Abstract
Major depressive disorder (MDD) has been associated with biased memory formation for mood-congruent information, which may be related to altered monoamine levels. The piccolo (PCLO) gene, involved in monoaminergic neurotransmission, has previously been linked to depression in a genome-wide association study. Here, we investigated the role of the PCLO risk allele on functional magnetic resonance imaging (MRI) correlates of emotional memory in a sample of 89 MDD patients (64 PCLO risk allele carriers) and 29 healthy controls (18 PCLO risk allele carriers). During negative word encoding, risk allele carriers showed significant lower activity relative to non-risk allele carriers in the insula, and trend-wise in the anterior cingulate cortex and inferior frontal gyrus. Moreover, depressed risk allele carriers showed significant lower activity relative to non-risk allele carriers in the striatum, an effect which was absent in healthy controls. Finally, amygdalar response during processing new positive words vs. known words was blunted in healthy PCLO+ carriers and in MDD patients irrespective of genotype, which may indicate that signalling of salient novel information does not occur to the same extent in PCLO+ carriers and MDD patients. The PCLO risk allele may increase vulnerability for MDD by modulating local brain function with regard to responsiveness to salient stimuli (i.e. insula) and processing novel negative information. Also, depression-specific effects of PCLO on dorsal striatal activation during negative word encoding and the absence of amygdalar salience signalling for novel positive information further suggest a role of PCLO in symptom maintenance in MDD.
Introduction
Major depressive disorder (MDD) is a highly prevalent psychiatric disorder, with twin studies showing that up to 40% of MDD is genetically determined [1]. Phenotypically, depression is characterized by depressed mood and/or anhedonia (loss of interest in nearly all activities) and has been associated with attentional deficits, resulting in poor functioning in daily life [2], [3]. Symptoms of negative mood, lack of positive affect, and attentional impairments may ensue from, or be reinforced by, dysfunctional emotional memory processes. Phenotypic features of abnormal perception, encoding, and consolidation of emotional information, often seen in depression, may be moderated by altered monoamine levels. Much of the candidate gene literature has focused on genes from the monoaminergic neurotransmitter system, such as the serotonin transporter, monoamine oxidase A and tryptophan hydroxylase 2 [4]–[9]. A recent study by our consortium demonstrated that candidate genetic association studies are not well replicated [10], which suggests that a hypothesis-free approach is more useful to identify possible genetic variants that contribute to MDD. A genome-wide association study for MDD found the SNP rs2522833 located at position 82453708 (hapmap genome build 37.1) in the piccolo gene (PCLO), which is involved in monoaminergic neurotransmission, to be of particular interest in its genetic model [11]. This association was confirmed in a number of studies with similar and related phenotypes [12]–[17], but not in others [18]–[20]. The rs2522833 SNP alters the hydrophilic, uncharged aminoacid serine to the charged aminoacid alanine in the calcium-binding C2A domain of PCLO and may affect protein stability [21]. The PCLO protein is localized at the cytomatrix of the presynaptic active zone and is important in monoaminergic neurotransmission in the brain [13], [20]. Recently, we have shown that the risk allele on the piccolo gene in healthy controls and depressed patients was associated with abnormal processing of negative emotional faces rather than executive functioning [12]. However, whether the rs2522833 polymorphism in the PCLO gene can also affect emotional memory processing has not been studied yet.
On a cognitive-behavioural level, MDD has been associated with attentional bias towards mood-congruent (i.e. negative) information [22]. Using neuropsychological assessments [23], it has been shown that negative emotional processing bias may be predictive of depression symptoms and may represent a state marker of MDD. It is thought that this negative bias is associated with abnormal responsiveness of brain regions involved in emotion processing, as well as disruption of cortico-limbic connections that are important for regulating emotional responses [24]. Recently we found an association of memory processing of positive and negative information in MDD with altered activity in the amygdala, ventral striatum, insula, hippocampus, anterior cingulate cortex (ACC), and inferior frontal gyrus (IFG) [25]. We found left ventral insular activation specifically during processing of negative words [25], which may reflect a general increased sensitivity for negative information, as suggested by Surguladze et al. (2010) [26]. In another study, an abnormal response for recollecting negative faces was found in MDD patients, which may reflect activation of negative schemas [27]. MDD has also been associated with a mood-incongruent bias (i.e., away from positive information) [28]–[30] which may affect memory formation for positive as compared to negative and neutral stimuli. Using event related potentials (ERP), Shestyuk et al. observed smaller slow wave amplitudes to positive self-relevant words in MDD relative to controls, whereas group differences for negative or neutral stimuli were absent [30]. In summary, negative and positive biases may lead to abnormal memory formation, reinforcing negative mood and further contributing to a chronic course of the disorder [31], [32].
Recent studies have underscored the importance of PCLO in MDD [13]–[16], and in neural processes underlying memory formation [33]. Although we recently found evidence for the PCLO risk allele to be associated with emotional processing of negative faces, it remains unclear whether effortful classification of emotional words is characterized by a similar association. Moreover, it is unknown whether PCLO modulates negative bias and emotional memory in depression.
Over the last few years, imaging genetics has shown to be a powerful method for investigating neurobiological pathways in various psychiatric disorders [34], [35]. Using an intermediate phenotype, such as emotional memory processing, which is probably closer to the neurobiological substrate of MDD than the clinical diagnosis itself [36], may be helpful in identifying genetic risk alleles. Until now, imaging genetics studies on emotional memory have mainly been conducted in healthy controls and in psychiatric disorders other than MDD. We have recently studied emotional processing and executive function in the context of genetic association with PCLO in a group of MDD patients and healthy controls [12] and found an association increased amygdalar activity during the processing of negative faces. Considering that encoding and retrieval of emotional stimuli is a complex form of cognitive and emotional processing, we hypothesize that the PCLO risk allele will also modulate emotional, especially negative, memory processing. Focussing on the amygdala, ventral striatum, hippocampus, anterior cingulate cortex (ACC), insula, and inferior frontal gyrus (IFG), regions that are important for encoding and recognition of valenced semantic information, we studied functional MRI correlates of successful emotional word encoding and recognition, and focussed on activation patterns explained by PCLO genotype in these areas independent of psychopathology. Since the pathophysiology of MDD is complex and diverse, we also investigated whether PCLO genotype effects on the brain were different in the presence of MDD psychopathology. In addition, we controlled for the use of selective serotonin reuptake inhibitors (SSRI) and tested whether functional effects coincided with morphometric variations related to PCLO genotype.
Methods
Participants
The present study was part of a large imaging study (details described elsewhere [25]) included in the Netherlands Study of Depression and Anxiety (NESDA) [37]. After excluding participants due to missing PCLO genotype data, technical problems during scanning and/or insufficient task performance, our final sample consisted of 89 MDD patients and 29 healthy controls (see figure S1 in File S1 for a detailed flowchart of the numbers of participants included). Exclusion criteria were: presence of MRI contraindications, DSM-IV axis I disorder other than MDD, Panic Disorder (PD) or Social Anxiety Disorder (SAD) (except Generalized Anxiety Disorder/GAD) lifetime, or any DSM-IV disorder (for HC), dependence or recent abuse of alcohol and/or drugs, hypertension, major internal and/or neurological disorders, and use of psychotropic medication (other than stable use of a selective serotonin reuptake inhibitor [SSRI] or incidental use of benzodiazepines). To assess depressive symptom characteristics and severity scores, the Inventory of Depressive Symptomatology (IDS) [38], and the Montgomery-Åsberg Depression Rating Scale (MADRS) [39], were used. PCLO groups did not differ with regard to age, gender, education, MDD/HC ratio, depression severity, or SSRI use/duration (see Table 1). All participants provided written informed consent and the Ethics Committees at the VU University Medical Center, and Academic Medical Center, Amsterdam, the Leiden University Medical Center and at the University Medical Center Groningen approved this study.
