Abstract
PURPOSE
Recent studies have identified genetic variants associated with both increased serum PSA concentrations and prostate cancer risk, raising the possibility of diagnostic bias. By correcting for the effects of these variants on PSA levels, it may be possible to create a personalized PSA cutoff to more accurately identify individuals for whom biopsy is recommended. We therefore determined how many men would continue to meet common biopsy criteria after genetic correction of their measured PSA concentrations.
MATERIALS AND METHODS
The genotypes of 4 single nucleotide polymorphisms (SNPs) previously associated with serum PSA levels (rs2736098, rs10788160, rs11067228, and rs17632542) were determined in 964 healthy Caucasian volunteers without prostate cancer. Genetic correction of the PSA was performed by dividing an individual's PSA value by his combined genetic risk. Analyses were used to compare the percentage of men that would meet commonly used biopsy thresholds (≥2.5 or ≥4.0 ng/mL) before and after genetic correction.
RESULTS
Genetic correction of serum PSA results was associated with a significantly decreased frequency of men meeting biopsy thresholds. Genetic correction could lead to a 15% and 20% relative reduction in the total number of biopsies using a biopsy threshold of ≥2.5 or ≥4.0 ng/mL, respectively. In addition, genetic correction could result in an 18–22% reduction in the number of potentially unnecessary biopsies and a 3% decrease in potentially delayed diagnoses.
CONCLUSIONS
Our results suggest that 4 SNPs can be used to adjust a man's measured PSA concentration and potentially delay or prevent unnecessary prostate biopsies in Caucasian men.
Keywords: prostate cancer, genetic variants, PSA, prostate biopsy
Introduction
Prostate cancer is the most frequent non-cutaneous cancer in men, as well as the second-leading cause of cancer death in the United States1. Despite evidence showing a decline in prostate cancer-specific mortality in the prostate specific antigen (PSA) screening era2, 3, the routine measurement of serum PSA as a screening tool remains controversial. Although PSA is highly prostate-specific, it is not cancer-specific. For example, serum PSA levels may be increased with urinary tract infections, urinary tract instrumentation, and/or other benign conditions, including benign prostatic hyperplasia4. These conditions may confound the interpretation of PSA results and lead to many potentially unnecessary prostate biopsies. These confounders may help to explain why roughly one-third of men with serum PSA levels above 10 ng/mL have no evidence of prostate cancer at biopsy5. Furthermore, many conditions (e.g., obesity, 5-alpha-reductase inhibitors), may falsely lower serum PSA values below biopsy threshold and result in a delay of prostate cancer diagnosis. Similarly, not all prostate cancers produce elevated serum PSA concentrations6.
The traditional single-cutoff PSA screening test can be improved by using other clinical features to determine whether a man should be further evaluated with a prostate biopsy. For example, the American Urological Association Best Practice Statement recommends considering factors such as PSA kinetics (e.g. velocity), patient age, ethnicity, and family history of prostate cancer7. We hypothesized that genetic factors that are associated with increases or decreases in serum PSA concentrations may provide improved specificity for early detection and aid in determining whether a prostate biopsy is warranted.
Genome-wide association studies (GWAS) and linkage analyses have identified genetic variants, called single nucleotide polymorphisms (SNPs), that are associated with an increased risk of prostate cancer8. More than 40 SNPs have been identified that appear to have a cumulative association with prostate cancer risk9–11.
It has been estimated that 40–45% of the inter-individual variability in serum PSA concentrations can be explained by genetic factors12, 13. Recent studies have demonstrated that SNPs in or near the gene that encodes PSA [e.g. kallikrein-related peptidase 3 (KLK3)] can influence prostate cancer detection14–16. For example, a SNP near KLK3 was associated with elevated PSA concentrations in men without prostate cancer16. Similarly, the results of a recent GWAS found SNPs associated with serum PSA levels in or near the following six genes: telomerase reverse transcriptase (TERT; chromosome 5p15.33, SNP rs2736098); β microseminoprotein (MSMB; chromosome 10q11, rs10993994); fibroblast growth factor receptor 2 (FGFR2; chromosome 10q26, rs10788160); T-box transcription factor (TBX3; chromosome 12q24, rs11067228); hepatocyte nuclear factor 1B (HNF1B; chromosome 17q12, rs4430796); and KLK3 (chromosome19q13.33, rs17632542)17. Interestingly, 4 of these SNPs (hereafter referred to as “PSA-SNPs”) were found to be principally associated with serum PSA concentrations.
