Abstract
Nicotinamide N-methyltransferase (NNMT) is overexpressed in a variety of human cancers, where it contributes to tumorigenesis by a still poorly understood mechanism. Here, we show using metabolomics that NNMT impairs the methylation potential of cancer cells by consuming methyl units from S-adenosyl methionine to create the stable metabolic product 1-methylnicotinamide. As a result, NNMT-expressing cancer cells possess an altered epigenetic state that includes hypomethylated histones and other cancer-related proteins combined with heightened expression of pro-tumorigenic gene products. Our findings thus point to a direct mechanistic link between the deregulation of a metabolic enzyme and widespread changes in the methylation landscape of cancer cells.
Building on the classical studies by Warburg showing that cancer cells exhibit enhanced dependence on glycolysis1, researchers over the past decade have uncovered a diverse array of metabolic changes that support tumorigenesis2, 3, and altered metabolism is now considered a hallmark of cancer4. Metabolism is tightly linked with signaling and transcriptional networks, as well as cellular epigenetic machinery and the post-translational modification state of proteins5. Recent data support the concept that metabolic reprogramming in tumor cells is driven by a number of biochemical changes, including activation of oncogenes, inactivation of tumor suppressors, and pro-tumorigenic mutations in metabolic enzymes themselves5, 6. Reciprocally, signaling and transcriptional pathways can be regulated by metabolism5. Large-scale profiling experiments, including genomics7, 8 and proteomics9, have uncovered many metabolic enzymes that show altered expression in cancers. Nonetheless, the biochemical and cellular functions that these deregulated enzymes play in cancer often remain unknown. Here, we investigate the function of one such cancer-associated metabolic enzyme nicotinamide N-methyltransferase (NNMT) and show through metabolomics that it exerts specific control over the methylation potential of cancer cells and, through doing so, imparts a broad effect on their epigenetic state.
RESULTS
NNMT activity correlates with cancer aggressiveness
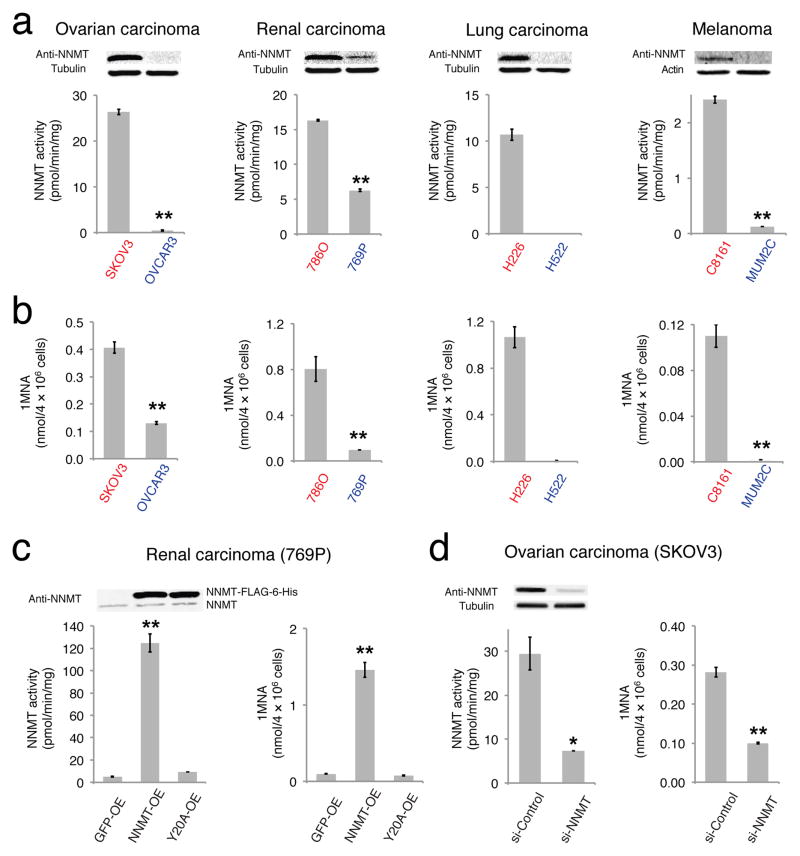
NNMT is a cytosolic enzyme that catalyzes the transfer of methyl group from S-adenosyl-L-methionine (SAM) to nicotinamide (NA), generating S-adenosylhomocysteine (SAH) and 1-methylnicotinamide (1MNA). NNMT is overexpressed in a variety of tumors, including cancers of the lung, liver, kidney, bladder, and colon, and has been shown to promote the migration, invasion, proliferation, and survival of cancer cells10–14. Despite considerable experimental evidence that NNMT supports tumorigenesis and may thus serve as a potential anticancer target, the actual metabolic functions played by this enzyme in cancer cells have not yet been determined. We set out to address this question by first profiling NNMT expression and activity across a panel of aggressive and non-aggressive human cancer cell lines from multiple tumors of origin, including ovarian (aggressive SKOV3, non-aggressive OVCAR3), kidney (aggressive 786O, non-aggressive 769P), lung (aggressive H226, non-aggressive H522) and uveal melanoma (aggressive C8161, non-aggressive MUM2C). Differences in the aggressive properties of these cancer cell lines have been previously established by measuring their respective in vitro migration/invasion and in vivo tumor-growth activities15 (also see Supplementary Results, Supplementary Fig. 1). Consistent with past studies11, 14, we found that NNMT expression and activity were highly elevated in aggressive cancer cells compared to their non-aggressive counterparts (Fig. 1a). We also found that aggressive cancer lines possessed higher levels of the NNMT-catalyzed product 1MNA (Fig. 1b). We next stably overexpressed NNMT in non-aggressive cancer lines (NNMT-OE cells) and generated two control cell populations that overexpressed GFP (GFP-OE cells) and an inactive NNMT mutant (Y20A16; Y20A-OE cells), respectively (Fig. 1c and Supplementary Fig. 2). NNMT-OE cells, but not GFP-OE or Y20A-OE cells showed dramatic increases in NNMT activity and cellular levels of 1MNA (Fig. 1c and Supplementary Fig. 2). NNMT-OE cells also displayed enhanced migration compared to control cells (Supplementary Fig. 3).
FIGURE 1. NNMT is elevated in aggressive human cancer cells.
(a) NNMT levels and activity are consistently elevated in aggressive human cancer cell lines (red) compared to non-aggressive cancer cell lines (blue), as determined by western blotting with anti-NNMT antibodies (upper panels) and nicotinamide (NA) substrate assays (bar graphs). NNMT activity in H522 cells was below detection limit. (b) Aggressive cancer cells possess higher levels of the NNMT product 1-methylnicotinamide (1MNA). 1MNA levels in H522 cells were below detection limit. (c) Overexpression of NNMT in non-aggressive 769P cells (NNMT-OE) confirmed by WB and NA substrate assay. GFP-OE and Y20A-OE cells correspond to control cancer cell models infected with GFP-vector and a catalytically inactive NNMT mutant (Y20A16), respectively. All exogenous proteins contained FLAG-6-His epitope tag and migrated slower on SDS-PAGE compared to endogenous proteins. NNMT-OE cells also showed elevated levels of 1MNA compared to GFP-OE or NNMT-Y20A cells. (d) SKOV3 cells were transiently transfected with the indicated siRNA constructs. A greater than 75% reduction in NNMT was confirmed by western blotting and activity assay in si-NNMT cells compared to si-Control cells transfected with nonsilencing siRNA that has no homology to any known mammalian gene. si-NNMT cells also showed decreased levels of 1MNA. All data shown as mean ± SEM; for metabolomics experiments N = 3–4/group; for substrate assay N = 2/group; * P < 0.05, ** P < 0.01 for NNMT-OE versus control (GFP-OE and Y20A-OE) groups or for si-NNMT versus si-Control groups. See Supplementary Fig. 13 for full gel images.