Table 1. Sample characteristics and task performance.
| Group; mean (SD) | |||||
| total (n = 118) | PCLO+ (n = 82) | PCLO− (n = 36) | p-value | ?2 | |
| Sample characteristics | |||||
| Gender (%female) | 61.9 (n = 73) | 62.2 (n = 51) | 61.1 (n = 22) | 1 | .012 |
| Age (years) | 38.1 (10.2) | 37.5 (10.1) | 39.3 (10.6) | .42 | |
| Education (years) | 12.6 (3.2) | 12.9 (3.3) | 12.0 (2.8) | .16 | |
| Scancenter (% A, L, G) | 28.8, 41.5, 29.7 | 26.8, 43.9, 29.3 | 33.3, 36.1, 30.6 | .69 | .747 |
| Diagnosis (MDD/HC) | 89/29 | 64/18 | 25/11 | .36 | .999 |
| IDS (score) | 18.8 (12.9) | 19.7 (12.4) | 16.9 (14.1) | .28 | |
| MADRS (score) | 12.5 (10.5) | 13.4 (10.4) | 10.3 (10.5) | .14 | |
| Duration SSRI use (months) | 18.9 (28.8) | 14.5 (19.8) | 32.0 (46.6) | .17 | |
| SSRI use (no/yes) | 90/28 | 61/21 | 29/7 | .64 | .525 |
| Memory performance (p) | |||||
| CREC.neg | .685 (.138) | .685 (.139) | .685 (.135) | .99 | |
| CREC.pos | .725 (.132) | .719 (.132) | .739 (.132) | .45 | |
| CREC.neu | .69 (.166) | .692 (.155) | .688 (.191) | .90 | |
| CREC.all | .70 (.122) | .697 (.122) | .70 (.124) | .83 | |
| FA.neg | .17 (.11) | .17 (.10) | .18 (.11) | .72 | |
| FA.pos | .12 (.11) | .13 (.11) | .11 (.11) | .35 | |
| FA.neu | .07 (.06) | .07 (.07) | .06 (.06) | .72 | |
| FA.all | .124 (.079) | .125 (.08) | .119 (.079) | .71 | |
| CREJ.neg | .66 (.141) | .66 (.137) | .67 (.151) | .623 | |
| CREJ.pos | .70 (.175) | .69 (.175) | .72 (.175) | .503 | |
| CREJ.neu | .81 (.13) | .81 (.13) | .82 (.131) | .699 | |
| CREJ.all | .729 (.13) | .72 (.127) | .74 (.137) | .54 | |
| Response time (sec) | |||||
| rt SCR.neg | 1.296 (.385) | 1.311 (.411) | 1.263 (.321) | .63 | |
| rt SCR.pos | 1.487 (.379) | 1.507 (.384) | 1.441 (.368) | .45 | |
| rt SCR.neu | 1.563 (.393) | 1.567 (.415) | 1.553 (.343) | .69 | |
| rt CREC.neg | 1.264 (.256) | 1.265 (.237) | 1.263 (.30) | .61 | |
| rt CREC.pos | 1.355 (.289) | 1.37 (.291) | 1.319 (.287) | .27 | |
| rt CREC.neu | 1.339 (.297) | 1.335 (.276) | 1.349 (.346) | .90 | |
| rt FA.neg | 1.50 (.45) | 1.48 (.42) | 1.53 (.50) | .65 | |
| rt FA.pos | 1.62 (.52) | 1.59 (.41) | 1.69 (.71) | .37 | |
| rt FA.neu | 1.58 (.53) | 1.60 (.51) | 1.53 (.57) | .56 | |
| rt CREJ.neg | 1.48 (.35) | 1.50 (.33) | 1.45 (.39) | .52 | |
| rt CREJ.pos | 1.52 (.34) | 1.54 (.34) | 1.50 (.35) | .53 | |
| rt CREJ.neu | 1.39 (.31) | 1.40 (.31) | 1.37 (.33) | .61 | |
Sample characteristics and task performance of the total sample. SD: standard deviation; n:number of participants; PCLO+: PCLO risk allele carriers; PCLO−: PCLO non-risk allele carries; A: Amsterdam; L: Leiden; G:Groningen; MDD: major depressive disorder; HC: healthy controls; IDS: Inventory of Depressive Symptomatology; MADRS: Montgomery-Åsberg Depression Rating Scale; SSRI: Selective Serotonin Reuptake Inhibitors; p: proportion correct answers; neg: negative (words); pos: positive (words); neu: neutral (words); sec: seconds; rt: response time.
Mean proportion correct and response times for encoding (subsequent hits) and recognition (hits, false alarms, correct rejection) indices. Correct RECognition (CREC): correct recognition of a previously encoded word; False Alarm (FA): incorrect indication of a newly presented word as a previously encoded word; Correct REJection (CREJ): correct recognition of a newly presented word as a new word; Subsequent Correct Recognition (SCR): consists of a word, presented during the encoding phase, that is correctly recognized during the subsequent recognition phase.
Genotyping
As described in detail elsewhere [11], genotyping was performed by Perlegen. Observed genotypes in our sample did not deviate from Hardy-Weinberg equilibrium (CC:AC:AA = 25∶57:36; χ2[1] = 0.08; p>0.05). All subjects reported Western European ancestry. We formed two groups based on the PCLO genetic association study in MDD. One group consisted of participants carrying the risk allele (AC/CC), and one group included participants not carrying the risk allele (AA). In the following, we will refer to these groups as PCLO+ (risk allele) carriers and PCLO− carriers, respectively.
Emotional Memory Task Paradigm
An event-related, (subject-paced), word encoding and recognition paradigm was used which has been described extensively elsewhere [25]. During the encoding part, participants were asked to classify 40 positive, 40 negative, and 40 neutral words according to their valence. Words were presented pseudo-randomized together with 40 baseline trials in 20 blocks of eight words. After a brief retention interval, participants were asked to complete a word recognition task. This task consisted of the 120 old encoding target words and 120 new distracter words (matched for valence), and 40 baseline trials, presented pseudo-randomized in 20 blocks of 14 words. Participants had to indicate whether they had ‘seen’ (i.e. remembered) the words previously, ‘probably seen’ (‘know’), or ‘not seen’ (rejection).
Magnetic Resonance Imaging Data Acquisition
The functional neuroimaging methods have been comprehensively reported elsewhere [40], [41]. In summary, T2*-weighted echo-planar images (EPI) sensitive to the blood oxygenation level–dependent (BOLD) effect were acquired using similar Philips 3T MR systems (repetition time [TR] = 2300 ms, echo time [TE] = 30.0 ms (UMCG: 28.0 ms), 35 slices (UMCG 39 slices)), situated at different locations (Amsterdam, Leiden, and Groningen, the Netherlands).
The EPI volumes were acquired at 35 slices (UMCG: 39 slices), interleaved axial acquisition, 3 mm thickness, matrix size 96×96 (UMCG: 64×64), in-plane resolution 2.29×2.29 mm (UMCG: 3×3 mm). A T1-weighted anatomical MRI was also acquired for each subject and included a sagittal 3-dimensional gradient-echo sequence (TR = 9 ms, TE = 3.5 ms, matrix 256×256, voxel size: 1×1×1 mm, 170 slices).
Statistical Analysis
Performance
Responses and response times were recorded and were used to calculate proportions (p) Hits, correct rejections (pCREJ), False Alarms (pFA), and old/new discriminant accuracy (d’ = pHits-pFA), overall and per valence (negative, neutral, and positive). Repeated-measures analyses of covariance (ANCOVAs) were performed to test for effects of PCLO genotype, genotype × diagnosis effects, and interaction effects of genotype, diagnosis and genotype × diagnosis with valence on task performance (pHits_all, pFalseAlarms_all, and d’_all) and response times during successful encoding and successful recognition. Significance for behavioural analyses was set at P<.05 and post hoc paired tests (T-test or Mann-Whitney [U] were Bonferroni-corrected for multiple comparisons (P Bonferroni).