In an Icelandic cohort, Gudmundsson et al. assessed whether the presence of the 4 PSA-SNPs could be used to genetically correct a man’s measured serum PSA. These genetically corrected PSA values significantly improved the performance of PSA as a screening tool (area under the curve; AUC=73.2%) compared to unadjusted values (AUC=70.9%). Similarly, a prior study from the Baltimore Longitudinal Study of Aging showed that the risk of prostate cancer on biopsy differed based on genotype for PSA-associated SNPs18.
The objective of the current study was to determine the effect of genetic correction of measured serum PSA results using the 4 PSA-SNPs in a U.S. Caucasian population. In addition, we sought to determine whether correction of a patient’s serum PSA concentration based on the presence of 4 PSA-SNPs could significantly lower the frequency of men who meet common serum PSA thresholds for biopsy.
Patients and Methods
Our study cohort consisted of 964 healthy Caucasian volunteers who enrolled between 2003 and 200919. The study was approved by Northwestern University’s Institutional Review Board, and all participants provided written informed consent as well as a blood sample used for genotype analysis. Clinical and pathologic features were recorded for all participants, including serum total PSA concentrations, first-degree family history of prostate cancer, and number of prostate biopsies. DNA was extracted from whole blood at deCODE® Genetics Inc., in Reykjavik, Iceland. Each sample was genotyped for the 4 PSA-SNPs as previously described17.
Genetic correction was performed by dividing the measured PSA concentration by a man’s combined genetic risk factor, as previously described17. Briefly, we calculated the genetic factor associated with any individual PSA-SNP allele using classical linear regression analysis. The relative genotypic effect on serum PSA concentrations for each SNP was calculated under a log additive model (i.e. risk/effect for heterozygous carriers is = r and the risk/effect for homozygous risk/effect-allele carriers is r2). The combined genotypic effect (using more than one SNP) was also determined by including all of the genetic variants into an additive logistic regression model. The combined genetic factor was determined in reference to the median frequency of the PSA-SNP alleles in the population. After genetic correction was applied, statistical analyses were used to compare the percentage of men who would meet commonly used PSA thresholds, ≥2.5 or ≥4.0 ng/ml. A p-value of <0.05 was considered significant. All statistical analyses were performed using SAS® 9.2.
Results
The baseline characteristics of the cohort of 964 Caucasian volunteers, some of whom may have had prostate cancer at the study initiation, are shown in Table 1. The genotypes of the 4 SNPs previously associated with PSA expression levels (rs2736098, rs10788160, rs11067228, and rs17632542) were determined. Based upon the distribution of genotype values, “controls” carried a median of 4 PSA-SNP alleles. We next genetically corrected the measured PSA values relative to the median in the population.
Table 1.
Demographics and Serum PSA Concentrations of 964 Healthy Caucasian Volunteers Without Prostate Cancer.
