Abstract
Ligand-binding dynamics control allosteric signaling through the estrogen receptor-α (ERα), but the biological consequences of such dynamic binding orientations are unknown. Here, we compare a set of ER ligands having dynamic binding orientation (dynamic ligands) with a control set of isomers that are constrained to bind in a single orientation (constrained ligands). Proliferation of breast cancer cells directed by constrained ligands is associated with DNA binding, coactivator recruitment and activation of the estrogen-induced gene GREB1, reflecting a highly interconnected signaling network. In contrast, proliferation driven by dynamic ligands is associated with induction of ERα-mediated transcription in a DNA-binding domain (DBD)-dependent manner. Further, dynamic ligands displayed enhanced anti-inflammatory activity. The DBD-dependent profile was predictive of these signaling patterns in a larger diverse set of natural and synthetic ligands. Thus, ligand dynamics directs unique signaling pathways, and reveals a novel role of the DBD in allosteric control of ERα-mediated signaling.
Introduction
The current dogma of ligand binding suggests that ligands have higher affinity for a subset of the ensemble of the dynamic conformations of their protein targets. In this scenario, binding drives the allosteric selection process, as the ligand forms energetically more stable complexes with these higher affinity conformers1. It is also generally thought that ligands bind in a single orientation, and dissociate if the protein dramatically changes conformation2. For example, the ER antagonist tamoxifen only binds to the inactive conformer of ER, and a switch to the active conformer would drive ligand dissociation because it has a pocket that is too small to accommodate the ligand3. In support of this notion, a large body of structural data suggests that ligands generally bind in a single, unique orientation.
We recently discovered that this dogma also reflects a selection process inherent to X-ray crystallography, which favors the conformer most likely to crystallize. To overcome this, we used a conformational trapping approach, adding surface hydrogen bonds to ER in order to stabilize the canonical active or inactive conformers4. This allows one to obtain structures of the same compound bound to two different protein conformers5. Surprisingly, different protein conformers select different binding orientations of the ligand, and notably, modifying the ligand alters the dynamics or options for its binding orientation5, indicating an unexpected reciprocity between ligand-binding orientation and receptor-conformer selection. We also demonstrated that modifying the ligand alters its preference for two different binding orientations, which are associated with different protein conformers and associated activity profiles5. This identified ligand dynamics as a novel mechanism to titrate the activity profile of an allosteric signaling system, and has since been verified by NMR in two other allosteric signaling proteins, PPARγ6 and dihydrofolate reductase7, suggesting that it is a general tenet of ligand-protein allostery.
ERα is a ligand-activated transcription factor that interacts with diverse binding partners. When activated by the ligand, ERα binds to the DNA and recruits transcriptional coregulators, which in turn control gene expression by mediating post-translational modification of histones and the associated transcriptional proteins and by recruiting the basal transcriptional machinery8. Further, ERα also binds to select interacting proteins via its N-terminal activation function 1 (AF1) domain, the DNA-binding domain (DBD), a short flexible hinge region, and the C-terminal ligand-binding domain (LBD). Although interdomain allosteric communication is well documented and bidirectional9, 10, it is not known how this occurs.
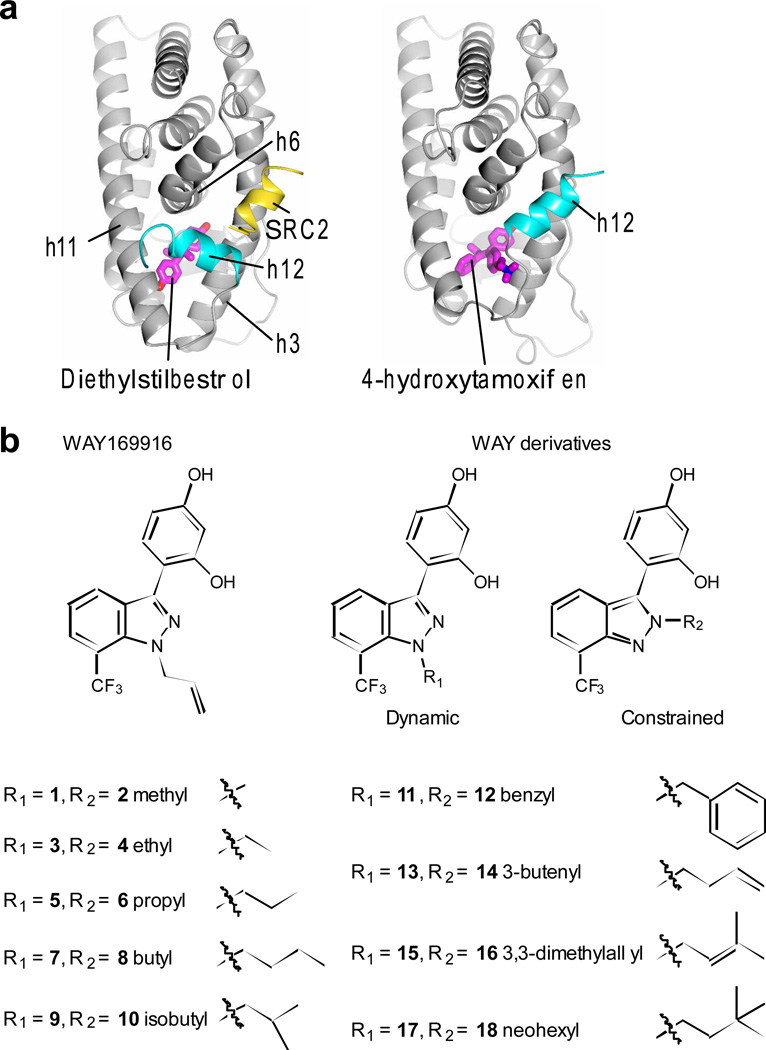
The LBD has a binding surface for transcriptional coregulators, called activation function 2 (AF2) that is allosterically regulated by ligand binding-induced positioning of the switch helix, helix 12 (h12), which in turn determines the relative recruitment of coregulators to coordinate a transcriptional response3, 11–14 Antagonists such as tamoxifen have a bulky pendent group that extends between h3 and h11 to directly reposition h12, forming what can be called an antagonist conformation (Fig. 1a, and Supplementary Results, Supplementary Fig. 1a,b). We also discovered a second epitope for bulky ligand substitution on the opposite side of the pocket from h12, facing h8 (Supplementary Fig. 1c). The h8 epitope accommodates this type of bulky substitution in a fashion that allows the receptor to retain full agonist activity15. In our initial work on WAY-166916, an anti-inflammatory ER ligand16, we found that the ligand could bind in two orientations (dynamic binding)5 – with the bulky side group in the hydrophobic side pocket towards h8, supporting an active conformer, and with the ligand flipped over and the bulky side group oriented towards h12, thereby disrupting h12 positioning from the active conformation (Supplementary Fig. 2a). Further, we discovered that the R group of WAY-166916 could be modified to favor binding orientations towards either the hydrophobic h8 side pocket or towards h12, thereby controlling the activity profile of the derivatives, via modulation of their ligand-binding dynamics. Here, we compared our original set of ligands that can bind in two different orientations (dynamic ligands) with a new control set of matching isomers, where placing the R groups on an adjacent atom restricts the ligand to one binding orientation (constrained ligands) (Fig. 1b), to systematically explore the biological consequences of ligand dynamics.
