Abstract
Staphylococcus epidermidis is an opportunistic bacterium that thrives as a commensal cutaneous organism and as a vascular pathogen. The S. epidermidis extracellular matrix binding protein (Embp) has been reported to be a virulence factor involved in colonization of medical device implants and subsequent biofilm formation. Here, we characterize the expression patterns of Embp in planktonic and biofilm cultures, as well as under high osmotic stresses that typify the commensal environment of the skin. Embp expression without osmotic stress was similar for planktonic and adherent cultures. Addition of osmotic stress via NaCl caused slight increases in embp expression in planktonic cultures. However, in adherent cultures a 100-fold increase in embp expression with NaCl versus controls occurred and coincided with altered biofilm morphology. Results suggest that the central role of Embp lies in commensal skin colonization, stabilizing the cell wall against osmotic stresses, rather than as a virulence factor promoting adhesion.
Introduction
Over half of hospital-acquired infections are associated with implanted devices with the case-to-fatality ratio of these infections as high as 40 % [1]. Even innocuous skin bacteria, such as Staphylococcus epidermidis, can adhere to implanted medical devices in the vasculature via specific and non-specific adhesion mechanisms. Biofilm colonies formed by adherent S. epidermidis enable the bacterium to evade both antibiotics and the host immune system, leading to infection, sepsis, and potentially death. Biofilms are microbial cell-produced mucilaginous matrices of extra-cellular DNA [2], proteins [3, 4], and polysaccharides [5]. The danger of biofilms arises from their secretion of virulence factors, their relatively large size within the vasculature, and the propensity for fragments to detach from the biofilm and occlude blood vessels downstream [6].
While expression of the icaADBC operon and its associated production of polysaccharide intracellular adhesin (PIA) is the most studied mechanism of S. epidermidis biofilm formation [7], a number of S. epidermidis strains lacking the icaADBC operon still possess the ability to form biofilms through alternative mechanisms. These alternative matrix molecules include proteins, such as the extracellular matrix binding protein (Embp) and the accumulation associated protein (Aap), and extracellular DNA [2–4].
Embp is a giant 1,100-kDa protein localized to the cell wall of S. epidermidis with potential functional similarity to large homologous proteins produced in other pathogenic bacteria such as Staphylococcus aureus and Abiotrophia defectiva. In A. defectiva, the Emb protein has been identified as a major fibronectin (FN) adhesion protein [8]. In S. aureus, however, while the 1,000-kDa extracellular matrix binding homolog (Ebh) is able to bind weakly to FN, its major function is proposed to be structural support of the cell wall [9, 10]. S. aureus exhibits strong binding to FN through other protein receptors, FnbpA and FnbpB, while Ebh-negative S. aureus mutants are unable to withstand hyper-osmotic pressure shifts.
The S. epidermidis Embp has been reported to be involved in both specific binding to FN-coated surfaces and in subsequent biofilm formation by Christner et al. [4]. However, significant specific receptor:ligand-mediated adhesion of S. epidermidis to FN adsorbed on a variety of such surfaces has been contradicted by other works [11, 12]. Embp-mediated cell:cell adhesion properties elucidated by the Christner et al. work could serve other beneficial roles in biofilm producing strains of S. epidermidis. Consequently, there is a need to further characterize the expression and purpose of Embp in both suspended and adherent biofilm cultures.
Materials and Methods
Cell Culture and Sample Collection
Staphylococcus epidermidis strain 1457, isolated from a patient with catheter-related sepsis, and strain CV2 (an isogenic agr- deletion mutant derived from 1457) were generously provided by Dr. Michael Otto (NIH, Bethesda MD) [13, 14]. The biofilm-negative strain S. epidermidis 12228 was purchased from ATCC. S. epidermidis strains SCH1.2, SCH1.3, and SCH1.5, cultured from intravascular catheter infections, were provided by Dr. Xuan Qin (Seattle Children’s Hospital) and further genotyped and characterized biochemically. All strains were kept in glycerol at −80 °C until use, then cultured on 10 g/L tryptic soy broth (TSB) agar plates. A single colony was then transferred into TSB medium and grown overnight at 37 °C. From this cell suspension, samples were diluted 1/50 into new medium for growth at 37 °C either as batch planktonic cell cultures under shaking conditions (200 rpm waterbath shaker), or as batch biofilm cultures under static fluid (minimal mixing) conditions.