Imaging data analysis
Image processing was performed using Statistical Parametric Mapping (SPM5; http://www.fil.ion.ucl.ac.uk/spm/software/spm5; software implemented in Matlab 7.5.0 (The Matlab Inc, Natick, MA, USA). Details of preprocessing and first-level single-subject analyses have been described elsewhere [25]. Briefly, following temporal and spatial preprocessing (final smoothing: 8 mm full-width at half-maximum [FWHM]), data were analyzed in the context of the General Linear Model. The subject-specific first-level models included regressors for encoding and recognition events. Due to the small proportion of recognition trials that were responded to with a ‘know’ response, these responses were treated as ‘remembered’ and consequently added to either correct recognized (CREC) or false alarms (FA). Activation maps associated with different valences were calculated per subject. To avoid inclusion of non-task related signal which might be expected when contrasting against the repetitive lower baseline, contrast images with visual input that only differed in its emotional content (‘encoding positive > encoding neutral’, ‘encoding negative > encoding neutral’ resulting from the encoding phase, and ‘recognized positive > recognized neutral’, and ‘recognized negative > recognized neutral’ were likewise included in a second-level random-effect analysis. Although our primary aim was to investigate valence effects on processes of word encoding and recognition, we also investigated specific effects of successful recognition by setting up the following contrast: ‘correct recognition (hits) > correct rejection’, resulting from the recognition phase) and included this contrast in a second-level random-effect analysis.
Based on our previous study [25] we included the following regions of interest for the emotional memory task, defined using the Automated Anatomical Labeling (AAL) atlas or Talairach Daemon (for the striatum) [42], implemented in the Wake Forest University (WFU) Pick Atlas toolbox: amygdala, hippocampus, ACC, IFG, insula, and striatum (includes caudate head, tail, body, putamen). Main effects of task are reported at a threshold of P<.05, whole-brain corrected for False Discovery Rate (FDR). We conducted a full factorial using genotype and diagnosis as between subject factors, and valence per encoding or recognition as within-subject factor, to test whether the PCLO+ was associated with altered activity in our ROIs for each valence for encoding or recognition. Scan location was entered as covariate by means of two dummy variables. Furthermore, to test for the specificity of valence for correctly recognized old versus new words, we conducted a 2×2×3 ANOVA with genotype (PCLO+, PCLO−) and diagnosis (MDD, HC) as between-subject factor, and valence (positive, negative, and neutral; e.g. CREC_positive > CREJ_positive) as within-subject factor. Scan location was entered as covariate by means of two dummy variables. Main effects of genotype and interaction of PCLO genotype with current psychopathology were reported at a voxel-wise threshold of P<.05 FDR corrected for the regions of interest, with an initial threshold of P<.001 uncorrected. In addition, to explore activity common to both negative and positive vs. neutral stimuli we post hoc computed the conjunction of these two contrasts, based on the global null hypothesis (k≥1) [43]. Each contrast had to meet a threshold of P<.001 uncorrected. To test whether between-group effects were related to volumetric differences, we conducted a two-by-two ANOVA for those regions that showed a between-group effect during the functional paradigm. Effects were reported at a threshold of P<.05 FDR corrected for the region of interest. To account for the number of a priori regions of interest, we corrected the critical corrected p-value for the number of regions (n = 6: amygdala, hippocampus, ACC, IFG, insula, and striatum). Using a standard Bonferroni correction would be too stringent, however, since the dependent variables were measured within the same individuals. Therefore, we took this interdependency into account and calculated the optimal threshold for positive vs. neutral and negative vs. neutral encoding and recognition using the Simple Interactive Statistical Analysis Bonferroni tool (www.quantitativeskills.com/sisa/calculations/bonfer.htm). Response in the six a priori regions of interest showed a mean correlation of r = .75 during positive encoding (>neutral encoding) across all participants, r = .63 (>neutral encoding) during negative encoding, r = .84 (>neutral recognition) during positive recognition, and r = .69 (>neutral recognition) during negative recognition, leading to a critical alpha of.026,.032,.037, and.029, respectively (Table S2 in File S1). Because SSRI use may alter regional brain function in psychiatric diseases [44], we repeated our analyses omitting SSRI users, to test for possible effect of SSRI use.
Results
Sample Descriptives
Table 1 lists the sample characteristics and behavioural statistics. Genotype groups were matched for MDD diagnosis, age, gender, and education.
Behavioural analyses of the emotional word encoding and recognition task revealed no significant main effect of genotype, no genotype × valence, and no genotype × valence × diagnosis interaction on accuracy and response time indices.
Table 2 lists the genotype × diagnosis sample and behavioural characteristics.
Table 2. Sample characteristics and task performance group × diagnosis.
| Group; mean (SD) | ||||
| PCLO+ | PCLO− | |||
| MDD (N = 64) | HC (N = 18) | MDD (N = 25) | HC (N = 11) | |
| Sample characteristics | ||||
| Gender (%female) | 65.6% (n = 42) | 50.0% (n = 9) | 60% (n = 15) | 63.6% (n = 7) |
| Age (years) | 35.9 (9.57) | 43.2 (10.05) | 39.0 (11.38) | 40.0 (9.12) |
| Education (years) | 12.4 (3.24) | 14.7 (2.78) | 11.3 (2.85) | 13.6 (2.16) |
| Scancenter (% A, L, G) | 23.4, 46.9, 29.7 | 38.9, 33.3, 27.8 | 24, 36, 40 | 54.5, 36.4, 9.1 |
| IDS (score) | 24.3 (10.03) | 3.9 (3.26) | 22.1 (13.29) | 3.7 (3.43) |
| MADRS (score) | 16.8 (9.12) | 1.5 (2.18) | 14.4 (9.82) | .27 (.65) |
| Duration SSRI use (months) | 14.5 (19.76) | N/A | 32.0 (46.59) | N/A |
| SSRI use (no/yes) | 43/21 | 18/0 | 18/7 | 11/0 |
| Memory performance (p) | ||||
| pCREC.neg | .693 (.143) | .658 (.127) | .700 (.118) | .652 (.170) |
| pCREC.pos | .724 (.129) | .700 (.145) | .728 (.143) | .764 (.103) |
| pCREC.neu | .702 (.163) | .656 (.114) | .717 (.175) | .621 (.219) |
| pFA.all | .123 (.084) | .132 (.067) | .129 (.078) | .099 (.079) |
| pCREJ.neg | .658 (.136) | .668 (.143) | .655 (.146) | .718 (.161) |
| pCREJ.pos | .696 (.165) | .701 (.212) | .713 (.165) | .739 (.205) |
| pCREJ.neu | .807 (.136) | .846 (.106) | .800 (.142) | .884 (.083) |
| Response time (sec) | ||||
| rt SCR.neg | 1.28 (.32) | 1.32 (.57) | 1.26 (.26) | 1.24 (.37) |
| rt SCR.pos | 1.53 (.39) | 1.35 (.31) | 1.43 (.36) | 1.45 (.26) |
| rt SCR.neu | 1.59 (.39) | 1.47 (.45) | 1.52 (.32) | 1.54 (.31) |
| rt CREJ.neg | 1.51 (.32) | 1.48 (.35) | 1.44 (.42) | 1.50 (.35) |
| rt CREJ.pos | 1.56 (.34) | 1.49 (.34) | 1.50 (.37) | 1.50 (.33) |
| rt CREJ.neu | 1.41 (.31) | 1.40 (.30) | 1.37 (.37) | 1.37 (.24) |
| rt CREC.neg | 1.26 (.21) | 1.33 (.30) | 1.23 (.30) | 1.29 (.25) |
| rt CREC.pos | 1.36 (.27) | 1.42 (.31) | 1.31 (.29) | 1.30 (.24) |
| rt CREC.neu | 1.31 (.25) | 1.43 (.30) | 1.31 (.31) | 1.40 (.35) |
Sample characteristics and task performance of the total sample, divided into genotype × diagnosis. SD: standard deviation; n:number of participants; PCLO+: PCLO risk allele carriers; PCLO−: PCLO non-risk allele carries; A: Amsterdam; L: Leiden; G:Groningen; MDD: major depressive disorder; HC: healthy controls; IDS: Inventory of Depressive Symptomatology; MADRS: Montgomery-Åsberg Depression Rating Scale; SSRI: Selective Serotonin Reuptake Inhibitors; p: proportion correct answers; neg: negative (words); pos: positive (words); neu: neutral (words); sec: seconds; rt: response time.
