Abstract
Background and Aims
Gloxinia (Sinningia speciosa) is a popular commercial plant for its attractive and colourful flowers. However, the genetic mechanism of flowering time regulation in gloxinia is largely unknown. Recent studies on model plants have elucidated that miR159 acts as a negative regulator of floral transition in short-day photoperiods. The aim of this study was to investigate whether genetic modification of miR159 expression can offer an effective approach for regulation of flowering characteristics in gloxinia.
Methods
Transgenic gloxinia plants were generated that over-express or suppress miR159 by means an efficient Agrobacterium-mediated transformation system in order to study the effect of miR159 on flowering time. In addition, the full-length cDNA of gloxinia GAMYB (SsGAMYB) was also cloned, and was verified to be a target of miR159 by modified RNA ligase-mediated 5′ rapid amplification of cDNA ends.
Key Results
Transgenic gloxinia plants that over-express or suppress miR159 exhibited significantly late or early flowering, respectively. During flower development, the expression level of miR159 was negatively correlated with SsGAMYB in gloxinia. MiR159-mediated SsGAMYB expression affected the expression levels of SsLEAFY (SsLFY) and three MADS-box genes (SsAP1, SsAP3 and SsAG), which regulated floral transition downstream of GAMYB. In addition, suppression of miR159 caused a conversion of petals and sepals in a few transgenic plants.
Conclusions
miR159-regulated GAMYB expression is an effective pathway of flowering time control in gloxinia. Transgenic manipulation of miR159 can be used as an applicable strategy to regulate flowering time in commercial ornamental plants.
Keywords: Flowering time; microRNA, miR159; GAMYB; ornamental; gloxinia; Sinningia speciosa
INTRODUCTION
Flowering time is a key trait in the breeding of commercial plants (Jung and Muller, 2009). Being able to induce flowering when desired could fit well with the need to get more flowers or fruits. For example, the ability to control flowering time can satisfy the increase in demand for fresh flowers during holidays, and can also meet the need for ornamental flowers at different seasons or all year round. Therefore, it will be of economic benefit to establish an applicable floral regulation system in ornamental plants.
Genetic modification by transformation of key flowering-related genes seems to offer an effective approach for manipulating flowering characteristics in plants (Samach, 2011; Srikanth and Schmid, 2011). Basic studies on the timing of flowering in model species have shed light on a complicated network of floral transition pathways (Levy and Dean, 1998; Blazquez, 2000). The photoperiod, gibberellin, ambient-temperature, light-quality and ageing pathways are currently implicated in promoting floral transition (Blazquez, 2000; Henderson and Dean, 2004). The five pathways constitute a complex gene-network, as a precise mechanism of regulating flowering time. The ultimate targets for the flowering-time pathways are the floral meristem identity genes, whose activity confers floral identity to newly emerging primordia (Blazquez, 2000). LEAFY (LFY) is a key player for integrating signals from all pathways, especially in the specification of floral meristem identity. Plants with severe LFY mutations fail to initiate floral meristems and instead produce secondary inflorescence branches (Weigel et al., 1992). Over-expression of exogenous LFY has been reported to cause early flowering in poplar (Weigel and Nilsson, 1995), rice (He et al., 2000) and gloxinia (Zhang et al., 2008). Taken together, current knowledge of flowering time control has been exploited through either over-expression or suppression of gene activity (Jung and Muller, 2009). However, it is not feasible to suppress a floral gene by RNA interference if its sequence is unavailable, especially in most ornamental plants with unsequenced genomes.
Recently, more and more miRNAs have been identified by bioinformatics prediction or advanced sequencing technologies. MiRNAs play pivotal roles in various aspects of plant development (Rubio-Somoza et al., 2009). Previous studies revealed that the majority of the targets of plant miRNAs are important transcription factors, such as APETALA2 (AP2), GAMYB, CUPSHAPED COTYLEDONS1 (CUC1) and CUC2, PHAVOLUTA (PHV) and PHABULOSA (PHB), which have key roles in meristem identity, hormone signalling, cell division, and formation of organ separation and polarity (Carrington and Ambros, 2003; Reyes and Chua, 2007). MiR159 has been identified to be one of eight highly conserved miRNA families in the common ancestor of all embryophytes (Cuperus et al., 2011). Targets of the miR159 family have been revealed to be the highly conserved GAMYB gene family, which play an important role in aleurone cells, flower and seed development as well as vegetative tissues growth (Allen et al., 2007). Over-expressing miR159 in transgenic arabidopsis causes delayed flowering and anther defects through down-regulation of AtMYB33 under short-day (SD) conditions (Achard et al., 2004). Similarly in rice, over-expression of miR159 results in delayed head formation mainly due to decreases in OsGAMYB and OsGAMYBL1 (Tsuji et al., 2006). In contrast, another piece of evidence showed that over-expressed miR159, down-regulating AtMYB101 but not MYB33 and MYB65, did not alter flowering time under the long-day photoperiod (Schwab et al., 2005). It also reported that a mir159ab double mutant did not have an altered flowering time under long-day conditions but delayed flowering under SD conditions (Alonso-Peral et al., 2010). In addition, this mir159ab mutant exhibited stunted growth with rounded/curled leaves (Allen et al., 2007; Alonso-Peral et al., 2010). These phenotypes of reduced stature and curled leaves were similar in MIM159 transgenic plants, which were used to reduce endogenous miR159 activity (Todesco et al., 2010). Based on these observations, the biological role of miR159 appears to be determined by a complex mechanism in plant growth development. Considering different reports about roles of miR159 and GAMYB in the promotion of floral transition, it seems to be necessary to investigate miR159-mediated GAMYB functions in flower development in a wider range of plant species. To date, it is largely unknown whether miR159 acts as a regulator of flowering time in ornamental plants.