Planktonic cell growth was quantified by optical density at 600 nm (OD600) and calibrated to approximate cell number (OD600 of 1.0 ≈ 109 cells). Adherent biofilm cells were removed from substrata using a microultrasonic cell disruptor with ASI tip (Kontes Co., Vineland, NJ) operated at mild sonication (30 % maximum setting for 15 s) before measuring the optical density of the resultant cell suspension. For mRNA and protein expression studies, ~109 cells were collected from either planktonic or adherent cultures and immediately added to an equal volume of a 1:1 mixture of ice cold acetone and ethanol and stored at −80 °C for at least twenty minutes or until further use.
Δembp S. epidermidis 1457 Mutation
Creation of an embp deletion mutant (Δembp) was attempted by four separate methods. Homologous recombination using the pKOR1 shuttle vector was first attempted to remove the entire embp gene. Insertional inactivation was performed to interrupt the expression of embp by replacing the first 1,000 bp with an antibiotic cassette. A suicide vector (pCIV2) was used to prevent transmission of non-recombined plasmid to daughter cells. Finally, a temperature sensitive vector (pBT2) containing erythromycin resistance was used to prevent embp expression. Summary of results related to these attempts is provided in the Supplementary Information.
Western Blotting
Proteins, expressed by planktonic or adherent cells, were collected by centrifuging 109 cells, rinsing twice in PBS (pH 7.4), followed by re-suspending cells in 0.5 ml Laemmli sample buffer and boiling at 95 °C for 5 min. Proteins were then separated by SDS-PAGE and analyzed via Western blot. Because of its large size, Embp migrates only into the first 2 mm of a 4–15 % acrylamide gel (BioRad, Carlsbad, CA) and must be transferred to the PVDF membrane without methanol while using high voltage (60 V max for 16 h). The membranes were blocked with 1 % BSA in TBS overnight and then probed for two hours with a 1/5,000 dilution of anti-rEmbp2588 antiserum donated by Dr. Holger Rodhe (Universitätsklinikum Hamburg-Eppendorf, Germany). Blots were rinsed and then incubated with a secondary alkaline phosphatase-conjugated goat anti-rabbit antibody (Pierce, Rockford, IL) at a dilution of 1/10,000, rinsed again, and detected by incubating with BCIP/NBT (MP Bio, Solon, OH).
Cell Lysis and mRNA Purification
Frozen cell suspensions of either planktonic or adherent cells were thawed on ice, centrifuged at 10,000×g for 10 min at 4 °C, and the supernatant removed. Cell pellets were dried for five minutes on ice and then re-suspended in 500 μl Tris–EDTA buffer (pH 7.5). The cell suspension was transferred to a Bio101 FastProtein™ Blue tube with Lysing Matrix B (1-μm silica beads) and processed in a Mini-Beadbeater-16 for one minute, then cooled on ice for one minute. The process was repeated for a total of four homogenization cycles. Suspensions were centrifuged and the supernatant was removed to a new tube for RNA purification. RNA purification was performed using an RNeasy™ Mini Kit (QIAgen) following the manufacturers protocol including the optional on-column RNase-free DNase I treatment. RNA purity was assessed on a UV spectrophotometer and samples with an OD280/OD260 ratio between 1.8 and 2.2 were used for further analysis.
Two-Step Reverse Transcription Quantitative PCR
QIAgen’s QuantiTect™ RT Kit was used to synthesize cDNA for qRT-PCR according to the manufacturer’s instructions with products stored at −20 °C until use (Table 1). qPCR was performed on cDNA using Quanti-Tect™ SYBR™ Green PCR Kit. Samples were assayed in triplicate and qPCR was performed on a Rotor-Gene 3000™ Real Time Thermal Cycler (Corbett Research/QIAgen) followed by a melt curve of PCR products. Genomic DNA standards were amplified with each reaction to calculate the relative copy number of each gene of interest. A housekeeping gene encoding gunaylate kinase, gmk, was used as a reference control [15].
Table 1.