| Total men | 964 |
| European Ancestry | 100 |
| Median Age (years) | 57 |
| Age (years) in Quartiles | n (% of total) |
| ≤50 | 203 (21.1%) |
| 51–57 | 263 (27.3%) |
| 58–65 | 263 (27.3%) |
| ≥66 | 235 (24.4%) |
| Median Serum PSA (ng/ml) | 0.8 |
| Serum PSA Concentration | n (% of total) |
| 0.0 – < 2.5 ng/mL | 869 (90.2%) |
| 2.5 – 4.0 ng/mL | 40 (4.2%) |
| 4.0 – 10.0 ng/mL | 50 (5.2%) |
| >10.0 ng/mL | 5 (0.5%) |
| Number of Men Undergoing Prostate Biopsies (to Date) | 59 (6.1%) |
| Number of Men Diagnosed with Prostate Cancer After Study Initiation (to Date) | 8 (0.8%) |
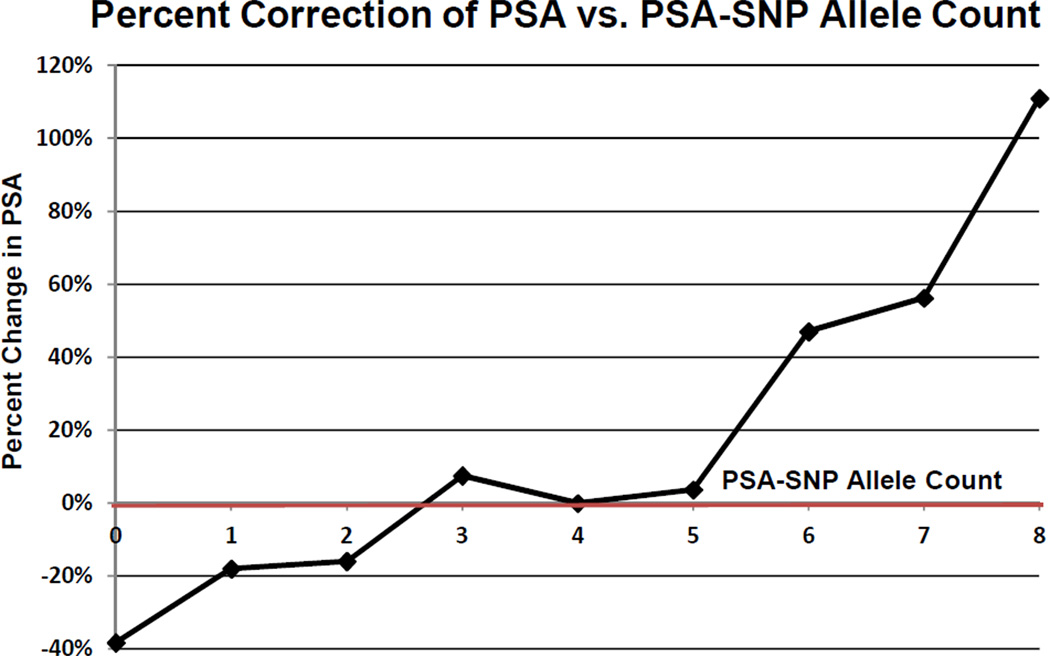
Figure 1 demonstrates the percent change in PSA after genetic correction compared with the PSA-SNP allele count. For example, if a man was found to be a carrier of all 8 PSA-SNP alleles, then his PSA level would be 110% higher than the population median. His PSA would have to be decreased by this percentage to correct for the presence of all 8 PSA-SNP alleles. In contrast, if another man was found to be a carrier of none of the PSA SNP alleles, then his PSA would be 38% lower than the population. His PSA would have to be increased by this percentage to correct for the absence of the PSA SNP alleles.
Figure 1. Percent Change in Serum PSA Concentrations Based on PSA-SNP Allele Count Relative to Median in the Population.
*It should be noted that not all of the PSA SNPs influence PSA expression equally. Therefore, the allele count represents the average percent change in PSA (correction) based upon all combinations of PSA-SNP alleles for that count.
Next, we compared the frequency of men who would meet common biopsy threshold criteria before and after genetic correction of the measured PSA values (Table 2). Prior to genetic correction, 9.7% of men had a PSA value ≥2.5ng/mL and thus may have been eligible for a prostate biopsy. In comparison, only 8.2% of men met this biopsy threshold criterion after genetic correction for the PSA SNPs; representing a significant 15.5% relative risk reduction for biopsy (Table 2, p<0.0001). Interestingly, there was an 18.3% reduction in the number of men who initially had a measured serum PSA above biopsy criteria but fell below it after genetic correction (i.e. potentially unnecessary biopsies) and a 3.4% reduction in the number of men who had a measured serum PSA below biopsy criteria but went above it after genetic correction (i.e. potentially delayed biopsies). Based upon these values, one of every 57 men undergoing PSA screening could be spared a biopsy using genetic testing at a threshold of 2.5ng/ml. Similarly, a significantly decreased number of subjects met a biopsy threshold criteria ≥ 4.0 ng/ml after genetic adjustment (5.3% corrected vs. 6.6% uncorrected, 19.7% relative risk reduction, p<0.0001; Table 2). This was associated with a 21.8% and 3.3% reduction in the number of men who would have undergone potentially unnecessary and delayed biopsies, respectively (Table 2). Using this biopsy threshold of 4.0ng/ml, one of every 69 Caucasian men undergoing PSA screening could be spared a potentially unnecessary biopsy using genetic testing.
Table 2.