Gloxinia (Sinningia speciosa), belonging to Gesneriaceae, is cultivated as a popular houseplant for its large and colourful flowers. Our previous study established an effective Agrobacterium-mediated transformation system in gloxinia and showed that over-expression of exogenous LFY promotes early flowering (Zhang et al., 2008). In the present study, we generated transgenic gloxinia that over-expresses or suppresses miR159, leading to late or early flowering, respectively. Meanwhile, a full-length cDNA of gloxinia GAMYB (SsGAMYB) was cloned and identified to be a target of miR159. In addition, altered expression levels of miR159a caused up- or down-regulation of SsGAMYB during flower development, resulting in changed transcript levels of endogenous LFY and MADS-box genes in transgenic lines. Our results suggest that miR159-mediated GAMYB expression takes part in the control of flowering time in gloxinia. Our aim is to establish an applicable floral regulation system that will allow flowering to be either accelerated or delayed, depending on the demand for ornamental plants.
MATERIALS AND METHODS
Plant materials and growth conditions
In our study, all the materials were derived from tissue culture, including wild-type and transgenic plants. For tissue culture, leaves of Sinningia speciosa were cut aseptically from seedlings and maintained in Murashige and Skoog (MS) culture medium, pH 5·8. The cultures were grown in a chamber maintained in SD conditions (8 h light/16 h dark; 28·8 µmol m−2 s−1) at 24 °C. For plant regeneration, we cut the same size leaves from tissue cultures, and put them on root-inducing medium for 1 month. Then, plantlets were transferred and grown in perlite–peat mix in a greenhouse maintained at a temperature of 24 ± 1 °C (SD, 8 h light/16 h dark) and humidity ≥80 %.
Plasmid construction
The miR159 precursor fragment was amplified by PCR from wild-type arabidopsis (Columbia-0, Ws ecotype) cDNA using a forward primer (5′-GGGGTACCCACGTTCTCATCAAAACTTTC-3′) and a reverse primer (5′-GCTCTAGAACACGCTAAACATTGCTTCG-3′), containing a KpnI site and an XbaI site at their 5′ ends. Artificial target mimics were generated as previously described (Franco-Zorrilla et al., 2007). All the fragments were cloned into pCAMBIA 13011, consisting of a CaMV 35S promoter and a hygromycin resistance gene. Then all the binary constructs were transferred into Agrobacterium tumefaciens strain EHA105.
Plant transformation and selection
Transgenic gloxinia was generated using Agrobacterium-mediated transformation (Zhang et al., 2008). Leaf pieces and Agrobacterium were co-cultivated on basic MS medium for 3 d, washed with sterile water and then transferred to the selection medium containing 20 mg L−1 hygromycin. After four cycles of 2-week selection culture, hygromycin-resistant plantlets were obtained, and then transferred to root-inducing medium for regeneration. Meanwhile, same-size plantlets of untransformed cultures were chosen to grow under the same medium as a wild-type control.
Total RNA isolation
Total RNA was isolated from different tissues of gloxinia using TRIZOL Regent (TaKaRa, Dalian, China) according to the manufacturer's instructions. After addition of isopropanol, the sample was incubated at –20 °C >10 h to get more RNAs. Following an ethanol wash, the total RNA was dissolved in RNase-free water and then treated with DNase I (RNase-free) (TaKaRa) at 37 °C for 15 min to get rid of genomic DNA. DNase I was removed by chloroform and the RNA sample was precipitated by 2·5 volumes absolute ethanol and 1/10 volume of 3 m sodium acetate at –20 °C overnight. The precipitation was resuspended in RNase-free water. The concentration and quality of RNA was tested by BioSpectrometer (Eppendorf, Hamburg, Germany).
Reverse transcription polymerase chain reaction analysis (RT-PCR)
The first-strand cDNA synthesis was performed using a cDNA synthesis kit (TaKaRa) according to the manufacturer's instructions, using 1 µg of RNA as template for reverse transcription.
The expression levels of pre-AtmiR159a and MIM159 in transgenic gloxinias were analysed by semi-quantitative RT-PCR (sqRT-PCR), and PCR cycles were optimized as described previously (Zhang et al., 2008).
Quantitative real-time RT-PCR (qRT-PCR) was used to analyse the expression levels of mature miR159a and flowering genes in wild-type and transgenic gloxinia. SYBR Green was used as the reporter dye (TaKaRa) and β-actin (GenBank accession number EF428182) was used to normalize expression levels of mRNAs. The mature miR159a level was detected by an efficient and reliable stem-loop RT-PCR procedure (Varkonyi-Gasic and Hellens, 2010) and miR159a level was standardized with U6. U6 is a class of metabolically stable small noncoding RNAs presented in the nuclei of eukaryotic cells, which are about 100 nucleotides long, showing a high degree of conservation. So we used U6 as a control in RT-PCR analysis of microRNA as previously reported (Allen et al., 2007; Xia et al., 2012). For each qRT-PCR, 0·4 µL cDNA was used in a 10-μL PCR system, containing 200 nm of each primer, 4·2 µL tri-distilled water and 5 µL SYBR Green. PCRs were carried out using Mastercycler ep realplex (Eppendorf) with three biological replications of the following programme; pre-denaturation at 94 °C for 5 min, then 40 cycles at 94 °C for 15 s, 60 °C for 15 s, 72 °C for 15 s and a dissociation stage generated by the software to analyse the melting curve. All primers used for RT-PCR were designed by Primer Premier5 and listed in Supplementary Data Table S1.
Cloning full-length cDNA of SsGAMYB and sequence analysis
Total RNA was isolated from wild-type gloxinia buds as described above. Poly(A) mRNA was enriched by the Oligotex mRNA purification Kit (TaKaRa). Previous research suggested that GAMYB proteins contain a conserved domain of R2R3 and three special regions (BOX1, BOX2 and BOX3). Based on these features, degenerate primers (forward primer: 5′-CAYGGYGWGGGBAACTGGAAY-3′ and reverse primer: 5′-TTGGAVTGAAGGGRGCTCSAKCTTC-3′) were designed according to sequences in arabidopsis, rice, barley, maize and other homologous genes to be able to amplify partial cDNA from gloxinia. Then we employed SMARTer RACE cDNA amplification kit (Clontech, CA, USA) to clone the full length of SsGAMYB (5′-TGCCTGGACGCACGGATAATGAGAT-3′ for 3′-RACE and 5′-GCGCTAGCAGGCTACTTGCAGGAAT-3′ for 5′-RACE). The RACE products were cloned into T-simple vector (TaKaRa) and six independents inserts were determined. The software of Omiga2·0 was used to determine the open reading frame of SsGAMYB.