RT-PCR and qRT-PCR primers used in RNA expression studies
| Gene | RT-PCR type | Forward primer | Reverse primer | PCR product size (bp) |
|---|---|---|---|---|
| gmk (SE0776) | Standard and quantitative | tcaggtgttggaaagggaac | cgcctcaaattcttcctttg | 153 |
| embp (SE1011) | Quantitative | tgacggttccggaagtattg | Ttagctccctgccatctttc | – |
| icaA (SERP2293) | Quantitative | ttatcaatgccgcagttgtc | Accgttggatattgcctctg | – |
qRT-PCR Analysis
Initial analysis was performed on the Rotor-Gene™ RG-3000 (Corbett Life Sciences/QIAgen, Valencia, CA) to insure that melt curves showed only one peak per primer set. Samples that failed to amplify were excluded from further analysis. 2−ΔΔCt analysis was performed according to the method of Livak and Schmittgen [16]. Relative gene copy expression analysis was performed to determine the differences in expression between multiple genes being analyzed to compare across multiple times/conditions (as described in Supplementary Information).
Biofilm Staining and Confocal Microscopy
An overnight culture of S. epidermidis 1457 cells was rinsed, adjusted to a concentration of 5 × 104 cells/ml in PBS, and added to wells of a sterile 6-well polystyrene plate. The plate was then incubated at 37 °C for 1 h to allow cells to attach. The supernatant was then replaced with TSB or TSB + 1.5 M NaCl and plates were incubated for an additional 48 h. Biofilm samples were then removed with a cell scraper and washed with PBS. Non-specific binding was blocked by incubating the cell suspension in goat serum before antibody binding. Cell suspensions were then incubated in a 1:250 dilution of rabbit anti-rEmbp2588 antibody before being re-suspended in a 1:200× dilution of DyLight 488™-conjugated goat anti-rabbit secondary antibody solution (Pierce, Rockford, IL). SYTO 60 red fluorescent nucleic acid stain (Invitrogen, Carlsbad, CA) was used as counterstain. Cell suspensions were then filtered onto a 0.22-μm Cyclopore™ membrane (Whatman, Piscataway, NJ). Five random fields of each membrane were imaged using a 100× oil immersion lens on an epifluorescent microscope (Carl Zeiss Microimaging, Thornwood, NY). These images were analyzed by Image J software (NIH, Bethesda, MD) to determine % of cells staining for Embp from the total number of cells. Cells incubated with isotype matched rabbit anti-Pseudomonas aeruginosa as a negative control were also analyzed for each sample type to confirm specific binding of anti-rEmbp2588 to Embp.
Cultivation of biofilms under varying conditions of osmotic pressures was performed as above, but used 4-well Lab-Tek™ chamber slides (Thermo Scientific) instead of 6-well plates. After 48-h incubation, the supernatant was aspirated without disturbing the biofilms and the samples were incubated with Live/Dead BacLight™ bacterial stain (Invitrogen) for visualization. The medium chamber was then removed and biofilms samples were covered with coverslip for microscopy. All microscopic image acquisitions were carried out by Confocal Laser Scanning Microscopy (Nikon A1R-Si Confocal/RIRF imaging system, Melville, NY) with a 100× oil objective.
Biofilm Quantitation
COMSTAT software (Center for System Microbiology, Arne Heydorn, Oxford, England) [17], a powerful tool for classifying biofilm 3-D structure, was used to quantify the architecture of biofilms (defined by different COMSTAT-generated features of the three-dimensional biofilms). Biofilm thickness, roughness, and surface area to volume ratio were calculated for each of the biofilm conditions tested.
To determine the roughness coefficient, R, Eq. 1 was used:
| (1) |
where Lfi is the ith individual biofilm thickness measurement, L̄f is the mean biofilm thickness, and N is the number of thickness measurements. Surface area-to-volume ratio is equivalent to the bilofilm’s surface area divided by the volume of the biofilm. Three independent experiments and three image stacks for each condition were analyzed to acquire mean values for each condition.
Statistical Analysis
Unpaired Student’s T-tests were performed to determine significance between controls and sample types, as well as between osmotic conditions with and without NaCl supplementation.