We also produced a complementary cell model where NNMT was knocked-down by > 75% in the aggressive ovarian cancer cell line SKOV3 using siRNA methods and found that these si-NNMT cells exhibited decreased NNMT activity and reduced 1MNA levels compared to SKOV3 cells treated with a scrambled siRNA control probe (si-Control cells) (Fig. 1d).
NNMT impairs the methylation potential of cancer cells
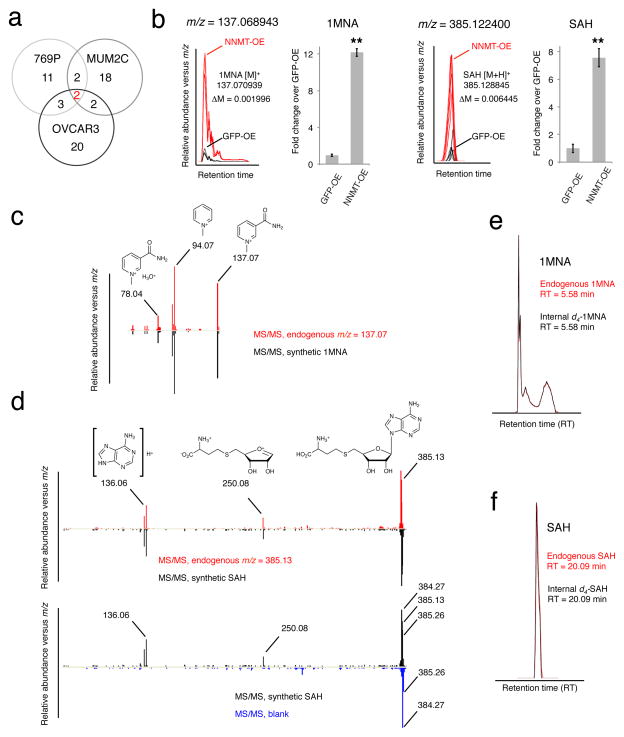
Next, we tested whether the NNMT product 1MNA might itself be responsible for the enhanced migratory activity of NNMT-OE cells. However, treatment of non-aggressive cancer cells with 1MNA did not affect their migration (Supplementary Fig. 4). This result motivated us to consider more broadly the impact of NNMT on the metabolic state of cancer cells, which we examined by performing a global analysis of polar metabolites17 in NNMT-OE and control cells. We were particularly interested in identifying metabolic changes caused by NNMT in multiple cancer cell types, as we considered such consistent alterations as having the highest probability of reflecting a conserved functional role for NNMT in cancer metabolism. To address this problem, we employed an untargeted metabolomics approach18, where metabolomes from NNMT-OE and GFP-OE renal carcinoma (769P), ovarian cancer (OVCAR3), and melanoma (MUM2C) cells were comparatively analyzed by an HPLC-Q-TOF-MS system operating in the broad mass scanning mode (m/z range of 50–1200 Da). Metabolites with significantly differing levels in NNMT-OE and GFP-OE cells were identified using the XCMS analyte profiling software19, which aligns and quantifies the relative signal intensities of mass peaks from multiple LC-MS traces (Supplementary Dataset 1). We then clustered the changing metabolites into groups based on whether they appeared in one, two, or all three pairs of NNMT-OE /GFP-OE comparisons. The resulting Venn diagram revealed only two metabolites that were consistently deregulated across all three types of cancer (Fig. 2a). These metabolites, which were both significantly elevated in NNMT-OE cells, were identified by a combination of high-resolution MS (Fig. 2b and Supplementary Fig. 5), tandem MS analysis (Fig. 2c,d), and LC-migration (Fig. 2e,f) as 1MNA (m/z measured = 137.068943; m/z predicted = 137.070939) and SAH (m/z measured = 385.122400; m/z predicted = 385.128845).
FIGURE 2. Structural assignment of 1MNA and SAH as deregulated metabolites in NNMT-OE cells.
(a) Comparative metabolomics of NNMT-OE and GFP-OE cancer lines. Venn diagram showing the number of metabolites commonly and significantly deregulated in three NNMT-OE cancer lines compared to their respective GFP-OE control cells. Two metabolites were identified as significantly different, based on showing a > 2-fold change in level in NNMT-OE compared to GFP-OE cells with a P -value for this difference of < 0.01. (b–f) These two metabolites were identified as 1MNA and SAH by searching their exact masses (b) against the Human Metabolome Database and comparing their fragmentation patterns (c, d) and LC-migration times (e, f) with synthetic standards. Part b shows bar graphs of quantified values for 1MNA and SAH in a representative pair (769P) of NNMT-OE and GFP-OE cancer cells. Note that for part d, synthetic SAH and endogenous m/z = 385.13 showed identical fragmentation spectra producing 250.08 and 136.06 daughter ions (upper panel). Ions with m/z values of 384.27 and 385.26 observed in SAH fragmentation spectrum are contamination signals as they were also observed in a control (blank) run (bottom panel). (b), data are presented as mean values ± SEM; N = 4 experiments/group; ** P < 0.01 for NNMT-OE versus control GFP-OE group.
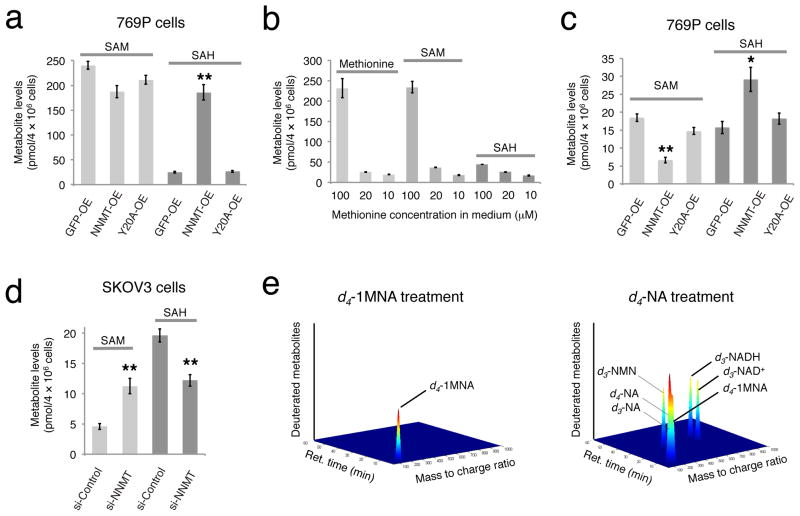
Targeted LC-MS profiling with a deuterated internal standard confirmed elevated SAH levels in NNMT-OE cells compared to both GFP-OE and Y20A-OE control cells (Fig. 3a and Supplementary Fig. 5). On the other hand, SAM levels were relatively unchanged (fold < 1.5) in NNMT-OE versus control cells (Fig. 3a). We wondered whether the lack of effect of NNMT expression on SAM might be due to the high endogenous concentrations of this metabolite in cancer cells grown in standard culture medium, which contains 100 μM methionine, a concentration that is 5–10 times higher than blood levels of this amino acid in humans20. Lowering the methionine concentration in media to 10–20 μM caused a corresponding dramatic reduction in cellular methionine and SAM levels, while SAH levels were less affected (Fig. 3b). NNMT-OE 769P cells grown in 20 μM methionine continued to show significantly elevated SAH levels, as well as a modest, but significant reduction in SAM levels, compared to control cells (Supplementary Fig. 6). Growth in 10 μM methionine media produced a more dramatic shift in metabolic profile such that NNMT-OE 769P cells now showed both a significant reduction in SAM levels and significant increase in SAH levels compared to control cells (Fig. 3c). Similar changes in SAM and SAH were observed in NNMT-OE 769P cells when processed by one of multiple metabolite extraction protocols (harvesting cells in cold PBS followed by metabolite extraction versus direct extraction of metabolites from plated cells without any PBS washes; Supplementary Fig. 7). NNMT-OE MUM2C cells maintained high SAH and unchanged SAM levels at all tested methionine concentrations (Supplementary Fig. 5 and 8). Importantly, in all human cancer cell types and at all methionine concentrations tested, elevated levels of NNMT resulted in more than a twofold reduction in cellular methylation potential (MP), defined as the ratio of SAM:SAH (Supplementary Table 1; see below for more discussion of MP). Conversely, si-NNMT SKOV3 cells showed elevated SAM and reduced SAH levels to produce a net four-fold increase in MP compared to si-Control cells (Fig. 3d).
