Abstract
Background and Aims
Plants growing at high densities perceive a decrease in the red to far-red (R/FR) ratio of incoming light. These changes in light quality trigger a suite of responses collectively known as the shade-avoidance syndrome (SAS) including hypocotyl and stem elongation, inhibition of branching and acceleration of flowering.
Methods
Quantitative trait loci (QTLs) were mapped for hypocotyl length to end-of-day far-red (EOD), a simulated shade-avoidance response, in recombinant inbred line (RIL) populations of Arabidopsis thaliana seedlings, derived from Landsberg erecta (Ler) and three accessions (Columbia, Col; Nossen, No-0; and Cape Verde Islands, Cvi-0).
Key Results
Five loci were identified as being responsible for the EOD response, with a positive contribution of Ler alleles on the phenotype independently of the RIL population. Quantitative complementation analysis and transgenic lines showed that PHYB is the candidate gene for EODRATIO5 in the Ler × Cvi-0 RIL population, but not for two co-localized QTLs, EODRATIO1 and EODRATIO2 mapped in the Ler × No-0 and Ler × Col RIL populations, respectively. The ERECTA gene was also implicated in the SAS in a background-dependent manner. For hypocotyl length EOD response, a positive contribution of erecta alleles was found in Col and Van-0, but not in Ler, Cvi-0, Hir-1 or Ws. Furthermore, pleiotropic effects of ERECTA in the EOD response were also detected for petiole and lamina elongation, hyponastic growth, and flowering time.
Conclusions
The results show that the analysis of multiple mapping populations leads to a better understanding of the SAS genetic architecture. Moreover, the background- and trait-dependent contribution of ERECTA in the SAS suggest that its function in shaded natural environments may be relevant for some populations in different phases of plant development. It is proposed that ERECTA is involved in canalization processes buffering the genetic variation of the SAS against environmental light fluctuations.
Keywords: ERECTA, shade-avoidance syndrome (SAS), natural genetic variation, phytochrome B (phyB), far-red end-of-day response, EOD response, quantitative trait loci, QTL
INTRODUCTION
Individual plant species thrive in specific environments, some growing well across a wide area, others being restricted to survival in very narrow ecological niches. Within a niche, a species must compete for space, light and other resources with the same and other species. The shade-avoidance syndrome (SAS) is a strategy of major adaptive significance to plants (Schmitt, 1997). The SAS is highly widespread in most species growing in open habitats and depends on the ability of the plant to perceive the presence of neighbours to anticipate competition for scarce resources such as light in crowded stands (Schmitt, 1997). A plant canopy reduces the ratio of red to far-red light (R/FR) by the efficient absorption of red light by photosynthetic pigments and the relative increase of FR photons (Smith, 1982). Plants perceive this light quality change through the phytochrome system and respond very rapidly, enhancing SAS, including via vegetative structure elongation, hyponastic response and acceleration of flowering (Casal, 2012).
In terms of molecular mechanisms involved in the SAS, the reduction of R/FR in shaded environments leads to rapid transcriptional changes principally mediated by phytochrome B (phyB). Shade light signals stabilize PIF4 and PIF5 proteins promoting elongation growth by the synthesis and redirection of auxin and favouring the degradation of DELLA proteins (Djakovic-Petrovic et al., 2007; Lorrain et al., 2008). PIF4 and PIF5 promote SAS by directly binding to G-boxes present in the promoter of early shade genes. To avoid an exaggerated SAS, some transcription factors such as HFR1, PAR1 and PAR2, expression of which is transcriptionally induced by shade, directly interact with PIF4 and PIF5 forming non-DNA-binding heterodimers that prevent their ability to induce the expression of elongation genes (Hornitschek et al., 2009; Galstyan et al., 2011). In addition, B-box-containing zinc finger transcription factors (BBX) can promote or inhibit hypocotyl growth in simulated and natural shade light (Crocco et al., 2011). In low R/FR, bbx21 and bbx22 mutants show an exaggerated elongation response and the bbx21bbx22 double mutant restores the SAS response in the cop1 mutant background, indicating that these genes are also part of the feedback inhibition of shade-avoidance responses downstream of COP1 (Crocco et al., 2010).
Phenotypes within most species are not fixed and show significant levels of natural genetic variation between individuals. In the model plant Arabidopsis thaliana, extensive natural variation in the SAS has been explored for seedling hypocotyl growth and flowering acceleration (Botto and Smith, 2002; Botto and Coluccio, 2007; Jiménez-Gómez et al., 2010; Coluccio et al., 2011). An SAS causal gene has recently been identified by analysing the genetic variation in the recombinant inbred line (RIL) population between Bay and Sha genotypes (Jiménez-Gómez et al., 2010; Coluccio et al., 2011). In both of these studies, the authors demonstrated that the ELF3 gene is a component of SAS signalling involved in the elongation and flowering responses, the Sha-ELF3 allele being less functional than the Bay-ELF3 allele in low R/FR. The causal mechanism of ELF3 genetic variation appears to be a single amino acid change in the Sha-ELF3 allele (Coluccio et al., 2011). Besides, natural alleles of PHYD and PIF4 contribute differentially to SAS (Aukerman et al., 1997; Brock et al., 2010). Using a genome-wide association analysis, it was suggested that polymorphisms with small effects could be responsible for the natural variation in hypocotyl growth under shade (Filiault and Maloof, 2012).
