Abstract
Human epidermal growth factor receptor (HER)-2 overexpression occurs in 15–20% of all breast cancers and is associated with increased metastatic potential and poor patient survival. Abnormal HER2 activation, either through HER2 overexpression or heregulin (HRG):HER3 binding, elicits the formation of potent HER2-HER3 heterodimers and drives breast cancer cell growth and metastasis. In a previous study, we found that fibroblast growth factor-inducible 14 (Fn14), a member of the TNF receptor superfamily, was frequently overexpressed in human HER2+ breast tumors. We report here that HER2 and Fn14 are also co-expressed in mammary tumors that develop in two different transgenic mouse models of breast cancer. In consideration of these findings, we investigated whether HER2 activation in breast cancer cells could directly induce Fn14 gene expression. We found that transient or stable transfection of MCF7 cells with a HER2 expression plasmid increased Fn14 protein levels. Also, HRG1-β1 treatment of MCF7 cells transiently induced Fn14 mRNA and protein expression. Both the HER2- and HRG1-β1-induced increase in Fn14 expression in MCF7 cells as well as basal Fn14 expression in HER2 gene-amplified AU565 cells could be reduced by HER2 kinase inhibition with lapatinib or combined HER2 and HER3 depletion using siRNA. We also report that Fn14-depleted, HER2-overexpressing MCF7 cells have reduced basal cell migration capacity and reduced HRG1-β1-stimulated cell migration, invasion and matrix metalloproteinase (MMP)-9 expression. Together, these results indicate that Fn14 may be an important downstream regulator of HER2/HER3-driven breast cancer cell migration and invasion.
Keywords: Fn14, breast cancer, HER2, HER3, heregulin, invasion, MMP-9
Introduction
Breast cancer is the most common malignancy in women world-wide and in the USA ~40,000 women are predicted to die from this disease in 2012 (1). HER2, a member of the epidermal growth factor receptor (EGFR) family, is overexpressed in 15–20% of invasive breast cancers and is associated with aggressive tumor behavior, decreased time to relapse and poor clinical outcome (2). HER3, a kinase-inactive member of the EGFR family (3), is often expressed in HER2+ breast tumors (4) and it is the preferred heterodimerization partner for HER2 (5). Indeed, HER3 has been shown to play a pivotal role in mediating both HER2 oncogenesis (6) and the resistance of breast cancers to HER2-targeted therapies (7). Heregulin-1 (HRG1) (also called neuregulin-1), a member of the EGF family (8), is a HER3 and HER4 ligand that elicits the formation of potent HER2-HER3 heterodimers (9). HRG1 is expressed in breast tumors (8) and has been implicated in modulating breast cancer cell invasion (10–12) and metastasis (13). Thus, the HER2-HER3 receptor pair forms a very potent mitogenic and transforming unit that promotes breast tumorigenesis.
Cancer cell migration and invasion is controlled by complex signaling events that regulate cytoskeletal changes, cell-cell and cell-extracellular matrix interactions, and extracellular proteolytic activity (14). HER2/HER3 heterodimer-driven cellular migration and invasion has been previously shown to require PI3K activity(15) and a variety of downstream mediators have been implicated in these cellular processes (e.g., FAK-Src (16) and matrix metalloproteinase (MMP)-9 (17)). In regard to the MMPs, they are known to play an important role in the tumor microenvironment by mediating effects such as remodeling of the extracellular matrix, tissue invasion and intravasation, inflammation and angiogenesis (18). HER2/HER3 signaling has been reported to up-regulate MMP production and activity (17, 19, 20) but the biological basis of this effect remains poorly understood.
Tumor necrosis factor (TNF)-like weak inducer of apoptosis (TWEAK) and fibroblast growth factor (FGF)-inducible 14 (Fn14) are a TNF superfamily ligand-receptor pair involved in many cellular processes associated with wound repair, including inflammation and angiogenesis (21, 22). TWEAK:Fn14 binding promotes Fn14 association with adaptor molecules called TNF receptor-associated factors (TRAFs) which couple to various signaling pathways such as the NF-κB, MAPK and PI3K/Akt pathways (21). The TWEAK-Fn14 signaling axis plays an important role in regulating various aspects of tumor behavior such as growth, survival, invasion and angiogenesis (23–29). Importantly, Fn14 is highly expressed in many solid tumors, and in some tumor types elevated Fn14 levels have been shown to correlate with disease progression and poor patient outcome (23, 25, 28). There is also evidence that Fn14 overexpression in tumor cells can activate signaling pathways and stimulate cellular processes; for example, ectopic Fn14 expression promotes glioma cell invasion by activating Rac1 and NF-κB(25).
In a previous study we showed that elevated Fn14 expression strongly correlated with the HER2+/ER- intrinsic subtype of breast cancer and with clinical indicators of poor prognosis(26). However, the mechanisms underlying preferential Fn14 expression in HER2+ breast tumors, as well as the potential contributions of Fn14 to HER2-mediated disease, have yet to be elucidated. Here we report that Fn14 expression is increased in MMTV-c-Neu and MMTV-polyoma middle T antigen (PyMT) transgenic mouse breast tumors with elevated Neu (HER2) levels. Also, both HER2 overexpression in MCF7 breast cancer cells and HRG1-β1 treatment of MCF7 cells induces Fn14 gene expression and these effects are dependent on HER2/HER3 signaling. Finally, we show that stable knockdown of Fn14 in HER2-overexpressing MCF7 cells decreases basal cell migration capacity and HRG1-β1-stimulated migration, invasion and MMP-9 expression.
Materials and Methods
Transgenic mouse models
MMTV-c-Neu mice (FVB/N-Tg(MMTV-neu)202Mul/J) (30) were purchased from Jackson Laboratories (Bar Harbor, ME, USA). These mice were bred and mammary tissue samples isolated as previously described (31). All MMTV-c-Neu animal studies were approved by the Case Western Reserve University Institutional Animal Care and Use Committee. The MMTV-PyMT mice (FVB/N-Tg(MMTV-PyVT)634Mul/J) (32,33) were also purchased from Jackson Laboratories. Male hemizygous transgenic mice were bred to FVB/N females and at various time points wild-type and hemizygous littermates were selected, euthanized and five mammary fat pad pairs were isolated and then frozen until use. All MMTV-PyMT animal studies were approved by the University of Maryland School of Medicine Institutional Animal Care and Use Committee.
Cell culture and treatments
Cell lines were obtained from the following sources: MCF7, BT474, SKBR3, MDA-MB-453, AU565 and NIH3T3 (ATCC; Manassas, VA, USA), MCF7/HER2 (Dr. Dihua Yu, University of Texas MD Anderson Cancer Center), MCF7/HER2-18 (Dr. Anne Hamburger, University of Maryland School of Medicine), NIH3T3/HER2 (Dr. Peter Choyke, NIH), MCF7 Ca/LTLT-Ca (Dr. Angela Brodie, University of Maryland School of Medicine). MCF7, MCF7/HER2, BT474, SKBR3 and MDA-MB-453 cells were maintained in DMEM (Cellgro, Manassas, VA, USA) and AU565, NIH3T3, NIH3T3/HER2 and MCF7/HER2-18 cells were maintained in RPMI 1640 (Cellgro). Both cell mediums were supplemented with 10% FBS (HyClone, Logan, UT, USA), 2 mM L-glutamine and 1% penicillin-streptomycin. MCF7/HER2 and MCF7/HER2-18 cells were additionally maintained in 750 or 500 μg/ml G418 (Cellgro), respectively. Lentivirus-infected MCF7/HER2-18 cells were additionally maintained in 0.5 μg/ml puromycin (Cellgro). Fn14 shRNA-448 cells expressing myc epitope-tagged Fn14 were additionally maintained in 1 μg/ml blasticidine (Sigma, St. Louis, MO, USA). MCF7 Ca and LTLT-Ca cells were grown as previously described (34). Cells were treated with the indicated concentrations of U0126, wortmannin (both from Cell Signaling Technology, Beverly, MA, USA), lapatinib (LC Laboratories, Woburn, MA, USA), MMP-2/MMP-9 Inhibitor IV (SB-3CT) (Calbiochem, La Jolla, CA, USA), MK-2206 (Alexis Corporation), EGF, HB-EGF, BTC, HRG1-α or HRG1-β1 (all from R & D Systems, Minneapolis, MN, USA).