FIGURE 3. NNMT regulates SAM and SAH levels in cancer cells.
(a) Targeted LC-MS with a deuterated internal standard confirmed elevated SAH levels in NNMT-OE cells compared to GFP-OE and Y20A-OE control cells. Data are shown for 769P cells; see Supplementary Fig. 5 for similar data on MUM2C and OVCAR3 cells. (b) Cellular levels of methionine and SAM were much lower in 769P cells cultured for 48 h in 10–20 μM versus 100 μM concentrations of methionine, while SAH levels were less affected. (c) NNMT-OE 769P cells cultured with 10 μM methionine possess significantly lower SAM levels and elevated SAH levels compared to GFP-OE and Y20A-OE cells. (d) si-NNMT cells grown on low (10 μM) methionine exhibit elevated SAM and lower SAH levels compared to si-Control cells. (e) LC-MS profiling of 769P cells treated with d4-labeled 1MNA (d4–1MNA) for 24 h revealed accumulation of d4–1MNA, but no other deuterated metabolites were detected. In contrast, d4-labeled-nicotinamide (d4-NA) was converted by cancer cells into d4–1MNA and several deuterated intermediates in the NAD+ biosynthetic pathway, including NAD+, NADH, and nicotinamide mononucleotide (NMN). For illustration purposes, signal intensities of each metabolite were arbitrarily set to a value of 1; see Supplementary Fig. 9 for actual signal intensity values for each metabolite. For (a–d), data are presented as mean values ± SEM; N = 3–4 experiments/group; * P < 0.05, ** P < 0.01 for NNMT-OE versus control (GFP-OE and Y20A-OE) groups or for si-NNMT versus si-Control groups.
1MNA is a stable metabolic product in cancer cells
Taken together, our metabolomic findings revealed that the NNMT-catalyzed conversion of nicotinamide to 1MNA exerted a more global impact on the MP of cancer cells. This result, combined with the lack of effect of 1MNA on the migratory activity of cancer cells, led us to consider that this metabolite, rather than functioning as a pro-tumorgenic signal, might instead act as a sink for storing methylation units in cancer cells that express high levels of NNMT. To further investigate such a model, we performed metabolic labeling studies using deuterated 1MNA and found that this metabolite is remarkably stable in cancer cells, as we did not detect its conversion to other deuterated metabolites even after incubation with cancer cells for 24 h (Fig. 3e and Supplementary Fig. 9). In contrast, deuterated nicotinamide (NA) was transformed by cancer cells into numerous metabolic products, including NAD+, NADH, nicotinamide mononucleotide (NMN), and 1MNA (Fig. 3e and Supplementary Fig. 9). These data indicate that cancer cells have a limited capacity to recover the methylation units that accumulate in 1MNA as a result of NNMT catalysis, offering a satisfying biochemical mechanism to explain the effect of this enzyme on cellular MP.
NNMT regulates protein methylation in cancer cells
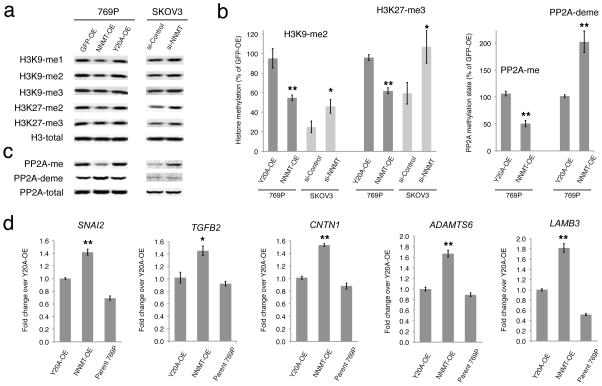
The MP of cells has the potential to influence a variety of important pathways and processes, including the modification states of proteins, DNA, RNA, and lipid metabolites, which are regulated by methyltransferases that utilize SAM as a universal methyl donor21–24. SAH is a product of these transmethylation reactions and acts as a competitive inhibitor of methyltransferases25. For this reason, MP is an important metabolic indicator of cellular methylation status, and a decrease in this parameter has been associated with a global hypomethylation state in cells22. We therefore next tested whether NNMT-induced changes in MP affect protein methylation, focusing on methylation events that have been linked to cancer, such as those that occur on histones26, 27. When grown in media containing 10 and 20 μM methionine, NNMT-OE cells showed a significant decrease in many (e.g., H3K4, H3K9, H3K27 and H4K20), but not all (e.g., H3R17) of the tested histone methylation events compared to control (GFP-OE or Y20A-OE) cells (Fig. 4a,b and Supplementary Fig. 6). This effect was most dramatic in 769P cells (Fig. 4a,b and Supplementary Fig. 6), but also observed in MUM2C cells (Supplementary Fig. 8), correlating with the greater change in MP caused by NNMT expression in 769P cells (four-fold change, compared to a two-fold change in MUM2C cells). Conversely, si-NNMT cells showed increased levels of histone methylation events compared to si-Control cells (Fig. 4a,b and Supplementary Fig. 8). We also noted that the siControl-SKOV3 cells exhibited lower basal levels of histone methylation compared to the control (GFP-OE, Y20-OE) 769P cells (Fig. 4b), consistent with the higher endogenous levels of NNMT (Fig. 1c,d) controlling histone methylation in SKOV3.
FIGURE 4. NNMT regulates the methylation state of histones and other signaling proteins in cancer cells.
(a, b) NNMT-OE 769P cells show reductions in histone 3 methylation events compared to GFP-OE and Y20A-OE control cells as determined by Western blotting. Conversely, si-NNMT SKOV3 cells show increases in most of these histone methylation events compared to si-Control cells. (b) Bar graphs quantifying representative methylation changes shown in (a) and (c). (c) NNMT-OE 769P cells show reduced levels of PP2A methylation compared to GFP-OE and Y20A-OE control cells as determined by Western blotting. Conversely, si-NNMT SKOV3 cells show increased levels of PP2A methylation compared to si-Control cells. Lane assignment is defined as in (a). (d) Real-time (RT)-PCR analysis showing upregulation of five cancer-related genes in NNMT-OE compared to Y20A-OE or parental 769P cells. Gene changes were normalized to the average of three housekeeping genes, including ACTB, GAPDH and HPRT1. For all studies, cells were cultured in low (10 μM) methionine medium. All data are presented as mean values ± SEM; For (b), N = 4–6 experiments/group. Western blot band intensity was first normalized to total histone 3 levels, followed by normalization of all samples to GFP-OE control group. * P < 0.05, ** P < 0.01 for NNMT-OE versus control Y20A-OE groups, or for si-NNMT versus si-Control groups. For (d), N = 4 experiments/group. * P < 0.05, ** P < 0.01 for NNMT-OE versus control (Y20A-OE and parental 769P cells) groups. See Supplementary Fig. 14 for full gel images.
The histone methylation changes observed in NNMT-OE cells were mostly blunted when these cells were grown on high methionine (100 μM, Supplementary Fig. 10) despite their altered MP (Supplementary Table 1). This result suggests that the methylation state of cells may be tied to more than just their relative MP values, but also to the absolute SAM and SAH concentrations that form the basis for this ratiometric measurement. We further found that 1MNA (0.5 mM) did not alter histone methylation when added to GFP-OE 769P cells grown on low (10 μM) methionine (Supplementary Fig. 11), providing additional support that NNMT affects protein methylation by altering SAM and SAH rather than through a direct action of its product 1MNA.