Figure 1. ERα structure and ligand sets.
a) The LBD is shown as ribbon, with h12 bound to either full-agonist diethylstilbesterol (PDB ID: 3ERD) or tamoxifen (PDB ID: 3ERT), demonstrating how tamoxifen positioning of h12 blocks binding of SRC2.
b) Ligand isomer pairs were designed with the R group at a position that allows for ligand dynamics or at an adjacent atom, such that it only binds to ER in one constrained orientation.
Results
Structure-class analysis of ligand dynamics
We have developed an approach coined “structure-class analysis”, where analyses of groups of structures and/or ligands enables one to identify structural and conformational features not apparent in the noise of a single structure, and thereby to identify patterns not visible from comparison of individual compounds. This approach allowed definition of the structural basis of differential ligand recognition by ERα versus ERβ17, partial agonist/graded signaling4 and its control by ligand dynamics5 and allostery in JNK family kinases18. While a comparison of two paired ligands could be informative, it is not possible to differentiate between class-specific phenotypes and attributes specific to the ligand. As we describe below, the different signaling phenotypes of dynamic versus constrained ligands are only apparent using structure-class analysis.
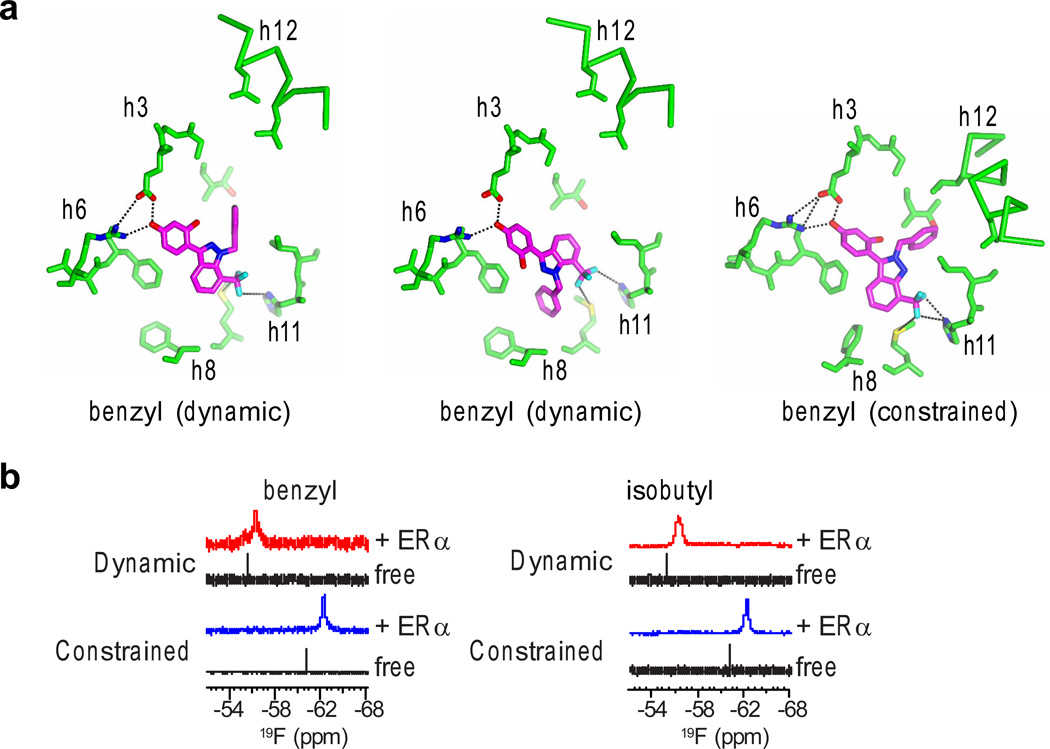
Molecular modeling, crystal structures and NMR analysis of our newly generated constrained ligands bound to ERα confirmed they bound in a single orientation. Indeed, modeling clearly shows that even the methyl-substituted constrained compound could not bind with the R group facing away from h12, as it is not able to reach the hydrophobic side pocket, but rather clashes (Supplementary Fig. 2b). In the eight new and several published5 crystal structures of ERα bound to WAY-169916 or the derivatives, we found that the constrained ligands only bound with the R group directed towards h12, whereas the dynamic ligands bound in orientations with the R group either facing the hydrophobic side pocket or in both the h8 and h12 orientations (Fig. 2a, Supplementary Fig. 2a,c and Supplementary Table 1). As a more direct test of the ligand dynamics, NMR data were obtained from two of the isomer sets, probing the NMR-active fluorine nuclei in the CF3 group when bound to ERα. For the benzyl isomers, the constrained ligand shows a single resonance (peak), while the dynamic isomer had two distinct resonances, suggesting there were two distinct chemical environments around the CF3 group for the dynamic isomer (Fig. 2b). Similarly for the isobutyl isomers, the constrained ligand shows a single NMR resonance, while the matching dynamic isomer displayed a broader, shifted peak (Fig. 2b). Deconvolution of this broad peak revealed two overlapping resonances that recapitulated the shape of the measured broad peak (Supplementary Fig. 3), again supporting the idea of a more dynamic local chemical environment for the dynamic isomer. Having verified the multiple poses with docking, X-ray crystallography and NMR, these structurally well-characterized compounds were then used as a test set for how ligand dynamics impacts cellular signaling.
Figure 2. Structural validation of ligand binding.
a) The ERα LBD was crystallized with both of the benzyl isomers. Select ligand-binding pocket residues that lie in the indicated helices are shown.
b) The CF3 group in the ligand provided signal for NMR determination of the local chemical environment of the indicated ligands, data for ligand alone in solution (free) or interacting with ERα.
Chemical biology phenotyping
We developed a platform to mechanistically characterize the major ERα signaling pathways in a high throughput fashion, with robotic pintool dispensing in a 384-well format, yielding highly robust assays. Gene expression, biochemical and phenotypic assays were used to probe the underlying differences in structure that might contribute to class-specific signaling outcomes. The data for the WAY-169916 derivatives are shown in Supplementary Figure 4, and the assays are described in detail in Supplementary Table 2 and Online methods, while the estradiol (E2) and tamoxifen control curves for each assay are shown in Supplementary Figure 5a–o.
