Abstract
Microbial fermentation process development is pursuing a high production yield. This requires a high throughput screening and optimization of the microbial strains, which is nowadays commonly achieved by applying slow and labor-intensive submerged cultivation in shake flasks or microtiter plates. These methods are also limited towards end-point measurements, low analytical data output, and control over the fermentation process. These drawbacks could be overcome by means of scaled-down microfluidic microbioreactors (μBR) that allow for online control over cultivation data and automation, hence reducing cost and time. This review goes beyond previous work not only by providing a detailed update on the current μBR fabrication techniques but also the operation and control of μBRs is compared to large scale fermentation reactors.
INTRODUCTION
Recently, there is a great attention to the development of novel microbial cell cultivation technologies for high-throughput screening and industrial bioprocess follow-up, to accelerate our scientific insights in the functioning of microorganisms at molecular and genetic levels. Instrumented stirred-tank bioreactors effectively control temperature, pH, and dissolved oxygen (DO) levels by regulating mixing and the gas sparging rate, resulting in valuable physiological and metabolic information at different stages of the fermentation process. Conventional fermentation by stirred-tank bioreactors are, however, typically expensive and labor intensive for screening purposes. Cultivations performed in bench scale bioreactors, on the other hand, result in reliable and information rich data, because these bioreactors are equipped with sophisticated measurement and control devices. Still, such bench scale bioreactors remain expensive and time consuming to operate, limiting the number of experiments that can be carried out within a certain time frame. Bench-scale stirred-tank bioreactors have been enhanced by reducing the reactor volume and increasing the number of parallel operating reactors,1 but more efforts are still required, to overcome the problems of sterilization, assembly, cleaning, and calibration of reactors’ sensors. Test tubes, shake flasks, and microtiter plates are widely used as simple bioreactors for fermentation and cell culture experiments. These bioreactors can easily be operated with small volumes, but they have weak control over bioprocess conditions and the data obtained are often limited to endpoint measurements. The main advantage of microtiter plates as a cell cultivation system is that they are designed for high-throughput and automated experimental set-ups. Microtiter plates for cell cultivation typically contain 24 or 96 parallel wells with working volumes typically ranging from 0.1 to 3 ml.2, 3 As normally only temperature is controlled, efforts have been made to implement on-line pH or DO measurements units in microtiter plates as well4, 5, 6 to obtain continuous process information during fermentation. Indeed, 24- and 96-well plates with integrated optical sensors (optodes) for the measurement of pH or dissolved oxygen concentration are already commercially available and will help to increase their use to study the physiological conditions of the fermentation processes.4, 5, 7, 8 A downside of the optodes is their small measurement range. Several industrially relevant species, e.g., yeasts, are generally cultivated at a pH around 5, which is outside the working range of the optodes. Therefore, the current challenge in the further development of high throughput microreactor systems is the increase of the measurement and control possibilities.9, 10, 11, 12, 13
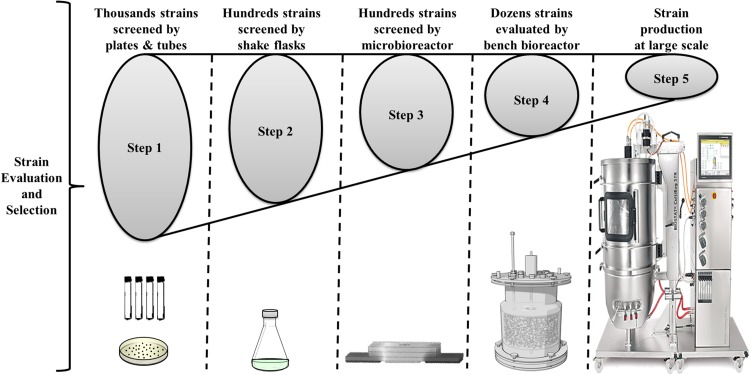
Microbioreactors with integrated sensors combine the small volumes of microtiter plates with the monitoring and control features found in bench-scale systems to form promising tools for rapid, high-throughput, and cost-effective screening. If a bioreactor could be miniaturized to the micrometer scale, numerous new applications would become possible, such as biosensors, micro-fermentor arrays, or microbiological assay kits. Micro-fermentor systems could also facilitate the process flow for strain screening and evaluation as illustrated in Fig. 1. To achieve such miniaturization, at least three requirements must be fulfilled. First, the reaction chamber itself should be reduced to micrometer dimensions. Second, the microorganism must be injected into the chamber without any damage or leakage. Finally, novel microfluidic mechanisms must be devised to deliver material to the tiny chamber and recover it again. Recently, there has been a growing concern to develop microfluidic lab-on-a-chip and micro-chemical reactors able to accomplish chemical and biological experiments and analysis. Current developments focus on relatively fast mixing, sufficient heat transfer, sensitive and efficient reaction control, and simple scaling up. Some of these micro-devices have already been produced on commercial production scales, but many more designs have been developed on a laboratory scale. In this review paper, applicability of the most recent integrated microfluidic microbioreactors (μBR) systems are described and compared to traditional bioreactors. The scope of this review is restricted to microbioreactors that allow fermentations with microorganisms growing in suspension, since the majority of industrial fermentations are based on this type of process. Moreover, in this review, we will only consider microbioreactors with an operating volume smaller than 1 ml, which are designed to mimic typical bench-scale reactors. Therefore, the term “μBR” will exclusively refer to this type of reactor.
Figure 1.
The process flow for the strain screening and evaluation with high throughput μBR positioned in the middle directly before scaling-up to the bioreactor.