We next tested whether the impact of NNMT on cellular methylation extended beyond histones to include other proteins and biomolecules. The tumor suppressor protein phosphatase 2A (PP2A), is regulated by methylation of the C-terminal Leu309 residue of its catalytic subunit28, and a decrease in PP2A methylation has been shown to promote basal ERK pathway activity required for efficient growth factor responses in cancer cells29. We found that NNMT-OE cells show a striking reduction in methylated PP2A and a corresponding increase in demethylated PP2A compared to control cells (Fig. 4b, c). si-NNMT cells conversely displayed higher methylated PP2A compared to si-Control cells (Fig. 4c). We then more globally assessed the effect of NNMT on arginine methylation of proteins using the ASYM25 antibody, which is specific for asymmetrical dimethylated arginines30, and found that several, but not all cellular proteins showed lower arginine methylation levels in NNMT-OE cells compared to control cells (Supplementary Fig. 12). si-NNMT cells showed more limited changes in arginine methylation, but the observed effects trended, as expected, toward increases in methylation levels (Supplementary Fig. 12). In contrast, NNMT overexpression or knockdown did not affect global DNA methylation as measured by total cellular 5-methyl-2′-deoxycytidine content (Supplementary Fig. 12).
We evaluated whether NNMT-induced changes in histone methylation correlated with changes in gene expression by performing DNA microarray experiments comparing NNMT-OE and Y20A-OE 769P cells (Supplementary Dataset 2). These global profiling studies identified several cancer-related genes with altered expression in NNMT-OE cells, including SNAI231, TGFB232, CNTN133, ADAMTS634 and LAMB335. We used quantitative RT-PCR to confirm higher expression levels for all five genes in NNMT-OE compared to Y20A-OE or parental 769P cancer cells (Fig. 4d). Notably, methylation of H3K27 has previously been shown to regulate the SNAI2 and CNTN1 genes in human endothelial cells36 and the LAMB3 gene in breast cancer cells37. Upregulation of SNAI2 and TGFB2 gene expression has also been observed in breast cancer cells treated with the global methyltransferase inhibitor 3-deazaneplanocin A, but not in cells treated with the DNA methylation inhibitor 5-azacytidine38. These past findings provide supportive evidence that the gene expression changes observed in our study are likely due to reductions in MP and histone methylation in NNMT-OE cells. Finally, we found that NNMT-regulated changes in protein methylation observed on low methionine medium correlated with altered cancer cell behavior, as NNMT-OE and si-NNMT cells showed enhanced migration and reduced migration/invasion, respectively, compared to their corresponding control cells (Supplementary Fig. 3). In contrast, si-NNMT cells grown on high methionine medium showed no change in migration capacity compared to si-Control cells (Supplementary Fig. 3).
DISCUSSION
Deregulated metabolic pathways can affect cancer cell biology in a number of ways that extend beyond simply providing energy and primary building blocks to support proliferation. Recent studies on mutant forms of isocitrate dehydrogenase-1 (IDH1) associated with glioma have, for instance, provided evidence that these IDH1 variants affect the methylation state of histones and DNA39 possibly by generating the novel metabolite 2-hydroxyglutarate40, which can competitively inhibit α-ketoglutarate-dependent demethylase enzymes41. Here, we show that NNMT, which is overexpressed in a diverse set of cancers, regulates the protein methylation state of tumor cells through a distinct mechanism that involves altering cellular ratios of SAM:SAH. In this model, 1MNA, the primary product of NNMT catalysis, serves principally as a stable sink to store methylation units in tumor cells rather than as a “bioactive” pro-tumorigenic metabolite, allowing NNMT to act in turn as a metabolic rheostat that can fine-tune the methylation state of cancer cells. A similar model has been proposed to explain the function of glycine-N-methyltransferase in normal liver, where this enzyme regulates MP42, although its downstream impact on liver protein methylation has not, to our knowledge, been examined. Interestingly, administration of nicotinamide to GNMT−/− mice can rectify their aberrantly high liver levels of SAM42, suggesting that, in the absence of GNMT, NNMT may regulate the MP of normal tissues. Metabolic pathways that are upstream of SAM biosynthesis may also influence MP and protein methylation, as has recently been demonstrated for threonine metabolism in mouse embryonic stem cells43. Finally, we should also emphasize that we observed an effect of NNMT on protein methylation when cancer cells were cultured in low (10–20 μM), but not high (100 μM) concentrations of methionine. These findings serve as a reminder that culturing conditions can exert a substantial influence over the metabolic and protein modification state of cancer cells. In this regard, it would be important, in future studies, to examine the impact of nicotinamide concentrations on NNMT-regulated methylation pathways in cancer cells. Nicotinamide levels have been reported to range from 0.4 to 400 μM in mammalian tissues and fluids44–46, and nicotinamide concentration was 8 μM in the RPMI media used in this study. Under conditions where nicotinamide is more limiting, it is possible that NNMT could alter additional metabolic parameters, such as NAD+/NADH ratios. Nicotinamide is also under clinical investigation as a sensitizer for radiotherapy in a variety of cancers47. Our data suggest that exogenously administered nicotinamide could have effects beyond radiosensitization and directly impact the metabolism and biology of tumors, especially if they express high levels of NNMT.
Projecting forward, it would be helpful to better understand how NNMT is regulated in cancer cells. We are not aware of reported somatic mutations or evidence of gene amplification for NNMT in cancer, but recent studies indicate that Ras48, Stat349, and NF-κB50 signaling pathways may be responsible for driving NNMT overexpression in cancer cells, where its heightened levels appear to correlate with epithelial-to-mesenchymal transition8, 48. We further found that NNMT does not regulate all histone methylation events or global DNA methylation, which suggests that the enzyme selectively impacts some, but not all cellular methylation pathways, possibly depending on the relative Km and Ki values of individual methyltransferase enzymes (including NNMT itself) for SAM and SAH, respectively. Consistent with this premise, we found that methylation events that are controlled by methyltransferases with higher Km values (and Ki or IC50 values) for SAM (and SAH) tended to be more sensitive to NNMT (Supplementary Table 2). Finally, we should emphasize that further studies are required to understand the functional relationship between the specific protein methylation and gene expression changes and pro-tumorigenic effects caused by NNMT in cancer cells. Our findings so far suggest that the methylation events regulated by NNMT can alter histone-dependent gene expression, but also extend beyond histones to include tumor suppressor proteins like PP2A. Given its apparently widespread influence over protein methylation patterns in tumor cells, NNMT could represent an attractive drug target to modulate the cancer epigenome for therapeutic benefit.
ONLINE METHODS
Materials
All chemical reagents were purchased from Sigma-Aldrich unless otherwise noted. Nicotinamide-2,4,5,6-d4 (d4-NA) and S-adenosyl-L-methionine-d3 (S-methyl-d3) (d3-SAM) were purchased from C/D/N isotopes (D-3457 and D-4093, respectively). L-methionine-(methyl-13C,d3) (13Cd3-methionine) was from Sigma-Aldrich (#299154). Serine-2,3,3-d3 (d3-serine) was purchased from Cambridge Isotope Laboratories (DLM-1073-1). S-(5′-deoxyadenosin-5′-yl)-L-homocysteine-d4 (d4-SAH) was purchased from Cayman Chemical Company.
General synthetic methods
1H NMR and 13C NMR spectra were recorded on Bruker DRX-600 spectrometers using residual solvent peak as an internal standard. NMR chemical shifts are reported in ppm using residual solvent peak as internal standard, and J values are reported in Hz. High resolution mass spectra were recorded on an Agilent 6520 mass spectrometer using ESI-TOF.