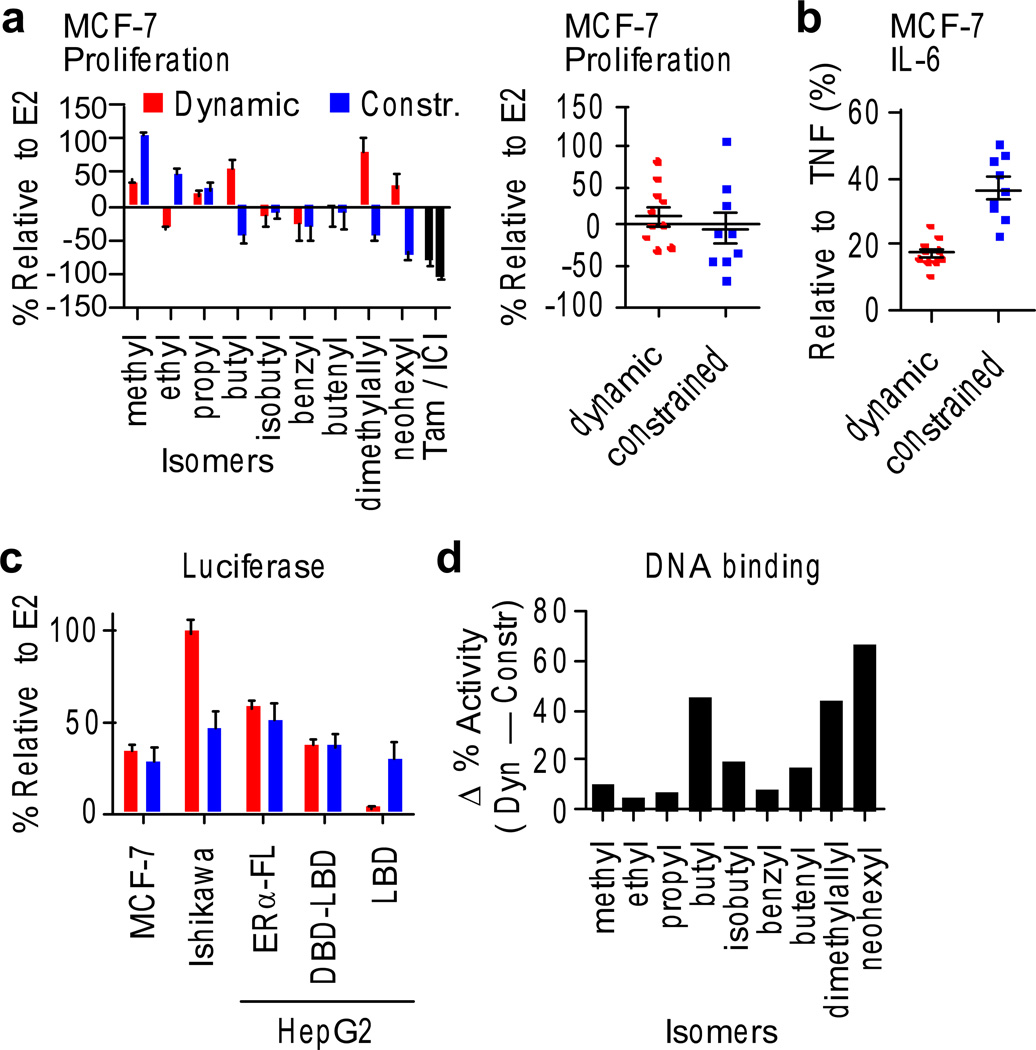
While individual compounds displayed unique assay profiles, we observed clear class-specific differences. Proliferation of MCF-7 breast cancer cells revealed great variability between the isomer pairs, though the class averages were identical (Fig. 3a), underscoring the need to perform the class analysis to assess characteristic receptor-conformational effects such as those induced by ligand dynamics. Ligands were also assessed for the suppression of TNFα-stimulated production of Interleukin-6 (IL-6) cytokine – a hallmark of the ERα-mediated anti-inflammatory response, which derives from ERα binding to the IL-6 promoter via indirect tethering and modulating coregulator recruitment4. In contrast to the proliferation data, IL-6 showed strikingly different class-specific responses, where the dynamic ligands displayed greater suppression of secreted IL-6 (Fig. 3b). To visualize the magnitude of differences between the pairs, we subtracted the dynamic from the constrained response for each pair, which highlighted that greater anti-inflammatory effects occurred with dynamic ligands for each pair (Supplementary Fig. 6a). These results demonstrate a novel principle in biology, that ligand dynamics can be exploited to modulate signaling specificity of an allosteric molecule, a concept that could also be applied more broadly to drive new therapeutic targeting strategies.
Figure 3. Ligand phenotyping.
a) MCF-7 breast cancer cells were switched to steroid-free serum and treated with 10 µM compound or control, including estradiol (E2), 4-hydroxytamoxifen (Tam) and the full ER antagonist ICI 182,780 (ICI). Growth was assayed after 7 days and normalized to the E2 response. Data represent mean values ± s.e.m.
b) MCF-7 cells were stimulated with 10 ng/ml TNFα + test compounds, and secreted IL-6 was quantitated after 24 hr, using AlphaLISA. Data represent mean values ± s.e.m.
c) MCF-7 and Ishikawa cells were transfected with 3xERE-luciferase reporter for assays of native ERα activity. Liver (HepG2) cells were transfected with the 3xERE- or Gal4-luciferase reporters, and the indicated ERα expression plasmids. Class averages for dynamic versus constrained ligands are shown. Data represent mean values ± s.e.m.
d) The DNA-binding profile was determined in HEK293T cells transfected with a fusion of ERα to the strong activation domain of VP16 on the 3xERE-luciferase reporter, which overrides the ER ligand pharmacology. Plotted is the difference in normalized data (% relative to E2) between the isomer pairs, showing that the dynamic ligands have greater DNA binding activity profile for each pair.
We developed a set of reporter-gene assays to determine which ERα domains are responsible for gene activation and to compare cells derived from different tissue types. In the breast, ERα activation requires AF2, while the liver and uterus have different coactivator profiles, enabling activity via the N-terminal AF1. This is why tamoxifen, an antagonist in the breast, mimics some of the effects of estradiol (E2) in the liver and uterus, thus acting as a selective estrogen-receptor modulator (SERM)19. The estrogen-response element (ERE) luciferase assays show identical class averages for the breast and liver cells, but the dynamic ligands showed dramatically enhanced stimulation in the uterine cell line (Fig. 3c). While the breast and liver cell assays showed variability between isomer pairs, the class-specific effects in the uterine cells were found for each pair (Fig. 3c and Supplementary Fig. 6b). This may reflect class differences in protein interaction profiles between these cell types, which are known to underlie tissue-specific effects of SERMs19.
Many eukaryotic proteins contain modular domains with distinct intrinsic activities that can be allosterically modified by interdomain communication; a good example is c-Src, whose kinase activity is regulated by interactions between its SH2 and SH3 domains20. With ERα, E2 drives similar 15-fold activation of the full-length ERα, or a construct lacking AF1 (ERα-DBD-LBD) (Supplementary Fig. 5a–c), while the isolated LBD (fused to the Gal4 DBD, a construct termed ERα-LBD) displays some 90-fold activation, likely due to very low basal activity. This demonstrates that deletion of AF1 or DBD/hinge region does not affect E2 response and hence E2 activates transcription via the ERα-LBD. This is consistent with a wealth of biochemical and structural data that E2 stabilizes the AF2 surface to mediate recruitment of coactivators via their nuclear-receptor interaction domain, LxxLL sequence motif3, 21. In contrast, tamoxifen agonist activity is completely absent in the context of the ERα-DBD-LBD or ERα-LBD constructs, demonstrating that tamoxifen’s agonist activity is dependent upon AF1 (Supplementary Fig. 5a,b)10. Further, in the mammalian two-hybrid assay, tamoxifen recruitment of steroid receptor coactivator-1 (SRC1) only occurred with full-length ERα (Supplementary Fig. 5f) and not with the isolated LBD (Supplementary Fig. 5i), consistent with the established role of AF1 in tamoxifen agonist activity22. It is not known how the LBD communicates structurally with other ERα domains, but the current model is that various ERα ligands use AF1, AF2 or a mixture of them for recruitment of coactivators and transcriptional activation.