Number of Men Meeting Biopsy Criteria (PSA ≥2.5 ng/mL or PSA ≥4.0 ng/mL) Before and After Genetic Correction of Serum PSA Concentrations.
| A. Biopsy Threshold: PSA ≥2.5 ng/mL | ||||
|---|---|---|---|---|
| Corrected PSA Concentration; n= (%) | ||||
| PSA <2.5 ng/mL | PSA ≥2.5 ng/mL | Total | ||
| Measured Serum PSA Concentration; n= (%) |
PSA <2.5 ng/mL | 868 (90.0) | 3 (0.3) | 871 (90.3) |
| PSA ≥2.5 ng/mL | 17 (1.8) | 76 (7.9) | 93 (9.7) | |
| Total | 885 (91.8) | 79 (8.2) | ||
| B. Biopsy Threshold: PSA ≥4.0 ng/mL | ||||
|---|---|---|---|---|
| Corrected PSA Concentration; n= (%) | ||||
| PSA <4.0 ng/mL | PSA ≥4.0 ng/mL | Total | ||
| Measured Serum PSA Concentration; n= (%) |
PSA <4.0 ng/mL | 897 (93.3) | 3 (0.3) | 900 (93.6) |
| PSA ≥4.0 ng/mL | 14 (1.5) | 50 (5.1) | 64 (6.6) | |
| Total | 911 (94.6) | 53 (5.3) | ||
If we limited our population to men with an unadjusted PSA ≥2.5 ng/mL, genetic correction could possibly prevent one of every 6 biopsies using a biopsy threshold of ≥ 2.5 ng/mL. Similarly, limiting our cohort to men with an unadjusted PSA ≥4.0 ng/mL, genetic correction also could prevent one of every 6 biopsies using a threshold of ≥ 4.0 ng/mL (Table 3).
Table 3.
Number of Men With Initial Uncorrected Serum PSA ≥2.5 ng/mL or ≥4.0 ng/mL Meeting Biopsy Criteria (PSA ≥2.5 ng/mL or ≥4.0 ng/mL) Before and After Genetic Correction of Serum PSA Concentrations
| A. Biopsy Threshold ≥ 2.5 ng/ml | ||||
|---|---|---|---|---|
| After Genetic Correction; n= (%) | ||||
| PSA <2.5 ng/ml | PSA ≥2.5 ng/ml | Total | ||
| Measured PSA Level ≥2.5 ng/ml |
16 (17.2) | 77 (82.8) | 93 (100.0) | |
| B. Biopsy Threshold ≥ 4.0 ng/ml | |||
|---|---|---|---|
| After Genetic Correction | |||
| PSA <4.0 ng/ml | PSA ≥4.0 ng/ml | Total | |
| Measured PSA Level ≥4.0 ng/ml |
11 (17.2) | 53 (82.8) | 64 (100.0) |
A total of 59 men in our cohort have undergone prostate biopsy and 15 have been diagnosed with prostate cancer based upon the initial PSA or subsequent screening PSA values (Table 1). Interestingly, amongst 17 men who fluctuated below the cutoff of 2.5ng/ml after genetic correction, 13 have been biopsied and none was found to have prostate cancer. In comparison, of the 76 men whose PSA remained above the threshold, 43 have been biopsied and 8 have biopsy proven cancer to date. Similarly, amongst 14 men who fluctuated below the 4.0ng/ml cutoff after genetic correction, 7 have been biopsied and none was found to have prostate cancer. In comparison, of the 50 men whose PSA remained above the 4.0ng/ml threshold, 32 have been biopsied and 7 have been diagnosed with cancer to date. None of the men whose PSA went above the threshold has been diagnosed with cancer as yet.