Results and Discussion
Low Expression of embp Gene During Planktonic and Biofilm Cultures
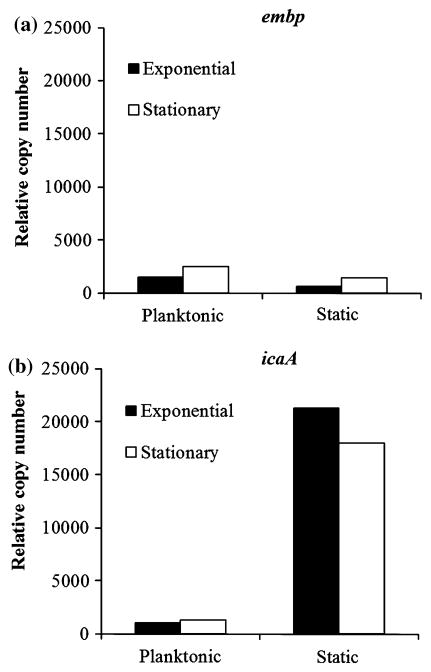
We hypothesized that embp expression levels would be increased during events (either adhesion or biofilm formation) that required Embp. Thus, we quantified embp expression in S. epidermidis over time in both planktonic and biofilm cultures and compared this to icaA expression, a gene known to be involved in biofilm formation but not in initial adhesion. The embp gene exhibited only low levels of expression throughout both planktonic and biofilm cultures (Fig. 1a). This was in stark contrast to the expression patterns of the icaA gene (Fig. 1b), which exhibited low levels of expression throughout planktonic growth but were dramatically increased in biofilm cultures.
Fig. 1.
Relative copy number for embp (a) and icaA (b) during exponential (4 h) and stationary (24 h) growth in planktonic and adherent cultures. Gene expression determined by qRT-PCR and normalized to guanylate kinase (gmk) housekeeping gene
At these later stages in biofilm cultures, quorum sensing and environmental stress responses would have been activated, as cell density had peaked and cells would have exhausted nutrient sources or dissolved oxygen. icaA gene expression is known to be regulated by both quorum sensing (LuxS) and stress response mechanisms (RsbU and σB) [5, 18]. Since expression of icaA, but not embp, is significantly different under biofilm forming conditions, it would appear that embp is not under the same control as icaA. Indeed, Yao et al. [19] reported no difference between embp expression in 24-hr planktonic cultures as compared to 24-hr adherent cultures in their genome wide analysis of S. epidermidis 1457.
Embp Protein Expression
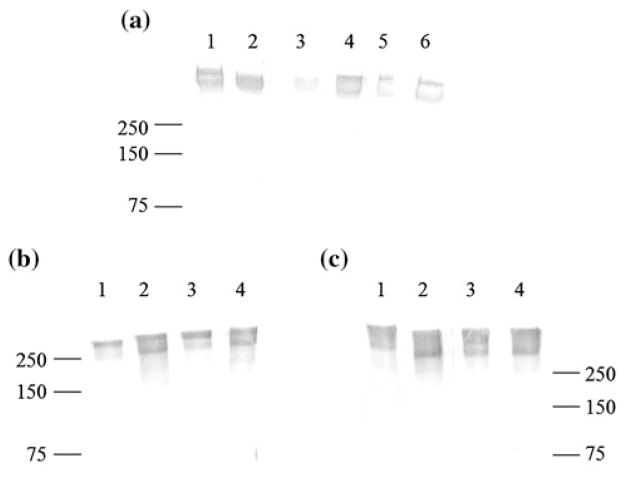
Western blots confirmed low level synthesis and production of Embp in cell wall extracts of various S. epidermidis strains (Fig. 2a). Biofilm-forming S. epidermidis (strain 1457), a non-biofilm forming cutaneous strain of S. epidermidis (ATCC 12228), and four separate clinical S. epidermidis strains isolated from catheter associated infections each produced Embp in TSB planktonic cultures although the clinical strains showed more variation in production. In addition, we confirmed that expression of Embp in S. epidermidis 1457 cells was not different over time compared to the CV2 strain (an isogenic agr- mutant) for planktonic or adherent biofilm cells at either 4 h (Fig. 2b) or 16 h (Fig. 2c) of culture. Samples of the only high MW (≥500 kDa) protein band visible in silver stained SDS-PAGE gels were sequenced at the University of Washington’s Mass Spectrometry Center and verified that the giant protein detected in Western Blots was Embp. Because agr plays an important role in cell survival during stationary growth, we would have expected a difference in Embp expression in any one or more of these culture conditions (early vs. late, planktonic vs. static biofilm growth) had Embp been controlled by the accessory regulator gene [20]. However, this was not the case, indicating that Embp is likely to be regulated by unrelated mechanisms.
