evidence and theoretical musings. Brain Res 1380: 78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Klei L, Sanders SJ, Murtha MT, Hus V, Lowe JK, et al. (2012) Common genetic variants, acting additively, are a major source of risk for autism. Mol Autism 3: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, et al. (2008) Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 9: 356–369. [DOI] [PubMed] [Google Scholar]
- 13. Liu PY, Zhang YY, Lu Y, Long JR, Shen H, et al. (2005) A survey of haplotype variants at several disease candidate genes: the importance of rare variants for complex diseases. J Med Genet 42: 221–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pritchard JK (2001) Are rare variants responsible for susceptibility to complex diseases? Am J Hum Genet 69: 124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pritchard JK, Cox NJ (2002) The allelic architecture of human disease genes: common disease-common variant…or not? Hum Mol Genet 11: 2417–2423. [DOI] [PubMed] [Google Scholar]
- 16. Gargus JJ (2009) Genetic calcium signaling abnormalities in the central nervous system: seizures, migraine, and autism. Ann N Y Acad Sci 1151: 133–156. [DOI] [PubMed] [Google Scholar]
- 17. Palmieri L, Papaleo V, Porcelli V, Scarcia P, Gaita L, et al. (2010) Altered calcium homeostasis in autism-spectrum disorders: evidence from biochemical and genetic studies of the mitochondrial aspartate/glutamate carrier AGC1. Mol Psychiatry 15: 38–52. [DOI] [PubMed] [Google Scholar]
- 18. Zacharias DA, DeMarco SJ, Strehler EE (1997) mRNA expression of the four isoforms of the human plasma membrane Ca(2+)-ATPase in the human hippocampus. Brain Res Mol Brain Res 45: 173–176. [DOI] [PubMed] [Google Scholar]
- 19. Stahl WL, Eakin TJ, Owens JJ, Breininger JF, Filuk PE, et al. (1992) Plasma membrane Ca(2+)-ATPase isoforms: distribution of mRNAs in rat brain by in situ hybridization. Brain Res Mol Brain Res 16: 223–231. [DOI] [PubMed] [Google Scholar]
- 20. Stauffer TP, Guerini D, Celio MR, Carafoli E (1997) Immunolocalization of the plasma membrane Ca2+ pump isoforms in the rat brain. Brain Res 748: 21–29. [DOI] [PubMed] [Google Scholar]
- 21. Ueno T, Kameyama K, Hirata M, Ogawa M, Hatsuse H, et al. (2002) A mouse with a point mutation in plasma membrane Ca2+-ATPase isoform 2 gene showed the reduced Ca2+ influx in cerebellar neurons. Neurosci Res 42: 287–297. [DOI] [PubMed] [Google Scholar]
- 22. Burette A, Rockwood JM, Strehler EE, Weinberg RJ (2003) Isoform-specific distribution of the plasma membrane Ca2+ ATPase in the rat brain. J Comp Neurol 467: 464–476. [DOI] [PubMed] [Google Scholar]
- 23. Kozel PJ, Friedman RA, Erway LC, Yamoah EN, Liu LH, et al. (1998) Balance and hearing deficits in mice with a null mutation in the gene encoding plasma membrane Ca2+-ATPase isoform 2. J Biol Chem 273: 18693–18696. [DOI] [PubMed] [Google Scholar]
- 24. Bortolozzi M, Brini M, Parkinson N, Crispino G, Scimemi P, et al. (2010) The novel PMCA2 pump mutation Tommy impairs cytosolic calcium clearance in hair cells and links to deafness in mice. J Biol Chem 285: 37693–37703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Spiden SL, Bortolozzi M, Di Leva F, de Angelis MH, Fuchs H, et al. (2008) The novel mouse mutation Oblivion inactivates the PMCA2 pump and causes progressive hearing loss. PLoS Genet 4: e1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Prasad V, Okunade GW, Miller ML, Shull GE (2004) Phenotypes of SERCA and PMCA knockout mice. Biochem Biophys Res Commun 322: 1192–1203. [DOI] [PubMed] [Google Scholar]
- 27. McCullough BJ, Tempel BL (2004) Haplo-insufficiency revealed in deafwaddler mice when tested for hearing loss and ataxia. Hear Res 195: 90–102. [DOI] [PubMed] [Google Scholar]