Western blot analysis
Western blotting was performed as previously described (35). The following primary antibodies were used: Fn14, p-HER2 (Tyr1248), p-HER3 (Tyr1289), p-Erk1/2 (Thr202/Tyr204), Erk1/2, p-Akt (S473), Akt, p-p90RSK (Ser380), p90RSK, p-p70 S6 Kinase (Thr389), p70 S6 Kinase, GAPDH (all from Cell Signaling Technology), Neu, ErbB3, ErbB4 (all from Santa Cruz Biotechnology, Santa Cruz, CA, USA), EGFR, Myc and tubulin (all from Millipore).
FACS analysis
Flow cytometry was conducted using phycoerythrin-labeled anti-Fn14 mAb ITEM-4 and IgG3 isotype control (eBioscience Inc., San Diego, CA, USA) as previously described (26).
RNA isolation and quantitative real-time RT-PCR assays
Total cellular RNA was extracted using the RNeasy kit (Qiagen, Valencia, CA, USA) as previously described (36). RNA was converted to cDNA using the ProtoScript AMV LongAmp Taq RT-PCR kit (New England Biolabs, Ipswich, MA, USA) according to manufacturer’s instructions. Fn14 and GAPDH mRNA levels were quantified using an ABI Prism 7900HT Real-time PCR system (Applied Biosystems, Beverly, MA, USA) and the following primers and probes: Fn14, Cat.# Hs00171993_m1; MMP-9, Cat.# Hs00234579_m1; GAPDH, Cat.# Hs99999905_m1 (Taqman Gene Expression Assay, Applied Biosystems).
Plasmid DNA and siRNA transfections
MCF7 cells were transiently transfected with 2 or 4 μg of the pcDNA6-HER2 expression plasmid (provided by Dr. Mien-Chie Hung (University of Texas MD Anderson Cancer Center)) per 60 mm dish using the Effectene transfection kit (Qiagen) according to the manufacturer’s instructions. Cells were harvested 24 h post transfection for Western blot analysis. In other experiments, Fn14 shRNA-448 cells were transfected with pcDNA6 vector or pcDNA6-Fn14-myc plasmid (37) as above and drug-resistant cell colonies were selected using blasticidine. Positively transfected clones were expanded and screened for Myc levels by Western blot analysis. For siRNA experiments, MCF7, MCF7/HER2, MCF7/HER2-18 or AU565 cells were plated at a density of 2 × 105 in 60 mm dishes and 24 h later the cells were transfected with either Allstars non-silencing control, luciferase control, HER4, HER2 and/or HER3 siRNA duplexes (Qiagen) at a concentration of 25 nM using the Lipofectamine 2000 transfection reagent as previously described(26). Cells were harvested 48 h post transfection for Western blot analysis.
Lentiviral transduction and isolation of Fn14-depleted cell lines
Lentiviral constructs encoding control, non-target shRNA or shRNAs targeting two different regions of the Fn14 transcript (Fn14 shRNA#448, Cat.# TRCN0000072448 and Fn14 shRNA#562, Cat.# TRCN0000222562) were purchased from Sigma. Lentiviral packaging was performed as previously described (27). MCF7/HER2-18 cells were transduced with the various lentiviruses and drug-resistant cell colonies were selected using puromycin. Positively transduced clones were expanded and screened for Fn14 levels by Western blot analysis.
Scratch wound assays
Parental MCF7, MCF7/HER2-18, MCF7/HER2-18 control shRNA, and MCF7/HER2-18 Fn14 shRNA cells were plated in triplicate in 6-well cluster dishes and allowed to grow to confluence in normal RPMI growth medium (10% FBS). The confluent cell monolayers were scratched in the shape of a cross using a 200 μl pipette tip and normal RPMI growth medium (10% FBS) or low serum medium (0.5%) containing HRG1-β1 (50 or 200 ng/ml as indicated) was added to the cells. Wound closure was monitored over a 24 h time period and photographs were taken at the four intersecting edges of the cross. Wound width at 0 and 24 h was measured and the difference plotted as percent wound closure.
Invasion assays
Parental MCF7, MCF7/HER2-18, MCF7/HER2-18 control shRNA, MCF7/HER2-18 Fn14 shRNA-562 and MCF7/HER2-18 Fn14 shRNA-448 cells (2 × 105), and MCF7/HER2-18 Fn14 shRNA-448 cells transfected with vector or Fn14-myc plasmid (1 × 105) were suspended in RPMI containing 0.5% FBS and seeded in the upper chamber of 24-well BD Biocoat Matrigel Invasion Chambers (BD Biosciences, San Jose, CA, USA). Lower chambers contained RPMI with 0.5% FBS and HRG1-β1 (50 or 200 ng/ml as indicated). After 48 h of incubation the cells were fixed and stained as previously described (27). Cells were counted from 25 high-power-fields per filter.
Gelatin zymography
MCF7/HER2-18 control shRNA cells, Fn14 shRNA-448 cells and Fn14 shRNA-448 cells transfected with vector or Fn14-myc expression plasmid were plated in 100-mm dishes, serum-starved for 24 h in serum-free, phenol red-free RPMI medium and then treated with 50 ng/ml HRG1-β1. Conditioned media was collected at 0, 12 and 24 h following treatment and concentrated using Amicon Ultra-15 Centrifugal Filter Units (Millipore). Protein concentrations were determined using the BCA protein assay (Thermo Fisher Scientific). Equal amounts of protein were subjected to gelatin zymography using Novex 10% Zymogram gels (Invitrogen, Carlsbad, CA, USA) according to manufacturer’s instructions.
Results
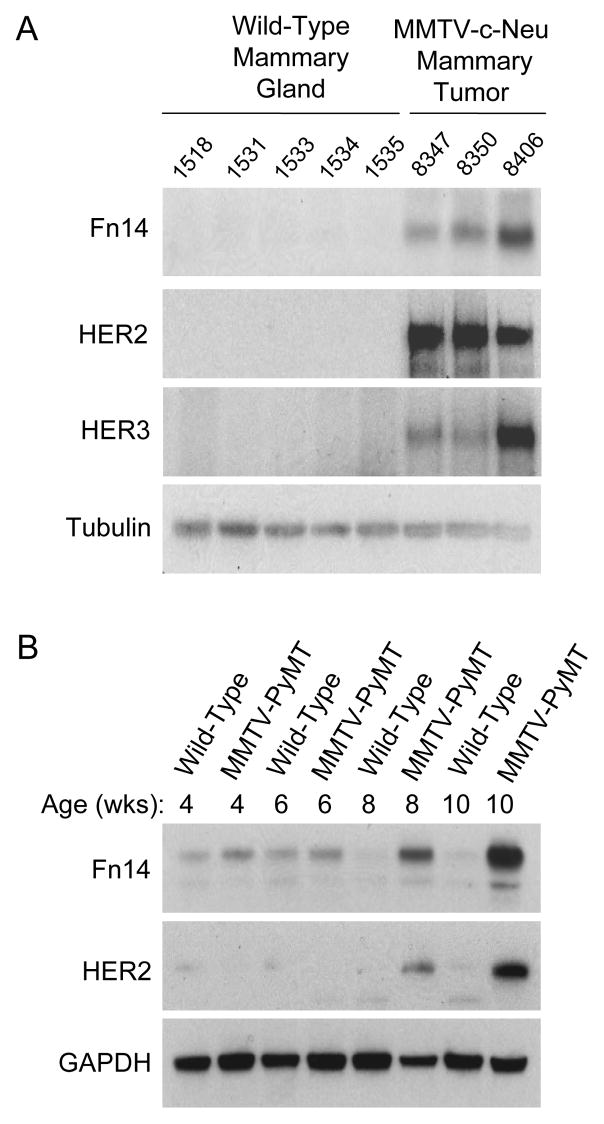
Fn14 and HER2 are co-expressed in MMTV-c-Neu and MMTV-PyMT transgenic mouse breast tumors
Previous reports have indicated that Fn14 is highly expressed in human breast tumors (26, 38, 39); and in particular, it has been shown that Fn14 is most frequently overexpressed in the HER2+ breast cancer subtype (26, 39). To investigate this further, we analyzed Fn14 levels in the mouse mammary tumor virus (MMTV)-c-Neu transgenic mouse model of Neu (HER2)-induced tumorigenesis. These mice express the rat c-Neu proto-oncogene under transcriptional control of the MMTV promoter/enhancer and develop spontaneous mammary tumors (30, 31). Fn14 expression was detected in all of the HER2+ mammary tumors examined with no expression seen in age-matched wild-type mammary glands (Fig. 1A). HER3 expression was also up-regulated in the tumors, consistent with a previous report (40). We also examined Fn14 levels in the MMTV-PyMT model of breast cancer. In this model, mammary hyperplasia is detected at ~4 weeks of age and advanced carcinoma at ~12 weeks (32, 33). It has been reported that Neu (HER2) expression is activated in PyMT-driven mammary tumors, with the highest HER2 expression levels in carcinoma stage tumors(33). We detected Fn14 expression in both wild-type mammary glands and PyMT tumors isolated from 4- to 10-week-old mice, but Fn14 levels were highest in the 8- and 10-week-old HER2+ mammary tumor samples (Fig. 1B).