Mean proportion correct and response times for encoding (subsequent hits) and recognition (hits, false alarms, correct rejection) indices. Correct RECognition (CREC): correct recognition of a previously encoded word; False Alarm (FA): incorrect indication of a newly presented word as a previously encoded word; Correct REJection (CREJ): correct recognition of a newly presented word as a new word; Subsequent Correct Recognition (SCR): consists of a word, presented during the encoding phase, that is correctly recognized during the subsequent recognition phase.
Imaging Results
Main effects of the encoding and recognition contrast across groups can be found in Table 3. Table S3 in File S1 describes the main effects of encoding and recognition per valence.
Table 3. Main effects of encoding of emotional words vs. neutral words.
| Emotional Encoding | ||||||||
| Side | BA | MNI coordinates | p (FDR) | ka | ||||
| Regions | x | y | z | Z | ||||
| Frontal | ||||||||
| Inferior Frontal Gyrus | R | 45 | 54 | 24 | 6 | 3.82 | 0.004 | 68 |
| R | 47 | 42 | 21 | −15 | 3.41 | 0.009 | 10 | |
| Medial Frontal Gyrus | L | 10 | −3 | 57 | −3 | 5.2 | 0.004 | 513 |
| L | 10 | −3 | 60 | 21 | 4.72 | 0.001 | 513 | |
| R | 6 | 3 | 48 | 39 | 3.16 | 0.014 | 455 | |
| Middle Frontal Gyrus | L | 10 | −33 | 42 | 30 | 2.83 | 0.025 | 11 |
| R | 6 | 48 | 3 | 48 | 3.82 | 0.004 | 317 | |
| R | 6 | 36 | −6 | 57 | 3.86 | 0.004 | 317 | |
| L | 9 | −33 | 33 | 39 | 2.71 | 0.031 | 11 | |
| Temporal/Parietal | ||||||||
| Precentral Gyrus | L | 6 | −30 | −9 | 54 | 5.29 | 0.001 | 4504 |
| Anterior Cingulate | L | 32 | −6 | 27 | 30 | 3.61 | 0.006 | 226 |
| R | 24 | 6 | 18 | 33 | 3.31 | 0.01 | 226 | |
| Posterior Cingulate | R | 30 | 30 | −72 | 9 | 3.68 | 0.005 | 1574 |
| Subcortical | ||||||||
| Amygdala | L | N/A | −24 | 3 | −24 | 2.74 | 0.032 | 10 |
| Putamen | L | N/A | −21 | 9 | −3 | 3.04 | 0.023 | 15 |
| Cuneus | L | 30 | −15 | −72 | 9 | 3.56 | 0.011 | 394 |
| Lingual Gyrus | L | 18 | −12 | −75 | −9 | 3.71 | 0.011 | 394 |
| Inferior Parietal Lobule | R | 40 | 45 | −33 | 42 | 4.29 | 0.002 | 1574 |
| Medial Dorsal Nucleus | L | thalamus | −3 | −15 | 9 | 4.04 | 0.003 | 265 |
| Middle Temporal Gyrus | L | 21 | −51 | 3 | −24 | 5.4 | 0.001 | 4504 |
| Superior Temporal Gyrus | R | 38 | 48 | 6 | −27 | 3.25 | 0.012 | 19 |
| Supramarginal Gyrus | R | 40 | 63 | −48 | 24 | 3.93 | 0.003 | 1574 |
Main effects of encoding of emotional words vs. neutral words. Main effects are reported at P FDR<.05, whole brain corrected with a minimum cluster size of 10. MNI: Montreal Neurological Institute; BA: Brodmann area; k: clustersize; L: left; R: right; a: clustersize at p<.05 FDR whole brain corrected.
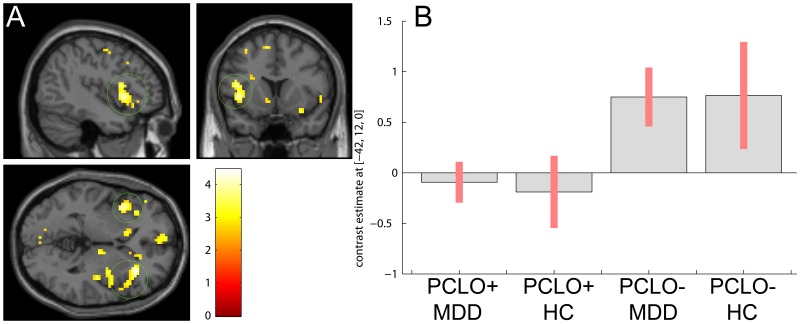
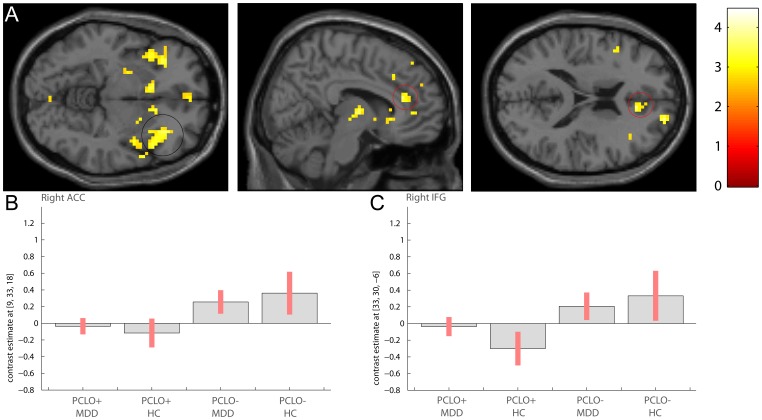
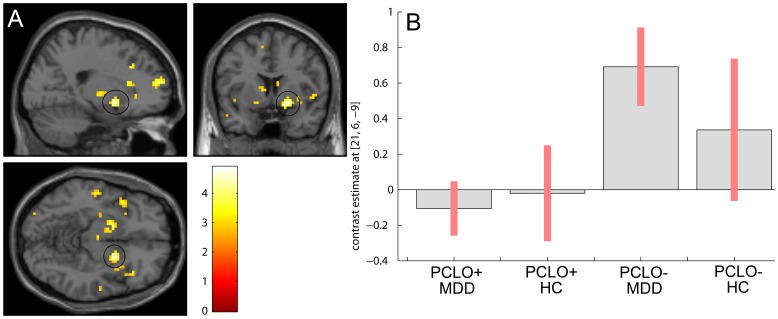
A significant main effect of genotype was observed during successful negative word encoding (‘encoding negative > encoding neutral’) in the insular cortex (left insula; MNI [−42 12 0]; Z = 3.92; right insula; P FDR = .008 corrected for small volume; MNI [33 27 −3]; Z = 4.26; P FDR = .007 corrected for small volume; Figure 1; Table S4 in File S1) and trend-wise in the dorsal part of the pregenual ACC and inferior frontal gyrus (left pregenual ACC; MNI [−3 33 30]; Z = 3.12; P FDR = .046 corrected for small volume; right pregenual ACC; MNI [9 33 18]; Z = 3.49; P FDR = .046 corrected for small volume; left IFG; MNI [−36 30 −3]; Z = 3.41; P FDR = .045 corrected for small volume; right IFG; MNI [33 30 −6]; Z = 3.42; P FDR = .045 corrected for small volume; Figure 2; Table S4 in File S1). This was due to lower activity in PCLO+ compared to PCLO−, which was observed independent of diagnostic status. An interaction of PCLO genotype and diagnosis was observed in the striatum (left ventral striatum (caudate head); MNI [−18 21 −3]; Z = 3.6; P FDR = .028 corrected for small volume; (caudate body); MNI [−9 3 9]; Z = 3.3; P FDR = .028 corrected for small volume; right ventral striatum (caudate body); MNI [9 0 15]; Z = 3.22; P FDR = .028 corrected for small volume; left dorsal putamen; MNI [−18 3 −9]; Z = 3.57; P FDR = .028 corrected for small volume; right dorsal putamen; MNI [21 6 −9]; Z = 4.65; P FDR = .002 corrected for small volume. In the MDD group we found reduced activity in the PCLO+ carriers relative to the PCLO− carriers in this region, which was absent in healthy controls (Figure 3).