The leucine-rich repeat receptor-like Ser/Thr kinase gene ERECTA modulates plant developmental programmes as well environmental and biotic responses (Torii et al., 1996; van Zanten et al., 2009; Fu et al., 2009). Among the most characteristic phenotypes associated with the erecta mutation are the short stature and the reduced internode and pedicel lengths of inflorescences (Torii et al., 1996). The ERECTA locus has been implicated in the control of hypocotyl growth under continuous blue light, far-red and red pulses, and darkness (Borevitz et al., 2002; Botto et al., 2003). In A. thaliana plants, ERECTA is involved in the ethylene-induced hyponastic response, allowing an upward leaf movement response through differential petiole growth (van Zanten et al., 2010). Moreover, ERECTA has been implicated in the control of leaf, flower and fruit development, resistance to pathogens, and the regulation of transpiration (Douglas et al., 2002; Godiard et al., 2003; Llorente et al., 2005; Masle et al., 2005). In spite of the extensive characterization of ERECTA functions in plant development, its participation in the SAS is largely unknown.
The aim of this work was to improve our understanding of the SAS genetic architecture of hypocotyl elongation to an end-of-day FR signal (EOD), a light treatment that simulates shade conditions in nature. First, we describe the natural genetic variation in EOD response among 108 accessions. Secondly, we conducted a quantitative trait loci (QTLs) analysis using three sets of A. thaliana RIL populations, having Landsberg erecta (Ler) as a common parent. Thirdly, we demonstrate that PHYB is the candidate gene for EODRATIO5, but not for two co-localized QTLs, suggesting PHYB background-dependent effects on shade. Finally, we focused on ERECTA as a candidate gene for EOD6. We found that the ERECTA gene is implicated in the SAS in a background-dependent manner. Furthermore, ERECTA polymorphic effects in EOD responses were detected for other SAS traits, suggesting that its role in shaded environments is relevant for some populations in different phases of plant development.
MATERIALS AND METHODS
Plant material
Arabidopsis thaliana accessions were obtained from the Arabidopsis Biological Resource Center (ABRC, Ohio State University, USA). RIL populations derived from crosses between Ler and No-0 (Magliano et al., 2005), Ler and Col (Lister and Dean, 1993), and Ler and Cvi-0 populations (Alonso Blanco et al., 1998) were also obtained from the ABRC. We also used near isogenic lines (NILs) polymorphic for ERECTA between Ler and Cvi-0 genotypes (Keurentjes et al., 2007); PHYB transgenic lines carrying 35S::PHYB-Cvi and 35S::PHYB-Ler alleles in phyB-9 mutant Col background (Filiault et al., 2008); ERECTA transgenic lines in Landsberg erecta (Ler), Vancouver-0 (Van-0) and Hiroshima (Hir-1) backgrounds carrying ERECTA-Col (van Zanten et al., 2009), and er-104 (Torii et al., 1996) and er-2 (CS3401), erecta mutants in Ws and Col backgrounds, respectively.
Experimental conditions
We cultivated plants at 23 °C under 16/8-h light/dark long-day conditions. Seeds were harvested and kept dry in darkness. For the experiments, 15 seeds of each genotype were sown on 0·8 % agar contained in clear plastic boxes and incubated in darkness at 5 °C for 4 d. Thereafter, the boxes with seeds were transferred to 22 °C and irradiated with a red light (R) for 3 h to promote homogeneous germination. After 24 h incubation in darkness, seedlings were exposed to WL and EOD treatments. The WL treatment consisted of growing the seedlings in a white light chamber with 8/16-h light/dark short-day photoperiod at 22 °C (WL, 85 µmol m−2 s−1, otherwise as indicated in the text). The EOD treatment consisted of adding 30 min of far-red light at the end of WL (FR, 40 µmol m−2 s−1). The FR light was provided by incandescent lamps in combination with an RG9 filter (Schott, Mainz, Germany). On day 4, the ten tallest of 15 seedlings of each box for each genotype were measured in at least three replicates.
For vegetative and flowering traits, we followed the experimental protocols indicated above. Five days after germination, seedlings were transplanted on a mixture of peat moss/vermiculite/perlite (1 : 3 : 3 ratio) contained in plastic pots. The pots were transferred and randomly distributed into a chamber under short day conditions (8/16-h light/dark) at 22 °C. The light treatment was done with mercury lamps (60 µmol m−2 s−1). At the end of the photoperiod, half of the pots received a pulse of 30 min of FR light (45 µmol m−2 s−1). Over 6 weeks, we measured petiole and lamina length, and leaf angle in the youngest leaf totally expanded. Leaf angle, defined as the angle between the petiole and a line perpendicular to the soil surface, was measured at the middle of the photoperiod with a protractor. Flowering time, determined as leaf number at bolting, was measured by counting only primary rosette leaves when the flowering stem reached 2 cm in length. All light measurements were made with a spectrophotometer (SpectroSense2/2 + ; Skye Instrument Ltd, Llandrindod Wells, UK).