Comparing the activity of each compound to the full agonist E2 allowed classification of compounds according to the ERα domains used for gene activation (Supplementary Fig. 7). Ligands from both the dynamic and constrained classes showed similar activity relative to E2 for the ERα-DBD-LBD and the full-length receptor; thus, AF1 is not necessary for activity (Fig. 3c). For the ERα-LBD, the constrained ligands again showed a similar activity relative to E2 as with the full-length receptor, suggesting a slight contribution to signaling from AF1 but primary utilization of AF2. However, dynamic ligands showed no activity on the LBD alone, indicating an absolute requirement for the DNA-binding domain or adjacent small hinge for activity (Fig. 3c). To test this, DNA binding was assessed in a high-throughput fashion by assaying luciferase activity of an ERα-VP16 fusion, where the strong activation function of VP16 overrides the pharmacology of the ER ligand23 and gives a proxy for DNA binding, which is constitutive in this overexpression system. As expected, the full antagonist ICI (ICI 182,780, also known was Faslodex or Fulvestrant) blocked activity in this assay (Supplementary Fig. 5o), consistent with its known role in redistributing ERα to an insoluble cellular fraction24. Notably, there were significant differences in this assay (Student’s t-test, P < 0.0001), with the dynamic ligands showing more robust activation of the reporter for each isomer pair (Fig. 3d), indicative of a different character of DNA binding or gene activation that we find to be significant. As described below, this DBD-dependent activity profile was also predictive of compound-signaling behavior in a larger, more diverse collection of ER ligands, demonstrating the power of this comparative domain analysis approach.
We also probed for other class-specific differences in ER biochemistry and signaling. The dogma for how ER induces proliferation is via dimerization, binding to DNA, recruiting transcriptional coactivators, including members of the Steroid Receptor Coactivator (SRC) family, and activating key proliferative genes, including Cyclin D1 (CCND1), c-MYC, GREB1 and pS225–29. The GREB1 and pS2 genes contain canonical EREs, while ER control of CCND1 and c-MYC involves indirect tethering to other transcription factor complexes28, 30. We used a bioluminescence resonance energy transfer (BRET) assay to measure dimerization of YFP-ERα-renilla-ERα, and also of renilla-ERα-YFP-ERβ dimers31. Association of full-length ERα with SRC family members was assayed by mammalian two-hybrid assay. No class-specific differences were identified in these assays, including dimerization, coactivator recruitment and native gene analyses (Supplementary Fig. 8a–c). However, structure-activity relationships that somewhat correlate with the size of the R group are apparent (Supplementary Fig. 9), for instance increasing the size of the R group enhances the ability of dynamic ligands to stimulate endogenous ER activity relative to the constrained counterparts. Thus, dynamic ligands induced a unique signaling profile through ER, with selective differences in transrepression of IL-6 and dependence on the DBD for gene activation.
Ligand dynamics and the proliferative response
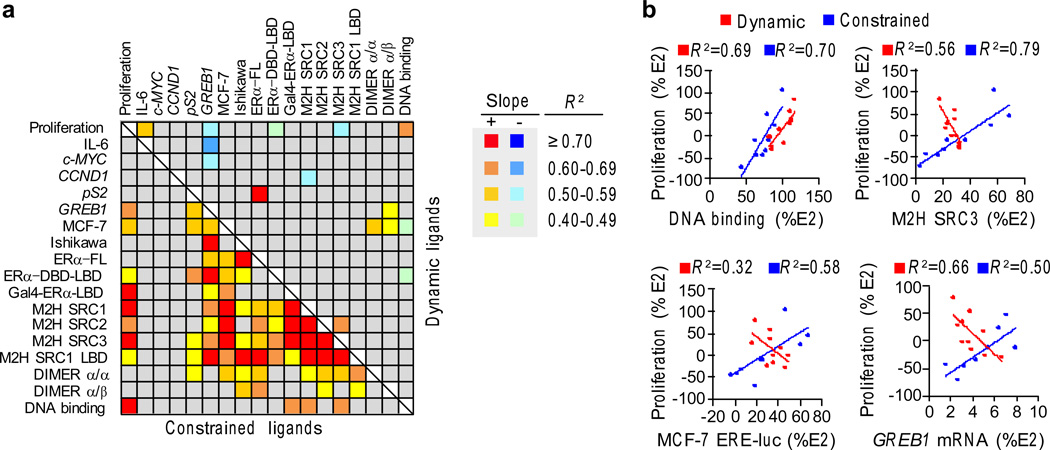
Although ligands for ERα are generally thought to behave as agonists or antagonists, it is evident that they can display a full range of graded signaling responses. This graded signaling is apparent in the biochemical assays used to probe the mechanism of ER action, but it is not known whether this graded response is reflected in the native gene activation that eventually impacts a physiological phenotype, such as proliferation. ERα uses multiple mechanisms, binding partners and effector genes to determine proliferation; therefore it is also of interest to understand the role of ligand dynamics in this complex relationship. To this end, we performed linear regression analyses assessing relationships between all of the screens, and the coefficient of determination, R2, was calculated to determine how the structural class, underlying biochemical mechanisms and transcriptional output correlate with the proliferative response. R2 was then plotted as a heat map (Fig. 4a and Supplementary Fig. 10).
Figure 4. Cellular correlates of ER proliferation.
a) Cross-correlation matrix for R2 between all the bioassays. Linear regression analysis was performed to identify relationships between the assays. The R2 ranges are shown as a heat map. The slope of the regression line indicates whether the associations are positively or negatively correlated with the dynamic-ligand data on the top and the constrained-ligand data on the bottom. (Proliferation – MCF-7 proliferation assay; IL-6 – TNFα induced IL-6 expression measured by AlphaLISA in MCF-7 cells; cMYC, CCND1, and pS2 – mRNA measured in MCF-7 cells; MCF-7 and Ishikawa – ERE-Luciferase driven by endogenous ERα; ERα-FL, ERα-DBD-LBD and GAL4-ERα-LBD – ERE-Luciferase assays in HepG2 cells; M2H SRC1, M2H SRC2 and M2H SRC3 – mammalian 2-hybrid assays with ERα as the bait and with either SRC1, SRC2 or SRC3 as prey; M2H SRC1 LBD – mammalian 2-hybrid assays with ERα-LBD as the bait and with SRC1 as prey; DIMER α/α and α/β – dimerization between ERα homodimers and ERα/β heterodimers measured using BRET assay). All assays are represented as percentage of E2 response, except for IL-6, which is represented as percentage of TNFα stimulated IL-6. The assays are described in detail in Supplementary Table 2 and online methods. The R2 values are shown in Supplementary Figure 10.
b) Data from the dynamic ligands are red, while the data from the constrained ligands are blue for the indicated assays. The linear regression plots are shown with the R2 values.
Constrained and dynamic ligands showed dramatic differences in the biological correlates of the proliferative signal. The constrained ligands showed the expected relationships, where the degree of ligand-specific proliferation was associated with binding to DNA, recruitment of coactivators and gene activation (Fig. 4a,b and Supplementary Fig. 11a). Further, these underlying biochemical relationships were also highly correlated with each other for the constrained ligands (Fig. 4a,b), suggesting a common structural mechanism that represents the canonical activation pathway for ER. Surprisingly, the degree of proliferation correlated specifically with GREB1 (Fig. 4b), but not the other critical proliferative genes c-MYC or CCND1 (Supplementary Fig. 11b). As expected, c-MYC is rapidly induced, while GREB1 slowly accumulates (Supplementary Fig. 12a), demonstrating that these genes were examined at peak expression levels. Accordingly, though its function is not resolved, GREB1 is necessary for hormone-directed proliferation of breast (Supplementary Fig. 12b)27, 32 and prostate cancer cells27, 32.
Notably, the R2 matrix of assays with the dynamic ligands shows a loss or inversion of almost all of the relationships between proliferation and the different assays, including reporter gene activation, coactivator recruitment and induction of GREB1 mRNA (Fig. 4a,b and Supplementary Fig. 11a). While DNA-binding correlated directly with the degree of proliferation induced by the different dynamic ligands (Fig. 4b), it correlated inversely with reporter-gene activation by dynamic ligands (Fig. 4a,b and Supplementary Fig. 11c). GREB1 correlations were also different and were inversely related to proliferation, c-MYC mRNA and IL-6 protein (Supplementary Fig. 11d). Thus, dynamic ligands provoke a dramatic rewiring of the canonical ER signaling pathway, an observation that is revealed only by comprehensive systems biology ligand-class analysis.