Figure 1. PCLO genotype effect during negative word encoding in the insula.
PCLO+ carriers show significant hypoactivation of the insula, relative to PCLO− carriers during negative word encoding. Panel A shows sagittal, coronal, and axial section at peak activation (green circle: insula; results shown at P<.005; Z = 3.92 (left), Z = 4.26 (right)). Panel B shows the cluster means for each peak voxel, with their standard errors for the different groups. PCLO: Piccolo genotype; PCLO+: PCLO risk allele carriers; PCLO−: PCLO non-risk allele carriers; MDD: Major Depressive Disorder; HC: Healthy controls; AU: arbitrary units.
Figure 2. PCLO genotype effect during negative word encoding in the inferior frontal gyrus and anterior cingulate cortex.
PCLO+ carriers show trend-wise hypoactivation of the inferior frontal gyrus and anterior cingulate cortex relative to PCLO− carriers during negative word encoding. Panel A shows sagittal, coronal, and axial section at peak activation (black circle: IFG, red circle: dorsal part of pgACC; results shown at P<.005; IFG, Z = 3.79 (left), Z = 3.42 (right); pgACC, Z = 3.12 (left), Z = 3.49 (right)). Panel B shows the cluster means for each peak voxel, with their standard errors for the different groups. PCLO: Piccolo genotype; PCLO+: PCLO risk allele carriers; PCLO−: PCLO non-risk allele carriers; MDD: Major Depressive Disorder; HC: Healthy controls; AU: arbitrary units.
Figure 3. Genotype × diagnosis interaction during negative word encoding in the striatum.
PCLO+ carriers within the MDD group show reduced striatal activity relative to the PCLO− carriers within the MDD group during negative word encoding. No effect of genotype is seen in the HC group. Panel A shows sagittal, coronal, and axial section at peak activation (results shown at P<.005; black circle: dorsal putamen, Z = 4.65 (right). Panel B shows the cluster means for each peak voxel, with their standard errors for the different groups. PCLO: Piccolo genotype; PCLO+: PCLO risk allele carriers; PCLO−: PCLO non-risk allele carriers; MDD: Major Depressive Disorder; HC: Healthy controls; AU: arbitrary units.
Effects of genotype during successful recognition of negative words did not meet the required threshold of P<.05 FDR corrected. Subtreshold at P<.001 uncorrected, increased activity in the insula in PCLO+ carriers relative to PCLO− carriers was observed (Table S4 in File S1).
During encoding of positive words or during recognition of positive words effects of genotype likewise did not meet the required a priori threshold. Exploration of these contrasts at a threshold of P<.001 uncorrected revealed decreased activity in frontal and limbic areas for encoding positive words (regions are listed in table S4 in File S1).
During correct recognition>correct rejection (in the recognition phase) of positive words we found a significant interaction of emotion × diagnosis × genotype: During rejection of positive new words, healthy PCLO− carriers showed increased left amygdalar activation, while no difference between processing positive old and new words was observed in PCLO+ carriers and in MDD patients, indicating blunting to novel positive information in PCLO+ carriers and patients (MNI [−27 −3 −24]; Z = 3.36; Figure S5 in File S1). No effect of negative or neutral words was found.
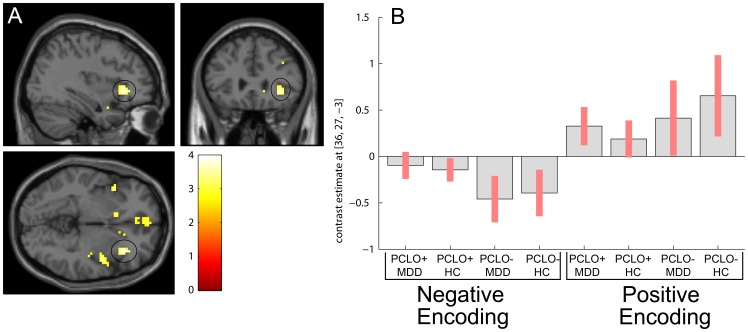
We tested post hoc for common valence effects by performing a conjunction analysis, using both the negative and positive word encoding contrasts. We found a PCLO genotype effect in regions including IFG, medial frontal, insula, and caudate head (Figure 4), reflecting reduced activity of the PCLO+ carriers relative to PCLO− carriers. It should be noted that our significant conjunction (although at an explorative threshold of p<.001 uncorrected) does not mean all the contrasts were individually significant (i.e., a conjunction of significance). It indicates that the contrasts were consistently high and jointly significant. This is equivalent to inferring one or more effects were present.
Figure 4. Valence specific effects during word encoding.
Panel A shows valence specific effects during word encoding (results shown at P<.005). Black circle: inferior frontal gyrus (right); MNI [x = 36 y = 27 z = −3], Z = 3.9. Panel B: Parameter estimates and 95% confidence intervals showing direction of effect for each valence during word encoding in peakvoxel (MNI [x = 36 y = 27 z = −3]). PCLO: Piccolo genotype; PCLO+: PCLO risk allele carriers; PCLO−: PCLO non-risk allele carriers; MDD: Major Depressive Disorder; HC: Healthy controls; AU: arbitrary units.
No volumetric differences were observed between groups in these regions.
Effects of SSRI
After excluding SSRI users (n = 28), PCLO effects observed for processing of emotional words memory (‘encoding negative vs. encoding neutral, encoding positive vs. encoding neutral, recognition negative vs. recognition neutral, and recognition positive vs. recognition neutral’) were similar to the main genotype analyses.
Discussion
In the present study, we examined the effects of the PCLO rs2522833 polymorphism on regional brain activation during performance of an emotional word encoding and recognition paradigm in MDD patients and healthy controls, while also controlling for SSRI use and volumetric differences. Just below threshold, results indicated that PCLO is associated with psychopathology-independent functional changes within the dorsal part of the pregenual (pg)ACC, predominantly during processing of novel, negative information. Whereas pgACC hypoactivation in PCLO+ carriers was specific for the processing of negative information, we found that the PCLO risk allele modulated both negative and positive information processing in the IFG and insula. In addition, PCLO was found to differentially affect striatal activation during negative encoding in health and disease, as genotype effects were observed in MDD patients but not in controls. Successful recognition of emotional words was not associated with significant PCLO effects. To avoid problematic interpretation of genotype effects on memory processing [45], [46], memory performance was modelled at first level. Moreover, we found a blunted effect in the amygdala in PCLO+ carriers and MDD patients of new positive words relative to old words, which may indicate that signalling of salient novel information does not occur to the same extent in PCLO+ carriers and MDD patients. During recognition of negative or neutral words, no difference between PCLO+ and PCLO− was found. We found no effect of PCLO genotype, or PCLO × MDD interaction during performance.