RNA ligase-mediated 5′ rapid amplification of cDNA ends (RLM 5′-RACE)
To validate a putative target of miR159, a modified RLM 5′-RACE was carried out, according to the protocol described before (Zhou et al., 2010). Total RNA was isolated from wild-type flower buds and ligated RNA was amplified with two nested special primers (outer: 5′-TCAAATCATCACAAATAAATGTCCCT-3′ and inner 5′-GAAGCGTGTGAAGTCTGCTGGAAAG-3′). PCR products were gel purified and cloned into T-simple vector (TaKaRa). Ten independent clones were sequenced.
RESULTS
Generation of transgenic gloxinia lines that over-express or suppress miR159a
MiR159 has been identified to be one of eight highly conserved miRNA families in plants (Cuperus et al., 2011). Sequence alignment indicated that miR159a shows very high conservation among species (Fig. 1A). Since the sequences of miR159 precursors in gloxinia are unavailable from the public database, we used the AtmiR159a precursor to over-express miR159a under the control of the cauliflower mosaic virus 35S (CaMV 35S) promoter. In addition, we carried out target mimicry to repress the activity of the endogenous miR159a. Target mimicry, in which an un-cleavable target is expressed in the plant, has been used to sequester specific miRNAs (Franco-Zorrilla et al., 2007). 35S:MIM159 was generated to harbour such a target-mimic construct for miRNA159a (for the sequence see Supplementary Data Table S2). Transgenic gloxinia lines were generated using Agrobacterium-mediated transformation, as described in our previous work (Zhang et al., 2008).
Fig. 1.

Alignment of mature miR159a and RT-PCR analysis of transgenic gloxinias. (A) Sequence alignment of mature miR159a in arabidopsis (At), tomato (Sl), barley (Hv), rice (Os), maize (Zm) and wheat (Ta). (B) Expression levels of Pre-AtmiR159a and MIM159 (35S:MIM159) in transgenic plants by RT-PCR analysis.
The transgenic plantlets were detected by RT-PCR with gene-specific primers. Distinct bands of expected size not found in wild-type plants were obtained from 32 plantlets of AtmiR159a over-expressing 35S:miR159a transgenic lines and 28 transgenic plants of MIM159 over-expressing 35S:MIM159 lines (Fig. 1B).
Altered flowering time in transgenic gloxinia
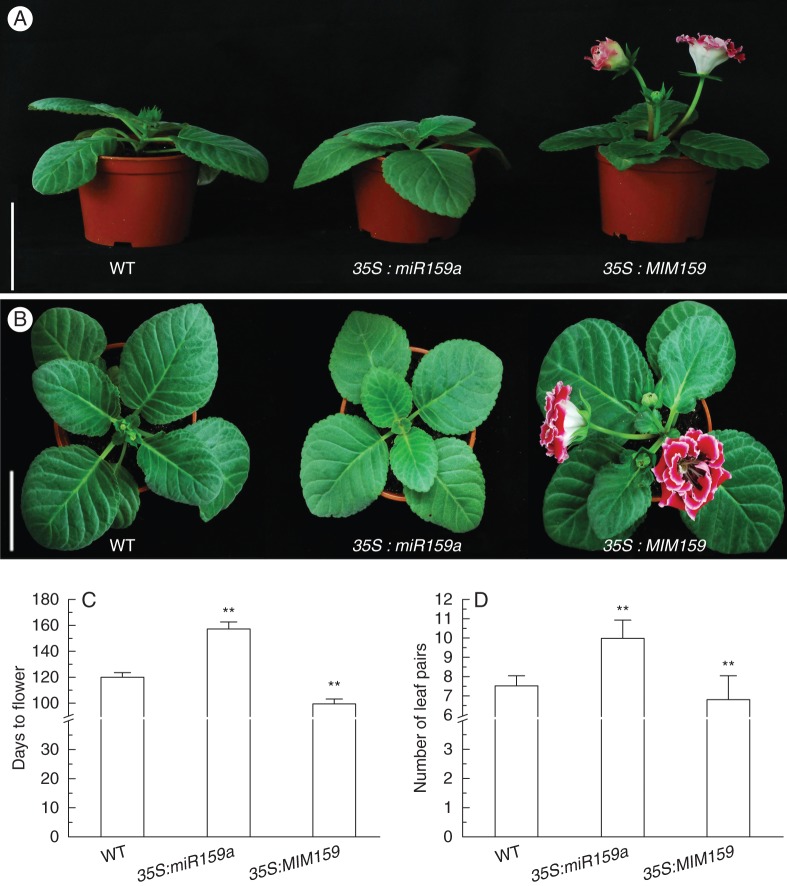
Transgenic gloxinias were grown under SD conditions (8 h light/16 h dark) in the greenhouse. Phenotypic analysis showed that the plant heights of transgenic lines were similar to that of wild type under the same cultivation period (Fig. 2A). As to vegetative tissues, 35S:miR159a displayed smaller leaves than either wild-type or 35S:MIM159 plants (Fig. 2B). Interestingly, compared with wild-type plants, 35S:miR159a plants exhibited a delay in flowering time, whereas 35S:MIM159 lines exhibited an increase in flowering during the reproductive stage (Fig. 2B). Wild-type plants flowered at day 120·2 (±3·15) after regeneration, while transgenic lines 35S:miR159a flowered at day 157·6 (±4·93) and lines 35S:MIM159 flowered at day 99·8 (±3·5) (Fig. 2C). In addition, at flowering, 35S:miR159a plants exhibited more leaves than wild type (10 ± 0·92 against 7·6 ± 0·5), whereas 35S:MIM159 lines exhibited fewer leaves than wild type (6·8 ± 1·22 against 7·6 ± 0·5; Fig. 2D). These results demonstrate that miR159-over-expression or inactivation in transgenic gloxinia can significantly alter flowering time.
Fig. 2.