Data collection and statistical analysis
We measured hypocotyl length in WL and EOD. We calculated an index of shade response as the ratio of EOD/WL following similar protocols used previously (Botto and Smith, 2002). Briefly, we estimated EOD response as the ratio of EOD/WLhypocotyl length. A similar criterion was used to calculate petiole and lamina length, and leaf angle. Because flowering time is accelerated by the EOD treatment, we estimated the EOD response for flowering as WL/EOD to maintain the index of shade response between a range of values similar to the other traits (Botto and Smith, 2002). Mean differences among treatments were tested by Student's t-test. Analyses were carried out with GraphPad Prism, Version 5·01 (GraphPad Sofware, Inc., La Jolla, CA, USA).
QTL mapping analysis
Forty-six, 66 and 99 markers obtained from Ler × No-0 (Magliano et al., 2005), Ler × Col (Lister and Dean, 1993) and Ler × Cvi (Alonso-Blanco et al., 1998) RIL populations were used for QTL analysis, respectively. MAPMAKER/EXP 3·0 was used to construct the linkage map (Lander et al., 1987). Linkage groups were verified with a minimum LOD = 2·5 and a maximum distance of 50 cM (Kosambi function). Both the linkage map data and the RIL phenotypic data were then imported to QTL Cartographer version 2·0 obtained from http://statgen.ncsu.edu/qtlcart/WQTLCart.htm (Wang et al., 2004). For the QTL analysis, we included 94 lines for the Ler × No-0 RIL population, 85 lines for the Ler × Col RIL population and 127 lines for the Ler × Cvi RIL population. We ran the data as absolute values in the WL and EOD responses. The likelihood, location, additive effect and percentage of variance explained by each QTL were calculated using model 6 based on the composite interval mapping (CIM) method (Zeng, 1994). QTL cofactors were initially selected by using forward–backward stepwise multiple regression. Mapping was conducted with a walking speed of 0·5 cM and a window size of 3 cM. For precise determination of significant QTLs, the LOD thresholds for each linkage group were calculated by a permutation test method (Doerge and Churchill, 1996) with 1000 permutations at the permutation significance level (P < 0·05). The support interval of each QTL was constructed using the 2-LOD rule with the confidence intervals being defined by all those values falling within 2-LOD score of the maximum value (Lynch and Walsh, 1998).
QTL confirmation assays
To reduce the QTL interval of EODRATIO1 in the Ler × No-0 RIL population, we generated heterogeneous inbred lines (HIF) using the advantage that some RILs are still individually segregating for one limited genomic region (Tuinstra et al., 1997). Using PCR, we screened F7 individuals of RIL152 that were still heterozygous for marker PLS5 to select the HIF152 marker polymorphic at PSL5 and with homogenous alleles in the rest of the genome. We then selected recombinant HIF (rHIF) by successive PCRs using seven polymorphic indel markers in the interval mapping between CIW3 and NGA1126 (Salathia et al., 2007). We also included two polymorphic markers at two positions of PHYB (sites 3 and 12, Filiault et al., 2008). For PHYB quantitative complementation analysis, we crossed No-0, Col, Cvi-0 and Ler parental lines with a PHYB + or phyB mutant in Ler (phyB-1) or Col (SALK_069700) following the protocols suggested by Mackay (2001). To evaluate the quantitative complementation with the phyB mutation, the phenotype of F1 individuals was analysed by a two-way ANOVA with tester genotypes as the ‘genotype’ factor and phyB mutation as the ‘phyB’ factor. Quantitative complementation of the phyB mutant alleles was detected using the non-significant ‘genotype’ by ‘phyB’ factor interaction (Mackay, 2001). Functional variation of PHYB alleles from Ler and Cvi-0 was confirmed by evaluating hypocotyl growth in at least three independent transgenic lines carrying 35S::PHYB-Cvi and 35S::PHYB-Ler alleles in a phyB-9 mutant Col background (Filiault et al., 2008).
RESULTS
Natural genetic variation for an EOD response among 108 Arabidopsis accessions
We analysed the phenotypic correlation for hypocotyl length in Arabidopsis seedlings grown under WL and EOD among 108 accessions distributed between 15 and 63 °N. Hypocotyl length varied between 1·55 and 5·86 mm in WL, and between 3·66 and 8·13 mm in EOD (Supplementary Data Fig. S1A, B). To identify the capacity of the accessions to respond to shade, we calculated different indexes for shade-avoidance elongation response. We obtained similar results using log-transformed and untransformed data, and for different ratio indexes of shade response (data not shown). Therefore, from here on we detail only the results from the absolute untransformed data and the ratio of hypocotyl length in EOD/WL (hereafter EOD response), a standardized shade response index used previosuly (Botto and Smith, 2002). The EOD response was between 1·30 and 2·73. Accessions with low EOD response included Got-7, Tsu-1, No-0 and Cvi-0, whereas Got-22 and Eden-1 appeared as the most responsive genotypes to the EOD response (Fig. 1). We found a slight positive tendency for the EOD response with an increase in latitude (Fig. 1, r = 0·33, P = 0·0004), which was negatively correlated with the pattern of response in WL (r = –0·31, P = 0·0011, Supplementary Data Fig. S1A). To confirm that the EOD treatment used in this work simulated natural shade conditions mediated principally by phyB, we studied the effects of EOD in phyB and phyAphyB double mutants in Ler and Col backgrounds (Supplementary Data Fig. S2A). As expected, the phyB mutants had a null EOD response, indicating that phyB is the phytochrome promoting the shade response (Supplementary Data Fig. S2B).