Discussion
The high survival rate of men with prostate cancer is largely a reflection of screening with serum PSA.20 However, support for the widespread use of PSA is questioned because of its limited specificity. This is due to other non-malignant conditions (e.g. benign prostatic hyperplasia, prostatitis, etc.) causing elevated serum PSA concentrations. In addition, recent studies suggest that heritable factors significantly contribute to variation in PSA expression17, 21–24. Specifically, men who carry an increased number of the 4 PSA-SNP alleles express more PSA and are considered to be “genetically high PSA producers”, while men who carry decreased numbers of the PSA-SNPs are considered to be “genetically lower PSA producers.” Thus, the number of PSA-SNP alleles carried by an individual directly affects his measured serum PSA concentrations. The present study demonstrates that genetic correction for the PSA-SNPs could decrease the number of men with an “abnormal” PSA based on commonly used biopsy thresholds. Our data suggest that the traditional single-cutoff PSA screening (e.g. ≥2.5 or ≥4.0 ng/mL) might be improved by genetic correction. If our results are validated, adjustment for the 4 PSA-SNPs could potentially prevent up to 15–20% of prostate biopsies. Since it has been estimated that more than 1 million biopsies are performed in the US annually25, this could translate into 150,000 to 200,000 potentially unnecessary biopsies every year. Because some prostate biopsy procedures result in infection, sepsis, and hospitalizations26, routine implementation of this genetic adjustment could have a substantial health impact. As sequencing technology advances, the associated cost continues to decrease. Assuming that a diagnostic laboratory has the available equipment, supplies and trained personnel, a panel of 4 SNPs would cost less than 60 cents. Therefore, although approximately 60 men need to be screened with both PSA and genetic testing to prevent an unnecessary biopsy, the cost to benefit ratio appears favorable.
Interestingly, genetic correction of the measured PSA result could decrease the number of potentially unnecessary biopsies by approximately 18–22%. In other words, genetic correction could significantly decrease the number of biopsies being performed in genetically high PSA producers without prostate cancer. Genetic correction also decreased the number of potentially delayed biopsies by approximately 3%. This would correspond to a significant decrease in the number of biopsies in genetically low PSA producers.
Limitations of our study include examining the relatively small study population of Caucasian men, and additional investigation is necessary in other ethnic groups. In addition, not all of the healthy volunteers who initially met biopsy criteria have been biopsied to date. Therefore, true confirmation of re-classification after genetic correction as potentially unnecessary or delayed biopsies remains to be determined prospectively. However, as mentioned in the results section, there is a significantly higher rate of prostate cancer diagnosis amongst men whose PSA was above the biopsy threshold and did not fluctuate after genetic correction compared to those who fell below after correction. Finally, the influence of genetic correction on clinical outcomes requires further prospective study in a large independent cohort.
Conclusion
Our results confirm that a personalized PSA value can be obtained by adjusting for the presence of 4 genetic variants that influence serum PSA concentrations. If confirmed, this approach could potentially be used to tailor PSA screening, possibly reduce unnecessary biopsies, and avoid delay in performing necessary biopsies.
Acknowledgments
Supported in part by the Urological Research Foundation, Prostate SPORE Grant (P50CA90386-05S2), and the Robert H. Lurie Comprehensive Cancer Center grant (P30 CA60553). SL is supported by The Louis Feil Charitable Lead Trust.
References
- 1.Jemal A, Siegel R, Xu J, et al. Cancer statistics, 2010. CA Cancer J Clin. 60:277. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Bartsch G, Horninger W, Klocker H, et al. Tyrol Prostate Cancer Demonstration Project: early detection, treatment, outcome, incidence and mortality. BJU Int. 2008;101:809. doi: 10.1111/j.1464-410X.2008.07502.x. [DOI] [PubMed] [Google Scholar]
- 3.Schroder FH, Hugosson J, Roobol MJ, et al. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 4.Nadler RB, Humphrey PA, Smith DS, et al. Effect of inflammation and benign prostatic hyperplasia on elevated serum prostate specific antigen levels. J Urol. 1995;154:407. doi: 10.1097/00005392-199508000-00023. [DOI] [PubMed] [Google Scholar]