Fig. 2.
Western Blot of rabbit-antiEmbp2588 binding to Embp expressed in various S. epidermidis strains and conditions. a Embp expression by S. epidermidis laboratory strains 1457 (lane 1) and 12228 (lane 2) plus four separate clinical isolates of S. epidermidis (lanes 3–6). b 4 h and c 16 h Embp expression by S. epidermidis strain 1457 (lanes 1 and 2) and agr- strain CV2 (lanes 3, 4) in planktonic cultures (lanes 1 and 3) versus adherent cultures (lanes 2 and 4). Molecular weight standards (in kDa) labeled for reference
Osmotic Pressure Effects on embp Gene Expression
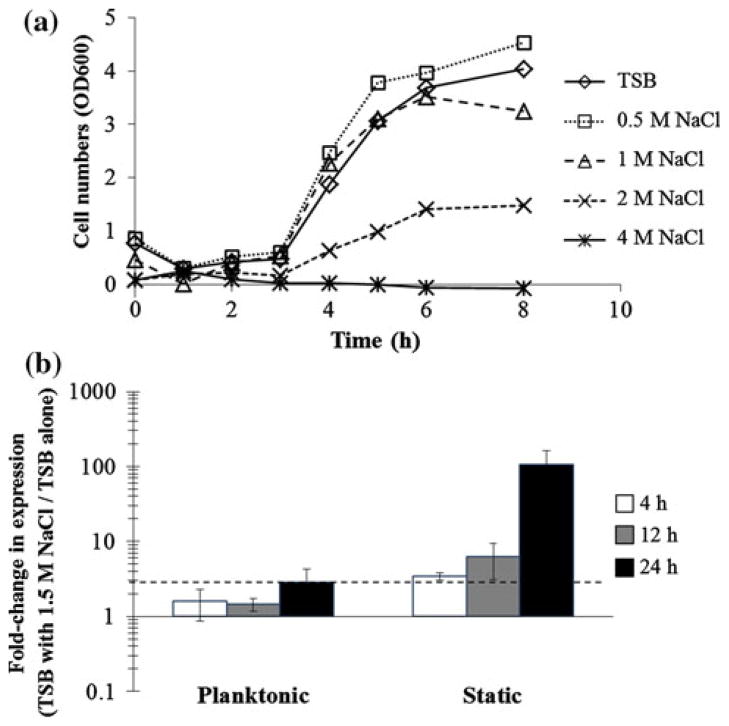
Since few changes in embp expression were seen comparing biofilm and planktonic cells, we hypothesized that S. epidermidis gene expression of embp, rather than being influenced by adhesion and/or biofilm formation would, instead, be altered under osmotic stresses. To determine the optimal concentration of NaCl for supplementation, a preliminary series of batch planktonic cultures were carried out at NaCl concentrations from 0 to 4 M NaCl (up to 23.2 % w/v NaCl). Figure 3a shows that S. epidermidis culture growth rates and extent were unaffected by NaCl concentrations ≤1 M (5.8 % w/v NaCl), but growth rate and extent were severely inhibited at 2 M NaCl and cells did not grow ≥4 M NaCl. Thus, subsequent osmotic stress experiments were carried out at 1.5 M NaCl to induce stress without severe growth inhibition.
Fig. 3.
S. epidermidis 1457 cell growth in TSB with increasing NaCl concentrations and relative embp gene expression by cells cultured with and without 1.5 M NaCl. a S. epidermidis 1457 planktonic growth in TSB alone or TSB with increasing NaCl. b Fold-change in embp expression from S. epidermidis cells grown in TSB with 1.5 M NaCl versus TSB only. Fold-change represents embp transcription in the presence of NaCl stress normalized expression levels in TSB alone. A twofold change was considered significant (dashed line). Error bars are standard error of the mean of 3 separate cultures and RNA preparations
To compare the gene expression of embp in the presence of osmotic stresses, cells were grown in planktonic and biofilm cultures using TSB supplemented with 1.5 M NaCl or TSB alone. As seen in Fig. 3b, planktonic cultures of S. epidermidis had similar embp expression in TSB and in TSB + 1.5 M NaCl. However, in biofilm cultures, the effect of NaCl on embp expression is pronounced with embp gene expression causing up-regulation throughout the course of the biofilm culture. The expression of embp is 3-times higher in 1.5 M NaCl supplemented TSB at 4 h growth, continues to increase to 6-times higher at 12 h, and reaches 100-times greater expression in osmotically stressed conditions at 24 h growth versus TSB alone. These experiments show that embp expression is induced by S. epidermidis cell stress under high salt concentrations, but that the effect is limited to adherent cells only.