Fig. 1.
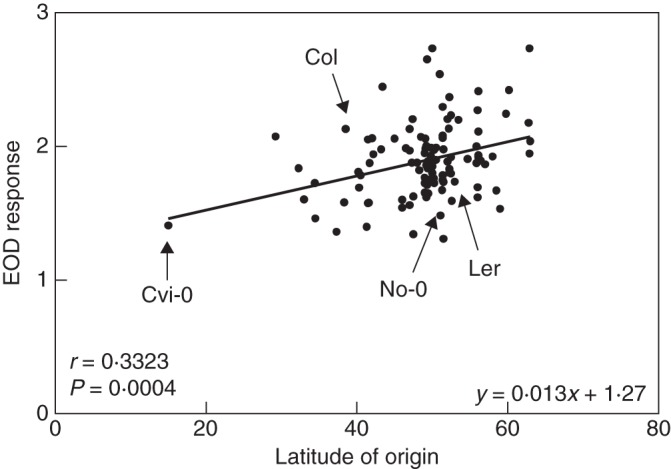
Natural variation in the EOD response for hypocotyl length in 108 accessions. The linear regression equation and the Pearson correlation index (r) with P value are also shown.
QTL mapping for EOD response in three RIL populations sharing Ler as the common parental genotype
We conducted a QTL quantitative approach using three sets of A. thaliana RIL populations, having Ler as a common parent. We used Ler × No-0, Ler × Col and Ler × Cvi-0 RIL populations (Lister and Dean, 1993; Alonso-Blanco et al., 1998; Magliano et al., 2005). We analysed the phenotypic variation of the three RIL populations in WL and EOD (Supplementary Data Fig. S3A, C, E, and Table S1). The four parental lines showed a range of variation for the EOD response between 1·13 for No-0 and 2·47 for Col (Supplementary Data Fig. S3B, D, F, and Table S1). The contribution of each parent to the EOD response was Col > Ler > Cvi-0 > No-0 (Supplementary Data Table S1). The average EOD response was 1·20 ± 0·02 for Ler × No-0, 1·75 ± 0·11 for Ler × Cvi-0 and 2·13 ± 0·12 for Ler × Col RIL populations (Supplementary Data Table S1). The genetic component of the phenotypic variation (heritability, H2) ranged between 0·54 and 0·84 for WL, 0·59 and 0·84 for EOD, and 0·30 and 0·63 for the EOD response (Supplementary Data Table S1).
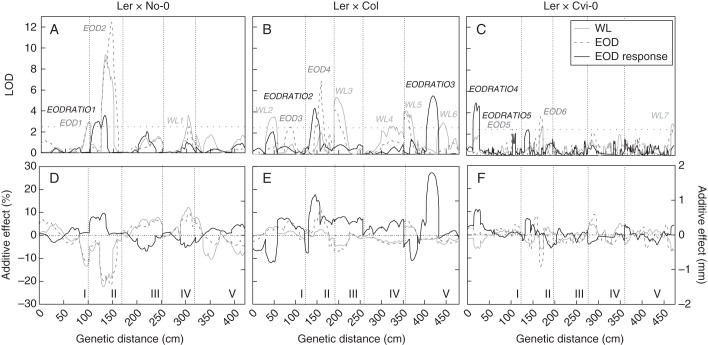
We mapped seven QTLs for WL, six QTLs for EOD and five QTLs for the EOD response (Supplementary Data Table S2). Most were specific to each trait, suggesting that the genetic architecture of the SAS elongation response depends on specific loci of small effects: WL2, WL4 and WL6 to WL; EOD2–EOD5 to EOD and EODRATIO2–EODRATIO5 to the EOD response (Supplementary Data Table S2; Fig. 2). On the other hand, WL1, WL3, WL5, WL7, EOD1 and EOD6 were common to WL and EOD conditions, but not significant for the EOD response, suggesting that these loci are involved in general elongation without effects in the EOD response (Supplementary Data Table S2; Fig. 2).
Fig. 2.
QTL mapping for the WL, EOD and EOD responses in three mapping populations sharing the Ler parent. LOD and additive effects are shown along accumulative distance (cM) of the five chromosomes of Arabidopsis (A, D for Ler × No-0; B, E for Ler × Col; and C, F for Ler × Cvi-0 RIL populations). WL, EOD and EODRATIO numbers of QTLs are arbitrary. Horizontal dotted lines represent significant thresholds for LOD scores. In the additive effect graphs, positive values indicate a positive contribution of Ler alleles and negative values indicate a positive contribution response of the alternative parent for each trait. Roman numbers at the bottom of the graphs indicate the chromosome number.