1-Methylnicotinamide
1-Methylnicotinamide was synthesized using a modification of a protocol reported by French et. al51. Briefly, nicotinamide (1 g, 8.2 mmol) was charged into flame-dried RBF under N2 and diluted in methanol (10 mL, 0.8 M). Methyl iodide (1.52 mL, 16.4 mmol) was then added dropwise at room temperature and the reaction allowed to stir overnight at room temperature. The resulting yellow precipitate was isolated by filtration, rinsed with methanol, and dried in vacuo. Recrystallization from methanol afforded a product (51% yield) whose spectral properties were consistent with previously reported values52: 1H-NMR (600 MHz, D2O) δ 9.30 (s, 1H), 8.98 (d, J = 6.0, 1H), 8.90 (d, J = 8.4, 1H), 8.19 (t, J = 7.2, 1H), 4.49 (s, 3H); 13C-NMR (600 MHz, D2O) δ 166.7, 148.1, 146.0, 144.4, 134.4, 128.8, 49.4. HRMS (ESI) (m/z): [M+] calculated for C7H9N2O+, 137.070939; found, m/z 137.071459.
1-Methylnicotinamide-2,4,5,6-d4
The same procedure as above was followed to produce d4-1MNAin 71% yield: 1H-NMR (600 MHz, D2O) δ 4.47 (s, 3H); 13C-NMR (600 MHz, D2O) δ 166.7, 147.8 (t, J = 119), 145.7 (t, J = 120), 144.1 (t, J = 110), 128.4 (t, J = 102), 49.4. HRMS (ESI) (m/z): [M+] calculated for C7H5D4N2O+, 141.096046; found, 141.097323.
Tissue culture
SKOV3 and OVCAR3 cell lines were obtained from NCI’s Developmental Therapeutics Program. 786O, 769P, H266 and H522 lines were purchased from ATCC. C8161 and MUM2C were provided by Mary Hendrix. All cell lines were cultured in RPMI-1640 medium (Cellgro) supplemented with 10% FBS, 2 mM glutamine and 10 mM HEPES buffer (complete RPMI-1640, 100 μM methionine). Unless otherwise noted, cells were seeded at 4 × 106 cells/150 mm dish in complete RPMI-1640 and allowed to proliferate for 16–24 h before experiments were performed. Low methionine medium was prepared by adding 10 and 20 μM methionine to a methionine-free RPMI-1640 (A14517-01, GIBCO), supplemented with 10% dialyzed FBS, 2 mM glutamine and 10 mM HEPES buffer. See overexpression and RNA interference studies for low methionine culture conditions.
Retroviral overexpression of wildtype and mutant NNMT in human cancer cell lines
The cDNA clone of human NNMT (BC000234, Open BioSystems) in pOTB7 was subcloned into pET-45b (+) vector for further manipulations. Catalytically inactive NNMT-Y20A mutant was generated by QuikChange site-directed mutagenesis using the primer 5′-atctaagccattttaaccctcgggatgccctagaaaaatattacaagtttg-3′ and its complement. Wildtype and mutant NNMT were cloned into modified pCLNCX retroviral vector 53. Retrovirus was prepared by taking 1.5 μg of both pCLNCX and pCL-Ampho vectors and 20 μl of FuGENE HD reagent (Roche) to transfect 80% confluent HEK293T cells. Virus containing supernatant from day 2 was collected and, in the presence of 8 μg/ml polybrene, used to stably infect cells for 72 h. Infection was followed by 7–14 days of selection in medium containing hygromycin B (100 μg/ml). Cells were expanded and cultured in complete RPMI-1640.
For all experiments except for studies in low methionine medium, cells were seeded at 4 × 106 cells/150 mm dish and were allowed to proliferate in complete RPMI for 16–24 h. For studies performed in low (10 μM and 20 μM) methionine medium, cells were washed with PBS, seeded at a concentration of 2 × 106 cells/150 mm dish, and cultured for 48h in methionine-free RPMI supplemented with indicated amounts of methionine.
NNMT activity assay
Cell pellets were resuspended in either 50 mM Tris-HCl, pH 8.0 or PBS (lung carcinoma lines), followed by sonication and centrifugation at 16,000 g for 10 min. Lysates (70–120 μg) were incubated with a reaction mixture (200 μM d4-NA, 50 μM S-adenosyl-L-methionine (SAM) and 2 mM DTT) at room temperature for 15–30 min in a volume of 20 μl. Reactions were quenched with equal amount of methanol, followed by a 10 min centrifugation at 16,000 g. Formation of d4-1MNA was followed by targeted LC-MS analysis. Briefly, 1MNA-d4 was separated with a Luna-NH2 column (5 μm, 100A, 50 × 4.6 mm, Phenomenex) together with a pre-column (NH2, 4 × 3.0 mm). Mobile phase A was composed of 100% CH3CN containing 0.1% formic acid, and mobile phase B was composed of 95:5 v/v H2O:CH3CN supplemented with 50 mM NH4OAc and 0.2% NH4OH. The flow rate started at 0.1 ml/min and the gradient consisted of 5 min 0% B, a linear increase to 100% B over 15 min at a flow rate of 0.4 ml/min, followed by an isocratic gradient of 100% B for 15 min at 0.5 ml/ml before equilibrating for 5 min at 0% B at 0.4 ml/min (40 min total). For each run the ejection volume was 20 μl. MS analysis was performed on an Agilent G6410B tandem mass spectrometer with ESI source. The dwell time for d4-1MNA was set to 100 ms. The capillary was set to 4 kV, the fragmentor was set to 100 V. The drying gas temperature was 350 °C, the drying gas flow rate was 11 l/min, and the nebulizer pressure was 35 psi. The mass spectrometer was run in MRM mode, monitoring the transition of m/z from 141 to 98 for d4-1MNA (positive ionization mode).
RNA interference studies in human cancer cell lines
Hs-NNMT-8 (si-NNMT, cagctactacatgattggtga) and Ctrl-AllStars-1 (si-Control, siRNA that has no homology to any known mammalian gene) were purchased from QIAGEN as FlexiTube siRNA premix. Cells were seeded at 0.25 × 106 cells/100 mm dish followed by treatment with siRNA premix reagent. Cells were cultured in complete RPMI for 72 h and tested for the loss of NNMT activity.
For studies performed in high and low methionine medium, 72 h after transfection complete RPMI medium was exchanged with RPMI containing indicated amount of methionine and cells were allowed to proliferate for additional 24 h. NNMT knockdown was confirmed in each experiment.
Untargeted metabolomic analysis of cancer cell lines
GFP-OE and NNMT-OE human cell lines were seeded at 4 × 106 cells/150 mm dish and cultured in complete RPMI medium for 24 h followed by 4 h serum starvation. Cells were scraped into ice-cold PBS and isolated by centrifugation at 1,400 × g at 4 °C. Water soluble cellular metabolites were extracted using methanol-water extraction protocol, essentially as previously described17. In brief, cell pellets were re-suspended in 100 μl of a 80:20 mixture of MeOH:H2O. For some experiments, internal deuterated standards, including 1 nmol d4-NA, 0.1 nmol d4-1MNA, 0.1 nmol 13Cd3-methionine, 0.1 nmol d3-SAM, 0.1 nmol d4-SAH, and 10 nmol d3-serine, were added to the extraction solution, for absolute quantification and sample normalization. The mixture was sonicated for 5 s followed by a 10 min centrifugation at 16,000 × g. The supernatant was collected and stored at −80° C or injected directly into mass spectrometer (30 μl). Metabolites were also extracted by an alternative protocol involving direct scrapping into organic solvent54 for the data shown in Supplementary Fig. 7. GFP-OE and NNMT-OE samples (3–4 replicates/line) were run sequentially. Water soluble cellular metabolites were separated by hydrophilic interaction chromatography17 with a Luna-NH2 column together with a pre-column. Mobile phase A was composed of 100% CH3CN, and mobile phase B was composed of 95:5 v/v H2O:CH3CN. Both solvents were supplemented with 0.1% formic acid to assist ion formation in a positive mode. For negative mode analysis, mobile phase B was supplemented with 50 mM NH4OAc and 0.2% NH4OH. The flow rate started at 0.1 ml/min for 5 min. The gradient started with 0% B for 5 min and increased linearly to 100% B over 40 min with a flow rate of 0.4 ml/min, followed by an isocratic gradient of 100% B for 10 min at 0.5 ml/min. Then column was equilibrated with 0% B for 5 min at 0.4 ml/min. MS analysis, scanning from m/z = 50–1200, was performed on Agilent 6520 Accurate Mass Q-TOF with ESI source. Untargeted LC-MS analysis was performed in both positive and negative ionization mode. The capillary was set to 4 kV. The drying gas temperature was 350 °C, the drying gas flow rate was 11 l/min, and the nebulizer pressure was 45 psi.