Osmotic Pressure Effects on Embp Expression and Biofilm Formation
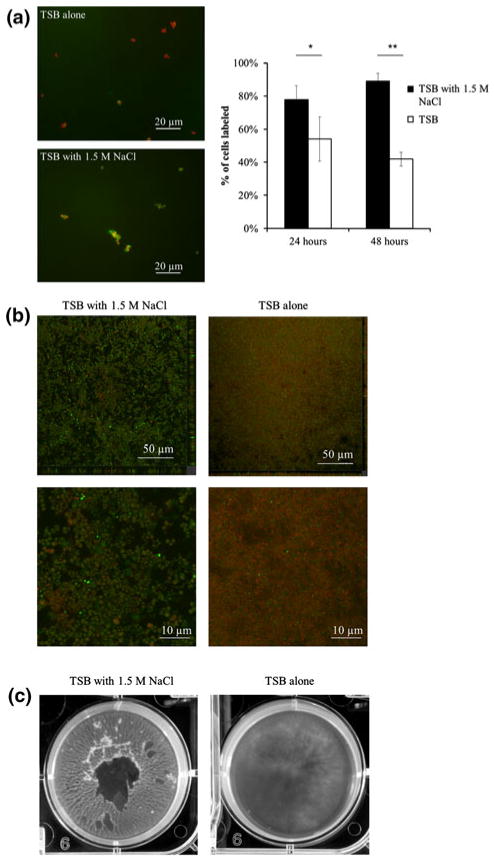
Because the addition of 1.5 M NaCl led to an upregulated gene expression in biofilm cultures, we then quantified the protein expression and morphological characteristics of the cells and biofilm. The addition of 1.5 M NaCl to TSB resulted in significantly increased cell wall-localized Embp as determined by immunofluorescence staining of adherent growth (Fig. 4a). This osmotic stress yielded 30 and 50 % increases in Embp protein expression at 24 and 48 h, respectively. Controls with isotype-matched antibodies against P. aeruginosa and with goat sera alone confirmed that binding was specific to Embp (data not shown). Confocal images also indicated a distinct difference in biofilm morphology due to the induced osmotic stress. As seen in Fig. 4b, confocal micrographs show biofilms to be thicker and less dense in TSB supplemented with 1.5 M NaCl compared to TSB only controls. Live/dead staining revealed many more live cells grown in 1.5 M NaCl + TSB culture after 24 h growth compared to the TSB only cultures. In addition, the size of cells grown under hyperosmotic stress was increased compared to the TSB alone. Table 2 displays the biofilm thickness, roughness, and surface area-to-volume ratios of S. epidermidis biofilms accumulated under the two osmotic pressure conditions. Compared to the control without NaCl, all three variables increased significantly under the osmotic stress induced by 1.5 M NaCl yielding a thicker, more heterogeneous biofilm.
Fig. 4.
Effects of high osmotic pressure conditions on biofilm formation. a Immunofluorescence microscopy of S. epidermidis cell Embp expression at 24 h (anti-rEmb25882 antibodies, green) and counterstained with SYTO9 (red) in TSB alone (Top) or TSB plus 1.5 M NaCl (bottom). Chart quantifies anti-Embp staining in TSB with 1.5 M NaCl (black bars) or TSB alone (white bars) at 24- and 48-h biofilm growth. Error bars standard deviations of the mean. *P <0.01, **P <0.00001. b Confocal micrograph of S. epidermidis biofilm formation in TSB with 1.5 M NaCl (left) versus TSB alone (right) with close ups of depicting cell morphology (below). Staining performed with BacLightLive/Dead stain. c Photograph of S. epidermidis biofilm formation at 48 h in TSB supplemented with 1.5 M NaCl (left) versus TSB alone (right)
Table 2.
