Abstract
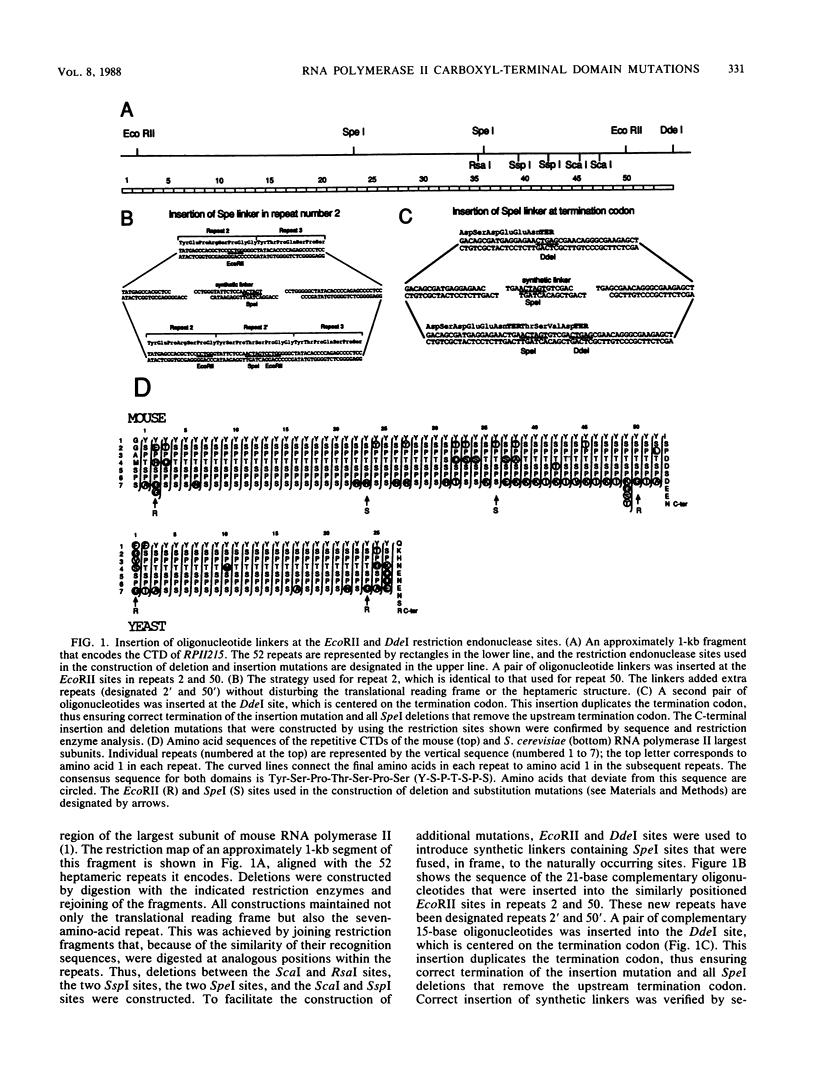
The carboxyl-terminal domain (CTD) of the mouse RNA polymerase II largest subunit consists of 52 repeats of a seven-amino-acid block with the consensus sequence Tyr-Ser-Pro-Thr-Ser-Pro-Ser. A genetic approach was used to determine whether the CTD plays an essential role in RNA polymerase function. Deletion, insertion, and substitution mutations were created in the repetitive region of an alpha-amanitin-resistant largest-subunit gene. The effects of these mutations on RNA polymerase II activity were assayed by measuring the ability of mutant genes to confer alpha-amanitin resistance after transfection of susceptible rodent cells. Mutations that resulted in CTDs containing between 36 and 78 repeats had no effect on the transfer of alpha-amanitin resistance, whereas mutations with 25 or fewer repeats were inactive in this assay. Mutations that contained 29, 31, or 32 repeats had an intermediate effect; the number of alpha-amanitin-resistant colonies was lower and the colonies obtained were smaller, indicating that the mutant RNA polymerase II was defective. In addition, not all of the heptameric repeats were functionally equivalent in that repeats that diverged in up to three amino acids from the consensus sequence could not substitute for the conserved heptamer repeats. We concluded that the CTD is essential for RNA polymerase II activity, since substantial mutations in this region result in loss of function.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahearn J. M., Jr, Bartolomei M. S., West M. L., Cisek L. J., Corden J. L. Cloning and sequence analysis of the mouse genomic locus encoding the largest subunit of RNA polymerase II. J Biol Chem. 1987 Aug 5;262(22):10695–10705. [PubMed] [Google Scholar]
- Allison L. A., Moyle M., Shales M., Ingles C. J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985 Sep;42(2):599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- Allison L. A., Wong J. K., Fitzpatrick V. D., Moyle M., Ingles C. J. The C-terminal domain of the largest subunit of RNA polymerase II of Saccharomyces cerevisiae, Drosophila melanogaster, and mammals: a conserved structure with an essential function. Mol Cell Biol. 1988 Jan;8(1):321–329. doi: 10.1128/mcb.8.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew B., Dahmus M. E., Meares C. F. RNA contacts subunits IIo and IIc in HeLa RNA polymerase II transcription complexes. J Biol Chem. 1986 Oct 25;261(30):14226–14231. [PubMed] [Google Scholar]
- Bartolomei M. S., Corden J. L. Localization of an alpha-amanitin resistance mutation in the gene encoding the largest subunit of mouse RNA polymerase II. Mol Cell Biol. 1987 Feb;7(2):586–594. doi: 10.1128/mcb.7.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Biggs J., Searles L. L., Greenleaf A. L. Structure of the eukaryotic transcription apparatus: features of the gene for the largest subunit of Drosophila RNA polymerase II. Cell. 1985 Sep;42(2):611–621. doi: 10.1016/0092-8674(85)90118-7. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyles S. S., Moss B. Homology between RNA polymerases of poxviruses, prokaryotes, and eukaryotes: nucleotide sequence and transcriptional analysis of vaccinia virus genes encoding 147-kDa and 22-kDa subunits. Proc Natl Acad Sci U S A. 1986 May;83(10):3141–3145. doi: 10.1073/pnas.83.10.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K. W., Khalili K., Zandomeni R., Weinmann R. The gene encoding the large subunit of human RNA polymerase II. J Biol Chem. 1985 Dec 5;260(28):15204–15210. [PubMed] [Google Scholar]