CHALLENGES OF μBRs
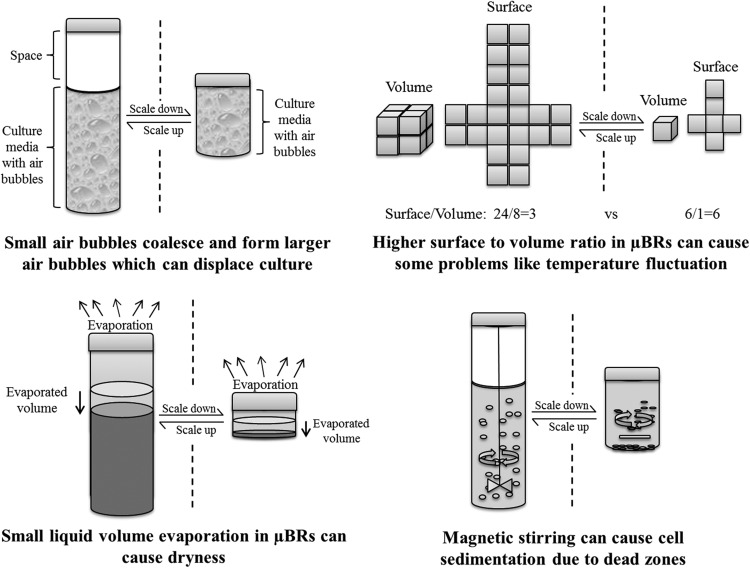
μBR indeed have clear advantages, like small volume, little or no need for cleaning (one time usage), and high throughput (multiple μBRs in parallel). Even though μBRs have many advantages, it is important to bear in mind that they also have issues related to their size and handling. Some of these issues are illustrated in Fig. 2. For example, continuous cultivations pose several difficulties to the design of a μBR system as the reactor volume has to stay constant by equalizing the volumetric inflow and the volumetric outflow of the reactor. Moreover, bioreactions can only truly be regarded as continuous culture reactions if the culture medium composition of the outflow is the same as that of the reactor content, i.e., if the assumption of an ideally mixed reactor holds. This in turn requires the mixing system to be efficient both in achieving a uniform distribution of nutrients and oxygen and in keeping the cells in suspension without the formation of dead zones. In μBRs, liquid motion in the reactor becomes predominantly laminar, so mixing in this case has to rely on minimizing diffusion distances.14 An additional problem is the fast sedimentation of some cells if there are any zones without uplift in the reactor. Also, due to the small size of μBRs, sparging of air is not an option. Small air bubbles coalesce and form larger air bubbles which can displace culture liquid or block fluidic ports and disturb optical measurements. Currently, cultivations are run with a fixed concentration of oxygen in the aeration gas without the flexibility to change the amount of DO via an input channel during the fermentation. Temperature fluctuations, due large surface to volume ratios, could significantly affect growth. Also, the limited pH range compatible with current polymer μBRs will not suit all kinds of organisms as many will require a more acidic pH. Also, evaporation becomes an issue at this reactor scale as it can reach relatively high rates. Boccazzi et al.15 measured an evaporation rate of 4.3 ± 0.4 μl/h in 100 μl volume polydimethylsiloxane (PDMS) μBR, which means that the reactor would thus completely dry out within 24 h. Leakage at the fluidic connections is another potential issue related to the pressures needed for the cultivation of cells. Finally, signal collection of different process variables in μBR is not straightforward. It relies on analytical methods which are not sufficiently developed at the moment.
Figure 2.
Some of the complications that are facing scaling down process of bioreactors.
μBR STATE OF ART
Fabrication methods
Fabrication materials
μBRs prototypes are typically made from polymethylmethacrylate (PMMA) and PDMS.16, 17, 18, 19 These two polymers are the most commonly used substrates for the fabrication of μBR prototypes due to their low material cost, easy handling, biocompatibility, solvent and chemical compatibility, and durability. Additionally, these materials also exhibit a high optical transmission in the visible wavelength range, which facilitates optical measurements.20, 21 Also, from processing perspective, PMMA and PDMS substrates are often preferred materials. It is possible to fabricate two- or three-dimensional (2D or 3D) microfluidic geometries using rather straightforward micromachining processes.22, 23 Their low material cost coupled with their cheap fabrication processes is ideal characteristics for cost-effective mass production and rapid prototyping. This also allows for the fabrication of disposable μBR, a useful advantage, as disposable reactors prevent sample contamination and reduce the effort involved in reactor preparation or cleaning.
PMMA is a synthetic thermoplastic polymer of methyl methacrylate which sometimes called acrylic glass and often used as an alternative to glass due to its light weight, transparency, and break-resistance. PMMA is often preferred because of its moderate properties, easy handling and processing, and low cost but behaves in a brittle manner when loaded, especially under an impact force, and is more prone to scratching than conventional inorganic glass.
PDMS is one of the polymeric organo-silicon compounds that are generally referred to as silicones.24 PDMS is the most extensively used silicon-based organic polymer that is known for its supreme rheological properties. An important advantage of PDMS substrates over other thermoplastic polymers like PMMA, PEEK, or PC is that PDMS enables the integration of microfluidic devices such as microvalves, micropumps, and mixers as an integral part of the reactor.16, 25 While the integration of microfluidic devices may complicate the reactor design, it—more importantly—decreases the cost of the entire μBR setup because fewer macro-scale devices are needed to drive the system. Furthermore, PDMS also exhibits high permeability to oxygen and carbon dioxide, which makes it highly suitable for cell-based systems.22 Despite all of these advantages, PDMS also suffers from several drawbacks. One drawback is that the gas permeability of PDMS can lead to unwanted levels of evaporation.22, 26 PDMS also swells in most organic solvents,22 although this is not really an issue for standard microbial fermentations because most fermentation media are prepared using water as a solvent. However, the swelling could become a problem for novel reactor designs where a solvent is added to the fermentation broth, for example, to remove product or inhibitory compounds from the water phase in an attempt to increase the productivity.27, 28
Polycarbonate (PC) is a more expensive alternative, when greater strength is required, but it contains potentially harmful bisphenol-A subunits. A yet other alternative polymer material is polyetheretherketone (PEEK), which is an expensive, but largely inert material. Glass or even silicon can also be used as substrates for the fabrication of μBR due to the easier integration of sensors and standard high throughput processing possibilities at foundries. Naturally, these materials are less suited for standard prototyping.29
Fabrication technology
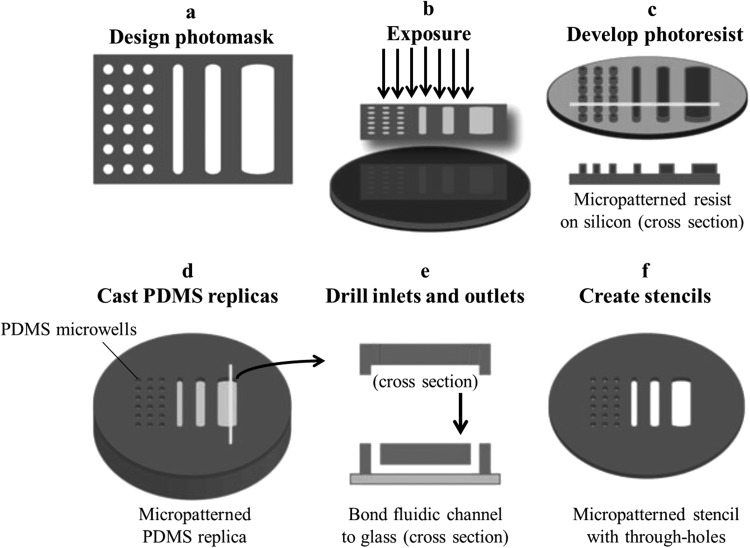
Microfabrication technology was initially developed for the semiconductor industry to create integrated circuits, and it offers precise reproducible spatial and temporal control. For building microfabricated cell-based devices, soft lithography is the most commonly used technique.30 In this technique, a photomask is depicted by a computer-aided design (CAD) tool and printed with a high-resolution printer with minimum feature sizes of 10 μm. Photolithography is then executed by using thin film of photosensitive material, such as SU-8 epoxy, to spin-coat a polished silicon wafer, and then the film was exposed through the printed photomask and developed to generate stable raised microstructured designs on the silicon substrate. Following this method, microstructure heights are settled by thin-film thickness (1-1000 μm), and the residual measurements are determined by the photomask. Once the silicon master mold is fabricated, elastomeric replicas can be repeatedly formed by mixing PDMS pre-polymer (resin plus a curing agent), casting or spin-coating it on the microstructured master, thermally curing the polymer, and peeling the resulting flexible and transparent films. Because the uncured pre-polymer flows and conforms to the underlying microstructures, patterns are transferred with high fidelity from silicon masters to the polymer replicas. The resulting microstructured PDMS films can then be used directly in different forms, i.e. stamps, open microwells, or three-walled microchannels. When PDMS templates with full thickness through-holes are required, the pre-polymer can be spin-coated onto the microstructures, and a rigid non-adhesive flat plate used to apply downward force during curing. Finally, stable microfluidic networks with closed-channel can be fabricated by permanently bonding the PDMS three-walled microchannel networks onto glass slides by treatment with oxygen plasma surface. This method is summarized pictorially in Fig. 3.31
Figure 3.