Effect of osmotic pressure on S. epidermidis biofilm structure analysis by means of COMSTAT (mean ± standard deviation)
| Osmotic stress | Thickness (μm) | Roughness coefficient | Surface area to volume ratio (μm2 μm−3) |
|---|---|---|---|
| None | 6.16 ± 0.75 | 0.32 ± 0.06 | 0.75 ± 0.08 |
| 1.5 M NaCl | 13.16 ± 2.28 | 0.85 ± 0.12 | 3.01 ± 0.40 |
| P value | P <0.005 | P <0.0005 | P <0.0001 |
We also noticed marked macroscale differences in biofilms formed under high osmotic pressure conditions compared to the controls. Biofilms cultured in TSB supplemented with 1.5 M NaCl appeared much more filamentous and thicker than biofilm formed in TSB alone (Fig. 4c). Osmotically stressed biofilms appeared to adhere less to the polystyrene itself and were easily dislodged from the plate surface. Rather, these S. epidermidis cells were bound to each other by thick filaments. Individual cells grown under higher osmotic stress were larger than their control counterparts. This phenomena was also reported by Vijaranakul et al. [21] for S. aureus cells grown in high NaCl and KCl concentrations though not in those grown in the presence of glycerol or sucrose. These authors concluded that the high ionic strength of the salt solution and not the osmotic effect alone was responsible for the larger size of the cells. Kuroda et al. [10] postulated that the Ebh protein of S. aureus might be involved in tolerance to hyperosmotic pressures as seen by commensal organisms on human skin. Extreme fluctuations in water activity and salt concentrations are common due to interactions between the skin’s sebaceous secretions, and the external environment [10, 22]. Our results suggest that there is a role for Embp in cell survival under such osmotic stresses.
Conclusion
In this work, we have characterized the expression patterns of the embp gene in planktonic and biofilm cultures, and in response to changes in NaCl-induced osmotic stress. The embp gene and protein were found to be expressed by various clinical and laboratory S. epidermidis strains at low levels in both planktonic and biofilm cultures cultivated under normal growth conditions. Embp gene and protein regulation was not correlated to either icaA or agr expression, genes related to biofilm and stationary phase growth, respectively. However, under biofilm culture conditions of sustained hyperosmotic stress, embp gene and protein expression were highly upregulated, and both cell and biofilm morphology were substantially altered at the micro- and macroscales. Results suggest that embp is upregulated by increases in osmotic pressure and may be involved in protein mediated resistance to plasmolysis due to osmotic stresses, such as those found on the skin. This upregulation results in increased Embp production, cell size, and survival, and correlates with altered biofilm morphology indicating a possible role for commensal biofilm maintenance through cell:cell adhesion properties.
Supplementary Material
Acknowledgments
This work was funded by the National Institute of Health/National Institute of Allergy and Infectious Disease grant (1R03AI079461-01) and NSF/BES (0102139). Additional graduate support was provided by the National Science Foundation Graduate Research Fellowship Program.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00284-013-0316-7) contains supplementary material, which is available to authorized users.
References
- 1.Darouiche RO. Treatment of infections associated with surgical implants. N Engl J Med. 2004;350(14):1422–1429. doi: 10.1056/NEJMra035415350/14/1422. [DOI] [PubMed] [Google Scholar]
- 2.Qin Z, Ou Y, Yang L, Zhu Y, Tolker-Nielsen T, Molin S, Qu D. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology. 2007;153(Pt 7):2083–2092. doi: 10.1099/mic.0.2007/006031-0. [DOI] [PubMed] [Google Scholar]
- 3.Sun D, Accavitti MA, Bryers JD. Inhibition of biofilm formation by monoclonal antibodies against Staphylococcus epidermidis RP62A accumulation-associated protein. Clin Diagn Lab Immunol. 2005;12(1):93–100. doi: 10.1128/CDLI.12.1.93-100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christner M, Franke GC, Schommer NN, Wendt U, Wegert K, Pehle P, Kroll G, Schulze C, Buck F, Mack D, Aepfelbacher M, Rohde H. The giant extracellular matrix-binding protein of Staphylococcus epidermidis mediates biofilm accumulation and attachment to fibronectin. Mol Microbiol. 2010;75(1):187–207. doi: 10.1111/j.1365-2958.2009.06981.x. [DOI] [PubMed] [Google Scholar]