The five EODRATIO QTLs were mapped on chromosomes 1, 2 and 5 and explained between 7 and 33 % of the total phenotypic variation (Fig. 2; Supplementary Data Table S2). The Ler alleles of the five EODRATIO QTLs contributed positively to the EOD response, having the smallest effects in EODRATIO5 and the largest effects in EODRATIO3 (0·07 and 0·27, respectively; Supplementary Data Table 2). Interestingly, the EODRATIO1, 2 and 5 QTLs identified in the three RIL populations co-localized on chromosome 2, intervals of which contained PHYB but not ERECTA (Supplementary Data Table 2; Fig. S2). Finally, EODRATIO3 mapped to the middle of chromosome 5 in the Ler × Col RIL population and EODRATIO4 mapped to the top of chromosome 1 in the Ler × Cvi-0 RIL population (Supplementary Data Table S2; Fig. 2B, C).
Confirmation of EODRATIO1 into an interval of 1·2 Mb
In the Ler × No-0 RIL population, EODRATIO1 was mapped for EOD response and WL, suggesting that this QTL might underlie variation in the shade-avoidance response primarily by controlling elongation in sun conditions with high R/FR. For EODRATIO1 confirmation, we selected a heterogeneous inbred family (HIF152) polymorphic for the PSL5 marker on chromosome 2 (Fig. 3). The No-0 alleles at PSL5 in the HIF152 reduced the EOD response significantly. This accounted for a higher hypocotyl length in WL and also in EOD compared with Ler alleles (Supplementary Data Fig. S4A, B). By successive PCRs with polymorphic markers in the interval between CIW3 and NGA1126 of EODRATIO1, we selected rHIF. While rHIF13 and rHIF88 did not show EOD response differences, rHIF44 and rHIF16 showed the EOD response, confirming EODRATIO1 to an interval of 1·2 Mb, between 8148 and 9354 kb, a region that includes the PHYB gene (Fig. 3C). To evaluate whether PHYB is the candidate gene for EODRATIO1, we selected the rHIF36 with polymorphic alleles for the PHYB gene but with homozygous alleles in the confirmed interval of this QTL. Polymorphic alleles at rHIF36 did not display a significant EOD response, suggesting that PHYB is not responsible for EODRATIO1 (Fig. 3A).
Fig. 3.
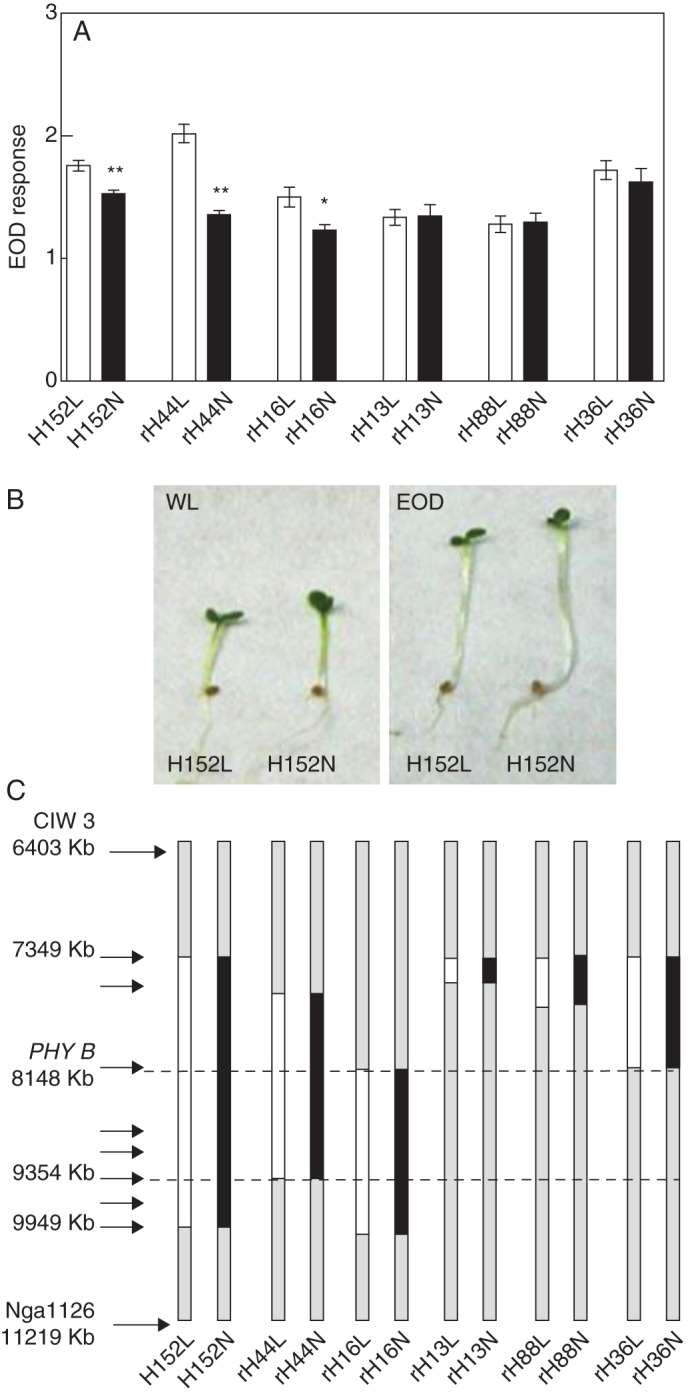
EODRATIO1 is confirmed to an interval of 1·2 Mb on chromosome 2. (A) Hypocotyl length for HIF and rHIF obtained from RIL152 polymorphic at the PSL5 marker. Each bar represents mean ± s.e.m. Significant differences between means were identified by Student's t-test and are indicated: *P < 0·05 and **P < 0·001. (B) Photographs of HIF152 grown under WL or EOD. (C) Diagram of the segregation area for HIF152 and rHIF152 lines on chromosome 2. The segregation area for each line is shown as heterozygous (Ler = white and No-0 = black) or homozygous (grey). Marker positions are indicated by arrows.