To our knowledge, this is the first genetic neuroimaging association study in MDD and healthy controls that shows an effect of PCLO genotype related hypoactivation of insula, and trend-wise of ACC and IFG during emotional memory processing. In a previous study, we showed that the PCLO risk allele is associated with abnormal involvement of limbic regions in response to negative stimuli (i.e. emotional faces), but not with altered prefrontal recruitment during an executive control task [12]. In the present study, we extended these findings by showing that in PCLO+ carriers, processing of negative information (words) is characterized by reduced insula, and trend-wise pgACC and IFG activation. These regions are considered important regions in the production and effortful regulation of mood states, and have been consistently associated with the psychopathology of MDD [47]. Moreover, the insula has been implicated in the salience network [48], where it is considered an important hub for processing salient events for action to be initiated, including calling on attentional resources and regulating autonomic activity in reaction to salient stimuli [49]. Although near-significant only, the present results demonstrate that PCLO impacts on processing of negative information in the dorsal part of the pgACC in both healthy controls and depressed patients. Our finding that PCLO genotype modulates the processing of negative novel information specifically in the pgACC replicates results obtained in healthy controls [50]–[52]. Therefore, these findings indicate that biases towards negative stimuli, as reflected in altered pgACC activation, may represent not only a feature of MDD, but also a vulnerability factor for developing mood disorders [53].
Taken together, this suggests that PCLO genotype may increase the risk for developing or maintaining a depressive disorder by affecting the mood regulating capacity of the brain.
In contrast to these psychopathology-independent findings, decreased activity in the ventral striatum during processing of negative stimuli was found in MDD patients only. MDD has been associated with altered striatal function [54], and with reward processing [55]. Since reduced serotonergic pathway signalling in the striatum was recently associated with MDD [56], it suggests a mediating effect of PCLO in MDD. Also, these findings are consistent with a recent proposition that PCLO may be particularly relevant for reward processing deficits in the context of stress [57].
We found that processing of emotional words, irrespective of valence, was modulated by the PCLO risk allele in regions including insula, IFG, caudate head, and medial frontal gyrus, as shown when performing a conjunction analysis. Low caudate activity has previously been associated with both altered inhibition of negative stimuli in subjects at risk for MDD [58] and motivational pathway dysfunction, or the inability to experience pleasure or engage in rewarding activities [59]. We propose that lower insula, and IFG activity in PCLO+ carriers may reflect a general inadequacy for regulating emotional responsiveness, either to enhance or induce a positive emotional mood state, or to down regulate negative mood states, increasing vulnerability for MDD in PCLO+ carriers.
In the present study, no effect of genotype during successful recognition was found. However, we found a blunted activity in the amygdala in healthy PCLO+ carriers and in MDD patients irrespective of genotype during recognition of new positive words, relative to old words, which underlines that new positive information results in less salience signalling in the amygdala in PCLO risk allele-carriers. This neural variation in processing novel positive information may further contribute to development of MDD symptomatology, as novel positive information appears to go undetected and may therefore further contribute to a negative biased orientation towards the world. Nevertheless, results suggested that PCLO may play a modest role in biased processing of familiar information: in PCLO+ carriers, processing of negative familiar information was associated with insular hyper-activation, as well as hypo-activation in a network implicated in reward processing and learning (including the inferior and medial frontal, hippocampal, caudate head, insula, and putamen), compared to non-risk carriers. However, given that these latter results were subtreshold only, we may conclude that PCLO predominantly affects deep processing during of semantic classification and successful encoding of novel information, which is indirect supported by animal studies showing that novel information contributes to fear aspects of depressive-like behaviour (i.e. the depression phenotype) [60].
In the present study, similar results were obtained when repeating our main analyses after excluding SSRI users, which is in line with previous findings in an emotional face processing task [12]. The recent proposal that PCLO enhances the neurophysiologic response to SSRIs in MDD patients [16] is not supported by our study.
Some limitations should be noted. First, although we used similar 3T systems at each site in this multicenter study, no systematic scanning site × diagnosis bias occurred. However, variability in image acquisition may have occurred due to minor differences in hardware (receiver coil), imaging parameters, and timing of software upgrades. Second, depression severity in our MDD patients was only mild to moderate, due to recruitment from the general population, general practitioners, and outpatient mental health organizations, but not from inpatient clinics. Consequently, we do not know whether our interaction findings would have been more robust when inpatients had also been included. Third, cell sizes were small when testing for genotype × group interactions, which may have biased our results. To increase cell sizes in neuroimaging genetic studies using GWAS as genotypic factor, correction for multiple testing requires very large sample sizes (including healthy controls), which is only feasible in a multicenter meta-analysis approach [61]. However, the present study was a follow-up of a previous GWAS for the clinical phenotype of MDD, testing only a single promising polymorphism in the PCLO gene.
Although this study provides evidence for modulation of negative word encoding related activity by PCLO genotype, its role in the serotonergic pathway remains unclear and should be the focus of future research. A promising approach is likely to be the use of positron emission tomography (PET) tracers to study radioligand binding to receptors that interact with PCLO, as shown when studying genes associated with serotonin transporter function [62]. In addition, the use of longitudinal MRI designs may be helpful to investigate whether PCLO+ carriers continue to show a negative bias reflected in lower frontostriatal activity and therefore may, indeed, be more vulnerable to develop MDD.
Conclusion
Our findings indicate that the presence of the PCLO risk allele may increase vulnerability for MDD by affecting the mood regulating capacity of the brain and by influencing dysfunctional reward processing in MDD. It further increases vulnerability for MDD by contributing to a general inadequacy for regulating emotional responsiveness. The interaction between the PCLO genotype and MDD reflected in decreased activity in the ventral striatum and amygdala also suggests that the pathophysiology of MDD is complex and may interact with the PCLO genotype. Moreover, we have found similar regions as MDD studies during emotional encoding, which indicates that the PCLO risk allele plays an important role in the mediation between MDD and altered brain activity.
Supporting Information
Figure S1: Flowchart of participants included in this study. NESDA: Netherlands Study of Depression and Anxiety; PCLO+: PCLO risk allele carriers; PCLO−: PCLO non-risk allele carriers; n: number of participants. Table S2: Optimal threshold calculation for multiple comparison correction. Table S3: Main effect of encoding and recognition, specified per valence. Table S4: PCLO genotype effect during task. Figure S5: Parameter estimates of correctly recognized old versus new words. Effect is shown at the amygdala (MNI [−27 −3 −24]: during rejection of positive new words, healthy PCLO− carriers showed increased left amygdalar activation, while no difference between processing positive old and new words was observed in PCLO+ carriers and in MDD patients, indicating blunting to novel positive information in PCLO+ carriers and patients. PCLO: Piccolo genotype; PCLO+: PCLO risk allele carriers; PCLO−: PCLO non-risk allele carriers; MDD: Major Depressive Disorder; HC: Healthy controls; AU: arbitrary units.
(DOC)
Acknowledgments
We would like to thank Anita Sibeijn, Edith Liemburg, and Ramona Demenescu for help with patient management and scanning.
Funding Statement
The infrastructure for the NESDA study (www.nesda.nl) is funded through the Geestkracht program of the Netherlands Organisation for Health Research and Development (Zon-Mw, grant number 10-000-1002) and is supported by participating universities and mental health care organizations: VU University Medical Center, GGZ inGeest, Arkin, Leiden University Medical Center, GGZ Rivierduinen, University Medical Center Groningen, Lentis, GGZ Friesland, GGZ Drenthe, Scientific Institute for Quality of Healthcare (IQ healthcare), Netherlands Institute for Health Services Research (NIVEL), Netherlands Institute of Mental Health and Addiction (Trimbos Institute), the Center for Medical Systems Biology (CSMB, NWO Genomics), Biobanking and Biomolecular Resources Research Infrastructure (BBMRI –NL), and VU University’s institutes for Health and Care Research (EMGO+) and Neuroscience Campus Amsterdam (NCA). The genotyping of the samples was provided through the Genetic Association Information Network (GAIN). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Sullivan PF, Neale MC, Kendler KS (2000) Genetic Epidemiology of Major Depression: Review and Meta-Analysis. Am J Psychiatry 157: 1552–1562. [DOI] [PubMed] [Google Scholar]
- 2.American Psychiatric Association (2002) Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, Text Revision. American Psychiatric Association.