Figure 1.
Fn14 is expressed in MMTV-c-Neu and MMTV-PyMT transgenic mouse breast tumors. (A) Five normal mammary glands and three mammary tumors isolated from 12-month-old wild-type or MMTV-c-Neu transgenic mice, respectively, were analyzed for Fn14, HER2, HER3 and tubulin expression by Western blotting. (B) Normal mammary gland and mammary tumor samples isolated from age-matched wild-type or MMTV-PyMT transgenic mice, respectively, were analyzed for Fn14, HER2 and GAPDH expression by Western blotting.
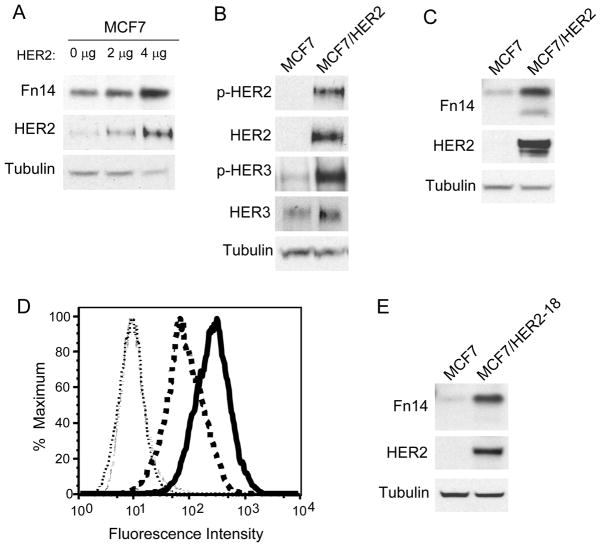
HER2 overexpression in human breast cancer cell lines and murine fibroblasts increases Fn14 levels
To investigate whether HER2 overexpression in breast cancer cells could directly up-regulate Fn14 gene expression, we used MCF7 cells since they normally express relatively low levels of HER1 and HER2, moderate levels of Fn14 and HER4, and high levels of HER3 (Supplementary Fig. S1). We found that transient transfection of these cells with a HER2 expression plasmid increased Fn14 protein levels (Fig. 2A). We then compared Fn14 levels in parental MCF7 cells and a HER2-overexpressing stably-transfected MCF7 cell line (MCF7/HER2). HER2 overexpression in this cell line activated both HER2 and HER3 as shown by Western blotting with phospho(p)-HER2 and p-HER3 antibodies (Fig. 2B). Fn14 expression was elevated in the MCF7/HER2 cells, as measured by both Western blot (Fig. 2C) and FACS (Fig. 2D) analysis. In this Western blot and in several others to follow, two Fn14 species can be detected. These species represent full-length Fn14 and an N-terminal truncated Fn14 isoform (unpublished results). We determined whether HER2 overexpression in MCF7 cells was increasing Fn14 mRNA expression by quantitative real-time RT-PCR analysis, and found that Fn14 mRNA levels were not significantly increased in MCF7/HER2 cells compared to MCF7 cells (Supplementary Fig. S2). HER2 overexpression also increased Fn14 levels in another stably-transfected MCF7 cell line (MCF7/HER2-18) (Fig. 2E). Finally, we compared Fn14 levels in MCF7 cells stably-transfected with an aromatase expression plasmid versus their HER2-overexpressing letrozole-resistant counterpart (34) and in parental murine NIH3T3 fibroblasts versus a HER2-overexpressing stably-transfected NIH3T3 cell line. In both cases, HER2 overexpression increased Fn14 levels (Supplementary Fig. S3).
Figure 2.
Both transient and stable HER2 overexpression in MCF7 breast cancer cells increases Fn14 protein expression. (A) MCF7 cells were transiently transfected with the indicated amount of a HER2 expression plasmid. Cells were harvested at 24 h post-transfection and Fn14, HER2 and tubulin expression analyzed by Western blotting. (B and C) MCF7 and MCF7/HER2 cells were analyzed for HER2, HER3, p-HER2, p-HER3, Fn14 and tubulin levels by Western blotting. (D) MCF7 and MCF7/HER2 cells were subjected to FACS analysis using an anti-Fn14 mAb (MCF7 cells: dark dashed line; MCF7/HER2 cells: dark solid line) or isotype control IgG (MCF7 cells: light dashed line; MCF7/HER2 cells: light dotted line). (E) MCF7 and MCF7/HER2-18 cells were analyzed for Fn14, HER2 and tubulin expression by Western blotting.
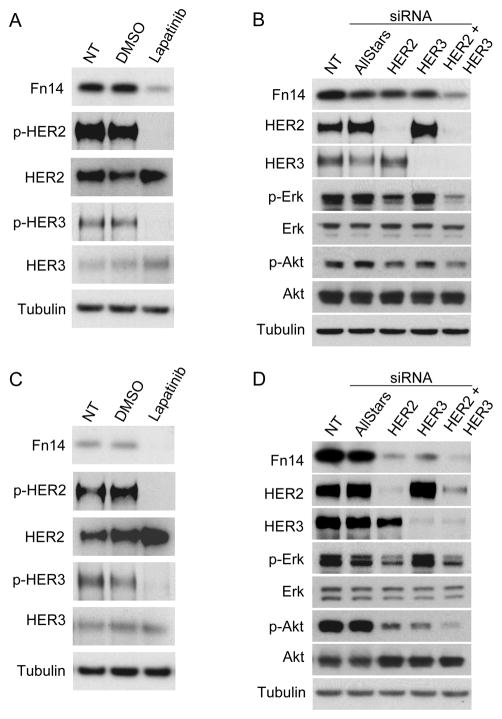
Inhibition of HER2 kinase activity, combinatorial HER2-HER3 depletion, and inhibition of MEK1/2 and PI3K signaling down-regulates Fn14 expression in HER2-overexpressing MCF7 and AU565 breast cancer cells
We tested whether HER2 tyrosine kinase activity was critical for HER2-mediated Fn14 up-regulation using lapatinib, a tyrosine kinase inhibitor that targets both HER1 and HER2 (41). Lapatinib treatment of MCF7/HER2 cells inhibited HER2 and HER3 phosphorylation and significantly downregulated Fn14 expression (Fig. 3A). We also used RNA interference to deplete HER2 and HER3 expression in MCF7/HER2 cells. Cells were transiently transfected with control siRNA or HER2 and HER3 siRNA alone or in combination. Combined HER2-HER3 depletion, but not HER2 or HER3 depletion alone, markedly inhibited Fn14 expression (Fig. 3B). Lapatinib treatment and combined HER2-HER3 depletion also decreased Fn14 expression in the MCF7/HER2-18 cell line (Supplementary Fig. S4). AU565 is a breast cancer cell line with HER2 gene amplification that normally expresses high levels of HER2 and HER3 (Supplementary Fig. S1) and constitutive HER2 and HER3 tyrosine phosphorylation can be detected in these cells (Fig. 3C). Lapatinib treatment and HER2, HER3 or both HER2 and HER3 depletion significantly decreased Fn14 expression in AU565 cells (Fig. 3C, D). HER4 depletion in MCF7/HER2 or AU565 cells did not alter Fn14 expression (data not shown).
Figure 3.
Lapatinib treatment or HER2/HER3 siRNA co-transfection of MCF7/HER2 and AU565 cells decreases Fn14 expression levels. (A) MCF7/HER2 cells were serum-starved overnight (0.5% FBS) and then either left untreated (NT, no treatment) or treated with DMSO vehicle or lapatinib (100 nM) for 8 h. Cells were harvested and Fn14, p-HER2, HER2, p-HER3, HER3 and tubulin levels analyzed by Western blotting. (B) MCF7/HER2 cells were either left untreated (NT) or transiently transfected with Allstars control, HER2 and/or HER3 siRNA duplexes. Cells were harvested 48 h later and analyzed for Fn14, HER2, HER3, p-Erk, Erk, p-Akt, Akt and tubulin levels by Western blotting. (C and D) AU565 cells were treated in the same manner as the MCF7/HER2 cells as described above in A and B.