- Corden J. L., Cadena D. L., Ahearn J. M., Jr, Dahmus M. E. A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc Natl Acad Sci U S A. 1985 Dec;82(23):7934–7938. doi: 10.1073/pnas.82.23.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenburg D., Dworniczak B., Faust D. M., Bautz E. K. RNA polymerase II of Drosophila. Relation of its 140,000 Mr subunit to the beta subunit of Escherichia coli RNA polymerase. J Mol Biol. 1987 Jun 20;195(4):929–937. doi: 10.1016/0022-2836(87)90496-7. [DOI] [PubMed] [Google Scholar]
- Frost E., Williams J. Mapping temperature-sensitive and host-range mutations of adenovirus type 5 by marker rescue. Virology. 1978 Nov;91(1):39–50. doi: 10.1016/0042-6822(78)90353-7. [DOI] [PubMed] [Google Scholar]
- Greenberg M. E., Ziff E. B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984 Oct 4;311(5985):433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- Gross-Bellard M., Oudet P., Chambon P. Isolation of high-molecular-weight DNA from mammalian cells. Eur J Biochem. 1973 Jul 2;36(1):32–38. doi: 10.1111/j.1432-1033.1973.tb02881.x. [DOI] [PubMed] [Google Scholar]
- Ingles C. J., Himmelfarb H. J., Shales M., Greenleaf A. L., Friesen J. D. Identification, molecular cloning, and mutagenesis of Saccharomyces cerevisiae RNA polymerase genes. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2157–2161. doi: 10.1073/pnas.81.7.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M., Sweetser D., Young R. A. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell. 1987 Sep 11;50(6):909–915. doi: 10.1016/0092-8674(87)90517-4. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Riva M., Memet S., Micouin J. Y., Huet J., Treich I., Dassa J., Young R., Buhler J. M., Sentenac A., Fromageot P. Isolation of structural genes for yeast RNA polymerases by immunological screening. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1554–1558. doi: 10.1073/pnas.83.6.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M., Elledge S., Davis R. W. Rapid mapping of antigenic coding regions and constructing insertion mutations in yeast genes by mini-Tn10 "transplason" mutagenesis. Proc Natl Acad Sci U S A. 1986 Feb;83(3):730–734. doi: 10.1073/pnas.83.3.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers D. G., Pearson M. L., Ingles C. J. Isolation and characterization of an alpha-amanitin-resistant rat myoblast mutant cell line possessing alpha-amanitin-resistant RNA polymerase II. J Biol Chem. 1975 Jul 10;250(13):4825–4831. [PubMed] [Google Scholar]
- Somers D. G., Pearson M. L., Ingles C. J. Regulation of RNA polymerase II activity in a mutant rat myoblast cell line resistant to alpha-amanitin. Nature. 1975 Jan 31;253(5490):372–374. doi: 10.1038/253372a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Southern P. J., Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1(4):327–341. [PubMed] [Google Scholar]
- Sweetser D., Nonet M., Young R. A. Prokaryotic and eukaryotic RNA polymerases have homologous core subunits. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1192–1196. doi: 10.1073/pnas.84.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yura T., Ishihama A. Genetics of bacterial RNA polymerases. Annu Rev Genet. 1979;13:59–97. doi: 10.1146/annurev.ge.13.120179.000423. [DOI] [PubMed] [Google Scholar]
- van der Eb A. J., Graham F. L. Assay of transforming activity of tumor virus DNA. Methods Enzymol. 1980;65(1):826–839. doi: 10.1016/s0076-6879(80)65077-0. [DOI] [PubMed] [Google Scholar]