- 3. Keedwell PA, Andrew C, Williams SC, Brammer MJ, Phillips ML (2005) The neural correlates of anhedonia in major depressive disorder. Biol Psychiatry 58: 843–853. [DOI] [PubMed] [Google Scholar]
- 4. Dannlowski U, Ohrmann P, Bauer J, Kugel H, Baune BT, et al. (2007) Serotonergic genes modulate amygdala activity in major depression. Genes Brain Behav 6: 672–676. [DOI] [PubMed] [Google Scholar]
- 5. Dannlowski U, Ohrmann P, Bauer J, Deckert J, Hohoff C, et al. (2007) 5-HTTLPR Biases Amygdala Activity in Response to Masked Facial Expressions in Major Depression. Neuropsychopharmacology 33: 418–424. [DOI] [PubMed] [Google Scholar]
- 6. Heinz A, Braus DF, Smolka MN, Wrase J, Puls I, et al. (2005) Amygdala-prefrontal coupling depends on a genetic variation of the serotonin transporter. Nat Neurosci 8: 20–21. [DOI] [PubMed] [Google Scholar]
- 7. Lau JYF, Goldman D, Buzas B, Fromm SJ, Guyer AE, et al. (2009) Amygdala Function and 5-HTT Gene Variants in Adolescent Anxiety and Major Depressive Disorder. Biol Psychiatry 65: 349–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee BT, Ham BJ (2008) Monoamine oxidase A-uVNTR genotype affects limbic brain activity in response to affective facial stimuli. NeuroReport 19: 515–519. [DOI] [PubMed] [Google Scholar]
- 9. Zill P, Baghai TC, Zwanzger P, Schule C, Eser D, et al. (2004) SNP and haplotype analysis of a novel tryptophan hydroxylase isoform (TPH2) gene provide evidence for association with major depression. Mol Psychiatry 9: 1030–1036. [DOI] [PubMed] [Google Scholar]
- 10. Bosker FJ, Hartman CA, Nolte IM, Prins BP, Terpstra P, et al. (2011) Poor replication of candidate genes for major depressive disorder using genome-wide association data. Mol Psychiatry 16: 516–532. [DOI] [PubMed] [Google Scholar]
- 11. Sullivan PF, de Geus EJC, Willemsen G, James MR, Smit JH, et al. (2009) Genome-wide association for major depressive disorder: a possible role for the presynaptic protein piccolo. Mol Psychiatry 14: 359–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Woudstra S, Bochdanovits Z, van Tol MJ, Veltman DJ, Zitman FG, et al. (2012) Piccolo genotype modulates neural correlates of emotion processing but not executive functioning. Trans Psychiatry 2: e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Choi KH, Higgs BW, Wendland JR, Song J, McMahon FJ, et al. (2011) Gene Expression and Genetic Variation Data Implicate PCLO in Bipolar Disorder. Biol Psychiatry 69: 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hek K, Mulder CL, Luijendijk HJ, van Duijn CM, Hofman A, et al. (2010) The PCLO gene and depressive disorders: replication in a population-based study. Hum Mol Genet 19: 731–734. [DOI] [PubMed] [Google Scholar]
- 15. Minelli A, Scassellati C, Cloninger CR, Tessari E, Bortolomasi M, et al. (2012) PCLO gene: Its role in vulnerability to major depressive disorder. J Affect Disord 139: 250–255. [DOI] [PubMed] [Google Scholar]
- 16. Schuhmacher A, Mössner R, Höfels S, Pfeiffer U, Guttenthaler V, et al. (2011) PCLO rs2522833 modulates HPA system response to antidepressant treatment in major depressive disorder. Int J Neuropsychopharmacol 14: 237–45. [DOI] [PubMed] [Google Scholar]
- 17. Furukawa-Hibi Y, Nitta A, Fukumitsu H, Somiya H, Furukawa S, et al. (2010) Overexpression of piccolo C2A domain induces depression-like behavior in mice. NeuroReport 21: 1177–1181. [DOI] [PubMed] [Google Scholar]
- 18.Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium (2012) A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry in press. [DOI] [PMC free article] [PubMed]
- 19. Shyn SI, Shi J, Kraft JB, Potash JB, Knowles JA, et al. (2011) Novel loci for major depression identified by genome-wide association study of Sequenced Treatment Alternatives to Relieve Depression and meta-analysis of three studies. Mol Psychiatry 16: 202–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wray NR, Pergadia ML, Blackwood DH, Penninx BW, Gordon SD, et al. (2012) Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned. Mol Psychiatry 17: 36–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Betts MJ, Russell RB (2007) Amino-Acid Properties and Consequences of Substitutions, in Bioinformatics for Geneticists: A Bioinformatics Primer for the Analysis of Genetic Data, Second Edition (ed M. R. Barnes), John Wiley & Sons, Ltd, Chichester, UK. In: Barnes M.R., editors. Bioinformatics for Geneticists: A Bioinformatics Primer for the Analysis of Genetic Data. Chichester, UK: John Wiley & Sons, Ltd. 289–316.
- 22. Stuhrmann A, Suslow T, Dannlowski U (2011) Facial emotion processing in major depression: a systematic review of neuroimaging findings. Biol Mood Anxiety Disord 1: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rude SS, Wenzlaff RM, Gibbs B, Vane J, Whitney T (2002) Negative processing biases predict subsequent depressive symptoms. Cogn Emot 16: 423–440. [Google Scholar]
- 24. Leppanen JM (2006) Emotional information processing in mood disorders: a review of behavioral and neuroimaging findings. Curr Opin Psychiatry 19: 34–39. [DOI] [PubMed] [Google Scholar]
- 25. van Tol MJ, Demenescu LR, van der Wee NJA, Kortekaas R, Marjan MA, et al. (2012) Functional Magnetic Resonance Imaging Correlates of Emotional Word Encoding and Recognition in Depression and Anxiety Disorders. Biol Psychiatry 71: 593–602. [DOI] [PubMed] [Google Scholar]
- 26. Surguladze SA, El-Hage W, Dalgleish T, Radua J, Gohier B, et al. (2010) Depression is associated with increased sensitivity to signals of disgust: A functional magnetic resonance imaging study. J Psychiatr Res 44: 894–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Wingen GA, van Eijndhoven P, Tendolkar I, Buitelaar J, Verkes RJ, et al. (2011) Neural basis of emotion recognition deficits in first-episode major depression. Psychol Med 41: 1397–1405. [DOI] [PubMed] [Google Scholar]
- 28. Burt DB, Zembar MJ, Niederehe G (1995) Depression and Memory Impairment: A Meta-Analysis of the Association, Its Pattern, and Specificity. Psychol Bull 117: 285–305. [DOI] [PubMed] [Google Scholar]
- 29. Heller AS, Johnstone T, Shackman AJ, Light SN, Peterson MJ, et al. (2009) Reduced capacity to sustain positive emotion in major depression reflects diminished maintenance of fronto-striatal brain activation. Proc Natl Acad Sci U S A 106: 22445–22450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shestyuk AY, Deldin PJ, Brand JE, Deveney CM (2005) Reduced Sustained Brain Activity During Processing of Positive Emotional Stimuli in Major Depression. Biol Psychiatry 57: 1089–1096. [DOI] [PubMed] [Google Scholar]
- 31. Wagner AD, Schacter DL, Rotte M, Koutstaal W, Maril A, et al. (1998) Building memories: Remembering and forgetting of verbal experiences as predicted by brain activity. Science 281: 1188. [DOI] [PubMed] [Google Scholar]
- 32. Elliott R, Rubinsztein JS, Sahakian BJ, Dolan RJ (2002) The Neural Basis of Mood-Congruent Processing Biases in Depression. Arch Gen Psychiatry 59: 597–604. [DOI] [PubMed] [Google Scholar]
- 33. Ibi D, Nitta A, Ishige K, Cen X, Ohtakara T, et al. (2010) Piccolo knockdown-induced impairments of spatial learning and long-term potentiation in the hippocampal CA1 region. Neurochem Int 56: 77–83. [DOI] [PubMed] [Google Scholar]