Schematic steps of microfabrication by soft lithography. (a) Using computer-aided design tool to draw photomasks, which then printed on transparency films. (b) Photoresist-coated silicon wafers are exposed through the photomask. (c) The photomask design is translated to the silicon surface. (d) The silicon then acts as a master mold for casting replicas of transparent PDMS polymer. (e) PDMS can be drilled to create inlets and outlets and bonded to a glass substrate to create closed microfluidic devices or (f) used to create stencils with micro-patterned through-holes. Reprinted with permission from M. L. Yarmush and K. R. King, Annu. Rev. Biomed. Eng. 11, 235 (2009). Copyright 2009 Annual Reviews.
Different types of μBRs
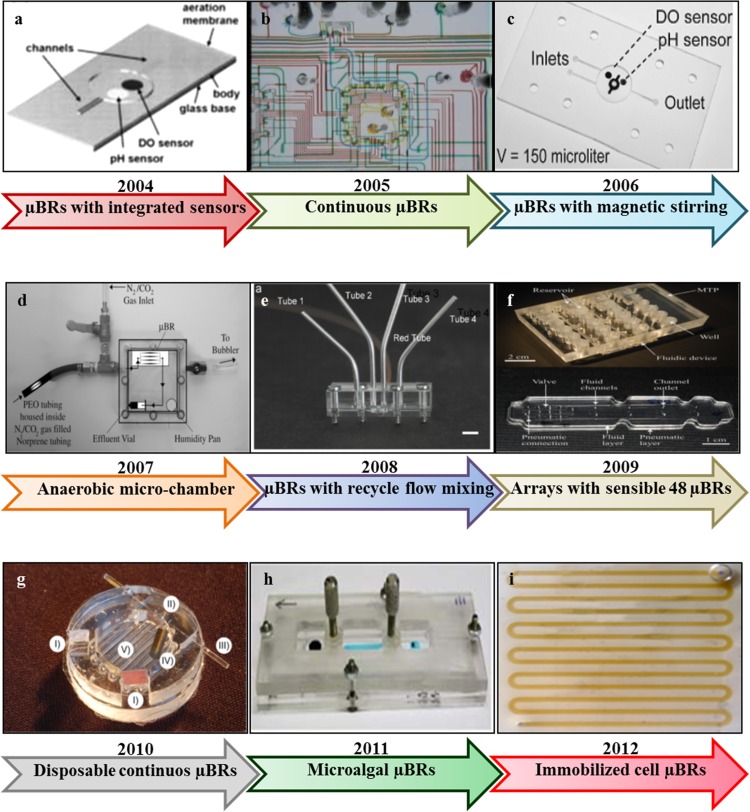
Microfluidic approaches using small channels and wells typically fabricated in PMMA and PDMS have been implemented to generate an inexpensive solution for microreactors. Applications being considered mainly include strain screening and examining growth parameters. Table TABLE I. highlights the main specifications of currently fabricated μBRs aiming to reproduce lab scale microbial batch processes and achieve reasonable biomass levels, while Fig. 4 illustrates the pictorial chronological milestones of μBRs progress. In the following paragraphs we will describe μBRs based on their type (single or array μBR) as well as on their level of integrated components for a better control of the microbial microenvironment.
TABLE I.
Fabricated μBRs devices and their main characteristics.
| Bioreactor type | Material | Volume (μl) | Dimensions H a × Ø b (mm) | Microorganism | Measurements and controls | Reference |
|---|---|---|---|---|---|---|
| μBR array | Polystyrene | 250 | N/A | Escherichia coli MG1655 | DO-pH-T-OD | 36 |
| Membrane-aerated μBR | PDMS/Glass | 50 | 0.3 × N/A | Escherichia coli FB21591 | DO-pH-OD | 18 |
| μBR | PDMS c/PMMA d | 150 | 1 × 10 | Escherichia coli FB21591 | DO e-pH-OD f-M g | 44 |
| Integrated μBR | PDMS | 50 | N/A | Escherichia coli strains DPD2276 and DPD2417 | DO-pH-luminescence-fluorescence-OD | 48 |
| μBR | PDMS/PMMA | 150 | 2 × 10 | Saccharomyces cerevisiae ATCC 4126 | DO-pH-OD-M | 15 |
| Anaerobic μBR | PDMS | 1 | 0.1 × 1.5 | Methanosaeta concilii GP6 | Anaerobic | 52 |
| μBR | PDMS | 30 | 1.5 × 5 | ··· | M | 47 |
| μBR | PMMA/PDMS | 1000 | 20 × 12 | Escherichia coli K12 | pH-OD | 37 |
| μBR | PDMS/PMMA | 190 | 3 × 9 | Saccharomyces cerevisiae CEN.PK 113–7D | DO-pH-OD-T-M | 46 |
| Microfluidic reactor | PDMS | 8 | N/A | Saccharomyces cerevisiae DSM 2155 | DO-OD | 33 |
| Parallel μBR | Polystyrene | 100 | 12.2 × 6.5 | Candida utilis CBS621 | DO-pH-OD-M | 38 |
| Microturbidostat | PDMS | 0.2 × 0.2 | Escherichia coli MG1655 | Pneumatic valve controller and fluorescence intensity of bacterial proteins | 45 | |
| Batch and continuous μBR | PDMS | 100 | 4.2 × 14 | Saccharomyces cerevisiae CEN.PK 113-7D | DO-pH-OD-T h-M | 32 |
| Parallel μBR | PDMS | 9 × 5 chambers | 21 × 39 | Aspergillus ochraceus Wilhelm DSM 63304 | ··· | 54 |
| Microfluidic device | PDMS/Glass/PMMA | 2.4 | 17.5 × 2.5 | Tetraselmis chuii LB 232 and Neochloris oleabundans UTEX 1185 | ··· | 53 |
| Microfluidic chemostat & turbidostat | PDMS/PC i | 1000 | N/A | Escherichia coli FB21591 | DO-pH-OD-M | 34 |
| BAY μBR | Parylene-C/Teflon-AF | 70 | 9.5 × 10.5 | Escherichia coli DH5α/Saccharomyces cerevisiae BY4741/Cyclotella cryptica (CCMP 332) | OD | 51 |
| Microchannel bioreactor | Quartz + PEI j | N/A | N/A k | Saccharomyces cerevisiae ATCC 204679 | ··· | 35 |
H: Height.