Flowering time alterations in transgenic gloxinias under short-day conditions. (A) Side view of wild-type (WT), 35S:miR159a and 35S:MIM159 transgenic plants. Scale bar = 10 cm. (B) Top view of WT, 35S:miR159a and 35S:MIM159 transgenic plants. Scale bar = 8 cm. All lines in (A, B) are 135 d old after transplantation to pots. (C) Days to flowering after regeneration. (D) Number of leaf pairs at flowering. Data represent means ± s.d. (n > 20). Double asterisks indicate a significant (ANOVA, P < 0·01) difference between transgenic lines and the wild type.
Identification and sequence analysis of SsGAMYB
The complex function of GAMYB under the regulation of miR159 (Murray et al., 2003; Millar and Gubler, 2005; Aya et al., 2011) prompted us to investigate the possible role of GAMYB in gloxinia during flower development. To obtain the full-length cDNA of GAMYB in gloxinia, the fragments with conserved MYB domain were cloned by RACE and sequences of six independent inserts were determined. The sequencing results showed that the length of the coding sequence was 1650 bp, putatively encoding a previously uncharacterized polypeptide of 549 amino acids that we named SsGAMYB (Supplementary Data Fig. S1). Phylogenetic analysis showed that the polypeptide of SsGAMYB shared significant similarity with those of both tomato and arabidopsis (Fig. 3A). A comparative analysis of SsGAMYB demonstrated that the homologue contained a conserved R2R3 DNA-binding domain and three typical regions (BOX1, BOX2 and BOX3), similar to the domains in GAMYBs of other plant species (Fig. 3B). A putative complementary site of miR159a exists in the region (964–985 bp) between BOX1 and BOX2, in which there are only two mismatched nucleotides (Fig. 3C).
Fig. 3.
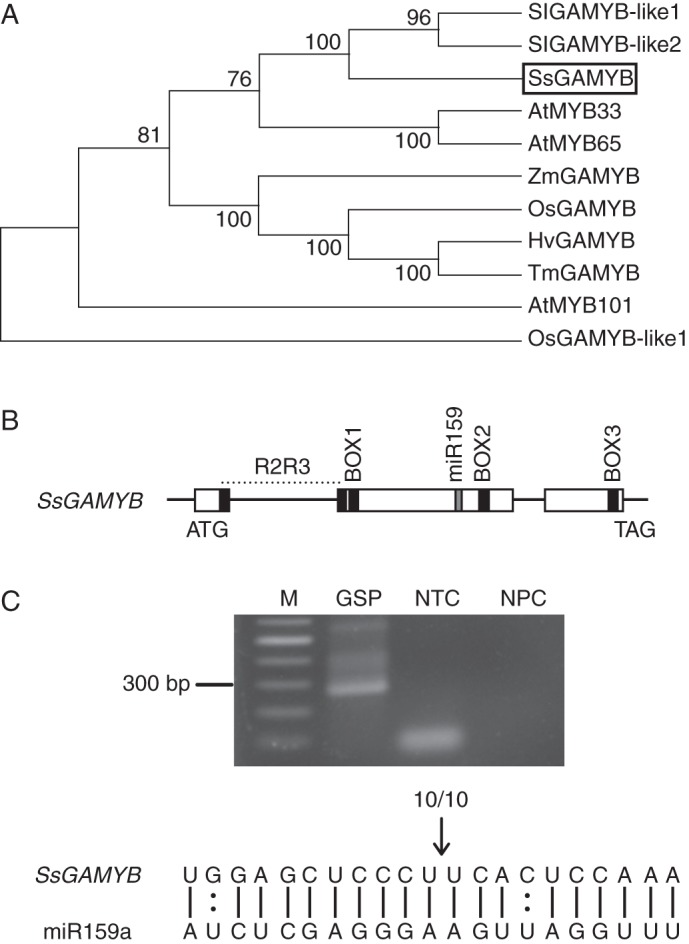
Analysis of SsGAMYB protein and validation of miR159-guided cleavage of SsGAMYB. (A) Phylogenetic analysis of SsGAMYB proteins in S. lycopersicum, A. thaliana, Z. mays, O. sativa, H. vulgare and T. aestivum. The phylogenetic tree was generated with the computer program MEGA4. Bootstrap values are shown. (B) Structure of SsGAMYB and location of the complementary sequence between SsGAMYB and miR159. The coding regions are indicated by boxes: first black box, R2R3 DNA-binding domain; second to the fourth black boxes, BOX1–3 domains; grey box, putative miR159 target site (C). Experimental validation of miR159 cleavage site in SsGAMYB. The cleavage site was determined by RLM 5′-RACE. M, Marker; GSP, gene specific primer; NTC, no template control; NPC, no primer control. As shown, miR159a sequence is complementary to the partial sequence of SsGAMYB. The arrow indicates the cleavage site of SsGAMYB and the number above the sequence indicates the frequency of 5′-RACE clones.
To test whether SsGAMYB is a direct target of miR159, we performed RLM 5′-RACE to detect the cleavage site of SsGAMYB. A band of predicted size was cloned in the flower buds sample (Fig. 3C). The amplified fragment was cloned into T-simple vector and ten independent inserts were determined by sequencing. The results revealed that ten out of ten of the cloned fragments were cleaved at the position of the tenth nucleotide (Fig. 3C), which confirms that SsGAMYB is a target of miR159a in gloxinia.
Expression patterns of miR159a and SsGAMYB in gloxinia
MiR159a, a main member of the miR159 family, predominantly accumulated in inflorescence and floral tissues in arabidopsis and rice (Tsuji et al., 2006; Allen et al., 2007). To investigate expression patterns of miR159a and GAMYB in gloxinia, we determined accumulation levels of mature miR159a and SsGAMYB mRNA in various tissues by qRT-PCR. In gloxinia, mature miR159a accumulated highly in young leaves (leaf 1) and sepals, while its level was relatively less abundant in old leaves (leaf 3) and other tissues of flowers (petals, stamens and carpels), and was especially lowest in stamens (Fig. 4A). The spatial expression pattern of miR159a in gloxinia was consistent with that in arabidopsis and rice, implying the conservation of miR159 in plants.
Fig. 4.

Expression patterns of SsmiR159a and SsGAMYB in various tissues using quantitative RT-PCR analysis. (A) Relative expression levels of mature SsmiR159a in various tissues of gloxinia. Leaf1, seedling leaf; leaf2, adult seedling leaf; leaf3, reproduction period leaf. (B) Relative mRNA levels of SsGAMYB. Mean values were obtained from three independent samples. Error bars represent s.d.