- 5.Knobloch JK, Bartscht K, Sabottke A, Rohde H, Feucht HH, Mack D. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J Bacteriol. 2001;183(8):2624–2633. doi: 10.1128/JB.183.8.2624-2633.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang R, Khan BA, Cheung GY, Bach TH, Jameson-Lee M, Kong KF, Queck SY, Otto M. Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J Clin Invest. 2011;121(1):238–248. doi: 10.1172/JCI42520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruckner R. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol Lett. 1997;151(1):1–8. doi: 10.1111/j.1574-6968.1997.tb10387.x. S0378-1097(97)00116-X. [DOI] [PubMed] [Google Scholar]
- 8.Manganelli R, van de Rijn I. Characterization of emb, a gene encoding the major adhesin of Streptococcus defectivus. Infect Immun. 1999;67(1):50–56. doi: 10.1128/iai.67.1.50-56.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke SR, Harris LG, Richards RG, Foster SJ. Analysis of Ebh, a 1.1-megadalton cell wall-associated fibronectin-binding protein of Staphylococcus aureus. Infect Immun. 2002;70(12):6680–6687. doi: 10.1128/IAI.70.12.6680-6687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuroda M, Tanaka Y, Aoki R, Shu D, Tsumoto K, Ohta T. Staphylococcus aureus giant protein Ebh is involved in tolerance to transient hyperosmotic pressure. Biochem Biophys Res Commun. 2008;374(2):237–241. doi: 10.1016/j.bbrc.2008.07.037. [DOI] [PubMed] [Google Scholar]
- 11.Linnes JC, Mikhova K, Bryers JD. Adhesion of Staphylococcus epidermidis to biomaterials is inhibited by fibronectin and albumin. J Biomater Res A. 2012;100(8):1990–1997. doi: 10.1002/jbm.a.34036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dexter SJ, Pearson RG, Davies MC, Camara M, Shakesheff KM. A comparison of the adhesion of mammalian cells and Staphylococcus epidermidis on fibronectin-modified polymer surfaces. J Biomed Mater Res. 2001;56(2):222–227. doi: 10.1002/1097-4636(200108)56:2<222::aid-jbm1087>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 13.Mack D, Siemssen N, Laufs R. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect Immun. 1992;60(5):2048–2057. doi: 10.1128/iai.60.5.2048-2057.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vuong C, Gerke C, Somerville GA, Fischer ER, Otto M. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J Infect Dis. 2003;188(5):706–718. doi: 10.1086/377239. [DOI] [PubMed] [Google Scholar]
- 15.Vandecasteele SJ, Peetermans WE, Merckx R, Van Eldere J. Expression of biofilm-associated genes in Staphylococcus epidermidis during in vitro and in vivo foreign body infections. J Infect Dis. 2003;188(5):730–737. doi: 10.1086/377452. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth. 2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146(Pt 10):2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 18.Xu L, Li H, Vuong C, Vadyvaloo V, Wang J, Yao Y, Otto M, Gao Q. Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect Immun. 2006;74(1):488–496. doi: 10.1128/IAI.74.1.488-496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao Y, Sturdevant DE, Otto M. Genomewide analysis of gene expression in Staphylococcus epidermidis biofilms: insights into the pathophysiology of S. epidermidis biofilms and the role of phenol-soluble modulins in formation of biofilms. J Infect Dis. 2005;191(2):289–298. doi: 10.1086/426945. [DOI] [PubMed] [Google Scholar]
- 20.Yao Y, Vuong C, Kocianova S, Villaruz AE, Lai Y, Sturdevant DE, Otto M. Characterization of the Staphylococcus epidermidis accessory-gene regulator response: quorum-sensing regulation of resistance to human innate host defense. J Infect Dis. 2006;193(6):841–848. doi: 10.1086/500246. [DOI] [PubMed] [Google Scholar]
- 21.Vijaranakul U, Nadakavukaren MJ, de Jonge BL, Wilkinson BJ, Jayaswal RK. Increased cell size and shortened peptidoglycan interpeptide bridge of NaCl-stressed Staphylococcus aureus and their reversal by glycine betaine. J Bacteriol. 1995;177(17):5116–5121. doi: 10.1128/jb.177.17.5116-5121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willson M. The indigenous microbiota of the skin. In: Willson M, editor. Bacteriology of humans: an ecological perspective. Blackwell; Oxford: 2008. pp. 56–94. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.