HER2 overexpression in MCF7 (Supplementary Fig. S5A, Fig. 3B) and AU565 (Fig. 3D) cells leads to constitutive activation of the MEK/ERK and PI3K/Akt signaling cascades. Therefore, we analyzed the effects of HER2 and/or HER3 depletion on p-Erk and p-Akt levels in order to investigate why HER2 or HER3 depletion alone decreased Fn14 levels in AU565 cells but not MCF7/HER2 or MCF7/HER2-18 cells. We found that HER2 or HER3 depletion alone had a much greater effect on p-Akt levels in the AU565 cells compared to the MCF7/HER2 cells (Fig. 3B, D). To determine the relative contribution of the MEK/ERK and PI3K/Akt pathways in regulating Fn14 expression in these two cell lines we treated cells with either the MEK1/2 inhibitor U0126 or the PI3K inhibitor wortmannin. Each of these inhibitors reduced Fn14 expression levels in both cell lines, but Fn14 expression in AU565 cells was more sensitive to wortmannin treatment (Supplementary Fig. S5B and C). Treatment of AU565 cells with the Akt-specific inhibitor MK-2206 also reduced Fn14 expression levels (data not shown).
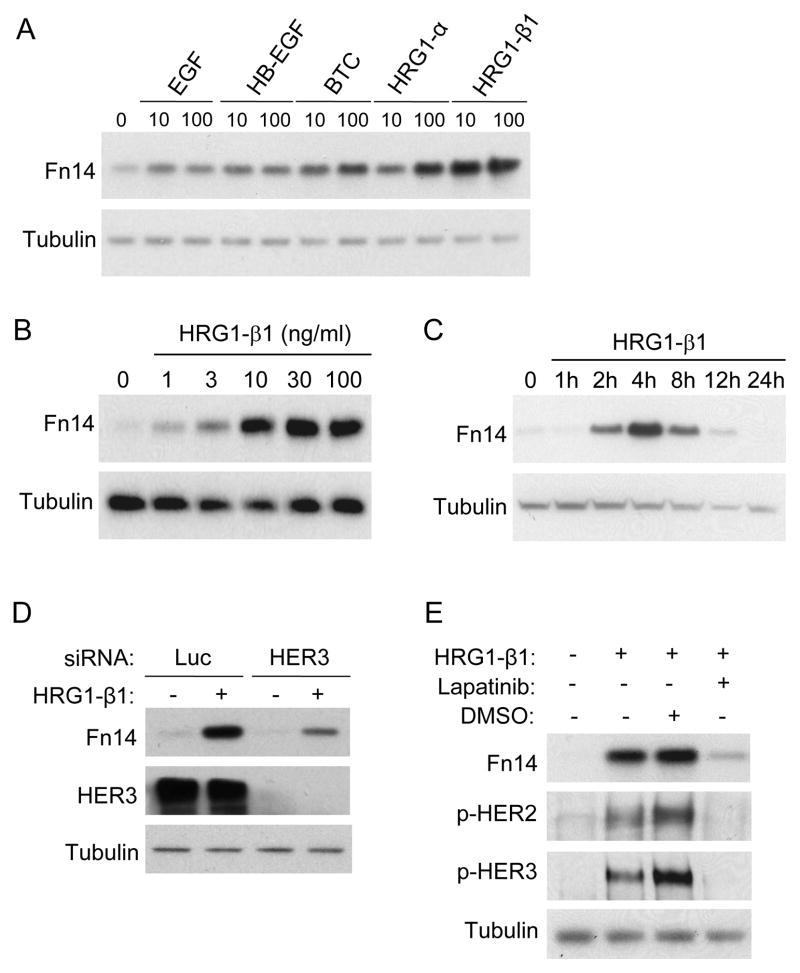
HRG1-β1 stimulation of MCF7 cells induces Fn14 gene expression
HER2 signaling in breast cancer cells can be activated by both HER2 overexpression and by ligand engagement of other EGFR family members; and in particular, by HRG1 binding to HER3 (5, 42). Therefore, we next evaluated the effects of HRG1 and other EGF family ligands on Fn14 expression in MCF7 cells. Cells were treated with either the HER1 ligand EGF, the HER1/HER4 ligands heparin-binding EGF-like growth factor (HB-EGF) and betacellulin (BTC), or the HER3/HER4 ligands HRG1-α and HRG1-β1. All ligands increased Fn14 expression, but the maximal effect was seen with HRG1-β1 (Fig. 4A). The HRG1-β1 stimulatory effect was both dose-dependent (Fig. 4B) and time-dependent (Fig. 4C). Also, HRG1-β1 treatment of MCF7 cells transiently increased Fn14 mRNA levels (Supplementary Fig. S6). HRG1-β1-mediated Fn14 up-regulation requires HER3 binding and/or signaling since HER3 depletion in MCF7 cells attenuated this effect (Fig. 4D). Lapatinib pretreatment of MCF7 cells also inhibited the HRG1-β1 stimulatory effect, indicating that HER2 signaling, most likely via HER2-HER3 heterodimer formation, is required for Fn14 gene induction (Fig. 4E). HRG1-β1 stimulation of AU565 cells also up-regulated Fn14 expression levels (data not shown).
Figure 4.
HRG1-β1 treatment of MCF7 cells induces Fn14 gene expression. (A) MCF7 cells were serum-starved overnight (0.5% FBS) and either left untreated or treated with 10 or 100 ng/ml of the indicated growth factors for 4 h. Cells were harvested and Fn14 and tubulin expression analyzed by Western blotting. (B) Serum-starved MCF7 cells were either left untreated or treated with the indicated doses of HRG1-β1 for 4 h. Cells were harvested and Fn14 and tubulin expression analyzed by Western blotting. (C) Serum-starved MCF7 cells were either left untreated or treated with HRG1-β1 (10 ng/ml) for the indicated time periods. Cells were harvested and Fn14 and tubulin expression analyzed by Western blotting. (D) MCF7 cells were transiently transfected with either luciferase control siRNA or HER3 siRNA. After 48 h of incubation the cells were serum-starved overnight (0.5% FBS) and then either left untreated or treated with HRG1-β1 (10 ng/ml) for 4 h. Cells were harvested and analyzed for Fn14, HER3 and tubulin expression by Western blotting. (E) Serum-starved MCF7 cells were either left untreated or treated with HRG1-β1 (10 ng/ml) for 4 h. In some cases the cells were first pretreated for 30 min with DMSO vehicle or lapatinib (100 nM). Cells were harvested and Fn14, p-HER2, p-HER3 and tubulin expression analyzed by Western blotting.
Fn14 depletion decreases MCF7/HER2-18 cell basal migration capacity
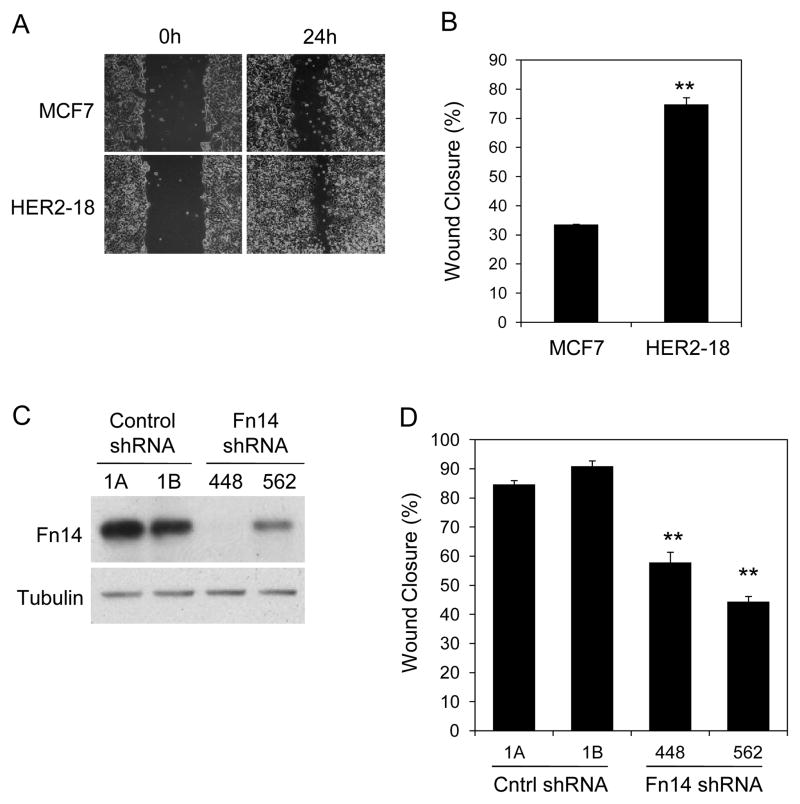
We compared the migratory potential of parental MCF7 cells and MCF7/HER2-18 cells using scratch wound assays. The HER2-overexpressing cells had an ~2.2-fold greater basal migration capacity (Fig. 5A, B). To determine the functional significance of Fn14 expression in HER2-overexpressing or HRG1-β1-stimulated breast cancer cells, we established MCF7/HER2-18 clonal cell lines stably transduced with either a control shRNA or two shRNAs targeting different regions of the Fn14 transcript. Effective Fn14 knockdown was confirmed by Western blot analysis (Fig. 5C). We analyzed the migratory potential of the four cell lines using the scratch wound assay. Fn14 depletion significantly decreased the ability of MCF7/HER2-18 cells to close the scratch wound (Fig. 5D).