Ø: Diameter.
PDMS: Polydimethylsiloxane.
PMMA: Poly(methylmethaacrylate).
DO: Dissolved oxygen.
OD: Optical density.
M: Mixing.
T: Temperature.
PC: Polycarbonate.
PEI: Polyethylenimine.
N/A: Not available.
Figure 4.
The main chronological milestones of microfluidic cell bioreactors development. (a) Reprinted with permission from Zanzotto et al., Biotechnol. Bioeng. 87, 243 (2004). Copyright 2004 Wiley. (b) Reprinted with permission from Balagadde et al., Science 309, 137 (2005). Copyright 2005 The American Association for the Advancement of Science (AAAS). (c) Reprinted with permission from Boccazzi et al., Biotechnol. Prog. 22, 710 (2006). Copyright 2006 Wiley. (d) Reprinted with permission from Steinhaus et al., Appl. Environ. Microbiol. 73, 1653 (2007). Copyright 2007 American Society for Microbiology (ASM). (e) Reprinted with permission from Li et al., Chem. Eng. Sci. 63, 3036 (2008). Copyright 2008 Elsevier. (f) Reprinted with permission from Buchenauer et al., J. Micromech. Microeng. 19, 8 (2009). Copyright 2009 IOPscience. (g) Reprinted with permission from Schäpper et al., Chem. Eng. J. 160, 891 (2010). Copyright 2010 Elsevier. (h) Reprinted with permission from Holcomb et al., Anal. Bioanal. Chem. 400, 245 (2011). Copyright 2011 Springer. (i) Reprinted with permission from Seo et al., Process Biochem. 47, 1011 (2012). Copyright 2012 Elsevier.
Batch/continuous μBRs
Continuous fermentation is the process in which cells are maintained in the exponential growth phase by the regular addition of fresh medium that is exactly balanced by the exclusion of cell suspension from the bioreactor. Research has been extended to evaluate the applicability of μBRs in continuous mode fermentation. Schäpper et al.32 presented a single-use μBR device for both batch and continuous cultivation of suspended cells. The device combined the small working volumes known from micro-well plates with the versatility of bench-scale reactors. Homogeneous mixing of the broth was achieved with a free-floating stirrer bar which also provided the updraft necessary to keep cells in suspension. Temperature and pH could be tightly controlled to the desired set point; DO and cell were also measured on-line. Aeration was provided through a semipermeable membrane which separates the gas from the liquid phase. The addition of CO2 and NH3 gases through the same membrane was used to control pH. Validation of the device was carried out by cultivations of Saccharomyces cerevisiae. The resulting growth curves and DO over time were similar to those seen for bench-scale reactors. Another diffusion-based microreactor system with a reaction volume of 8 μl was fabricated, operated, and characterized by Edlich et al.33 in order to increase the understanding of micro-scale cultivations and its potential as screening tool for biological processes. The advantage of this latter continuous cultivation μBR was the integrated online measurement technique for DO and optical density (OD). Degassed medium and moistened air flow were used above the gas permeable PDMS membrane to reduce undesirable bubble formation before and during operation. Because of the air-permeable PDMS membrane, a passive oxygen supply of the culture medium in the device was ensured by diffusion. The oxygen supply itself was monitored online via integrated DO sensors based on a fluorescent dye complex. Results of culturing experiments with S. cerevisiae DSM 2155 showed reproducible concentration courses of DO, glucose, ethanol, and dry weight of biomass. Although mixing was performed, biofilm formation was observed resulting in low oxygen transfer rates and limited growth in suspension.
Since chemostats are frequently used to study cell metabolic rates, then sampling of growth chamber fluid for offline analysis is essential to measure chemical products. While micro-scale cultures reduce cost by reducing media usage, volume reduction also directly affects sampling time negatively. For example, a typical 50 μl sample volume for high-performance liquid chromatography (HPLC) taken from a 100 nl bioreactor requires 500 turnovers of the chip volume. For bacterial cells with a 1 h−1 growth rate, a single sample can take 500 h. Based on this fact, Lee et al.34 demonstrated long term micro-scale continuous culture of Escherichia coli for 500 h. By developing a device platform that incorporated rigid materials with PDMS membranes, a novel strategy for evaporation prevention and reliable flow control was demonstrated allowing long term operation without concentration drift. An integrated peristaltic pump enabled online flow control, and multiple inputs allowed on-chip media preparation and concentration control. Both cell density control in chemostat mode using glucose and cell density control in turbidostat mode using flow rate were demonstrated. In order to observe detailed cell metabolism, HPLC analysis of chemical concentrations in different steady and dynamic states of operation was compared in both 1 ml volume and previous devices. From chemostat experiments, steady state production of different acids or products could be characterized. From turbidostat experiments, maximum cell growth rates could be directly measured, and observation of overflow metabolism could be analyzed to determine acid yields. Research was extended to apply continuous mode to the immobilized form of microbial cells. The immobilized biocatalysts have been extensively investigated during last few decades. Immobilization of microbial cells showed certain technical and economical advantages over free cell system. Microchannel bioreactor was designed by Seo et al.35 to test continuous fermentation with immobilized cell. The fermentation system included a hydrophilic quartz channel in the bottom to immobilize cells using 0.4 wt. % polyethyleneimine and a channel designed to continuously remove metabolically produced carbon dioxide in the top using hydrophobic polypropylene. Ethanol fermentation was executed using S. cerevisiae and Pichia stipitis to evaluate fermentation characteristics of immobilized cells and to identify long-term activity of immobilized S. cerevisiae cells. The continuous flow microchannel bioreactor was operated steadily over a period of 1 month.