Predictive power in larger test sets
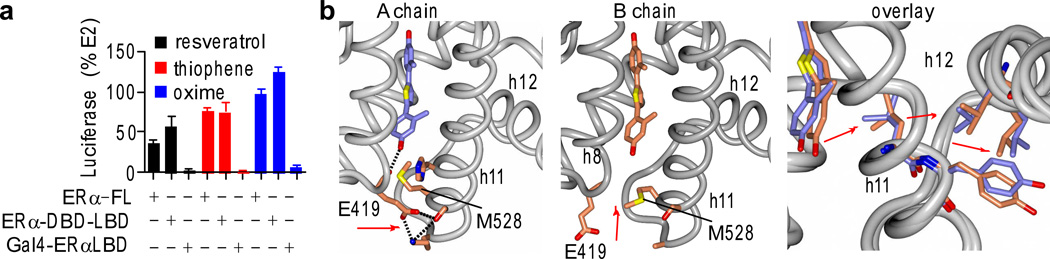
To assess if DBD-specific signaling correlates with distinct signaling mechanisms more generally, we tested our hypothesis on a diverse set of 80+ ligands representing 11 scaffolds (Supplementary Table 3), and from these complexes we obtained an additional 33 co-crystal structures bound to ERα. In our larger test set, three compounds had the same profile as the WAY dynamic ligands, where their transcriptional activity is dependent upon the DBD. These included the phytoestrogen resveratrol and two synthetic estrogens. For these compounds, the LBD alone had less than 10% activity, while deleting the N-terminal AF1 (ERα-DBD-LBD) had no effect on the activity profile, showing an absolute requirement for the DBD/hinge region for activity (Fig. 5a).
Figure 5. Identification of dynamic ligands based on DBD profile.
a) Three compounds from a larger test set of 82 compounds showed DBD-specific activity profiles that match the data profiles from the dynamic WAY-169916 derivatives. Assays were performed as in Figure 3a. Data represent mean values ± s.e.m.
b) Structure of ERα LBD bound to a thiophene ligand shows two different binding orientations on the two sides of the dimer, protein chain A and B, including differences in a ligand associated hydrogen bonding network found only chain A. An overlay of the models highlights the ligand-induced differences in ligand binding and the packing between h11 and h12.
The hypotheses generated with the dynamic WAY-169916 derivatives were predictive, as the three compounds with the DBD-dependent activity profiles were the only ligands to show dynamic binding out of the set of 33 crystal structures. Both resveratrol and the oxime compound (Supplementary Fig. 13a) showed clear evidence for two overlapping binding modes in the electron density maps. For the thiophene, the ligand bound differently in each subunit of the dimer (Fig. 5b): In chain A, one of the phenols forms a hydrogen bond to the loop between h7 and h8 (Fig. 5b), whereas in chain B, the phenol is shifted towards h11; this moves the loop between h7 and h8 more than 3 Å away from h11, allowing Met528 to exit the pocket, representing an unusual and significant perturbation from the typical agonist conformation. By superimposing the two LBD chains, it is clear that these differences are transmitted to the h11-h12 interface (Fig. 5b), which should impact h12 dynamics and receptor activity. Thus, a “DBD only” activity profile predicts ligand dynamics in a wider test set, and is apparent in both natural and synthetic ligands.
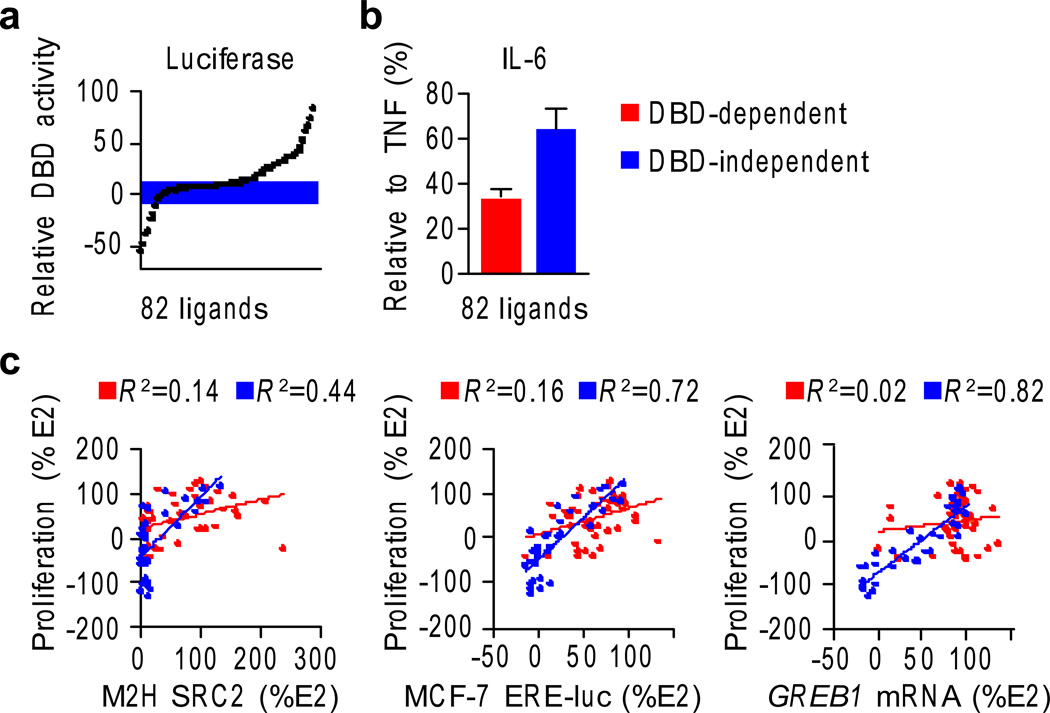
The three newly identified dynamic ligands showed strong anti-inflammatory effects, with resveratrol being the most anti-inflammatory compound in our larger screen (Supplementary Fig. 14a), effects that were clearly ER-mediated (Supplementary Fig. 14b). This suggested that the DBD-dependent activity profile might have a wider impact on cellular signaling. We therefore generated a DBD-dependent activity profile by subtracting the signal from the ERα-LBD assay from the ERα-DBD-LBD reporter data for all ligands from the 11 scaffolds (Supplementary Fig. 15), allowing a comparison of signaling relative to E2, with and without the DBD. The compounds were sorted by DBD-dependent activity, and the inflection points in the graph were used to define a set of 42 DBD-dependent and 40 DBD-inactive compounds (Fig. 6a) suitable for comparison.
Fig. 6. DBD-activity profile predicts signaling outcomes.
a) A DBD-activity profile was calculated for the larger test set by subtracting the ERα-Gal4-LBD luciferase data from the ERα-DBD-LBD luciferase data in HepG2 liver cells, indicating the relative contribution of the DBD to the gene activation signal. The compounds were binned into DBD-independent (i.e. relative DBD activity is close to zero) versus DBD-dependent profiles.
b) The IL-6 assay was performed for the larger test set, and results examined with respect to the DBD activity profiles. Data represent mean values ± s.e.m.
c) The larger ligand test set was assayed for the indicated screens as described for the WAY-169916 derivatives, and was analyzed based on their DBD-activity profile. R2 values obtained from linear regression analysis are indicated. Data are colored as in panel b.