Figure 5.
Fn14 depletion reduces MCF7/HER2-18 cell basal migration capacity. (A) Confluent monolayers of parental MCF7 and MCF7/HER2-18 cells were wounded using a pipette tip. Wound closure was monitored by microscopy and representative photomicrographs (20X magnification) are shown. (B) Wound width was calculated at 0 and 24 h and the difference plotted as percent wound closure. The values shown are mean +/− SEM of three experiments. **P< 0.01 compared to MCF7 cells by Students t test. (C) MCF7/HER2-18 clonal cell lines stably transduced with a control shRNA lentivirus (1A and 1B) or two different Fn14 mRNA-targeted shRNA lentiviruses (448 and 562) were analyzed for Fn14 and tubulin expression by Western blotting. (D) Confluent monolayers of the four cell lines described above were wounded using a pipette tip. Wound width was calculated at 0 and 24 h and the difference plotted as percent wound closure. The values shown are mean +/− SEM of three experiments. **P< 0.01 compared to control shRNA-1A cells by Students t test.
MCF7/HER2-18 cells are poorly invasive in Matrigel assays using serum as the stimulus
We next tried to determine the effects of Fn14 depletion on MCF7/HER2-18 cell basal invasive capacity using modified Boyden chambers coated with basement membrane extract (Matrigel). The MCF7/HER2-18 control shRNA cell lines did not invade through Matrigel when either 10% or 20% serum-containing medium was used in the bottom chamber as the chemoattractant and the incubation period was as long as 72 hours (data not shown); consequently, we could not evaluate if Fn14 depletion in these cells would reduce basal invasion capacity.
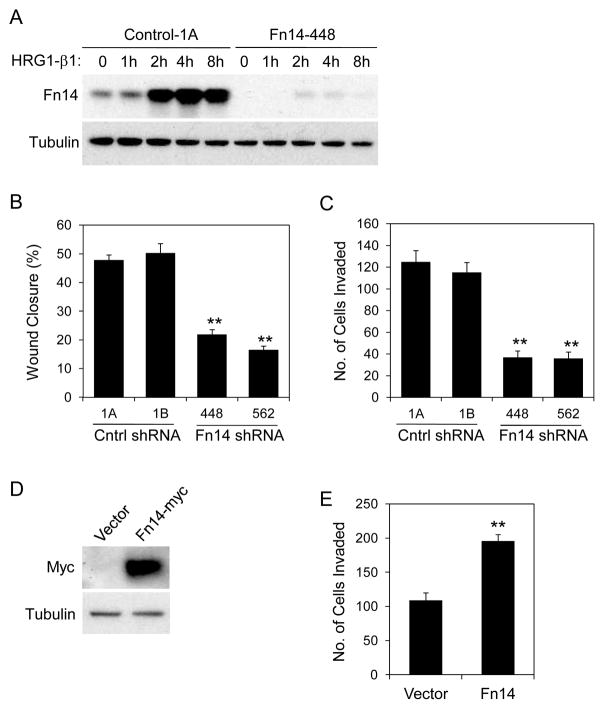
HRG1-β1-stimulated cell migration and invasion is attenuated in Fn14-depleted MCF7/HER2-18 cells
HRG1-β1 treatment of MCF7 cells increases Fn14 gene expression (shown above) and HRG1-β1 is a potent pro-migratory and pro-invasive factor for these cells (12). We found that HRG1-β1 treatment of MCF7/HER2-18 cells also stimulated migration and invasion, and this effect was ~2.9- and ~4.9-fold greater, respectively, than the effect on parental MCF7 cells (Supplementary Fig. S7). Since HRG1-β1 treatment of control shRNA, but not Fn14 shRNA MCF7/HER2-18 cells, increases Fn14 expression (Fig. 6A and data not shown), we examined whether Fn14 up-regulation was required for HRG1-β1-driven migration and invasion using our four cell lines. We found that Fn14 depletion attenuated both HRG1-β1-stimulated cell migration (Fig. 6B) and invasion (Fig. 6C). We next investigated whether re-introduction of Fn14 into Fn14-depleted MCF7/HER2-18 cells could rescue the invasion deficit. Fn14 shRNA-448 cells were stably transfected with vector control or a plasmid encoding myc epitope-tagged human Fn14. This plasmid did not include the Fn14 3′-UTR and thus ectopic Fn14 expression should not be inhibited by Fn14 shRNA co-expression, and this was confirmed by Western blot analysis (Fig. 6D). Ectopic Fn14 expression in the Fn14-depleted MCF7/HER2-18 cells resulted in an ~1.8-fold increase in HRG1-β1-stimulated cell invasion (Fig. 6E).
Figure 6.
Fn14 depletion in MCF7/HER2-18 cells reduces HRG1-β1-stimulated cell migration and invasion. (A) The MCF7/HER2-18 control shRNA-1A and Fn14 shRNA-448 cell lines were serum-starved overnight (0.5% FBS) and then either left untreated or treated with HRG1-β1 (50 ng/ml) for the indicated time periods. Cells were harvested and Fn14 and tubulin expression analyzed by Western blotting. (B) MCF7/HER2-18 control and Fn14 shRNA cell lines were treated with HRG1-β1 (200 ng/ml) and migration capacity was compared using the scratch wound assay. Wound width was calculated at 0 and 24 h and the difference plotted as percent wound closure. The values shown are mean +/− SEM of three experiments. **P< 0.01 compared to control shRNA-1A cells by Students t test. (C) MCF7/HER2-18 control and Fn14 shRNA cell lines were treated with HRG1-β1 (200 ng/ml) and invasive capacity was compared using the Matrigel invasion assay. HRG1-β1-stimulated cell invasion was quantitated as described in Materials and Methods. The values shown are mean +/− SEM of three experiments. **P< 0.01 compared to control shRNA-1B cells by Students t test. (D) Fn14 shRNA-448 cells, stably transfected with vector or Fn14-myc plasmid, were analyzed for Fn14-myc and tubulin expression by Western blotting. (E) Vector or Fn14-transfected Fn14 shRNA-448 cells were treated with HRG1-β1 (200 ng/ml) and invasive capacity was quantitated as described in Materials and Methods. The values shown are mean +/− SEM of three experiments. **P< 0.01 by Students t test.