Next, we examined the expression level of SsGAMYB in various tissues of gloxinia. SsGAMYB was highly expressed in stamens and carpels, while its expression level was relatively low in sepals and petals (Fig. 4B). In particular, the mRNA level of SsGAMYB was almost undetectable in sepals, which was opposite to the expression level of miR159a in sepals. Thus, the expression levels of SsGAMYB and miR159a are negatively correlated in gloxinia.
Effects of over-expressed or suppressed miR159a on SsGAMYB expression in transgenic gloxinia
To evaluate the effects of miR159a expression on SsGAMYB, we examined the expression level of mature miR159a and SsGAMYB in transgenic lines by qRT-PCR analysis. Three lines of 35S:miR159a (#2, #28 and #41) and 35S:MIM159 (#19, #21 and #23) transgenic plants were chosen for further study. Total RNA was extracted from vegetative leaves and flower buds at the same growth stage. Compared with wild type, mature miR159a level was significantly increased in 35S:miR159a lines and decreased in 35S:MIM159 lines in leaves and flower buds (Fig. 5A). Thus, we have successfully generated transgenic gloxinia with varying level of miR159a in either vegetative or floral tissues.
Fig. 5.
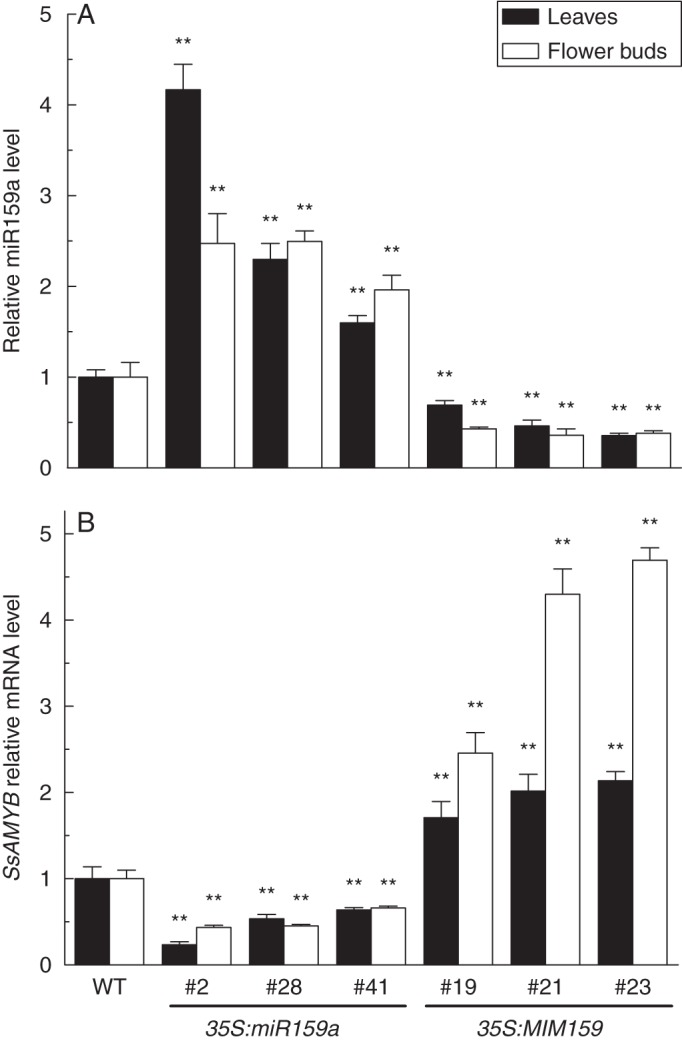
Transcriptional levels of mature miR159a and SsGAMYB in wild-type (WT) and transgenic gloxinia. (A) Transcriptional level of mature miR159a in adult seedling leaves and flower buds. (B) Expression levels of SsGAMYB mRNA in adult seedling leaves and flower buds. Mean values were obtained from three independent samples. Error bars represent s.d. ** P < 0·01, significant difference between transgenic lines and the wild type (ANOVA).
Furthermore, the steady-state mRNA level of SsGAMYB was also detected in these transgenic lines. Compared with the wild-type control, a low level of SsGAMYB was observed in the leaves and flower buds of 35S:miR159a lines, whereas the SsGAMYB level was significantly elevated in 35S:MIM159 lines. However, in the 35S:MIM159 lines, the relative mRNA level of SsGAMYB in flower buds was higher than those in leaves (Fig. 5B). This is consistent with the expression level of SsGAMYB being negatively regulated by miR159a in wild-type gloxinia (Fig. 4B). These results revealed that the accumulation level of miR159 exhibited an opposite trend to the mRNA level of SsGAMYB in transgenic lines. It suggested that the expression level of SsGAMYB is negatively regulated by miR159 in transgenic gloxinia.
Expression of downstream flowering genes in transgenic gloxinia
LFY and LFY-like genes control the initiation of floral meristems and regulate MADS-box genes in flowering plants (Weigel and Meyerowitz, 1994). Thus, it is necessary to determine whether the miR159 target SsGAMYB regulates expression of LFY during floral initiation in transgenic gloxinia. We isolated a partial cDNA fragment of LFY in gloxinia (SsLFY) using primers designed based on its homologues in tomato and arabidopsis (Supplementary Data Fig. S2). Three MADS-box genes are present in gloxinia, SsAG (GenBank accession number EF428183), SsAP1 (EF428184) and SsAP3 (EF428185) (Zhang et al., 2008). Quantitative RT-PCR analysis of flower buds showed that over-expression of miR159 resulted in a significant decline in SsLFY transcript level in 35S:miR159a lines, while suppression of miR159 caused an increase in SsLFY in 35S:MIM159 lines under SD conditions (Fig. 6A). Meanwhile, the mRNA levels of SsAG, SsAP1 and SsAP3 were decreased dramatically in 35S:miR159a lines but increased in 35S:MIM159 lines (Fig. 6B–D). These results revealed that the altered flowering time correlates with the changed expression levels of SsLFY in transgenic gloxinia, in which SsLFY expression promotes the expression of SsAG, SsAP1 and SsAP3. Therefore, the miR159-mediated flowering control pathway appears to be effective in gloxinia.