PHYB is the candidate gene for EODRATIO5 but not for EODRATIO1 or EODRATIO2
By quantitative complementation analysis, we confirmed that PHYB gene variation did not account for EODRATIO1 (Supplementary Data Fig. S5A). F1 individuals of PHYB + and phyB showed that No-0 and Ler alleles were able to complement the dysfunction of the PHYB mutant allele (P genotype × phyB = 0·11). Because PHYB was also mapped to the intervals of EODRATIO2 and EODRATIO5, we investigated whether PHYB is the candidate gene for these QTLs. By quantitative complementation analysis we confirmed that PHYB did not accounted for EODRATIO2 (P genotype × phyB = 0·43; Supplementary Data Fig. S5B). However, two pieces of evidence suggest that PHYB is the candidate gene for EODRATIO5. First, the PHYB + Ler allele failed to complement phyB mutation in F1 individuals (P genotype × phyB = 0·0006, Fig. 4A). Secondly, transgenic lines carrying PHYB alleles of Ler or Cvi-0 in the phyB-9 mutant background showed that the PHYB-Ler allele has a higher contribution to the EOD response than the PHYB-Cvi allele (Fig. 4B).
Fig. 4.
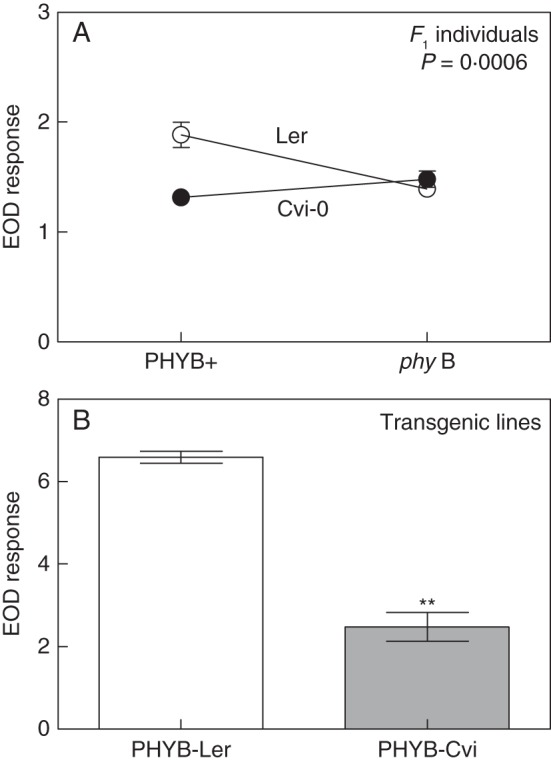
PHYB is the candidate gene for EODRATIO5. (A) Quantitative complementation test for the EOD response. The P value of the interaction between tester genotype (‘genotype’ factor) and phyB mutation in Ler background (‘phyB’ factor) is given. Each point represents the mean ± s.e.m. (B) EOD response in transgenic lines carrying 35S::PHYB-Cvi and 35S::PHYB-Ler alleles. Each bar represents mean ± s.e.m. Significant differences between means were identified by Student's t-test and are indicated: **P < 0·01.
ERECTA is the candidate gene for EOD6
While No-0, Col and Cvi-0 carry wild-type ERECTA alleles, Ler contains an induced mutation in the leucine-rich repeat receptor-like Ser/Thr kinase ERECTA gene (Torii et al., 1996). In our analysis, three QTLs were mapped at the ERECTA marker: EOD2 and EOD4 for EOD in Ler × No-0 and Ler × Col, respectively; and EOD6 for WL and EOD in Ler × Cvi-0 RIL populations (Supplementary Data Table S2). As expected for the ERECTA function in plant development, the additive effects of Ler carrying the erecta mutation at EOD2 and EOD6 were lower than No-0 and Cvi-0, respectively. However, ERECTA appears not to be responsible for EOD4 because opposite allelic effects were found (Supplementary Data Table S2). Using polymorphic NILs at the ERECTA region, we confirmed the positive contribution of Cvi-ERECTA alleles at EOD6 for the hypocotyl elongation in WL and EOD (Fig. 5). Furthermore, the EOD response was not significantly affected by ERECTA polymorphisms between Ler and Cvi-0, confirming the output of the QTL analysis (Fig. 5; Supplementary Data Table S2).