To identify metabolites with differential levels in NNMT-OE versus GFP-OE cells, we employed XCMS analyte profiling software. In brief, XCMS identifies features whose relative intensity varies between sample groups (group 1: NNMT-OE replicates; group2: GFP-OE replicates) and calculates fold changes, as well as P-values. XCMS software allows quick access to the quality of each feature by generating extracted ion chromatograms display panels (see Fig. 2b and Supplementary Fig. 5). Obtained data sets were first filtered based on P-value (P < 0.01) and fold change (fold > 2). Significant peak changes between samples were confirmed by manually extracting MS1 signals and by calculating the area under the peak from MS1 chromatograms. We next clustered the changing metabolites into groups based on whether they appear in one, two, or all three (769P, MUM2C and OVCAR3) pairs of NNMT-OE/GFP-OE. Prioritization was given to those metabolites that were found to change in all three cancer cell line sets. Two metabolites identified as 1MNA and SAH (see Fig. 2) were consistently deregulated across all three cell lines (Supplementary Dataset 1).
In addition to ions corresponding to endogenous 1MNA and SAH, an ion with m/z value of 152.074417 was also consistently elevated in all NNMT-OE lines compared to GFP-OE lines. This ion was identified as 3-methoxycarbonyl-1-methylpyridinium by using a combination of high-resolution MS (observed m/z = 152.074417, calculated m/z = 152.070605) and co-elution with authentic sample of 3-methoxycarbonyl-1-methylpyridinium. Elevated levels of 3-methoxycarbonyl-1-methylpyridinium in NNMT-OE metabolomes compared to GFP-OE metabolomes are likely due to the slow alcoholysis of NNMT product 1MNA in the methanolic extracts, as no 152.074417 ion was detected in metabolomes from GFP-OE and NNMT-OE cells when cell pellets were extracted with a 50:50 mixture of CH3CN:H2O. In addition, formation of this ester from synthetic 1MNA was also observed in methanolic solutions containing residual amounts of PBS without cellular extracts, confirming the non-metabolic origin of the compound. Interestingly, this ester is not formed in pure 80:20 MeOH:H2O mixture, suggesting that certain components of residual PBS catalyze the solvolysis. Considering the non-metabolic nature of this compound, it was excluded from the list of deregulated metabolites identified by metabolomic analysis.
LC-MS co-migration studies
Metabolomes from GFP-OE and NNMT-OE 769P cells were prepared and analyzed as described above. The identity of endogenous m/z = 137.07 was confirmed by overlapping its extracted ion chromatogram with MS1 ion chromatogram of d4-1MNAinternal standard (m/z = 141.10). Similarly, the identity of endogenous m/z = 385.13 was confirmed by overlapping its extracted ion chromatogram with MS1 ion chromatogram of d4-SAHinternal standard (m/z = 389.16).
MS/MS fragmentation studies
LC-MS/MS analysis was performed on an Agilent 6520 as just described in positive ionization mode. MS and MS/MS data were collected in scanning mode from m/z = 50–2000 and m/z = 50–2500, and a rate of 1.03 spectra/s. The capillary voltage was set to 4 kV, and the fragmentor voltage was set to 100 V. The drying gas temperature was 350 °C, the drying gas flow rate was 11 l/min, and the nebulizer pressure was 45 psi. The collision energy for 1MNA and SAH was 20 V and 5 V, respectively.
Metabolic labeling studies
769P cells were seeded at 1.5 × 106 cells/150 mm dish and were cultured overnight in complete RPMI medium. The next day, the medium was replaced with serum-free medium containing either d4-1MNA (100 μM) or d4-NA (100 μM). Control samples were prepared by incubating cells with the same concentration of authentic compound (either 1MNA or NA). After an additional 4 or 24 h, cellular metabolomes were prepared and analyzed in the untargeted scanning mode as described above. The resultant chromatograms were analyzed by extracting relative m/z values and quantified by calculating the area under the peak. The following deuterated metabolites were detected: d4-1MNA (m/z = 141.10, RT = 5.6 min, pos. mode), d4-NA (m/z = 127.08, RT = 6.1 min, pos. mode), d3-NA (m/z = 126.07, RT = 6.1 min, pos. mode), d3-NMN(m/z = 338.08, RT = 28.2 min, pos. mode), d3-NAD+ (m/z = 665.12, RT = 28.1 min, neg. mode), and d3-NADH(m/z = 667.14, RT = 33.9 min, neg. mode). These metabolites were absent in control cells and their identity was confirmed by co-elution with corresponding authentic metabolites in control cells and authentic standards (1MNA, m/z = 137.07, RT = 5.6 min, pos. mode; NA, m/z = 123.06, RT = 6.1 min, pos. mode; NMN, m/z = 335.06, RT = 28.2 min, pos. mode; NAD+, m/z = 662.10, RT = 28.1 min, neg. mode). The formation of d3-labeled metabolites in NAD+ biosynthetic pathway could be explained by the oxidation/reduction of NAD(H) which would result in the loss of deuterium. Flux of metabolites through the NAD+ pathway would then generate the steady state levels of d3-labeled metabolites that are measured in this assay. Consistent with this model, we observed the time-dependent increase in the ratio of d3-NA/d4-NA in our metabolic labeling studies.
Targeted MRM measurements of intracellular metabolites
Cells were collected by scraping into ice-cold PBS followed by centrifugation at 1,400 × g. Cellular metabolites were extracted with 100 μl of a 80:20 mixture of MeOH:H2O, containing the following deuterated standards: 0.1 – 1 nmol d4-NA, 0.1 nmol d4-1MNA, 0.1 nmol 13Cd3-methionine, 0.1 nmol d3-SAM, 0.1 nmol d4-SAH, and 10 nmol d3-serine. The mixture was sonicated for 5 s followed by a 10 min centrifugation at 16,000 × g. Supernatant was collected, and 30 μl was subjected to LC-MS analysis. LC separation was performed as described above for metabolomics experiments. MS analysis was performed on Agilent G6410B tandem mass spectrometer with ESI source as described in NNMT activity assay. Mass spectrometer was running in MRM mode. Metabolites were quantified by measuring the area under the peak in comparison to the deuterated standards. Cellular SAH levels were calculated by subtracting the residual amount of SAH present in internal standard d3-SAM from total SAH levels. Amount of SAH coming from d3-SAM constituted less than 30% of total SAH. The following MS transitions and retention time (RT) were used to measure the indicated metabolites: 1MNA (m/z 137 →94, RT = 5.6 min), d4-1MNA (m/z 141 →98, RT = 5.6 min), SAM (m/z 399 →250, RT = 16.2 min), d3-SAM (m/z 402→250, RT = 16.2 min), SAH (m/z 385→136, RT = 20.1 min), d4-SAH (m/z 389→136, RT = 20.1 min), methionine (m/z 150→133, RT = 17.8 min), and 13Cd3-methionine (m/z 154 →137, RT = 17.8 min). For easy comparison between cell lines, absolute concentrations of cellular metabolites are normalized to 4 × 106 cells.