- 5.Aus G, Damber JE, Khatami A, et al. Individualized screening interval for prostate cancer based on prostate-specific antigen level: results of a prospective, randomized, population-based study. Arch Intern Med. 2005;165:1857. doi: 10.1001/archinte.165.16.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGuire BB, Helfand BT, Loeb S, et al. Outcomes in patients with Gleason score 8–10 prostate cancer: relation to preoperative PSA level. BJU Int. doi: 10.1111/j.1464-410X.2011.10628.x. [DOI] [PubMed] [Google Scholar]
- 7.Greene KL, Albertsen PC, Babaian RJ, et al. Prostate specific antigen best practice statement: 2009 update. J Urol. 2009;182:2232. doi: 10.1016/j.juro.2009.07.093. [DOI] [PubMed] [Google Scholar]
- 8.Ishak MB, Giri VN. A systematic review of replication studies of prostate cancer susceptibility genetic variants in high-risk men originally identified from genome-wide association studies. Cancer Epidemiol Biomarkers Prev. 20:1599. doi: 10.1158/1055-9965.EPI-11-0312. [DOI] [PubMed] [Google Scholar]
- 9.Helfand BT, Fought AJ, Loeb S, et al. Genetic prostate cancer risk assessment: common variants in 9 genomic regions are associated with cumulative risk. J Urol. 184:501. doi: 10.1016/j.juro.2010.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun J, Chang BL, Isaacs SD, et al. Cumulative effect of five genetic variants on prostate cancer risk in multiple study populations. Prostate. 2008;68:1257. doi: 10.1002/pros.20793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng SL, Sun J, Wiklund F, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358:910. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 12.Bansal A, Murray DK, Wu JT, et al. Heritability of prostate-specific antigen and relationship with zonal prostate volumes in aging twins. J Clin Endocrinol Metab. 2000;85:1272. doi: 10.1210/jcem.85.3.6399. [DOI] [PubMed] [Google Scholar]
- 13.Pilia G, Chen WM, Scuteri A, et al. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2006;2:e132. doi: 10.1371/journal.pgen.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kote-Jarai Z, Amin Al Olama A, Leongamornlert D, et al. Identification of a novel prostate cancer susceptibility variant in the KLK3 gene transcript. Hum Genet. 129:687. doi: 10.1007/s00439-011-0981-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Penney KL, Schumacher FR, Kraft P, et al. Association of KLK3 (PSA) genetic variants with prostate cancer risk and PSA levels. Carcinogenesis. 32:853. doi: 10.1093/carcin/bgr050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eeles RA, Kote-Jarai Z, Giles GG, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 17.Gudmundsson J, Besenbacher S, Sulem P, et al. Genetic correction of PSA values using sequence variants associated with PSA levels. Sci Transl Med. 2:62ra92. doi: 10.1126/scitranslmed.3001513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loeb S, Carter HB, Walsh PC, et al. Single nucleotide polymorphisms and the likelihood of prostate cancer at a given prostate specific antigen level. J Urol. 2009;182:101. doi: 10.1016/j.juro.2009.02.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le BV, Griffin CR, Loeb S, et al. [-2]Proenzyme prostate specific antigen is more accurate than total and free prostate specific antigen in differentiating prostate cancer from benign disease in a prospective prostate cancer screening study. J Urol. 183:1355. doi: 10.1016/j.juro.2009.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schroder FH, Hugosson J, Roobol MJ, et al. Prostate-cancer mortality at 11 years of follow-up. N Engl J Med. 2012;366:981. doi: 10.1056/NEJMoa1113135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savblom C, Giwercman A, Malm J, et al. Association between polymorphisms in the prostate-specific antigen (PSA) promoter and release of PSA. Int J Androl. 2009;32:479. doi: 10.1111/j.1365-2605.2008.00882.x. [DOI] [PubMed] [Google Scholar]
- 22.Pal P, Xi H, Sun G, et al. Tagging SNPs in the kallikrein genes 3 and 2 on 19q13 and their associations with prostate cancer in men of European origin. Hum Genet. 2007;122:251. doi: 10.1007/s00439-007-0394-3. [DOI] [PubMed] [Google Scholar]
- 23.Chiang CH, Chen KK, Chang LS, et al. The impact of polymorphism on prostate specific antigen gene on the risk, tumor volume and pathological stage of prostate cancer. J Urol. 2004;171:1529. doi: 10.1097/01.ju.0000116538.15995.93. [DOI] [PubMed] [Google Scholar]
- 24.Cramer SD, Chang BL, Rao A, et al. Association between genetic polymorphisms in the prostate-specific antigen gene promoter and serum prostate-specific antigen levels. J Natl Cancer Inst. 2003;95:1044. doi: 10.1093/jnci/95.14.1044. [DOI] [PubMed] [Google Scholar]
- 25.Bostwick DG, Meiers I. Prostate biopsy and optimization of cancer yield. Eur Urol. 2006;49:415. doi: 10.1016/j.eururo.2005.12.052. [DOI] [PubMed] [Google Scholar]
- 26.Loeb S, van den Heuvel S, Zhu X, et al. Infectious Complications and Hospital Admissions After Prostate Biopsy in a European Randomized Trial. Eur Urol. doi: 10.1016/j.eururo.2011.12.058. [DOI] [PubMed] [Google Scholar]