Fig. 5.
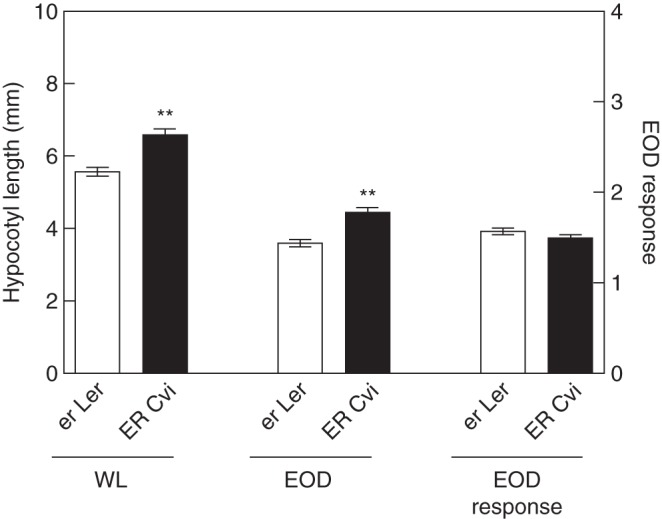
ERECTA is the candidate gene for EOD6. Hypocotyl length in WL and EOD, and EOD response of NIL carrying polymorphic ERECTA allele originating from the Ler × Cvi-0 RIL population. Each bar represents mean ± s.e.m. Significant differences between means were identified by Student's t-test and are indicated: **P < 0·001.
The effects of ERECTA depends on the genetic background
To have a better understanding of the ERECTA function in shade, we analysed the EOD response in polymorphic lines at the ERECTA gene in five genetic backgrounds. Using point mutations at the ERECTA gene in Col and Ws, and ERECTA transgenic lines in Ler, Van-0, Ws and Hir-1 accessions, we demonstrated that ERECTA effects in the EOD response are dependent on the genetic background (Fig. 6C). The erecta allele in Col and Van-0 showed a significantly higher EOD response compared with those genotypes carrying the ERECTA allele. Although the ERECTA allele did not have effects on the EOD response in Ler, Ws and Hir-1, the seedlings of these genotypes showed a higher elongation in WL and EOD as expected for the ERECTA function in cell expansion (Fig. 6A, B).
Fig. 6.
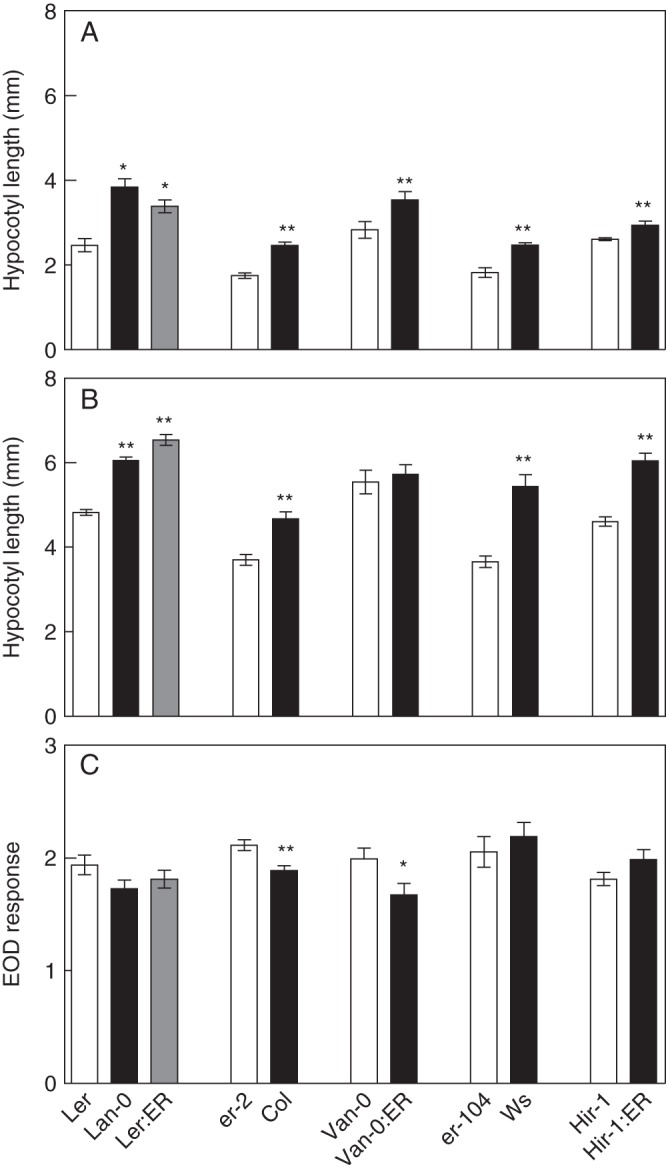
ERECTA effects on shade-avoidance responses depend on the genetic background. Hypocotyl length in WL (A), EOD (B) and EOD response (C). Each bar represents mean ± s.e.m. Significant differences between means were identified by Student's t-test and are indicated: *P < 0·05 and **P < 0·001.