Western blotting
Cell pellets were resuspended in lysis buffer followed by sonication and 10 min centrifugation at 16,000 g. Lysates were separated by SDS-PAGE, transferred to nitrocellulose membrane and blocked in 5% milk in TBST. The primary antibodies used were: anti-NNMT (Abcam, ab58743), anti-H3K9-me1 (Abcam, ab8896), anti-H3K9-me2 (Cell Signaling, 9753), anti-H3K9-me3 (Abcam, ab8898), anti-H3-total (Abcam, ab1791), anti-H3K4-me1 (Abcam, ab8895), anti-H3K4-me2 (Abcam, ab32356), anti-H3K4-me3 (Abcam, ab8580), anti-H3K27-me2 (Abcam, ab24684), anti-H3K27-me3 (Millipore, 07-449), anti-H3K79-me2 (Abcam, ab3594), anti-H4K20-me2 (Cell Signaling, 9759), anti-PP2A-total (Millipore, 07-324), anti-PP2A-me (Millipore, 04-1479), anti-PP2A-deme (Millipore, 05-577), anti-dimethyl-arginine, asymmetric or ASYM25 (Millipore, 09-814), anti-H3R17-me2a (Abcam, ab8284).
Migration and invasion studies
Migration and invasion assays were performed as described previously8, 15. For 1MNA treatment studies, 0.5 mM 1MNA was added to GFP-OE 769P cells during serum starvation and to the upper and bottom chamber during migration assay.
DNA methylation assay
Genomic DNA was isolated from 769P cells using DNeasy blood & tissue kit from QIAGEN. 1 μg of DNA was degraded into nucleosides using DNA Degradase Plus (Zymo Research). LC separation was achieved with a Sinergy Fusion-RP column (4 μm, 80A, 50 × 4.6 mm, Phenomenex) together with a pre-column (Fusion-RP, 4 × 3.0 mm). Mobile phase A was composed of 100% H2O, and mobile phase B was composed of 100% MeOH. Both solvents were supplemented with 0.1% formic acid to assist ion formation in a positive mode. The flow rate started at 0.1 ml/min for 5 min. The gradient started with 0% B for 5 min and increased linearly to 100% B over 20 min with a flow rate of 0.4 ml/min, followed by an isocratic gradient of 100% B for 2 min at 0.5 ml/min. Then column was equilibrated with 0% B for 3 min at 0.4 ml/min. MS analysis, scanning from m/z = 50–1200, was performed on Agilent 6520 Accurate Mass Q-TOF. 5-Methyl-2′-deoxycytidine (5mdC) content was calculated as [5mdC]/[dG] using external calibration curve as described55.
DNA microarray
mRNAs were isolated (RNeasy Mini Kit, QIAGEN) from NNMT-OE and Y20A-OE 769P cells, reversed transcribed, and hybridized to Affymetrix Human 1.0 ST microarray. Data were then filtered for genes that were upregulated or downregulated (>1.4-fold) in NNMT-OE versus Y20A-OE cells for further analysis.
Real-time (RT)-PCR analysis
mRNAs were isolated using RNeasy Mini Kit (QIAGEN). The cDNAs were synthesized by reverse transcription using the SABiosciences RT2 kit. Obtained cDNAs were added to RT2 SYBR Green mastermix followed by RT-PCR using custom RT2 Profiler PCR Arrays (SABiosciences). RT-PCR was performed on ABI 7900HT cycler (384-well block, Applied Biosystems). Results were normalized to the average of three housekeeping genes, including ACTB, GAPDH and HPRT1. Gene list is shown in table below.
| Gene | Catalog # | Refseq # | Official Full Name |
|---|---|---|---|
| SNAI2 | PPH02475 | NM_003068 | Snail homolog 2 (Drosophila) |
| TGFB2 | PPH00524 |
NM_003238 NM_001135599 |
Transforming growth factor, beta 2 |
| CNTN1 | PPH00632 | NM_001843 | Contactin 1 |
| ADAMTS6 | PPH15788 | NM_197941 | ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| LAMB3 | PPH20656 |
NM_000228 NM_001017402 NM_001127641 |
Laminin, beta 3 |
| ACTB | PPH00073 | NM_001101 | Actin, beta |
| GAPDH | PPH00150 | NM_002046 | Glyceraldehyde-3-phosphate dehydrogenase |
| HPRT1 | PPH01018 | NM_000194 | Hypoxanthine phosphoribosyltransferase 1 |
Statistical analysis
Data are shown as mean ± SEM. P-values were calculated using unpaired, two-tailed Student’s t-test. A P-value of < 0.05 was considered significant.
Supplementary Material
Acknowledgments
This work was supported by the NIH (CA132630), a postdoctoral fellowship from Bayer (O.A.U), and an NSF predoctoral fellowship (A.M.Z.).
Footnotes
Author Contributions. O.A.U. and B.F.C. designed the experiments, analyzed the data and wrote the manuscript. A.M.Z. synthesized 1MNA and d4-1MNA. O.A.U. performed all other experiments.
Competing Financial Interests Statement.
The authors have no competing financial interest.
References
- 1.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The Biology of Cancer: Metabolic Reprogramming Fuels Cell Growth and Proliferation. Cell Metabolism. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Hsu PP, Sabatini DM. Cancer Cell Metabolism: Warburg and Beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg Robert A. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Wellen KE, Thompson CB. A two-way street: reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol. 2012;13:270–276. doi: 10.1038/nrm3305. [DOI] [PubMed] [Google Scholar]
- 6.DeBerardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Current Opinion in Genetics & Development. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Possemato R, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nomura DK, et al. Monoacylglycerol lipase exerts dual control over endocannabinoid and fatty acid pathways to support prostate cancer. Chem Biol. 2011;18:846–856. doi: 10.1016/j.chembiol.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nomura DK, Dix MM, Cravatt BF. Activity-based protein profiling for biochemical pathway discovery in cancer. Nat Rev Cancer. 2010;10:630–638. doi: 10.1038/nrc2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roessler M, et al. Identification of nicotinamide N-methyltransferase as a novel serum tumor marker for colorectal cancer. Clin Cancer Res. 2005;11:6550–6557. doi: 10.1158/1078-0432.CCR-05-0983. [DOI] [PubMed] [Google Scholar]
- 11.Wu Y, Siadaty MS, Berens ME, Hampton GM, Theodorescu D. Overlapping gene expression profiles of cell migration and tumor invasion in human bladder cancer identify metallothionein 1E and nicotinamide N-methyltransferase as novel regulators of cell migration. Oncogene. 2008;27:6679–6689. doi: 10.1038/onc.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J, et al. Expression of nicotinamide N-methyltransferase in hepatocellular carcinoma is associated with poor prognosis. J Exp Clin Cancer Res. 2009;28:20. doi: 10.1186/1756-9966-28-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomida M, Mikami I, Takeuchi S, Nishimura H, Akiyama H. Serum levels of nicotinamide N-methyltransferase in patients with lung cancer. J Cancer Res Clin Oncol. 2009;135:1223–1229. doi: 10.1007/s00432-009-0563-y. [DOI] [PubMed] [Google Scholar]
- 14.Tang SW, et al. Nicotinamide N-methyltransferase induces cellular invasion through activating matrix metalloproteinase-2 expression in clear cell renal cell carcinoma cells. Carcinogenesis. 2011;32:138–145. doi: 10.1093/carcin/bgq225. [DOI] [PubMed] [Google Scholar]
- 15.Nomura DK, et al. Monoacylglycerol Lipase Regulates a Fatty Acid Network that Promotes Cancer Pathogenesis. Cell. 2010;140:49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng Y, et al. Structural basis of substrate recognition in human nicotinamide N-methyltransferase. Biochemistry. 2011;50:7800–7808. doi: 10.1021/bi2007614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bajad SU, et al. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J Chromatogr A. 2006;1125:76–88. doi: 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 18.Saghatelian A, et al. Assignment of Endogenous Substrates to Enzymes by Global Metabolite Profiling†. Biochemistry. 2004;43:14332–14339. doi: 10.1021/bi0480335. [DOI] [PubMed] [Google Scholar]