Fig. 6.
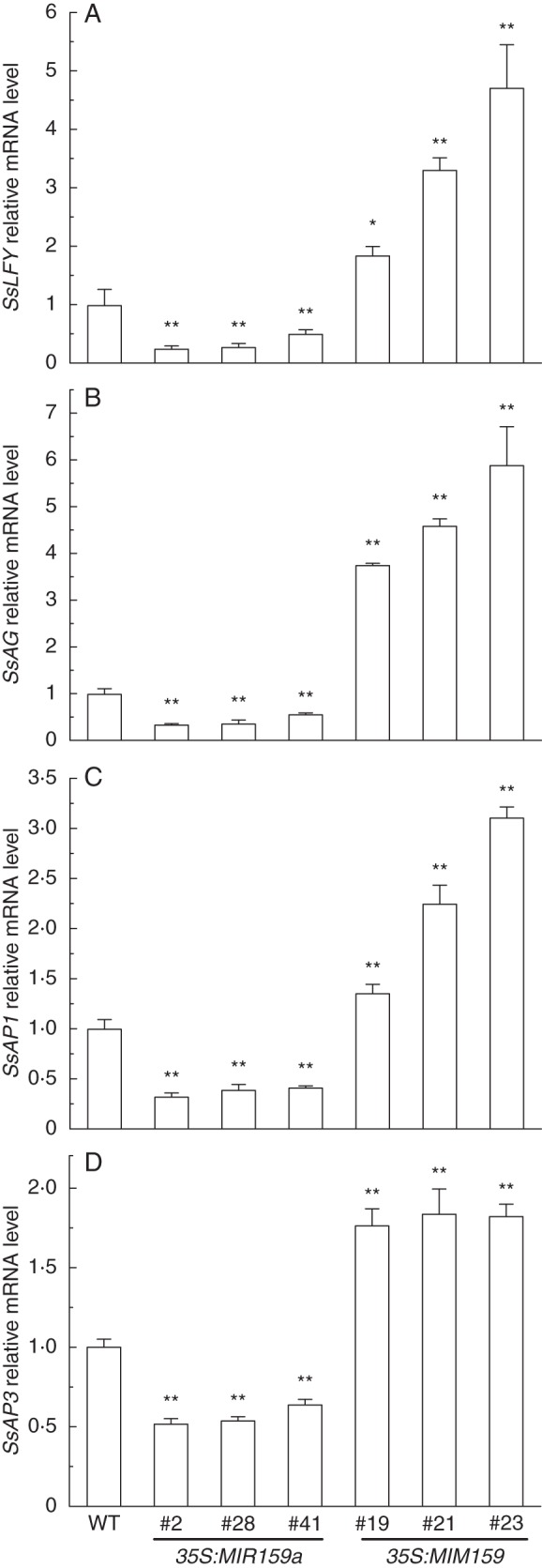
Expression patterns of flowering genes in wild-type (WT) and transgenic gloxinia. Quantitative RT-PCR analysis of (A) SsLFY, (B) SsAG, (C) SsAP1, and (D) SsAP3 in flower buds of WT and transgenic lines. Mean values were obtained from three independent samples. Error bars represent s.d. ** P < 0·01, significant difference between transgenic lines and the wild type (ANOVA).
Effects of altered miR159 expression on floral organ development
LFY plays a dual role in regulating floral meristem identity and floral organ patterning (Parcy et al., 1998). Our above results showed that elevated LFY expression promoted early flowering in transgenic plants. This prompted us to investigate whether alterations in LFY transcripts also affect floral organ patterning in gloxinia. Phenotype observation indicated that the floral morphology of 35S:miR159a appeared relatively normal compared with the wild-type (Fig. 7A, C, E). Nonetheless, abnormal morphology appeared in a few of 35S:MIM159 lines (3 of 28 lines). In the flower bud stage, sepals were converted partly into petals in the 35S:MIM159 lines, disturbing the normal development of sepals (Fig. 7B). When the flower buds opened completely, the red petals appeared partly green, displaying conversion of petals into sepals (Fig. 7D). Notably, when sepals were removed, obvious sepal-like petals were seen in 35S:MIM159 lines (Fig. 7F).
Fig. 7.

Morphological changes in 35S:MIM159 flowers. Floral organs shown are from wild-type (A, C, E) and 35S:MIM159 (B, D, F) plants: (A, B) flower buds; (C, D) front side of flowers; (E, F) underside of flowers with sepals removed.
The transgenic lines of 35S:MIM159 can be divided into two groups: mild (#19) and severe (#23) phenotypes. The altered floral organs can only be seen in three lines with severe phenotype where the expression level of SsGAMYB increased strongly (#23 in Fig. 5B). Thus, altered floral organs might be mainly due to the high expression level of LFY in 35S:MIM159 transgenic lines (Fig. 6A). According to our previous report (Zhang et al., 2008), over-expression of LFY caused morphological change and alteration in the expression of AP1 and AP3 in flower organs. Quantitative RT-PCR analysis of those abnormal flowers showed that AP1 transcript level was notably high in petals, while AP3 expression level was significantly high in sepals and dramatically low in petals compared with wild-type flowers (Supplementary Data Fig. S3). In wild-type flowers, AP1 was highly expressed in sepals and AP3 was accumulated in petals and stamens, while AG and GAMYB was high in stamens and carpels (Supplementary Data Fig. S4). However, the expression levels of LFY, AP1 and AP3 were not changed greatly in mild phenotype lines (#19) where floral organs are normal compared with wild type (Fig. 6A, C, D). It revealed that the conversion of petals and sepals might be caused by disturbed expression levels of AP1 and AP3 between the first and second whorls. In summary, during floral organ development in gloxinia, suppression of miR159 activity disrupted the formation of petals and sepals.