- 28. Empson RM, Akemann W, Knopfel T (2010) The role of the calcium transporter protein plasma membrane calcium ATPase PMCA2 in cerebellar Purkinje neuron function. Funct Neurol 25: 153–158. [PubMed] [Google Scholar]
- 29. Empson RM, Turner PR, Nagaraja RY, Beesley PW, Knopfel T (2010) Reduced expression of the Ca(2+) transporter protein PMCA2 slows Ca(2+) dynamics in mouse cerebellar Purkinje neurones and alters the precision of motor coordination. J Physiol 588: 907–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garside ML, Turner PR, Austen B, Strehler EE, Beesley PW, et al. (2009) Molecular interactions of the plasma membrane calcium ATPase 2 at pre- and post-synaptic sites in rat cerebellum. Neuroscience 162: 383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Empson RM, Garside ML, Knopfel T (2007) Plasma membrane Ca2+ ATPase 2 contributes to short-term synapse plasticity at the parallel fiber to Purkinje neuron synapse. J Neurosci 27: 3753–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McCauley JL, Li C, Jiang L, Olson LM, Crockett G, et al. (2005) Genome-wide and Ordered-Subset linkage analyses provide support for autism loci on 17q and 19p with evidence of phenotypic and interlocus genetic correlates. BMC Med Genet 6: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lauritsen MB, Als TD, Dahl HA, Flint TJ, Wang AG, et al. (2006) A genome-wide search for alleles and haplotypes associated with autism and related pervasive developmental disorders on the Faroe Islands. Mol Psychiatry 11: 37–46. [DOI] [PubMed] [Google Scholar]
- 34. Griswold AJ, Ma D, Cukier HN, Nations LD, Schmidt MA, et al. (2012) Evaluation of copy number variations reveals novel candidate genes in autism spectrum disorder-associated pathways. Hum Mol Genet 21: 3513–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Carayol J, Sacco R, Tores F, Rousseau F, Lewin P, et al. (2011) Converging evidence for an association of ATP2B2 allelic variants with autism in male subjects. Biol Psychiatry 70: 880–887. [DOI] [PubMed] [Google Scholar]
- 36. Prandini P, Pasquali A, Malerba G, Marostica A, Zusi C, et al. (2012) The association of rs4307059 and rs35678 markers with autism spectrum disorders is replicated in Italian families. Psychiatr Genet 22: 177–181. [DOI] [PubMed] [Google Scholar]
- 37. Krug DA, Arick J, Almond P (1980) Behavior checklist for identifying severely handicapped individuals with high levels of autistic behavior. J Child Psychol Psychiatry 21: 221–229. [DOI] [PubMed] [Google Scholar]
- 38. Schopler E, Reichler RJ, DeVellis RF, Daly K (1980) Toward objective classification of childhood autism: Childhood Autism Rating Scale (CARS). J Autism Dev Disord 10: 91–103. [DOI] [PubMed] [Google Scholar]
- 39. Jurinke C, van den Boom D, Cantor CR, Koster H (2002) The use of MassARRAY technology for high throughput genotyping. Adv Biochem Eng Biotechnol 77: 57–74. [DOI] [PubMed] [Google Scholar]
- 40. Storm N, Darnhofer-Patel B, van den Boom D, Rodi CP (2003) MALDI-TOF mass spectrometry-based SNP genotyping. Methods Mol Biol 212: 241–262. [DOI] [PubMed] [Google Scholar]
- 41.Paul Oeth MBCP (2007) Iplex Assay: Increased plexing efficiency and flexibility for massARRAY system through single base primer extension with mass-modified terminators. Sequenom Application note September 27:
- 42. O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63: 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rabinowitz D, Laird N (2000) A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Hum Hered 50: 211–223. [DOI] [PubMed] [Google Scholar]
- 44. Gauderman WJ (2002) Sample size requirements for association studies of gene-gene interaction. Am J Epidemiol 155: 478–484. [DOI] [PubMed] [Google Scholar]