The DBD-dependent activity profile demonstrated significant predictive power in the larger test set. Specifically, in the IL-6 assay (Fig. 6b), DBD-dependent compounds were significantly more robust at suppressing IL-6 induction by TNFα in breast cancer cells (Student’s t-test, P < 0.0001), reproducing the findings with the dynamic WAY-169916 derivatives (Fig. 3b). Only compounds that signaled via AF1 or AF2 (e.g., DBD-independent) showed the canonical relationships between proliferation and reporter genes, coactivator recruitment and GREB1 transcripts (Fig. 6c). Thus, allosterically induced signaling via the DBD interferes with the canonical ER-mediated proliferative signaling mechanisms, and is rather associated with a strong suppression of the inflammatory program.
Discussion
Our structural and chemical biology platform enables a systems-biology approach to defining nuclear receptor functions and their control by diverse ligands. Structure-class analyses revealed that ERα ligands differ widely in the domain(s) required to activate ERα-mediated transcription. For ligands that signal via the AF1 or LBD (i.e., DBD-independent), proliferation was strongly associated with ERE-reporter gene activation, coactivator recruitment and mRNA levels of the ERE-containing native gene GREB1. However, compounds with DBD-dependent activity profiles lack these canonical signaling pathways in a large and diverse set of ligands, and drives a strong anti-inflammatory response. Quite strikingly, ligands that bind dynamically drove the receptor to completely rely on the DBD to activate transcription, demonstrated a robust anti-inflammatory effect and altered signaling behavior with respect to proliferation. Furthermore, compounds having nominally similar activity profiles in simple-binding and reporter-gene assays, as are typically used in early stage drug discovery, can exhibit very different effects on target phenotypes in more complex cellular and in vivo biological contexts—a phenomenon that can be confounding in pharmaceutical development. Here, we show that ligand-class analysis enables the discernment of underlying mechanistic differences. Thus, ligand dynamics is revealed as a novel structural parameter that drives signaling and associated phenotypes for ERα. Finally, we believe that our results demonstrate a novel principle in biology—that ligand dynamics can be exploited to modulate signaling specificity of an allosteric molecule, a concept that could be applied more broadly to drive new therapeutic targeting strategies
Bidirectional allostery between the DBD and LBD has been observed in a number of different contexts, but little is known about the underlying structural mechanisms. Small changes in the DNA-response element sequence can alter gene activation and coactivator recruitment patterns9, 33, 34, while different ligands or coactivators can alter affinity for DNA35. Interestingly, a peptide that binds on the opposite side of the LBD from the AF2 surface displayed completely different ligand-mediated interaction profiles in the LBD versus the full-length receptor, thus defining an interaction site on the LBD that responds to interdomain allostery36, 37. Further, expression of this peptide in cells selectively blocked tamoxifen agonist activity, but not that of E2, suggesting that the binding site acts as a conduit of structural information from the LBD to AF1. In the only full-length nuclear receptor structure to date, there is a direct contact between the PPARγ LBD and the DBD of its dimer partner, RXR38, but this interaction was not observed in solution39, suggesting interdomain signaling is likely to be context dependent. For example, the DNA-repair enzyme MPG binds to both the ERα LBD and DBD, increases receptor affinity for DNA, and inhibits transcription, effects that likely require a unique ERα interdomain conformer40. Our work suggests that the opposite also occurs – that certain ligand-receptor interactions determine recruitment of transcriptional coregulators that depend upon the DBD to modulate ERα activity. We propose that nuclear receptors should best be viewed as allosteric signaling scaffolds41, not just for gene expression, but also for modulation of DNA repair42, rapid activation of kinases43 and many other emerging activities42, 44, 45. In this model, the nuclear receptor activity reflects an allosteric integration of receptor structure, DNA, ligand, post-translational modifications and bound proteins, where a change in any one of these interacting partners can alter the nuclear receptor scaffold interaction profile and the signaling output.
Biology is at an inflection point46, where in the nuclear receptor field, for example, most of the signaling pathways have been identified, but we still cannot explain why a set of ERα ligands will display such a wide variety of physiological phenotypes. Technological advances have now allowed for the application of systems-biology approaches across multiple disciplines, including X-ray crystallography, chemical biology and nuclear receptor pharmacology to enable our structure-class analyses and provide key insights into ligand dynamics and receptor function.
Online Methods
Protein production and X-ray crystallography
ERα protein was produced as previously described5. X-ray diffraction data for new ER LBD crystals (Supplementary Table 2) was collected at Advanced Photon Source (APS), Argonne National Laboratory (ANL) (beam-line 23-ID-B) and Stanford Synchrotron Radiation Lightsource (SSRL) (beam-line 11-1). Data reduction was performed using HKL-2000 software. All new ER LBD models were built via molecular replacement (MR) using PHENIX47. Coordinate files for ER ligands were generated using ChemDraw/Chem3D software suite (Cambridgesoft) or the PRODRG server (http://davapc1.bioch.dundee.ac.uk/prodrg/)48. Structures were refined using PHENIX and rebuild using COOT49. Model images were generated with CCP4MG50.
NMR spectroscopy
ERα protein was incubated with ligand and19F NMR spectra collected on a Bruker 700 MHz NMR instrument with a broadband observe (BBO) probe at 298K. Data were processed and analyzed, including peak deconvolution, using Bruker Topspin 3.0.
Cell culture
HepG2, MCF-7, HEK293-T and Ishikawa cells were cultured in Dulbecco’s minimum essential medium (DMEM) (Cellgro by Mediatech, Inc. Manassas, VA) supplemented with 10% fetal bovine serum (FBS) (Hyclone by Thermo Scientific, South Logan, UT), and 1% non-essential amino acids (Cellgro), Penicillin-Streptomycin-Neomycin antibiotic mixture and Glutamax (Gibco by Invitrogen Corp. Carlsbad, CA), were maintained at 37°C and 5% CO2.
Luciferase reporter assays
Cells were plated in 10 cm dish, and transfected next day with the respective vectors (HepG2 cells – 10 µg of 3X ERE-luciferase reporter and 1.6 µg of either ERα-FL or ERα-DBD-LBD expression vector, 10 µg GAL4- ERα-LBD and 1µg 5X UAS GAL4-Luc, MCF-7 cells – 10 µg 3X ERE-Luc, Ishikawa cells – 10 ug 3X ERE-Luc) using FugeneHD reagent (Roche Applied Sciences, Indianapolis, IN). The next day cells were trypsinized, plated onto 384 well plates (Greiner Bio-one, Manro, NC) in 25µl phenol red-free growth media supplemented with 10% charcoal-dextran sulfate-stripped FBS in triplicates and incubated overnight. After 24 hrs cells were stimulated with compounds dispensed using 100 nl pintool Biomeck NXP workstation (Beckman Coulter Inc.) Luciferase activity was measured after 24 hours using BriteLite reagent (PerkingElmer Inc., Shelton, CT) according to manufacturer’s protocol.
Mammalian two-hybrid assays
HEK293-T cells were transfected. To each well of a 384-well plate 15 µl OptiMEM containing 10 ng of ERα-VP16, 20 ng of either GAL4-SRC1, GAL4-SRC2 or GAL4-SRC3, 10 ng of 5X GAL4-UAS-Luc, 30 ng of pSPORT6 and 0.225 µl of Mirus LT1 transfection reagent was plated and incubated for 20 minutes. To this 15 µl of HEK293-T cells (10,000 cells per well) in phenol red free growth media with 10% charcoal-dextran sulfate-stripped FBS was added and incubated overnight. The next day cells were stimulated with compounds dispensed using 100 nl pintool Biomeck NXP workstation (Beckman Coulter Inc.) Luciferase activity was measured after 24 hours using BriteLite reagent (PerkingElmer Inc., Shelton, CT) according to manufacturer’s protocol.