μBR arrays
A scalable μBR array technology for parametric control of high-throughput cell cultivations has been regularly reported. The number of wells for such μBRs ranged from 2 to 48, and different technologies are used to control process parameters including optical sensor technology and electrochemical sensors. Maharbiz et al.,36 for example, applied printed circuit board (PCB) technology to continuously monitor the pH of 8 × 250 μl, whereas Buchenauer et al.37 monitored pH and biomass online in 48 μBRs by implementing a commercially available optical sensor technology. Using this latter technology, the pH could be sustained within 7–7.3 in a controlled culture of E. coli, while the pH in an uncontrolled culture ranged between 6.5 and 9. Even more parameters, namely, temperature, pH, DO, and viable biomass concentration, were controlled by an array of electrochemical sensors in a 2 × 100 μl μBRs array.38 The use of such online measurements resulted in very reproducible batch cultivations and was comparable to measurement obtained for a similar aerobic batch cultivation carried out in a conventional 4 L bench-scale bioreactor. For longtime experiments fouling issues were however observed, and further steps, for example the inclusion of electrochemical cleaning steps, might help in reducing the fouling problems on the electrochemical sensors.38
Control of microfluidics in μBRs
Modeling and simulation
There is a constant need for both strain and process improvement within the biotechnology industry to maintain competiveness. In this context, mechanistic models describing the different phenomena taking place in the μBRs are important tools for optimization and control of the process.39 The interplay between different metabolic pathways, regulation mechanisms, inhibition by substrates and products, and formation of intermediate substrates and products results in complex kinetic models. When it comes to modeling of the mass flow, the commonly used stirred tank bioreactors are most often assumed as ideal, and thus the existence of concentration gradients throughout the reactor is neglected. However, in large scale production bioreactors concentration gradients of substrates and products do exist as demonstrated by experimental work in combination with computational fluid dynamics (CFD) and may have a significant influence on the resulting yields and productivities of a process.40, 41
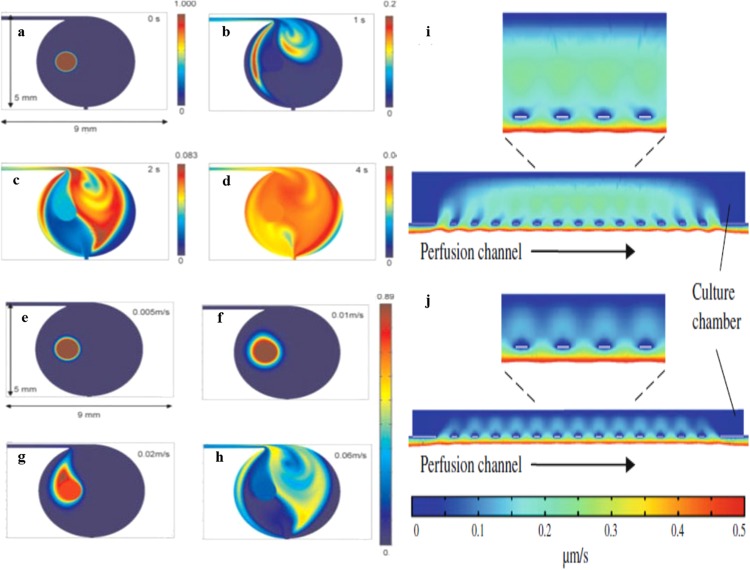
To optimally design the mixing performance of the μBR, comsol simulations were performed.42 Passive mixing was chosen, as it does not require the installation of moving parts. Two different parameters, namely velocity and oscillation, were taken into account in this simulation. According simulations, complete mixing could be achieved within 4 s for the specific μBR design. The mixing in the overall reactor system can be divided into two parts, a convection based dispersion (Figs. 5a, 5b, 5c, 5d) and a two-directional Taylor dispersion (Figs. 5e, 5f, 5g, 5h). Taylor dispersion arises from the velocity profile near the channel walls due to the no-slip flow condition at the solid-fluid interface. For another design, a 3D comsol finite element simulation was used based on the Navier–Stokes equation to study the distribution of the velocity field and shear stress.43 Different normal inflow velocities were applied to the inlet of the perfusion channel while the pressure of the outlet was set to zero (Figs. 5i, 5j). Simulations indicated that for this configuration diffusion was by far the dominant mass transport mechanism, while convective flows were significantly suppressed. It was also implied that a shallower chamber would result in further suppression of shear stress. In any case, these results clearly indicate that simulations help to make an effective reactor design ensuring an even flow field and leaving minimal room for dead zones within the μBR.
Figure 5.
Simulation result of the concentration profile of oscillation mixing in the reactor at different times: (a) t = 0 s, (b) t = 1 s, (c) t = 2 s, and (d) t = 4 s and with different pulling and pushing linear flow rates: (e) 0.005 m s−1, (f) 0.01 m s−1, (g) 0.02 m s−1, and (h) 0.06 m s−1. Reprinted with permission from Li et al., AIChE J. 55, 2725 (2009). Copyright 2009 Wiley. FEM simulation results of velocity fields around the piezoelectric transducer array and the culture chamber of models: (i) D100 and (j) D50, and arrows indicate the flow direction. Reprinted with permission from Y.-H. Hsu and W. Tang, Microfluid. Nanofluid. 11, 459 (2011). Copyright 2011 Springer.
μBRs with an integrated filter
Fermentation with obligate aerobes requires oxygen for aerobic cellular respiration to oxidize substrates (for example sugars and fats) in order to obtain energy. One of the most important cellular growth limitations in μBR is the depletion of oxygen. A lot of efforts have been made to overcome or control this problem. One suggested solution is the use of a thin layer of a gas-permeable PDMS for aeration through which oxygen can diffuse.18, 44 By changing the gaseous environmental conditions, it was demonstrated that oxygen levels within the μBR could be manipulated. Furthermore, it was demonstrated that the sensitivity and reproducibility of the μBR system were such that statistically significant differences in the time evolution of the OD, DO, and pH could be used to distinguish between different physiological states.18
Integrated filters were not only used for aeration purposes but also to prevent bacterial cells from being leaked and lost. In order to avoid bacterial leakage, the use of agarose filters has been described.45 The bacterial culture chamber was connected to a single inoculum input-output channel and to a nutrient channel in which the agarose filter safeguards the diffusion of nutrients and metabolites without bacterial leakage. Such a filter was also used for the supply of CO2 and NH3 gases to control pH.32
Besides filters to control aeration, control of evaporation is important. A novel strategy for evaporation prevention and reliable flow control was demonstrated using an integrated peristaltic pump. This approach enabled online flow control, while multiple inputs allowed on-chip media preparation and concentration control.
μBR with active mixer
Bioreactors include a complex three-phase flow (liquid, gas, and biomass), and the interaction between mixing and biological reactions is complicated by the fact that such interactions take place simultaneously. Mixing is one of the most important operations in a submerged cultivation; the mixing quality directly influences the distribution and suspension of substrate and microorganisms in the reactor and thus affects both the growth characteristics and the quality of the measurements. Also, optimal mixing ensures an even temperature profile throughout the reactor. Mixing becomes more difficult as the scale of the reactor decreases, as it becomes impossible to achieve the turbulent conditions associated with good mixing. It was also found that without stirring, gas bubbles may aggregate which could introduce unwanted variation in mass transfer characteristics.36
Several efforts to ensure an efficient mixing have been described. For some μBRs, mixing is achieved by means of a small magnetic stir bar.15, 32, 34, 38, 44, 46 The efficiency was examined by using fluorescent dyes mixing and computational fluid dynamics simulations. As an indication of the efficiency of mixing, the distribution of the dye in the μBR was a function of mixing time. The indicator color was uniformly distributed throughout the reactor indicating complete mixing within 30 s mixing at a stirring speed of 180 rpm. The simulation of the steady state flow at 180 rpm reveals a secondary vertical flow caused by the horizontal stir bar rotation. This vertical flow contributes to the oxygen mass transportation from the top membrane to the DO sensor located at the bottom of the reactor.44 Although such a solution is possible for experimental set-ups, integrating rotating magnetic fields clearly complicate the overall design of the μBRs and of the final device.