- 45. Nyholt DR (2004) A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet 74: 765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li J, Ji L (2005) Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity (Edinb) 95: 221–227. [DOI] [PubMed] [Google Scholar]
- 47. Clark AG (2004) The role of haplotypes in candidate gene studies. Genet Epidemiol 27: 321–333. [DOI] [PubMed] [Google Scholar]
- 48. Kruglyak L (1999) Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet 22: 139–144. [DOI] [PubMed] [Google Scholar]
- 49. Jorde LB (2000) Linkage disequilibrium and the search for complex disease genes. Genome Res 10: 1435–1444. [DOI] [PubMed] [Google Scholar]
- 50. Burette AC, Strehler EE, Weinberg RJ (2010) A plasma membrane Ca2+ ATPase isoform at the postsynaptic density. Neuroscience 169: 987–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fakira AK, Gaspers LD, Thomas AP, Li H, Jain MR, et al. (2012) Purkinje cell dysfunction and delayed death in plasma membrane calcium ATPase 2-heterozygous mice. Mol Cell Neurosci 51: 22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dodson HC, Charalabapoulou M (2001) PMCA2 mutation causes structural changes in the auditory system in deafwaddler mice. J Neurocytol 30: 281–292. [DOI] [PubMed] [Google Scholar]
- 53. Cline HT (2001) Dendritic arbor development and synaptogenesis. Curr Opin Neurobiol 11: 118–126. [DOI] [PubMed] [Google Scholar]
- 54. Moody WJ, Bosma MM (2005) Ion channel development, spontaneous activity, and activity-dependent development in nerve and muscle cells. Physiol Rev 85: 883–941. [DOI] [PubMed] [Google Scholar]
- 55. Kurnellas MP, Lee AK, Li H, Deng L, Ehrlich DJ, et al. (2007) Molecular alterations in the cerebellum of the plasma membrane calcium ATPase 2 (PMCA2)-null mouse indicate abnormalities in Purkinje neurons. Mol Cell Neurosci 34: 178–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Courchesne E, Yeung-Courchesne R, Press GA, Hesselink JR, Jernigan TL (1988) Hypoplasia of cerebellar vermal lobules VI and VII in autism. N Engl J Med 318: 1349–1354. [DOI] [PubMed] [Google Scholar]
- 57. Courchesne E, Saitoh O, Townsend JP, Yeung-Courchesne R, Press GA, et al. (1994) Cerebellar hypoplasia and hyperplasia in infantile autism. Lancet 343: 63–64. [DOI] [PubMed] [Google Scholar]
- 58. Hashimoto T, Tayama M, Murakawa K, Yoshimoto T, Miyazaki M, et al. (1995) Development of the brainstem and cerebellum in autistic patients. J Autism Dev Disord 25: 1–18. [DOI] [PubMed] [Google Scholar]
- 59. Fatemi SH, Halt AR, Realmuto G, Earle J, Kist DA, et al. (2002) Purkinje cell size is reduced in cerebellum of patients with autism. Cell Mol Neurobiol 22: 171–175. [DOI] [PubMed] [Google Scholar]
- 60. Ritvo ER, Freeman BJ, Scheibel AB, Duong T, Robinson H, et al. (1986) Lower Purkinje cell counts in the cerebella of four autistic subjects: initial findings of the UCLA-NSAC Autopsy Research Report. Am J Psychiatry 143: 862–866. [DOI] [PubMed] [Google Scholar]
- 61. Bailey A, Luthert P, Dean A, Harding B, Janota I, et al. (1998) A clinicopathological study of autism. Brain 121 (Pt 5): 889–905. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The demographic data of subjects affected with autism.
(DOC)
The information of primers and PCR-RFLP analysis of five SNPs in ATP2B2 .
(DOC)
Information of 5 SNPs in ATP2B2 and the genotype frequencies in 427 autism trios of Chinese Han descent.
(DOC)
Estimated haplotype frequencies and results of haplotype association analyses of ATP2B2 .
(DOC)
Comparison of the allele frequencies for the five SNPs in CHB and CEU from HapMap data.
(DOC)
Details for PCR-RFLP (polymerase chain reaction-restriction fragment length polymorphism) and direct sequencing.
(DOC)