Dimerization assays (BRET)
HEK293-T cells were plated on 6 well plates and transfected next day with a total DNA of 435 ng using Mirus LT1 as a transfection reagent at a 1:4 ratio of ERα-RLuc and ERα-YFP for ERα- ERα dimerization and with 1:4 ratio of ERα-RLuc and ERβ-YFP for ERα- ERβ dimerization. Cells transfected with 1:4 ratio of ERα-RLuc and pSPORT6 was used as a control. Next day the cells were trypsinized, resuspended with PBS and plated onto 384-well plates at 10,000 cells per well and stimulated with compounds dispensed using 100 nl pintool Biomeck NXP workstation (Beckman Coulter Inc.). After 45 minutes renilla luciferase substrate (Colentrazine-h, Promega) was added at 5 µM concentration and Bioluminescent Resonance Energy Transfer (BRET) was measured using an Envision Plate reader. The BRET ratio is calculated as described elsewhere (26).
Proliferation assays
MCF-7 cells were plated on 384-well plates in phenol red free growth media with 5% charcoal-dextran sulfate-stripped FBS and stimulated with compounds next day using 100 nl pintool Biomeck NXP workstation (Beckman Coulter Inc.). Treatment was repeated again after 3 days. 7 days after the initial treatment, 25 µl of CellTitre-Glo reagent (Promega) was added and luminescence was measured. Data was normalized using cell numbers estimated from a cell growth curve generated from cells plated on the 7th day.
DNA-binding assays
To each well of a 384-well plate 15 ul OPTIMEM containing 50 ng of ERα-VP16 and 50 ng of 3X ERE-Luc and 0.3 µl of Mirus LT1 transfection reagent was plated and incubated for 20 minutes. To this 15 µl of HEK293-T cells (12,000 cells per well) in phenol red free growth media with 10% charcoal-dextran sulfate-stripped FBS was added and incubated overnight. The next day cells were stimulated with compounds dispensed using 100 nl pintool Biomeck NXP workstation (Beckman Coulter Inc.) Luciferase activity was measured after 24 hours using BriteLite reagent (PerkingElmer Inc., Shelton, CT) according to manufacturer’s protocol.
IL-6 ELISA
MCF-7 cells were plated in 384-well plates in 25 µl phenol red-free growth media supplemented with 10% charcoal-dextran sulfate-stripped FBS at a density of 8,000 cells/well in triplicates. 48 hrs later cells were treated with vehicle, TNF (15 ng/ml) alone or TNF (15 ng/ml) and compounds at 10 µM dose. 24 hrs later, secreted IL-6 was measured using IL-6 AlphaLISA kit (Perkin Elmer) from 5 µl of media in 10 µl total reaction volume according to the manufacturer’s protocol.
Quantitative RT-PCR
MCF-7 cells were plated in 384-well plates in 25 µl phenol red-free growth media supplemented with 5% charcoal-dextran sulfate-stripped FBS at a density of 2500 cells/well in triplicates. Three days later cells were stimulated with compounds for 1 hr (c-MYC), 3 hr (CCND1) and 24 hr (GREB1 and pS2). Total RNA was extracted with RNAgem Tissue Plus RNA extraction kit (Zygem, New Zealand). Cells were washed once with PBS, lysed with 7.5 µl RNAgem extraction reagent at 75°C for 5 minutes. Genomic DNA was removed with DNASEI treatment for 5 minutes at 37°C and stopped by incubating at 75°C. 5ul of this mixture was used for cDNA synthesis in a total volume of 10 µl using High-Capacity cDNA Reverse Transcription Kit (Life Technologies, Grand Island, NY). The cDNA was diluted 3 times and 4 µl of diluted cDNA was used in qPCR reaction using TaqMan Gene Expression Master Mix (Life Technologies, Grand Island, NY) in a total reaction volume of 10 µl. The qPCR reaction was carried out in a duplex format using pre-designed real-time qPCR assays (Life Technologies, Grand Island, NY) with GAPDH primers with a VIC probe as an endogenous control and target gene (c-MYC, CCND1, GREB1 and pS2) primers with FAM probe. The reactions were run on ABI 7900 HT Real-Time PCR system and the fold change relative to vehicle treated samples were calculated using ΔΔCt method.
Data deposition
X-ray data and coordinates for new structures were deposited in the protein data bank (PDB) (www.pdb.org). These include the crystal structures of ERα in complex with WAY-derivatives 4 (PDB ID: 4IU7), 7 (4IUI), 9 (4IV2), 10 (4IV4), 12 (4IVW), 13 (4IVY), 14 (4IW6) and 17 (4IW8), and the dynamic thiophene (4IWC) and oxime (4IWF) ligands. The resveratrol-bound ERα structure will be published elsewhere.
Statistics
Data are represented as mean values ± s.e.m. Robustness of high-throughput assays was determined by Z-factor (Supplementary Table 2). An assay with a Z-factor of 0.5 or more is considered to be suitable for high-throughput format.
Supplementary Material
Acknowledgments
We thank Cleveland, J.L. for critically evaluating the manuscript. Research support from the National Institutes of Health (PHS 5R37 DK015556 to J.A.K.; 5R33CA132022 and 5R01DK077085 to K.W.N.) S.S. is supported by the Frenchman’s Creek Women for Cancer Research.