HRG1-β1-stimulated MMP-9 expression is attenuated in Fn14-depleted MCF7/HER2-18 cells
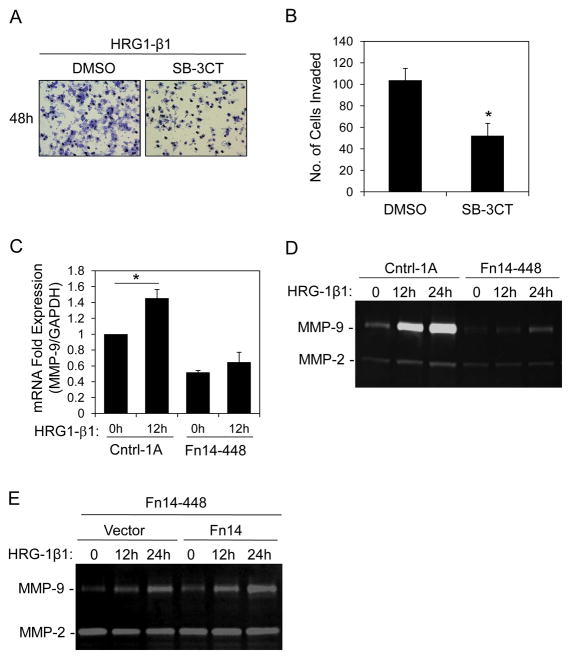
It has been previously reported that HRG1-β1 treatment of MCF7 cells stimulates MMP-9 expression and activity (17) and we found that HRG1-β1-stimulated MCF7/HER2-18 cell invasion is reduced by ~50% when cells are treated with SB-3CT, a specific inhibitor of MMP-2/MMP-9 activity (43) (Fig. 7A, B). Therefore, we determined whether Fn14 depletion had an effect on basal or HRG1-β1-stimulated MMP-9 gene expression. Quantitative real-time RT-PCR analysis indicated that basal as well as HRG1-β1-stimulated MMP-9 mRNA expression was lower in the Fn14 shRNA-448 cells compared to control shRNA cells (Fig. 7C). Gelatin zymography assays were then performed using conditioned media obtained from the two cell lines in order to examine MMP-9 protein levels. We found that the Fn14 shRNA-448 cells had reduced basal as well as HRG1-β1-stimulated MMP-9 gelatinolytic activity (Fig. 7D). Furthermore, forced expression of the Fn14 protein in the Fn14 shRNA-448 cells was able to increase HRG1-β1-stimulated MMP-9 expression (Fig. 7E).
Figure 7.
Fn14 depletion in MCF7/HER2-18 cells reduces HRG1-β1-stimulated MMP-9 expression. (A) MCF7/HER2-18 control shRNA-1A cells were plated into invasion chambers and treated with DMSO vehicle or MMP-2/MMP-9 Inhibitor 4 (SB-3CT) (20 μM) in the presence of HRG1-β1 (200 ng/ml). After 48 h of incubation the invasive cells were stained and representative photomicrographs (20X magnification) are shown. (B) HRG1-β1-stimulated cell invasion of vehicle- or drug-treated cells was quantitated as described in Materials and Methods. The values shown are mean +/− SEM of three experiments. *P< 0.05 by Students t test. (C) The MCF7/HER2-18 control shRNA-1 and Fn14 shRNA-448 cell lines were serum-starved overnight (0.5% FBS) and then either left untreated or treated with HRG1-β1 (50 ng/ml) for 12 h. Cells were harvested, RNA was isolated, and MMP-9 and GAPDH mRNA expression was quantitated using real-time RT-PCR. MMP-9 expression was normalized to GAPDH expression. The values shown are the mean +/− SEM of three experiments. *P< 0.05 by Students t test. (D) Serum-starved MCF7/HER2-18 control shRNA-1 and Fn14 shRNA-448 cells were either left untreated or treated with HRG1-β1 (50 ng/ml) for the indicated time periods. Conditioned media was collected and concentrated and MMP-2/MMP-9 activity analyzed using gelatin zymography. (E) Fn14 shRNA-448 cells stably transfected with vector or Fn14-myc plasmid were serum-starved overnight (0.5% FBS) and then either left untreated or treated with HRG1-β1 (50 ng/ml) for the indicated time periods. Conditioned media was collected and concentrated and MMP-2/MMP-9 activity analyzed using gelatin zymography.
Discussion
Overexpression of the EGFR family member HER2 occurs in 15–20% of all breast cancer cases and some HER2+ tumors also express the HER2-preferred binding partner HER3 (4). HRG1, a ligand for HER3, is also expressed in breast tumors (8). Both HER2 overexpression- and HRG1-mediated HER2/HER3 signaling likely contributes to breast cancer pathophysiology. In this study we first extended our earlier findings showing a correlation between HER2 and Fn14 expression in human breast tumors(26) by demonstrating their co-expression in mammary tumor specimens derived from MMTV-c-Neu and MMTV-PyMT transgenic mice. We then showed that (i) HER2 overexpression, achieved via plasmid transfection of MCF7 breast cancer cells or endogenous HER2 gene amplification in AU565 cells, activates HER2-HER3 signaling and increases Fn14 expression levels, (ii) HRG1-β1 treatment of MCF7 cells, which also activates HER2-HER3 signaling, can induce Fn14 gene expression, and (iii) HRG1-β1 treatment of HER2-overexpressing MCF7/HER2-18 cells further up-regulates Fn14 levels, and this “super-induction” is required for maximal cell migration, invasion and MMP-9 expression.
We identified a direct link between HER2 overexpression and elevated Fn14 levels in both MCF7 and AU565 breast cancer cells. First, we found that both transient and stable expression of HER2 in MCF7 cells increased Fn14 levels. Second, inhibition of HER2-HER3 signaling in MCF7/HER2 and AU565 cells using lapatinib significantly reduced Fn14 expression. Third, HER2 or HER3 depletion decreased Fn14 levels in AU565 cells, while combined HER2/HER3 depletion was required to see a similar effect in MCF7/HER2 and MCF7/HER2-18 cells. This cell line difference may be due to a more complete reduction of Akt activation in the AU565 cells following HER2 or HER3 depletion alone. This possibility is supported by our MEK, PI3K, and Akt inhibitor results indicating that Fn14 expression in AU565 cells is predominantly driven by PI3K/Akt signaling.
HER2 overexpression can activate gene transcription via several signaling pathways, including the MAPK (44), PI3K/Akt (44) and NF-κB (45) pathways, and the Fn14 gene promoter has potential transcription factor binding sites for both AP-1 and NF-κB (25). Therefore, we postulated that the increase in Fn14 expression in HER2-overexpressing cells was likely due to an increase in Fn14 gene transcription; however, we did not observe a significant difference in Fn14 mRNA levels between the MCF7 and MCF7/HER2 cells. Thus, it appears HER2 overexpression-mediated Fn14 up-regulation is occurring via a post-transcriptional mechanism.
The HER2 receptor has no known ligand, but it can be activated following ligand engagement of other EGFR family members via receptor heterodimerization (2, 3); therefore, we examined the effects of various EGF family members on Fn14 expression in MCF7 cells. HRG1-α and HRG1-β1, ligands for HER3 and HER4, were the two most potent inducers of Fn14 expression in MCF7 cells. HRG1-β1 treatment of MCF7 cells transiently increased Fn14 mRNA levels, suggesting that ligand engagement was inducing Fn14 gene transcription. Also, we found that both HER3 siRNA and lapatinib treatment prevented the HRG1-β1-stimulated increase in Fn14 expression observed in these cells, indicating that HRG1-β1-induced Fn14 expression was most likely due to HER2-HER3 heterodimerization and signaling.
We generated MCF7/HER2-18 cell lines with reduced Fn14 levels using Fn14 shRNA lentiviral constructs in order to investigate the potential role of Fn14 in HER2/HER3-activated breast cancer cells. First, we compared the basal migration capacity of the control and Fn14-depleted MCF7/HER2-18 cell lines and found that Fn14 depletion reduced migratory capacity. This finding is consistent with previous studies demonstrating that Fn14 depletion reduces lung (27) and prostate (28) cancer cell migration. Second, we attempted to examine the effect of Fn14 depletion on basal invasion capacity but found that the control MCF7/HER2-18 cell lines could not invade through a Matrigel barrier. Previous studies have also found that parental(26, 46) and HER2-overexpressing (47) MCF7 cells are poorly invasive. In our earlier report we showed that ectopic Fn14 expression in MCF7 cells increases invasive capacity (26). Our finding that HER2 overexpression in MCF7 cells increases Fn14 levels but not invasive capacity indicates that this degree of endogenous Fn14 up-regulation is not sufficient to increase MCF7 cell invasiveness under our experimental conditions.
Since HRG1-β1 treatment, like HER2 overexpression, can activate HER2/HER3 signaling and up-regulate Fn14 gene expression in parental MCF7 cells we investigated whether HRG1-β1 treatment of the control shRNA-expressing MCF7/HER2-18 cells might increase endogenous Fn14 levels over and beyond the level observed with HER2 overexpression alone. We found that this was the case; additionally, we showed that HRG1-β1 treatment of control MCF7/HER2-18 cells, like parental MCF7 cells (12), increased migration and invasion. These findings allowed us to test whether Fn14 up-regulation was required for HRG1-β-stimulated MCF7/HER2-18 cell migration and invasion, and this turned out to be the case. We confirmed that Fn14 levels could regulate invasion by showing that ectopic expression of Fn14 in Fn14-depleted MCF7/HER2-18 cells stimulated HRG1-β1-mediated cell invasion.