Other efforts for a simple active mixing method inside a μBR used a pressure-based recycle flow. Using pressure-driven recycle flow mixing, it appeared that both diffusion and convection contribute to the mixing; however, with a large recycle flow rate the contribution from convection predominates.47 A syringe pump was also used as an actuator to create recycle flow. The experimental results showed a reasonably good match with the calculated results. The mixing behavior in the prototype reactor was close to an ideally mixed system. Furthermore, because tubes and adaptors were used to connect syringe pump, reactor, and valves, the total volume of the system was larger than the reactor volume, leaving room for a more integrated system and even better mixing.
In yet another approach, active mixing inside a μBR was obtained using a pressure-based oscillating driving force.47 Good mixing performance was achieved under high oscillating flow conditions. The results showed a reasonably good match with the simulated results and the model-based calculated results. The mixing performance showed that both diffusion and convection contribute to mixing; however, with a large oscillating flow rate the contribution from convection predominates.
μBRs with integrated sensing modules
Online sensing and control in μBRs is often far from straightforward. In standard reactors chemostats and turbidostats are often used to control the cell metabolic rates and cell growth rates, respectively. For such offline measurements, regular sampling is, however, typically needed which may be problematic for the small volumes of μBRs. For example, a typical 50 μl sample volume for HPLC taken from a 100 nl bioreactor requires 500 turnovers of the chip volume. For bacterial cells with a 1 h−1 growth rate, a single sample can take 500 h. Based on this fact, Lee et al.34 demonstrated long term micro-scale continuous culture of E. coli for 500 h. Therefore, one fundamental requirement for milliliter- or microliter scale bioreactors is the ability to obtain OD, pH, and DO data in real time and thereby avoid the need for sample removal. Recent developments in electrochemical and optical sensor technologies have made such measurements possible in μBR. Various sensing techniques, i.e., electrochemical sensors, fluorescence sensors, quorum sensing, and bioluminescence sensors, were used in order to develop the control process over different fermentation parameters. Fluorescence lifetime-based sensors were integrated by many researchers in PMMA/PDMS μBRs to measure dissolved oxygen and pH,18, 44, 48 while quorum sensing was used by Balagadde et al.49 to regulate cell density through a feedback mechanism in order to observe the dynamics of E. coli. Also, integrated electrochemical sensors were tested for measurement of pH, temperature, DO, and viable biomass concentration for yeast cultivated under dynamic batch conditions as well as under prolonged continuous conditions.15, 50 All researchers compared fermentation results in the μBR with data from conventional bioreactors, test tubes, and shake flasks for benchmarking purpose. Both the time profile and the standard deviation of the OD curves obtained in the μBR were comparable with other conventional bioreactors. The similarity, also, included growth kinetics, dissolved oxygen profile within the vessel over time, pH profile over time, final number of cells, and cell morphology.
Microbial strain typing
Microorganisms such as bacteria, yeast, fungi, and algae are important for a wide range of applications. These types of cells are cultured in specialized growth media, often accompanied by active mixing and temperature control, and algae cultures have an added requirement of light as an energy source. So, there is urgent need for multiple μBR designs that can suit different microbial growth requirements. This was the aim of Au et al.51 research, where they introduced a μBR for automated culture and density analysis of microorganisms. The μBR was powered by digital microfluidics (DMF), and because it was used with bacteria, algae, and yeast, it was called the BAY μBR. Previous miniaturized bioreactors have relied on microchannels which often require valves, mixers, and complex optical systems. In contrast, the BAY μBR was capable of culturing microorganisms in distinct droplets on a format compatible with conventional bench-top analyzers without the use of valves, mixers, or pumps. Bacteria, algae, and yeast were grown till 5 days with automated semi-continuous mixing and temperature control. Growth profiles were shown to be comparable to those generated in conventional, macro-scale systems. Also, initial trials were made by Steinhaus et al.52 to setup anaerobic μBR. A μBR with 1 μl micro-channels was developed to study some optimum growth conditions for the methanogen Methanosaeta concilii. The μBR was enclosed in an anaerobic chamber designed to place it directly onto an inverted light microscope stage while retaining N2-CO2 environment. Under the ideal conditions for M. concilii of the shear stress and pH and at a temperature of 35 °C, gradients in a single microchannel were used to determine an optimum pH level of 7.6 and total NH4-N concentration of less than 1.1 g/l. The same trend was followed by Holcomb et al.,53 in which they developed a glass/PDMS microfluidic device capable of screening microalgal culturing and stress conditions. For culturing and stress experiments, the device contained power-free valves to isolate microalgae in a microfluidic growth chamber. The device was used to investigate lipid accumulation in the microalgae Neochloris oleabundans, and it was observed that it could be stressed to accumulate cytosolic lipids in a microfluidic environment, as evidenced with fluorescence lipid staining. Furthermore, the germination of fungal spores Aspergillus ochraceus was evaluated for two different key magnitudes (pH and temperature) in PDMS disposable μBR suitable for screening in batch or continuous mode.54 A grid structure was engraved on each chamber, allowing subsequent morphology imaging. Information on germination capacity with regard to interspecies’ variability allowed for optimization of industrial processes as optimal pH and temperature matched to the mesoscopic cultivation systems. The germination conditions therefore remained unaffected inside the μBR, while providing the advantages of dramatic reduction of medium consumption, submerged cultivation with constant oxygen supply, assured low cost and disposability, and possibility of a continuous cultivation mode.
CONCLUSIONS AND FUTURE PROSPECTS
There are several reasons to be interested in the development of new tools and technologies for improved microorganism cultivation. First, the current cultivation and bio-processing are mostly relying on shake flasks, which are not flexible for the microenvironment control. Furthermore, continuous-flow culture systems offer the advantages of constant environmental conditions which are impossible for batch based reactors.55 Second, the consummation of reagents is remarkably high for large volume systems, which is often not necessary for experimentations such as cultivation parameter determination. Third, it is difficult for the commonly used systems to perform parallel operation. Finally, many microorganisms found in nature are still unculturable using current techniques or conditions,36 and it is therefore important to develop new systems with an improved parameter control, reduced consumption, and increased parallelism and automation. The recent developments in the field of microliter-scale stirred bioreactors described in this review clearly demonstrate the usefulness of such systems for high-throughput bioprocess design (HTBD). The reduced reaction volume, the parallelization, and the automation of stirred tank bioreactors have the potential to significantly reduce process development times and assure a cost efficient bioprocess design. Current developments in lab-on-a-chip technologies offer a unique solution to these problems. Indeed, many groups have successfully demonstrated the relevance of fabricated microsystems in microorganism cultivation. In the last few years, an increasing number of alternative approaches for the cultivation of microorganisms in microfluidic devices on a microliter-scale were published. These approaches illustrate that μBR technology for microbial cultivations has reached a development stage where the operation of bench-scale reactors can be mimicked. The feasibility of pH and DO control has been demonstrated, and sufficient mixing has been proven at least for E. coli. However, no specifically designed μBR has yet been able to make the step from the laboratory environment to being a de-facto standard in industrial applications as the current μBR systems available on the market build on microtiter platforms. Additionally, scaling-up at different KLa values and for different types of organisms still has to be demonstrated. Islam et al.,56 for example, investigated the influence of three different scales of operation on the culture properties of E. coli and have suggested that KLa is a useful scale-up criterion for μBRs. However, the scale-up and scale-down capabilities of these reactors with respect to “technical” cultivations in standard stirred tank reactors (STRs) remain unclear and important engineering parameters are not known. To date, these systems therefore constitute a useful tool for automated screening tasks but seem less suitable for process development. The development of complementary miniaturized downstream processing technology is necessary as the number of parallel cultivations increases, and only little work on product recovery and purification has been published so far.57, 58 It also has to be stated that for many microliter-systems there is still a lack of knowledge of important variables related to engineering. The volumetric power input and maximum local energy dissipation have only been reported for a few systems. For a robust scale-up, the characteristics and the limits of parallel reaction systems must be known and taken into consideration. More scientific work on scale-up and/or scale-down issues and their application remains necessary.