Footnotes
Author Contributions
S.S. and J.C.N. designed and performed experiments, and wrote the manuscript; A.A.P, V.C., J.N. and T.S.H. performed experiments; D.J.K. designed experiments; J.A.K. and K.W.N. designed experiments and wrote the manuscript
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Hilser VJ, Wrabl JO, Motlagh HN. Structural and energetic basis of allostery. Annual review of biophysics. 2012;41:585–609. doi: 10.1146/annurev-biophys-050511-102319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll MJ, et al. Evidence for dynamics in proteins as a mechanism for ligand dissociation. Nature chemical biology. 2012;8:246–252. doi: 10.1038/nchembio.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shiau AK, et al. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 4.Nettles KW, et al. CBP Is a dosage-dependent regulator of nuclear factor-kappaB suppression by the estrogen receptor. Mol Endocrinol. 2008;22:263–272. doi: 10.1210/me.2007-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruning JB, et al. Coupling of receptor conformation and ligand orientation determine graded activity. Nature chemical biology. 2010;6:837–843. doi: 10.1038/nchembio.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hughes TS, et al. Ligand and receptor dynamics contribute to the mechanism of graded PPARgamma agonism. Structure. 2012;20:139–150. doi: 10.1016/j.str.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll MJ, et al. Direct detection of structurally resolved dynamics in a multiconformation receptor-ligand complex. Journal of the American Chemical Society. 2011;133:6422–6428. doi: 10.1021/ja2005253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 9.Meijsing SH, et al. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McInerney EM, Katzenellenbogen BS. Different regions in activation function-1 of the human estrogen receptor required for antiestrogen- and estradiol-dependent transcription activation. The Journal of biological chemistry. 1996;271:24172–24178. doi: 10.1074/jbc.271.39.24172. [DOI] [PubMed] [Google Scholar]
- 11.Nettles KW, Greene GL. Ligand control of coregulator recruitment to nuclear receptors. Annual review of physiology. 2005;67:309–333. doi: 10.1146/annurev.physiol.66.032802.154710. [DOI] [PubMed] [Google Scholar]
- 12.Darimont BD, et al. Structure and specificity of nuclear receptor-coactivator interactions. Genes & development. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-alpha. Nature. 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 14.Xu HE, et al. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha. Nature. 2002;415:813–817. doi: 10.1038/415813a. [DOI] [PubMed] [Google Scholar]
- 15.Nettles KW, et al. Structural plasticity in the oestrogen receptor ligand-binding domain. EMBO reports. 2007;8:563–568. doi: 10.1038/sj.embor.7400963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chadwick CC, et al. Identification of pathway-selective estrogen receptor ligands that inhibit NF-kappaB transcriptional activity. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2543–2548. doi: 10.1073/pnas.0405841102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nettles KW, et al. Allosteric control of ligand selectivity between estrogen receptors alpha and beta: implications for other nuclear receptors. Molecular cell. 2004;13:317–327. doi: 10.1016/s1097-2765(04)00054-1. [DOI] [PubMed] [Google Scholar]
- 18.Laughlin JD, et al. Structural Mechanisms of Allostery and Autoinhibition in JNK Family Kinases. Structure. 2012;20:2174–2184. doi: 10.1016/j.str.2012.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shang Y, Brown M. Molecular determinants for the tissue specificity of SERMs. Science. 2002;295:2465–2468. doi: 10.1126/science.1068537. [DOI] [PubMed] [Google Scholar]
- 20.Sicheri F, Kuriyan J. Structures of Src-family tyrosine kinases. Curr Opin Struct Biol. 1997;7:777–785. doi: 10.1016/s0959-440x(97)80146-7. [DOI] [PubMed] [Google Scholar]
- 21.Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 22.Berry M, Metzger D, Chambon P. Role of the two activating domains of the oestrogen receptor in the cell-type and promoter-context dependent agonistic activity of the anti-oestrogen 4-hydroxytamoxifen. The EMBO journal. 1990;9:2811–2818. doi: 10.1002/j.1460-2075.1990.tb07469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pham TA, et al. Antiestrogen can establish nonproductive receptor complexes and alter chromatin structure at target enhancers. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:3125–3129. doi: 10.1073/pnas.88.8.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lupien M, et al. Raloxifene and ICI182,780 increase estrogen receptor-alpha association with a nuclear compartment via overlapping sets of hydrophobic amino acids in activation function 2 helix 12. Mol Endocrinol. 2007;21:797–816. doi: 10.1210/me.2006-0074. [DOI] [PubMed] [Google Scholar]
- 25.Planas-Silva MD, Shang Y, Donaher JL, Brown M, Weinberg RA. AIB1 enhances estrogen-dependent induction of cyclin D1 expression. Cancer research. 2001;61:3858–3862. [PubMed] [Google Scholar]
- 26.Xu J, Wu RC, O'Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nature reviews. Cancer. 2009;9:615–630. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rae JM, et al. GREB 1 is a critical regulator of hormone dependent breast cancer growth. Breast cancer research and treatment. 2005;92:141–149. doi: 10.1007/s10549-005-1483-4. [DOI] [PubMed] [Google Scholar]
- 28.Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes & development. 2006;20:2513–2526. doi: 10.1101/gad.1446006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang YH, et al. Knockdown of c-Myc expression by RNAi inhibits MCF-7 breast tumor cells growth in vitro and in vivo. Breast cancer research : BCR. 2005;7:R220–R228. doi: 10.1186/bcr975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C, et al. Estrogen induces c-myc gene expression via an upstream enhancer activated by the estrogen receptor and the AP-1 transcription factor. Mol Endocrinol. 2011;25:1527–1538. doi: 10.1210/me.2011-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Powell E, et al. Identification of estrogen receptor dimer selective ligands reveals growth-inhibitory effects on cells that co-express ERalpha and ERbeta. PloS one. 2012;7:e30993. doi: 10.1371/journal.pone.0030993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rae JM, et al. GREB1 is a novel androgen-regulated gene required for prostate cancer growth. The Prostate. 2006;66:886–894. doi: 10.1002/pros.20403. [DOI] [PubMed] [Google Scholar]
- 33.Loven MA, Likhite VS, Choi I, Nardulli AM. Estrogen response elements alter coactivator recruitment through allosteric modulation of estrogen receptor beta conformation. The Journal of biological chemistry. 2001;276:45282–45288. doi: 10.1074/jbc.M106211200. [DOI] [PubMed] [Google Scholar]
- 34.Hall JM, McDonnell DP, Korach KS. Allosteric regulation of estrogen receptor structure, function, and coactivator recruitment by different estrogen response elements. Mol Endocrinol. 2002;16:469–486. doi: 10.1210/mend.16.3.0814. [DOI] [PubMed] [Google Scholar]
- 35.Wang JC, et al. Novel arylpyrazole compounds selectively modulate glucocorticoid receptor regulatory activity. Genes & development. 2006;20:689–699. doi: 10.1101/gad.1400506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bapat AR, Frail DE. Full-length estrogen receptor alpha and its ligand-binding domain adopt different conformations upon binding ligand. J Steroid Biochem Mol Biol. 2003;86:143–149. doi: 10.1016/s0960-0760(03)00262-0. [DOI] [PubMed] [Google Scholar]
- 37.Kong EH, et al. Delineation of a unique protein-protein interaction site on the surface of the estrogen receptor. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:3593–3598. doi: 10.1073/pnas.0407189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chandra V, et al. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature. 2008;456:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rochel N, et al. Common architecture of nuclear receptor heterodimers on DNA direct repeat elements with different spacings. Nature structural & molecular biology. 2011;18:564–570. doi: 10.1038/nsmb.2054. [DOI] [PubMed] [Google Scholar]
- 40.Likhite VS, Cass EI, Anderson SD, Yates JR, Nardulli AM. Interaction of estrogen receptor alpha with 3-methyladenine DNA glycosylase modulates transcription and DNA repair. The Journal of biological chemistry. 2004;279:16875–16882. doi: 10.1074/jbc.M313155200. [DOI] [PubMed] [Google Scholar]
- 41.Nwachukwu JC, Nettles KW. The nuclear receptor signalling scaffold: insights from full-length structures. The EMBO journal. 2012;31:251–253. doi: 10.1038/emboj.2011.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schultz-Norton JR, Ziegler YS, Nardulli AM. ERalpha-associated protein networks. Trends in endocrinology and metabolism: TEM. 2011;22:124–129. doi: 10.1016/j.tem.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 43.Hammes SR, Levin ER. Minireview: Recent advances in extranuclear steroid receptor actions. Endocrinology. 2011;152:4489–4495. doi: 10.1210/en.2011-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhan YY, et al. The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK. Nature chemical biology. 2012 doi: 10.1038/nchembio.1069. [DOI] [PubMed] [Google Scholar]
- 45.Sun Z, et al. Inhibition of beta-catenin signaling by nongenomic action of orphan nuclear receptor Nur77. Oncogene. 2012;31:2653–2667. doi: 10.1038/onc.2011.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.National Research Council (U.S.) A new biology for the 21st century. Washington, D.C.: National Academies Press; 2009. Committee on a New Biology for the 21st Century: Ensuring the United States Leads the Coming Biology Revolution., National Research Council (U.S.). Board on Life Sciences. & National Research Council (U.S.). Division on Earth and Life Studies. [PubMed] [Google Scholar]
- 47.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 49.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 50.McNicholas S, Potterton E, Wilson KS, Noble ME. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr D Biol Crystallogr. 2011;67:386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.