It has been reported that HRG1-β1 treatment and HER2/HER3 signaling in breast cancer cells can up-regulate MMP-9 production (17) and we found that maximal HRG1-β1-stimulated MCF7/HER2-18 cell invasion requires MMP-9 activity. Since TWEAK:Fn14 binding can up-regulate MMP-9 expression in various cell types, including glioma cells (48) and prostate cancer cells (28), we investigated whether Fn14 levels could also modulate MMP-9 expression. We found that in comparison to the control MCF7/HER2-18 cells, the Fn14-depleted cells had lower basal as well as HRG1-β1-stimulated MMP-9 expression. This finding is consistent with a recent report showing that transient Fn14 depletion in PC-3 prostate cancer cells can decrease basal MMP-9 expression (28).
In conclusion, we have demonstrated that ectopic HER2 overexpression in MCF7 cells, endogenous HER2 overexpression in AU565 breast cancer cells, and HRG1-β1 stimulation of parental as well as HER2-overexpressing MCF7 cells increases Fn14 gene expression. These effects are most likely mediated via HER2/HER3 activation and stimulation of the ERK and PI3K/Akt signaling pathways. We also found that HRG1-β1 treatment of HER2-overexpressing cells promotes cell migration, invasion and MMP-9 expression, and these effects are attenuated in Fn14-depleted cells. Our finding that HRG1-β1-mediated Fn14 up-regulation in MCF7/HER2-18 cells is required for maximal MMP-9 production may explain, at least in part, why HRG1-β1-stimulated invasion is impaired in the Fn14-depleted cell lines. Taken together, our results indicate that Fn14 may play a role in HER2/HER3-driven breast cancer cell migration and invasion. Furthermore, since Fn14 and HER2 are frequently co-expressed in human breast tumors (26, 39), agents targeting the Fn14 protein could be particularly beneficial to those HER2+ patients with intrinsic or acquired resistance to the current therapeutic agents trastuzumab (49) and lapatinib (50).
Supplementary Material
Acknowledgments
We thank Dr. Mark Williams (University of Maryland School of Medicine) for assistance with the FACS experiments. We also thank Dr. Mien-Chie Hung (University of Texas MD Anderson Cancer Center) for the HER2 expression plasmid, Dr. Dihua Yu (University of Texas MD Anderson Cancer Center) for the MCF7/HER2 cell line, Dr. Anne Hamburger (University of Maryland School of Medicine) for the MCF7/HER2-18 cell line, Dr. Peter Choyke (NIH) for the NIH3T3/HER2 cell line, and Dr. Angela Brodie (University of Maryland School of Medicine) for the MCF7-Ca and LTLT-Ca cell lines.
Grant Support
This work was supported in part by Susan G. Komen for the Cure Research Grant KG081095 (J.A.W.) and NIH grants R01 CA130940 (N.L.T.) and R01 CA090398 (R.A.K).
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Moasser MM. The oncogene HER2: its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene. 2007;26:6469–6487. doi: 10.1038/sj.onc.1210477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell MR, Amin D, Moasser MM. HER3 comes of age: new insights into its functions and role in signaling, tumor biology, and cancer therapy. Clin Cancer Res. 2010;16:1373–1383. doi: 10.1158/1078-0432.CCR-09-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naidu R, Yadav M, Nair S, Kutty MK. Expression of c-erbB3 protein in primary breast carcinomas. Br J Cancer. 1998;78:1385–1390. doi: 10.1038/bjc.1998.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Citri A, Skaria KB, Yarden Y. The deaf and the dumb: the biology of ErbB-2 and ErbB-3. Exp Cell Res. 2003;284:54–65. doi: 10.1016/s0014-4827(02)00101-5. [DOI] [PubMed] [Google Scholar]
- 6.Lee-Hoeflich ST, Crocker L, Yao E, Pham T, Munroe X, Hoeflich KP, et al. A central role for HER3 in HER2-amplified breast cancer: implications for targeted therapy. Cancer Res. 2008;68:5878–5887. doi: 10.1158/0008-5472.CAN-08-0380. [DOI] [PubMed] [Google Scholar]
- 7.Sergina NV, Rausch M, Wang D, Blair J, Hann B, Shokat KM, et al. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature. 2007;445:437–441. doi: 10.1038/nature05474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montero JC, Rodríguez-Barrueco R, Ocaña A, Díaz-Rodríguez E, Esparís-Ogando A, Pandiella A. Neuregulins and cancer. Clin Cancer Res. 2008;14:3237–3241. doi: 10.1158/1078-0432.CCR-07-5133. [DOI] [PubMed] [Google Scholar]
- 9.Agus DB, Akita RW, Fox WD, Lewis GD, Higgins B, Pisacane PI, et al. Targeting ligand-activated ErbB2 signaling inhibits breast and prostate tumor growth. Cancer Cell. 2002;2:127–137. doi: 10.1016/s1535-6108(02)00097-1. [DOI] [PubMed] [Google Scholar]
- 10.Xue C, Liang F, Mahmood R, Vuolo M, Wyckoff J, Qian H, et al. ErbB3-dependent motility and intravasation in breast cancer metastasis. Cancer Res. 2006;66:1418–1426. doi: 10.1158/0008-5472.CAN-05-0550. [DOI] [PubMed] [Google Scholar]
- 11.Hernandez L, Smirnova T, Kedrin D, Wyckoff J, Zhu L, Stanley ER, et al. The EGF/CSF-1 paracrine invasion loop can be triggered by heregulin beta1 and CXCL12. Cancer Res. 2009;69:3221–3227. doi: 10.1158/0008-5472.CAN-08-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kushima Y, Iida K, Nagaoka Y, Kawaratani Y, Shirahama T, Sakaguchi M, et al. Inhibitory effect of (−)-epigallocatechin and (−)-epigallocatechin gallate against heregulin beta1-induced migration/invasion of the MCF-7 breast carcinoma cell line. Biol Pharm Bull. 2009;32:899–904. doi: 10.1248/bpb.32.899. [DOI] [PubMed] [Google Scholar]
- 13.Atlas E, Cardillo M, Mehmi I, Zahedkargaran H, Tang C, Lupu R. Heregulin is sufficient for the promotion of tumorigenicity and metastasis of breast cancer cells in vivo. Mol Cancer Res. 2003;1:165–75. [PubMed] [Google Scholar]
- 14.Friedl P, Alexander S. Cancer invasion and the microenvironment: Plasticity and reciprocity. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 15.Adam L, Vadlamudi R, Kondapaka SB, Chernoff J, Mendelsohn J, Kumar R. Heregulin regulates cytoskeletal reorganization and cell migration through the p21-activated kinase-1 via phosphatidylinositol-3 kinase. J Biol Chem. 1998;273:28238–28246. doi: 10.1074/jbc.273.43.28238. [DOI] [PubMed] [Google Scholar]
- 16.Benlimame N, He Q, Jie S, Xiao D, Xu YJ, Loignon M, et al. FAK signaling is critical for ErbB-2/ErbB-3 receptor cooperation for oncogenic transformation and invasion. J Cell Biol. 2005;171:505–516. doi: 10.1083/jcb.200504124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao J, Xiong S, Klos K, Nguyen N, Grijalva R, Li P, et al. Multiple signaling pathways involved in activation of matrix metalloproteinase-9 (MMP-9) by heregulin-beta1 in human breast cancer cells. Oncogene. 2001;20:8066–8074. doi: 10.1038/sj.onc.1204944. [DOI] [PubMed] [Google Scholar]
- 18.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bosc DG, Goueli BS, Janknecht R. HER2/Neu-mediated activation of the ETS transcription factor ER81 and its target gene MMP-1. Oncogene. 2001;20:6215–6224. doi: 10.1038/sj.onc.1204820. [DOI] [PubMed] [Google Scholar]
- 20.Yuan G, Qian L, Song L, Shi M, Li D, Yu M, et al. Heregulin-beta promotes matrix metalloproteinase-7 expression via HER2-mediated AP-1 activation in MCF-7 cells. Mol Cell Biochem. 2008;318:73–79. doi: 10.1007/s11010-008-9858-6. [DOI] [PubMed] [Google Scholar]