There is also of course still room for improvement of the proposed μBR designs. In the current version of the reactor, the fluidic tubes are manually inserted into the reactor. Obviously, this procedure is not error-free both during insertion and reactor operation. A future version of the holder would therefore also include a guiding and clamping tunnel for the fluidic connections which would define position, direction, and insertion length of the tubes. This would then further facilitate the installation of the reactor. Currently cultivations are run with a fixed concentration of oxygen in the aeration gas. In a next step closed-loop DO-concentration control will be implemented using similar control algorithms and devices as for pH control. This will, for example, also allow investigating how cultivation reacts to stress due to lack of oxygen, by controlling the DO concentration at a low set point. Also, the pH range which can be controlled now will not suit all kinds of organisms. Many will require a more acidic pH (e.g., S. cerevisiae) which requires a different kind of sensor spot. Also, the comparison between this reactor design and larger scale cultivations, especially bench-scale or even industrial cultivation remains to be done. This is a very important part of the further investigations as it is essential to know how to translate μBR results into “real world” results. This comparison between reactor scales will be done by means of a wash-out curve where the maximal growth rate in the μBR and a bench-scale reactor are compared.
In the future more and more automated, fully monitored, and controlled microliter-scale reactors will be available, where almost the same process performances as in laboratory and pilot-scale reactors will be possible. The development and optimization of new micro-analytical methods for online or at-line measurement and consequently control of important state variables, for example, DO, pH, or optical density, especially enables cultivations comparable to conventional laboratory bioreactors. Highly advanced systems even offer the possibility to run and optimize fed-batch processes on a microliter-scale by, for example, combining the small-scale system with a liquid handler. This is important since the majority of industrial bioprocesses are run in fed-batch mode. Furthermore, the use of disposable miniaturized bioreactors becomes more and more popular because, especially on the small scale, cleaning can be a major obstacle to the whole process.
References
- Weuster-Botz D., Stevens S., and Hawrylenko A., Biochem. Eng. J. 11, 69 (2002). 10.1016/S1369-703X(02)00010-4 [DOI] [Google Scholar]
- Duetz W. A., Ruedi L., Hermann R. et al. , Appl. Environ. Microbiol. 66, 2641 (2000). 10.1128/AEM.66.6.2641-2646.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warringer J. and Blomberg A., Yeast 20, 53 (2003). 10.1002/yea.931 [DOI] [PubMed] [Google Scholar]
- John G. T., Goelling D., Klimant I. et al. , J. Dairy Res. 70, 327 (2003). 10.1017/S0022029903006344 [DOI] [PubMed] [Google Scholar]
- John G. T., Klimant I., Wittmann C. et al. , Biotechnol. Bioeng. 81, 829 (2003). 10.1002/bit.10534 [DOI] [PubMed] [Google Scholar]
- Weiss S., John G. T., Klimant I. et al. , Biotechnol. Prog. 18, 821 (2002). 10.1021/bp0200649 [DOI] [PubMed] [Google Scholar]
- Deshpande R. R. and Heinzle E., Biotechnol. Lett. 26, 763 (2004). 10.1023/B:BILE.0000024101.57683.6d [DOI] [PubMed] [Google Scholar]
- Kensy F., John G. T., Hofmann B. et al. , Bioprocess Biosyst. Eng. 28, 75 (2005). 10.1007/s00449-005-0010-7 [DOI] [PubMed] [Google Scholar]
- Betts J. I. and Baganz F., Microb. Cell Fact. 5, 21 (2006). 10.1186/1475-2859-5-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes P. and Cabral J. M. S., Biocatalysis Biotransform. 24, 237 (2006). 10.1080/10242420600667684 [DOI] [Google Scholar]
- Kumar S., Wittmann C., and Heinzle E., Biotechnol. Lett. 26, 1 (2004). 10.1023/B:BILE.0000009469.69116.03 [DOI] [PubMed] [Google Scholar]
- Micheletti M. and Lye G. J., Curr. Opin. Biotechnol. 17, 611 (2006). 10.1016/j.copbio.2006.10.006 [DOI] [PubMed] [Google Scholar]
- Sauer U., Curr. Opin. Biotechnol. 15, 58 (2004). 10.1016/j.copbio.2003.11.001 [DOI] [PubMed] [Google Scholar]
- Ottino J. M. and Wiggins S., Philos. Trans. R. Soc. London, Ser. A 362, 923 (2004) 10.1098/rsta.2003.1355. [DOI] [PubMed] [Google Scholar]
- Boccazzi P., Zhang Z., Kurosawa K. et al. , Biotechnol. Prog. 22, 710 (2006). 10.1021/bp0504288 [DOI] [PubMed] [Google Scholar]
- Lee H. L., Boccazzi P., Ram R. J. et al. , Lab Chip 6, 1229 (2006). 10.1039/b608014f [DOI] [PubMed] [Google Scholar]
- Szita N., Boccazzi P., Zhang Z. et al. , Lab Chip 5, 819 (2005). 10.1039/b504243g [DOI] [PubMed] [Google Scholar]
- Zanzotto A., Szita N., Boccazzi P. et al. , Biotechnol. Bioeng. 87, 243 (2004). 10.1002/bit.20140 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Boccazzi P., Choi H. G. et al. , Lab Chip 6, 906 (2006). 10.1039/b518396k [DOI] [PubMed] [Google Scholar]