- 34. Scharinger C, Rabl U, Pezawas L, Kasper S (2011) The genetic blueprint of major depressive disorder: Contributions of imaging genetics studies. World J Biol Psychiatry 12: 474–488. [DOI] [PubMed] [Google Scholar]
- 35. Bigos KL, Weinberger DR (2010) Imaging genetics - days of future past. NeuroImage 53: 804–809. [DOI] [PubMed] [Google Scholar]
- 36. Savitz JB, Drevets WC (2009) Imaging phenotypes of major depressive disorder: genetic correlates. Neuroscience 164: 300–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Penninx BWJH, Beekman AT, Smit JH, Zitman FG, Nolen WA, et al. (2008) The Netherlands Study of Depression and Anxiety (NESDA): rationale, objectives and methods. Int J Methods Psychiatr Res 17: 121–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rush A, Giles D, Schlesser M, Fulton C, Weissenburger J, et al. (1986) The inventory for depressive symptomatology (IDS): preliminary findings. Psychiatry Res 18: 65–87. [DOI] [PubMed] [Google Scholar]
- 39. Montgomery S, Åsberg M (1979) A new depression scale designed to be sensitive to change. Br J Psychiatry 134: 382–389. [DOI] [PubMed] [Google Scholar]
- 40. Demenescu LR, Renken R, Kortekaas R, van Tol MJ, Marsman JBC, et al. (2011) Neural correlates of perception of emotional facial expressions in out-patients with mild-to-moderate depression and anxiety. A multicenter fMRI study. Psychol Med 41: 2253–2264. [DOI] [PubMed] [Google Scholar]
- 41. van Tol MJ, van der Wee NJA, Demenescu LR, Nielen MMA, Aleman A, et al. (2011) Functional MRI correlates of visuospatial planning in out-patient depression and anxiety. Acta Psychiatr Scand 124: 273–284. [DOI] [PubMed] [Google Scholar]
- 42. Maldjian JA, Laurienti PJ, Kraft RA, Burdette JH (2003) An automated method for neuroanatomic and cytoarchitectonic atlas-based interrogation of fMRI data sets. NeuroImage 19: 1233–1239. [DOI] [PubMed] [Google Scholar]
- 43. Friston KJ, Penny WD, Glaser DE (2005) Conjunction revisited. NeuroImage 25: 661–667. [DOI] [PubMed] [Google Scholar]
- 44. Linden DEJ (2006) How psychotherapy changes the brain - the contribution of functional neuroimaging. Mol Psychiatry 11: 528–538. [DOI] [PubMed] [Google Scholar]
- 45. Dennis NA, Cabeza R, Need AC, Waters-Metenier S, Goldstein DB, et al. (2011) Brain-derived neurotrophic factor val66met polymorphism and hippocampal activation during episodic encoding and retrieval tasks. Hippocampus 21: 980–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van Wingen G, Rijpkema M, Franke B, van Eijndhoven P, Tendolkar I, et al. (2010) The brain-derived neurotrophic factor Val66Met polymorphism affects memory formation and retrieval of biologically salient stimuli. NeuroImage 50: 1212–1218. [DOI] [PubMed] [Google Scholar]
- 47. Fitzgerald PB, Laird AR, Maller J, Daskalakis ZJ (2008) A meta-analytic study of changes in brain activation in depression. Hum Brain Mapp 29: 683–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, et al. (2007) Dissociable Intrinsic Connectivity Networks for Salience Processing and Executive Control. J Neurosci 27: 2349–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Menon V, Uddin L (2010) Saliency, switching, attention and control: a network model of insula function. Brain Struct Funct 214: 655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Elliott R, Rubinsztein JS, Sahakian BJ, Dolan RJ (2000) Selective attention to emotional stimuli in a verbal go/no-go task: an fMRI study. NeuroReport 11: 1739–1744. [DOI] [PubMed] [Google Scholar]
- 51. Mak AKY, Hu Zg, Zhang JX, Xiao ZW, Lee TMC (2009) Neural correlates of regulation of positive and negative emotions: An fMRI study. Neurosci Lett 457: 101–106. [DOI] [PubMed] [Google Scholar]
- 52. Shafritz KM, Collins SH, Blumberg HP (2006) The interaction of emotional and cognitive neural systems in emotionally guided response inhibition. NeuroImage 31: 468–475. [DOI] [PubMed] [Google Scholar]
- 53. Chamberlain S, Sahakian B (2006) The neuropsychology of mood disorders. Curr Psychiatry Rep 8: 458–463. [DOI] [PubMed] [Google Scholar]
- 54. Hamilton JP, Gotlib IH (2008) Neural Substrates of Increased Memory Sensitivity for Negative Stimuli in Major Depression. Biol Psychiatry 63: 1155–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Smoski M, Felder J, Bizzell J, Green S, Ernst M, et al. (2009) fMRI of alterations in reward selection, anticipation, and feedback in major depressive disorder. J Affect Disord 118: 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Murrough J, Henry S, Hu J, Gallezot JD, Planeta-Wilson B, et al. (2011) Reduced ventral striatal/ventral pallidal serotonin1B receptor binding potential in major depressive disorder. Psychopharmacology 213: 547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bogdan R, Nikolova YS, Pizzagalli DA (2012) Neurogenetics of depression: A focus on reward processing and stress sensitivity. Neurobiol Dis in press. [DOI] [PMC free article] [PubMed]
- 58. Lisiecka DM, Carballedo A, Fagan AJ, Connolly G, Meaney J, et al. (2012) Altered inhibition of negative emotions in subjects at family risk of major depressive disorder. J Psychiatr Res 46: 181–188. [DOI] [PubMed] [Google Scholar]
- 59. Epstein J, Pan H, Kocsis JH, Yang Y, Butler T, et al. (2006) Lack of ventral striatal response to positive stimuli in depressed versus normal subjects. Am J Psychiatry 163: 1784–1790. [DOI] [PubMed] [Google Scholar]
- 60. Li Y, Zheng X, Liang J, Peng Y (2010) Coexistence of Anhedonia and anxiety-independent increased novelty-seeking behavior in the chronic mild stress model of depression. Behav Processes 83: 331–339. [DOI] [PubMed] [Google Scholar]
- 61. Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, et al. (2012) Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet 44: 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. David SP, Murthy NV, Rabiner EA, Munafó MR, Johnstone EC, et al. (2005) A functional genetic variation of the serotonin (5-HT) transporter affects 5-HT1A receptor binding in humans. J Neurosci 25: 2586–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Flowchart of participants included in this study. NESDA: Netherlands Study of Depression and Anxiety; PCLO+: PCLO risk allele carriers; PCLO−: PCLO non-risk allele carriers; n: number of participants. Table S2: Optimal threshold calculation for multiple comparison correction. Table S3: Main effect of encoding and recognition, specified per valence. Table S4: PCLO genotype effect during task. Figure S5: Parameter estimates of correctly recognized old versus new words. Effect is shown at the amygdala (MNI [−27 −3 −24]: during rejection of positive new words, healthy PCLO− carriers showed increased left amygdalar activation, while no difference between processing positive old and new words was observed in PCLO+ carriers and in MDD patients, indicating blunting to novel positive information in PCLO+ carriers and patients. PCLO: Piccolo genotype; PCLO+: PCLO risk allele carriers; PCLO−: PCLO non-risk allele carriers; MDD: Major Depressive Disorder; HC: Healthy controls; AU: arbitrary units.
(DOC)