- 19.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Analytical Chemistry. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 20.Zinellu A, et al. Plasma methionine determination by capillary electrophoresis –UV assay: Application on patients affected by retinal venous occlusive disease. Analytical Biochemistry. 2007;363:91–96. doi: 10.1016/j.ab.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 21.Lu SC. S-Adenosylmethionine. The International Journal of Biochemistry & Cell Biology. 2000;32:391–395. doi: 10.1016/s1357-2725(99)00139-9. [DOI] [PubMed] [Google Scholar]
- 22.Ulrey CL, Liu L, Andrews LG, Tollefsbol TO. The impact of metabolism on DNA methylation. Human Molecular Genetics. 2005;14:R139–R147. doi: 10.1093/hmg/ddi100. [DOI] [PubMed] [Google Scholar]
- 23.Jia G, et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo M. Current chemical biology approaches to interrogate protein methyltransferases. ACS Chem Biol. 2012;7:443–463. doi: 10.1021/cb200519y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cantoni GL. The role of S-adenosylhomocysteine in the biological utilization of S-adenosylmethionine. Prog Clin Biol Res. 1985;198:47–65. [PubMed] [Google Scholar]
- 26.Dawson Mark A, Kouzarides T. Cancer Epigenetics: From Mechanism to Therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 27.Varier RA, Timmers HTM. Histone lysine methylation and demethylation pathways in cancer. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 2011;1815:75–89. doi: 10.1016/j.bbcan.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 28.Eichhorn PJA, Creyghton MP, Bernards R. Protein phosphatase 2A regulatory subunits and cancer. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 2009;1795:1–15. doi: 10.1016/j.bbcan.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 29.Puustinen P, et al. PME-1 Protects Extracellular Signal-Regulated Kinase Pathway Activity from Protein Phosphatase 2A–Mediated Inactivation in Human Malignant Glioma. Cancer Research. 2009;69:2870–2877. doi: 10.1158/0008-5472.CAN-08-2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boisvert FM, Côté J, Boulanger MC, Richard S. A Proteomic Analysis of Arginine-methylated Protein Complexes. Molecular & Cellular Proteomics. 2003;2:1319–1330. doi: 10.1074/mcp.M300088-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Yang H, Menon L, Black P, Carroll R, Johnson M. SNAI2/Slug promotes growth and invasion in human gliomas. BMC Cancer. 2010;10:301. doi: 10.1186/1471-2407-10-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zavadil J, Bottinger EP. TGF-[beta] and epithelial-to-mesenchymal transitions. Oncogene. 24:5764–5774. doi: 10.1038/sj.onc.1208927. 0000. [DOI] [PubMed] [Google Scholar]
- 33.Su JL, et al. Knockdown of Contactin-1 Expression Suppresses Invasion and Metastasis of Lung Adenocarcinoma. Cancer Research. 2006;66:2553–2561. doi: 10.1158/0008-5472.CAN-05-2645. [DOI] [PubMed] [Google Scholar]
- 34.Wierinckx A, et al. A diagnostic marker set for invasion, proliferation, and aggressiveness of prolactin pituitary tumors. Endocrine-Related Cancer. 2007;14:887–900. doi: 10.1677/ERC-07-0062. [DOI] [PubMed] [Google Scholar]
- 35.Miyazaki K. Laminin-5 (laminin-332): Unique biological activity and role in tumor growth and invasion. Cancer Science. 2006;97:91–98. doi: 10.1111/j.1349-7006.2006.00150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dreger H, et al. Epigenetic Regulation of Cell Adhesion and Communication by Enhancer of Zeste Homolog 2 in Human Endothelial Cells / Novelty and Significance. Hypertension. 2012;60:1176–1183. doi: 10.1161/HYPERTENSIONAHA.112.191098. [DOI] [PubMed] [Google Scholar]
- 37.Tan J, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes & Development. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miranda TB, et al. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Molecular Cancer Therapeutics. 2009;8:1579–1588. doi: 10.1158/1535-7163.MCT-09-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turcan S, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu W, et al. Oncometabolite 2-Hydroxyglutarate Is a Competitive Inhibitor of α-Ketoglutarate-Dependent Dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Varela-Rey M, et al. Fatty liver and fibrosis in glycine N-methyltransferase knockout mice is prevented by nicotinamide. Hepatology. 2010;52:105–114. doi: 10.1002/hep.23639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shyh-Chang N, et al. Influence of Threonine Metabolism on S-Adenosylmethionine and Histone Methylation. Science. 2012 doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 45.Smythe GA, et al. Concurrent quantification of quinolinic, picolinic, and nicotinic acids using electron-capture negative-ion gas chromatography-mass spectrometry. Anal Biochem. 2002;301:21–26. doi: 10.1006/abio.2001.5490. [DOI] [PubMed] [Google Scholar]
- 46.Catz P, et al. Simultaneous determination of myristyl nicotinate, nicotinic acid, and nicotinamide in rabbit plasma by liquid chromatography-tandem mass spectrometry using methyl ethyl ketone as a deproteinization solvent. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;829:123–135. doi: 10.1016/j.jchromb.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 47.Wardman P. Chemical radiosensitizers for use in radiotherapy. Clin Oncol (R Coll Radiol) 2007;19:397–417. doi: 10.1016/j.clon.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 48.Joyce T, Cantarella D, Isella C, Medico E, Pintzas A. A molecular signature for Epithelial to Mesenchymal transition in a human colon cancer cell system is revealed by large-scale microarray analysis. Clin Exp Metastasis. 2009;26:569–587. doi: 10.1007/s10585-009-9256-9. [DOI] [PubMed] [Google Scholar]
- 49.Tomida M, Ohtake H, Yokota T, Kobayashi Y, Kurosumi M. Stat3 up-regulates expression of nicotinamide N-methyltransferase in human cancer cells. J Cancer Res Clin Oncol. 2008;134:551–559. doi: 10.1007/s00432-007-0318-6. [DOI] [PubMed] [Google Scholar]
- 50.Hsu S, et al. IKK-epsilon coordinates invasion and metastasis of ovarian cancer. Cancer Res. 2012;72:5494–5504. doi: 10.1158/0008-5472.CAN-11-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.French JB, et al. Characterization of Nicotinamidases: Steady State Kinetic Parameters, Classwide Inhibition by Nicotinaldehydes, and Catalytic Mechanism. Biochemistry. 2010;49:10421–10439. doi: 10.1021/bi1012518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Polkowska J, et al. A combined experimental and theoretical study of the pH-dependent binding mode of NAD+ by water-soluble molecular clips. Journal of Physical Organic Chemistry. 2009;22:779–790. [Google Scholar]
- 53.Martin BR, Giepmans BN, Adams SR, Tsien RY. Mammalian cell-based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nat Biotechnol. 2005;23:1308–1314. doi: 10.1038/nbt1136. [DOI] [PubMed] [Google Scholar]
- 54.Yuan J, Bennett BD, Rabinowitz JD. Kinetic flux profiling for quantitation of cellular metabolic fluxes. Nat Protocols. 2008;3:1328–1340. doi: 10.1038/nprot.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song L, James SR, Kazim L, Karpf AR. Specific Method for the Determination of Genomic DNA Methylation by Liquid Chromatography-Electrospray Ionization Tandem Mass Spectrometry. Analytical Chemistry. 2004;77:504–510. doi: 10.1021/ac0489420. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.