- 21.Winkles JA. The TWEAK-Fn14 cytokine-receptor axis: discovery, biology and therapeutic targeting. Nat Rev Drug Discov. 2008;7:411–425. doi: 10.1038/nrd2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burkly LC, Michaelson JS, Zheng TS. TWEAK/Fn14 pathway: An immunological switch for shaping tissue responses. Immunol Rev. 2011;244:99–114. doi: 10.1111/j.1600-065X.2011.01054.x. [DOI] [PubMed] [Google Scholar]
- 23.Kwon OH, Park SJ, Kang TW, Kim M, Kim JH, Noh SM, et al. Elevated fibroblast growth factor-inducible 14 expression promotes gastric cancer growth via nuclear factor-κB and is associated with poor patient outcome. Cancer Lett. 2012;314:73–81. doi: 10.1016/j.canlet.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 24.Tran NL, McDonough WS, Savitch BA, Sawyer TF, Winkles JA, Berens ME. The tumor necrosis factor-like weak inducer of apoptosis (TWEAK)-fibroblast growth factor-inducible 14 (Fn14) signaling system regulates glioma cell survival via NF-kappaB pathway activation and BCL-XL/BCL-W expression. J Biol Chem. 2005;280:3483–3492. doi: 10.1074/jbc.M409906200. [DOI] [PubMed] [Google Scholar]
- 25.Tran NL, McDonough WS, Savitch BA, Fortin SP, Winkles JA, Symons M, et al. Increased fibroblast growth factor-inducible 14 expression levels promote glioma cell invasion via Rac1 and nuclear factor-kappaB and correlate with poor patient outcome. Cancer Res. 2006;66:9535–9542. doi: 10.1158/0008-5472.CAN-06-0418. [DOI] [PubMed] [Google Scholar]
- 26.Willis AL, Tran NL, Chatigny JM, Charlton N, Vu H, Brown SAN, et al. The fibroblast growth factor-inducible 14 receptor is highly expressed in HER2-positive breast tumors and regulates breast cancer cell invasive capacity. Mol Cancer Res. 2008;6:725–734. doi: 10.1158/1541-7786.MCR-08-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whitsett TG, Cheng E, Inge L, Asrani K, Nathan M, Hostetter G, et al. Elevated expression of Fn14 in non-small cell lung cancer correlates with activated EGFR and promotes tumor cell migration and invasion. Am J Pathol. 2012;181:111–120. doi: 10.1016/j.ajpath.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang M, Narita S, Tsuchiya N, Ma Z, Numakura K, Obara T, et al. Overexpression of Fn14 promotes androgen-independent prostate cancer progression through MMP-9 and correlates with poor treatment outcome. Carcinogenesis. 2011;32:1589–1596. doi: 10.1093/carcin/bgr182. [DOI] [PubMed] [Google Scholar]
- 29.Ho DH, Vu H, Brown SAN, Donohue PJ, Hanscom HN, Winkles JA. Soluble tumor necrosis factor-like weak inducer of apoptosis overexpression in HEK293 cells promotes tumor growth and angiogenesis in athymic nude mice. Cancer Res. 2004;64:8968–8972. doi: 10.1158/0008-5472.CAN-04-1879. [DOI] [PubMed] [Google Scholar]
- 30.Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci USA. 1992;89:10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landis MD, Seachrist DD, Montañez-Wiscovich ME, Danielpour D, Keri RA. Gene expression profiling of cancer progression reveals intrinsic regulation of transforming growth factor-beta signaling in ErbB2/Neu-induced tumors from transgenic mice. Oncogene. 2005;24:5173–5190. doi: 10.1038/sj.onc.1208712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol. 2003;163:2113–2126. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jelovac D, Sabnis G, Long BJ, Macedo L, Goloubeva OG, Brodie AMH. Activation of mitogen-activated protein kinase in xenografts and cells during prolonged treatment with aromatase inhibitor letrozole. Cancer Res. 2005;65:5380–5389. doi: 10.1158/0008-5472.CAN-04-4502. [DOI] [PubMed] [Google Scholar]
- 35.Brown SAN, Ghosh A, Winkles JA. Full-length, membrane-anchored TWEAK can function as a juxtacrine signaling molecule and activate the NF-kappaB pathway. J Biol Chem. 2010;285:17432–17441. doi: 10.1074/jbc.M110.131979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yepes M, Brown SAN, Moore EG, Smith EP, Lawrence DA, Winkles JA. A soluble Fn14-Fc decoy receptor reduces infarct volume in a murine model of cerebral ischemia. Am J Pathol. 2005;166:511–520. doi: 10.1016/S0002-9440(10)62273-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown SAN, Richards CM, Hanscom HN, Feng SL, Winkles JA. The Fn14 cytoplasmic tail binds tumour-necrosis-factor-receptor-associated factors 1, 2, 3 and 5 and mediates nuclear factor-κ B activation. Biochem J. 2003;371:395–403. doi: 10.1042/BJ20021730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michaelson JS, Cho S, Browning B, Zheng TS, Lincecum JM, Wang MZ, et al. TWEAK induces mammary epithelial branching morphogenesis. Oncogene. 2005;24:2613–2624. doi: 10.1038/sj.onc.1208208. [DOI] [PubMed] [Google Scholar]
- 39.Chao DT, Su M, Tanlimco S, Sho M, Choi D, Fox M, et al. Expression of TweakR in breast cancer and preclinical activity of enavatuzumab, a humanized anti-TweakR mAb. J Cancer Res Clin Oncol. 2012 doi: 10.1007/s00432-012-1332-x. (epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siegel PM, Ryan ED, Cardiff RD, Muller WJ. Elevated expression of activated forms of Neu/ErbB-2 and ErbB-3 are involved in the induction of mammary tumors in transgenic mice: implications for human breast cancer. EMBO J. 1999;18:2149–2164. doi: 10.1093/emboj/18.8.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moy B, Kirkpatrick P, Kar S, Goss P. Lapatinib. Nat Rev Drug Discov. 2007;6:431–432. doi: 10.1038/nrd2332. [DOI] [PubMed] [Google Scholar]
- 42.Wallasch C, Weiss FU, Niederfellner G, Jallal B, Issing W, Ullrich A. Heregulin-dependent regulation of HER2/neu oncogenic signaling by heterodimerization with HER3. EMBO J. 1995;14:4267–4275. doi: 10.1002/j.1460-2075.1995.tb00101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin CW, Shen SC, Hou WC, Yang LY, Chen YC. Heme oxygenase-1 inhibits breast cancer invasion via suppressing the expression of matrix metalloproteinase-9. Mol Cancer Ther. 2008;7:1195–1206. doi: 10.1158/1535-7163.MCT-07-2199. [DOI] [PubMed] [Google Scholar]
- 44.Niu G, Carter WB. Human epidermal growth factor receptor 2 regulates angiopoietin-2 expression in breast cancer via AKT and mitogen-activated protein kinase pathways. Cancer Res. 2007;67:1487–1493. doi: 10.1158/0008-5472.CAN-06-3155. [DOI] [PubMed] [Google Scholar]
- 45.Merkhofer EC, Cogswell P, Baldwin AS. Her2 activates NF-kappaB and induces invasion through the canonical pathway involving IKKalpha. Oncogene. 2010;29:1238–1248. doi: 10.1038/onc.2009.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitra D, Brumlik MJ, Okamgba SU, Zhu Y, Duplessis TT, Parvani JG, et al. An oncogenic isoform of HER2 associated with locally disseminated breast cancer and trastuzumab resistance. Mol Cancer Ther. 2009;8:2152–2162. doi: 10.1158/1535-7163.MCT-09-0295. [DOI] [PubMed] [Google Scholar]
- 48.Winkles JA, Tran NL, Berens ME. TWEAK and Fn14: new molecular targets for cancer therapy? Cancer Lett. 2006;235:11–17. doi: 10.1016/j.canlet.2005.03.048. [DOI] [PubMed] [Google Scholar]
- 49.Pohlmann PR, Mayer IA, Mernaugh R. Resistance to trastuzumab in breast cancer. Clin Cancer Res. 2009;15:7479–7491. doi: 10.1158/1078-0432.CCR-09-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen FL, Xia W, Spector NL. Acquired resistance to small molecule ErbB2 tyrosine kinase inhibitors. Clin Cancer Res. 2008;14:6730–6734. doi: 10.1158/1078-0432.CCR-08-0581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.