DISCUSSION
A genetic and molecular understanding of floral transition in model systems has paved a way to breed plants through genetic modification by transformation. Current knowledge of flowering time control has been applied through either over-expression or suppression of an activity of a certain gene in crops (Jung and Muller, 2009). However, less progress has been made in the targeted genetic modification of flowering time in ornamental plants. The bottleneck is likely due to the relative unknown genomic information in most commercial species. Therefore, it is necessary to seek an applicable approach to produce novel varieties with altered flowering behaviour in commercial plants. In the present study, we provide a strategy for flowering time control by manipulation of miR159 expression in ornamental gloxinia. Over-expression or suppression of miR159 dramatically altered flowering time, suggesting that microRNA159-mediated flowering regulation appears effective and applicable in non-model plants.
First, miR159-mediated GAMYB functions in regulation of timing of flowering in gloxinia. The possible roles of miR159 prompted us to investigate how miR159-mediated GAMYB acts in gloxinia during flower development. Compared with wild-type gloxinia, under SD conditions, over-expression of miR159 delayed the timing of flowering to 37 d, whereas suppression of miR159 accelerated flowering to 20 d. Meanwhile, we identified a member of the GAMYB family in gloxinia with a complementary sequence to miR159. Consistent with the findings in arabidopsis, the steady-state mRNA level of SsGAMYB displayed a reverse correlation with accumulation of miR159 in vegetative and floral tissues in gloxinia. Our results suggested that GAMYB regulation by miR159 was a functionally effective pathway in triggering flowering under SD conditions. However, there are differing reports about roles of miR159 and GAMYB in the promotion of floral transition. In arabidopsis ecotype Landsberg erecta, over-expression of miR159 causes late flowering under short days due to down-regulation of target AtMYB33 (Achard et al., 2009). Under the long-day photoperiod, over-expression of miR159, down-regulating AtMYB101 but not MYB33 and MYB65, did not alter flowering time in transgenic lines (ecotype Columbia) (Schwab et al., 2005). Consistently, the flowering time of mir159ab double mutant is similar to the wild type under long-day conditions whereas mir159ab delayed flowering under SD conditions (Alonso-Peral et al., 2010). These reports suggested that miR159 regulation of flowering might be affected by photoperiod. Besides, it could be also explained by a difference in ecotypes which changed flowering time in the regulation of miR159 (Alonso-Peral et al., 2010). Thus, manipulation of miR159 expression might be an effective approach to control flowering time in ornamental plants.
Secondly, miR159 is highly conserved in sequence among all embryophytes (Cuperus et al., 2011). To date, more and more genomes and transcriptomes of plant species have been sequenced. However, genomic information is still far from being understood in most commercial plants. In our previous work, we introduced a LFY homologue (CFL) from cucumber (Cucumis sativus) into gloxinia due to lack of an endogenous LFY sequence, and this transgenic gloxinia over-expressed CFL which resulted in early flowering (Zhang et al., 2008). RNA interference is widely used to repress activity of a known gene in plants (Dunoyer et al., 2010). It is not feasible to suppress LFY activity by RNAi in a species where the LFY sequence is not known. To overcome this problem, we over-expressed miR159 instead to be able to repress the expression of its multiple targets. MiR159a has been shown to be a major identified member of the miR159 family (Cuperus et al., 2011). We used AtmiR159a precursor to generate over-expressing miR159a transgenic lines since the sequences of miR159 precursors in gloxinia are unavailable to date. Meanwhile, we also utilized the mimicry target approach to repress endogenous miR159 activity. Target mimicry has been used to sequester specific miRNAs, thus causing a repression of the normal miRNA activity (Franco-Zorrilla et al., 2007). Our results demonstrated that over-expression or repression of miR159 significantly altered the expression level of its target SsGAMYB, resulting in delayed or early flowering. In contrast to manipulating a single gene related to flowering, manipulating an miRNA expression level may completely affect the expression of its target genes. A recent study has shown that over-expression of a rice miR156 in switchgrass (Panicum virgatum) increases tiller number and improves biomass production (Fu et al., 2012). Therefore, it seems to be an efficient approach to elevate or reduce activity of target genes by genetic transformation of a conserved miRNA in a species with unknown genomic sequence.
Finally, conversion of petals and sepals due to miR159 repression could offer a new strategy for development of cultivars with various flower patterns. In spite of the effects on flowering time, over-expression of miR159 resulted in smaller leaves compared with wild type. This is consistent with the phenotype of miR159-over-expressing arabidopsis (Achard et al., 2004). Interestingly, conversion of petals and sepals was observed upon suppression of miR159 activity in gloxinia, which has not been reported yet even in the model species. Our results showed that suppression of miR159 activity caused an increase in the level of SsLFY in the flower buds. LFY plays a role in regulating not only floral meristem identity but also floral organ patterning (Parcy et al., 1998). Following the initial expression of LFY, then AP1, AP3 and AG are expressed throughout the floral meristem (Wagner, 2009). Based on the ABC model of floral organ identity specification, AP1 and AP2 are required for normal sepal and petal development, AP3 is expressed in the petals and stamens, AG and GAMYB are expressed in the stamens and carpels (Weigel and Meyerowitz, 1994; Weigel, 1998). Altered floral organs in 35S:MIM159 transgenic lines might be due to alterations in the expression of AP1 and AP3 in flower organs. In our study, abnormal sepals and petals were correlated with the significantly high expression levels of AP1 in the petals and AP3 in the sepals. As to ornamental plants, conversion of petals and sepals, due to novel floral organ patterning, might provide an alternative source for generating new cultivated species.
In conclusion, GAMYB regulation mediated by miRNA159 controls flowering time in gloxinia. Our study provides an applicable genetic modification approach to control the timing of flowering through manipulation of miR159 in commercial ornamental plants.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We sincerely thank Professor Jiliang Pang (Hangzhou Normal University) for kindly providing aseptic seedlings of gloxinia. This work was supported by the National Science Foundation of China (grant nos 30972016, 31171615 and 30571197).