- Fleger M. and Neyer A., Microelectron. Eng. 83, 1291 (2006). 10.1016/j.mee.2006.01.086 [DOI] [Google Scholar]
- Zhao Y.-G., Wu A., Lu H.-L. et al. , Appl. Phys. Lett. 79, 587 (2001). 10.1063/1.1389514 [DOI] [Google Scholar]
- Becker H. and Gartner C., Anal. Bioanal. Chem. 390, 89 (2008). 10.1007/s00216-007-1692-2 [DOI] [PubMed] [Google Scholar]
- Tsao C.-W. and DeVoe D., Microfluid. Nanofluid. 6, 1 (2009). 10.1007/s10404-008-0361-x [DOI] [Google Scholar]
- Lee J. N., Park C., and Whitesides G. M., Anal. Chem. 75, 6544 (2003). 10.1021/ac0346712 [DOI] [PubMed] [Google Scholar]
- Huang C.-W. and Lee G.-B., J. Micromech. Microeng. 17, 1266 (2007). 10.1088/0960-1317/17/7/008 [DOI] [Google Scholar]
- DeJong J., Ph.D. dissertation, University of Twente, Twente, 2008. [Google Scholar]
- Anvari M., Pahlavanzadeh H., Vasheghani-Farahani E. et al. , J. Ind. Microbiol. Biotechnol. 36, 313 (2009). 10.1007/s10295-008-0501-z [DOI] [PubMed] [Google Scholar]
- Zautsen R. R., Maugeri-Filho F., Vaz-Rossell C. E. et al. , Biotechnol. Bioeng. 102, 1354 (2009). 10.1002/bit.22189 [DOI] [PubMed] [Google Scholar]
- McDonald J. C. and Whitesides G. M., Acc. Chem. Res. 35, 491 (2002). 10.1021/ar010110q [DOI] [PubMed] [Google Scholar]
- Xia Y. and Whitesides G. M., Angew. Chem., Int. Ed. 37, 550 (1998). [DOI] [PubMed] [Google Scholar]
- Yarmush M. L. and King K. R., Annu. Rev. Biomed. Eng. 11, 235 (2009). 10.1146/annurev.bioeng.10.061807.160502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäpper D., Stocks S. M., Szita N. et al. , Chem. Eng. J. 160, 891 (2010). 10.1016/j.cej.2010.02.038 [DOI] [Google Scholar]
- Edlich A., Magdanz V., Rasch D. et al. , Biotechnol. Prog. 26, 1259 (2010). 10.1002/btpr.449 [DOI] [PubMed] [Google Scholar]
- Lee K. S., Boccazzi P., Sinskey A. J. et al. , Lab Chip 11, 1730 (2011). 10.1039/c1lc20019d [DOI] [PubMed] [Google Scholar]
- Seo T. Y., Eum K. W., Han S. O. et al. , Process Biochem. 47, 1011 (2012). 10.1016/j.procbio.2012.03.019 [DOI] [Google Scholar]
- Maharbiz M. M., Holtz W. J., Howe R. T. et al. , Biotechnol. Bioeng. 85, 376 (2004). 10.1002/bit.10835 [DOI] [PubMed] [Google Scholar]
- Buchenauer A., Funke M., Büchs J. et al. , J. Micromech. Microeng. 19, 074012 (2009). 10.1088/0960-1317/19/7/074012 [DOI] [Google Scholar]
- van Leeuwen M., Krommenhoek E. E., Heijnen J. J. et al. , Biotechnol. Prog. 26, 293 (2010). 10.1002/btpr.315 [DOI] [PubMed] [Google Scholar]
- Gernaey K. V., Lantz A. E., Tufvesson P. et al. , Trends Biotechnol. 28, 346 (2010). 10.1016/j.tibtech.2010.03.006 [DOI] [PubMed] [Google Scholar]
- Enfors S. O., Jahic M., Rozkov A. et al. , J. Biotechnol. 85, 175 (2001). 10.1016/S0168-1656(00)00365-5 [DOI] [PubMed] [Google Scholar]
- Lapin A., Müller D., and Reuss M., Ind. Eng. Chem. Res. 43, 4647 (2004). 10.1021/ie030786k [DOI] [Google Scholar]
- Li X., van der Steen G., van Dedem G. W. K. et al. , AIChE J. 55, 2725 (2009). 10.1002/aic.11880 [DOI] [Google Scholar]
- Hsu Y.-H. and Tang W., Microfluid. Nanofluid. 11, 459 (2011). 10.1007/s10404-011-0811-8 [DOI] [Google Scholar]
- Zhang Z., Szita N., Boccazzi P. et al. , Biotechnol. Bioeng. 93, 286 (2006). 10.1002/bit.20678 [DOI] [PubMed] [Google Scholar]
- Luo X., Shen K., Luo C. et al. , Biomed. Microdevices 12, 499 (2010). 10.1007/s10544-010-9406-5 [DOI] [PubMed] [Google Scholar]
- Alam M. N. H. Z., Schäpper D., and Gernaey K. V., J. Micromech. Microeng. 20, 055014 (2010). 10.1088/0960-1317/20/5/055014 [DOI] [Google Scholar]
- Li X., van der Steen G., Dedem G. W. K. van et al. , Chem. Eng. Sci. 63, 3036 (2008). 10.1016/j.ces.2008.02.036 [DOI] [Google Scholar]
- Zanzotto A., Boccazzi P., Gorret N. et al. , Biotechnol. Bioeng. 93, 40 (2006). 10.1002/bit.20708 [DOI] [PubMed] [Google Scholar]
- Balagadde F. K., You L., Hansen C. L. et al. , Science 309, 137 (2005). 10.1126/science.1109173 [DOI] [PubMed] [Google Scholar]
- Krommenhoek E. E., van Leeuwen M., Gardeniers H. et al. , Biotechnol. Bioeng. 99, 884 (2008). 10.1002/bit.21661 [DOI] [PubMed] [Google Scholar]
- Au S. H., Shih S. C., and Wheeler A. R., Biomed. Microdevices 13, 41 (2011). 10.1007/s10544-010-9469-3 [DOI] [PubMed] [Google Scholar]
- Steinhaus B., Garcia M. L., Shen A. Q. et al. , Appl. Environ. Microbiol. 73, 1653 (2007). 10.1128/AEM.01827-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcomb R. E., Mason L. J., Reardon K. F. et al. , Anal. Bioanal. Chem. 400, 245 (2011). 10.1007/s00216-011-4710-3 [DOI] [PubMed] [Google Scholar]
- Demming S., Sommer B., Llobera A. et al. , Biomicrofluidics 5, 14104 (2011). 10.1063/1.3553004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcott P. H., in Continuous Culture of Cells, edited by Calcott P. H. (CRC Press, Boca Raton, 1981), p. 127. [Google Scholar]
- Islam R. S., Tisi D., Levy M. S. et al. , Biotechnol. Bioeng. 99, 1128 (2008). 10.1002/bit.21697 [DOI] [PubMed] [Google Scholar]
- Jackson N. B., Liddell J. M., and Lye G. J., J. Membr. Sci. 276, 31 (2006). 10.1016/j.memsci.2005.09.028 [DOI] [Google Scholar]
- Shapiro M. S., Haswell S. J., Lye G. J. et al. , Biotechnol. Prog. 25, 277 (2009). 10.1002/btpr.99 [DOI] [PubMed] [Google Scholar]