LITERATURE CITED
- Achard P, Herr A, Baulcombe DC, Harberd NP. Modulation of floral development by a gibberellin-regulated microRNA. Development. 2004;131:3357–3365. doi: 10.1242/dev.01206. [DOI] [PubMed] [Google Scholar]
- Achard P, Gusti A, Cheminant S. Gibberellin signaling controls cell proliferation rate in Arabidopsis. Current Biology. 2009;19:1188–1193. doi: 10.1016/j.cub.2009.05.059. [DOI] [PubMed] [Google Scholar]
- Allen RS, Li J, Stahle MI, Dubroué A, Gubler F, Millar AA. Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proceedings of National Academy of Sciences of the USA. 2007;104:16371–16376. doi: 10.1073/pnas.0707653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Peral MM, Li J, Li Y. The microRNA159-regulated GAMYB-like genes inhibit growth and promote programmed cell death in Arabidopsis. Plant Physiology. 2010;154:757–771. doi: 10.1104/pp.110.160630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aya K, Hiwatashi Y, Kojima M. The gibberellin perception system evolved to regulate a pre-existing GAMYB-mediated system during land plant evolution. Nature Communications. 2011;2:544. doi: 10.1038/ncomms1552. [DOI] [PubMed] [Google Scholar]
- Blazquez M. Flower development pathways. Journal of Cell Science. 2000;113:3547–3548. doi: 10.1242/jcs.113.20.3547. [DOI] [PubMed] [Google Scholar]
- Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- Cuperus JT, Fahlgren N, Carrington JC. Evolution and functional diversification of MIRNA genes. The Plant Cell. 2011;23:431–442. doi: 10.1105/tpc.110.082784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P, Brosnan CA, Schott G. An endogenous, systemic RNAi pathway in plants. EMBO Journal. 2010;29:1699–1712. doi: 10.1038/emboj.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Franco-Zorrilla JM, Valli A, Todesco M. Target mimicry provides a new mechanism for regulation of microRNA activity. Nature Genetics. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- Fu C, Sunkar R, Zhou C. Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnology Journal. 2012;10:443–452. doi: 10.1111/j.1467-7652.2011.00677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Zhu Q, Dabi T, Li D, Weigel D, Lamb C. Transformation of rice with the Arabidopsis floral regulator LEAFY causes early heading. Transgenic Research. 2000;9:223–227. doi: 10.1023/a:1008992719010. [DOI] [PubMed] [Google Scholar]
- Henderson IR, Dean C. Control of Arabidopsis flowering: the chill before the bloom. Development. 2004;131:3829–3838. doi: 10.1242/dev.01294. [DOI] [PubMed] [Google Scholar]
- Jung C, Muller AE. Flowering time control and applications in plant breeding. Trends in Plant Science. 2009;14:563–573. doi: 10.1016/j.tplants.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Levy YY, Dean C. Control of flowering time. Current Opinion in Plant Biology. 1998;1:49–54. doi: 10.1016/s1369-5266(98)80127-1. [DOI] [PubMed] [Google Scholar]
- Millar AA, Gubler F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. The Plant Cell. 2005;17:705–721. doi: 10.1105/tpc.104.027920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray F, Kalla R, Jacobsen J, Gubler F. A role for HvGAMYB in anther development. The Plant Journal. 2003;33:481–491. doi: 10.1046/j.1365-313x.2003.01641.x. [DOI] [PubMed] [Google Scholar]
- Parcy F, Nilsson O, Busch MA, Lee I, Weigel D. A genetic framework for floral patterning. Nature. 1998;395:561–566. doi: 10.1038/26903. [DOI] [PubMed] [Google Scholar]
- Reyes JL, Chua NH. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. The Plant Journal. 2007;49:592–606. doi: 10.1111/j.1365-313X.2006.02980.x. [DOI] [PubMed] [Google Scholar]
- Rubio-Somoza I, Cuperus JT, Weigel D, Carrington JC. Regulation and functional specialization of small RNA-target nodes during plant development. Current Opinion in Plant Biology. 2009;12:622–627. doi: 10.1016/j.pbi.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Samach A. Control of flowering. In: Altman A, Paul Michael Hasegawa PM, editors. Plant biotechnology and agriculture: prospects for the 21st century. New York, NY: Academic Press; 2011. pp. 387–404. [Google Scholar]
- Schwab R, Palatnik JF, Riester M. Specific effects of microRNAs on the plant transcriptome. Developmental Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Srikanth A, Schmid M. Regulation of flowering time: all roads lead to Rome. Cellular and Molecular Life Science. 2011;68:2013–2037. doi: 10.1007/s00018-011-0673-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genetics. 2010;6 doi: 10.1371/journal.pgen.1001031. e1001031. http://dx.doi.org/10.1371/journal.pgen.1001031 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H, Aya K, Ueguchi-Tanaka M. GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers. The Plant Journal. 2006;47:427–444. doi: 10.1111/j.1365-313X.2006.02795.x. [DOI] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Hellens RP. qRT-PCR of Small RNAs. Methods in Molecular Biology. 2010;631:109–122. doi: 10.1007/978-1-60761-646-7_10. [DOI] [PubMed] [Google Scholar]
- Wagner D. Flower morphogenesis: timing is key. Developmental Cell. 2009;16:621–622. doi: 10.1016/j.devcel.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Weigel D. From floral induction to floral shape. Current Opinion in Plant Biology. 1998;1:55–59. doi: 10.1016/s1369-5266(98)80128-3. [DOI] [PubMed] [Google Scholar]
- Weigel D, Meyerowitz EM. The ABCs of floral homeotic genes. Cell. 1994;78:203–209. doi: 10.1016/0092-8674(94)90291-7. [DOI] [PubMed] [Google Scholar]
- Weigel D, Nilsson O. A developmental switch sufficient for flower initiation in diverse plants. Nature. 1995;37:495–500. doi: 10.1038/377495a0. [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. Leafy controls floral meristem identity in arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]
- Xia K, Wang R, Ou X. OsTIR1 and OsAFB2 downregulation via Osmir393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS ONE. 2012;7:e30039. doi: 10.1371/journal.pone.0030039. http://dx.doi.org/10.1371/journal.pone.0030039 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MZ, Ye D, Wang LL. Overexpression of the cucumber LEAFY homolog CFL and hormone treatments alter flower development in gloxinia (Sinningia speciosa) Plant Molecular Biology. 2008;67:419–427. doi: 10.1007/s11103-008-9330-8. [DOI] [PubMed] [Google Scholar]
- Zhou M, Gu LF, Li PC. Degradome sequencing reveals endogenous small RNA targets in rice (Oryza sativa L. ssp. Indica) Frontiers in Biology. 2010;5:67–90. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.