ERECTA is pleiotropic to other SAS traits
The SAS is present along the life cycle of the plants. To understand the genetic-dependent function of ERECTA in the SAS, we analysed the effects of ERECTA in the shade response of vegetative and flowering traits. The erecta allele increased the EOD response in some traits depending on the genetic background, increasing leaf angle in Ws, petiole and lamina length in Van-0, and flowering shade response in Ws (Fig. 7). However, the ERECTA allele in the Ler background increased the EOD response for petiole and lamina length (Fig. 7B, C). The ERECTA allele in Ws also increased the EOD response for petiole elongation (Fig. 7B). Depending on the genetic background, the ERECTA allele increased petiole and lamina elongation significantly, and petiole angle in WL and EOD conditions, while the erecta allele in the Ws background promoted the number of leaves at flowering in WL (Supplementary Data Fig. S6).
Fig. 7.
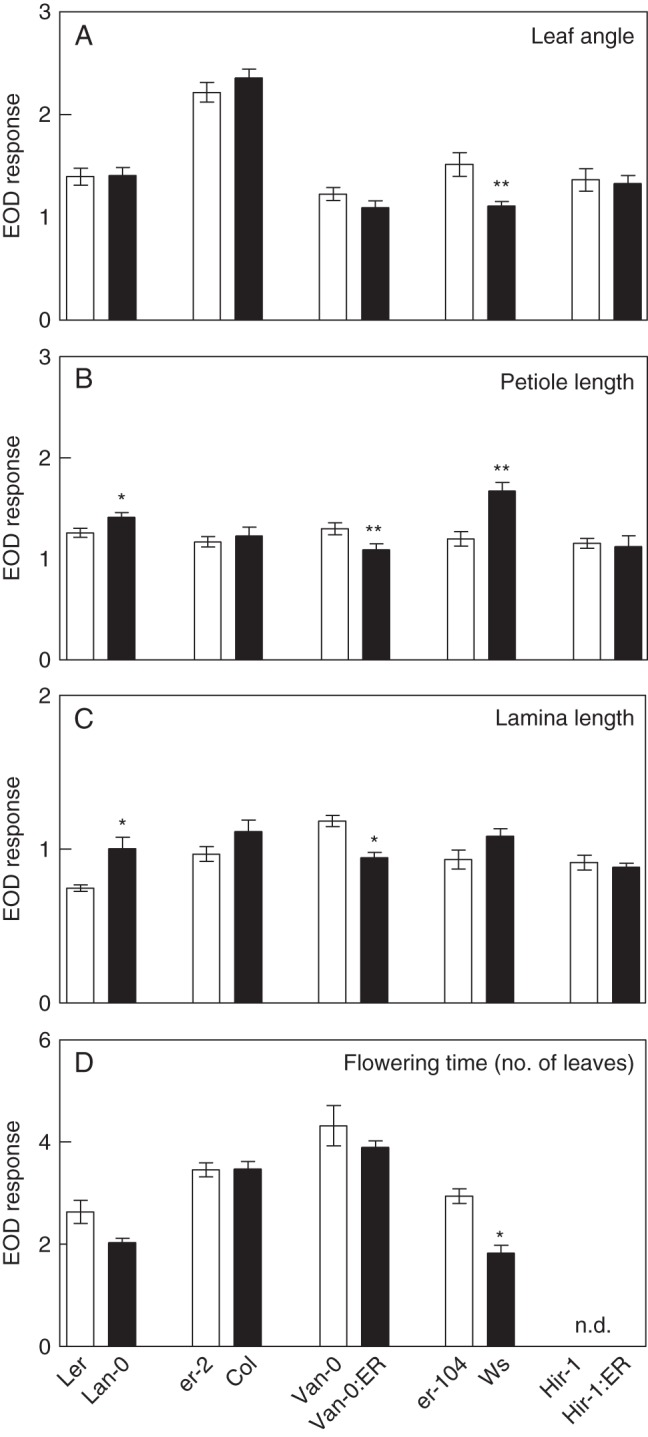
ERECTA function in the shade-avoidance syndrome is pleiotropic. EOD response for (A) leaf angle, (B) petiole length, (C) lamina length, and (D) number of leaves at flowering. Each bar represents mean ± s.e.m. Significant differences between means were identified by Student's t-test and are indicated: *P < 0·05 and **P < 0·001. nd, not determined.
DISCUSSION
In the present study, we exploited natural variation for shade-avoidance responses mapping QTLs in three RIL populations sharing Ler as a common parental genotype. SAS genetic variation was explained by several QTLs of small effect that fell into two groups. Six QTLs appeared in both WL and EOD light conditions. These loci could be responsible for the variation in general elongation without causing variation in the SAS. The second group of QTLs explained a more specific SAS genetic variation. In this group, three and two QTLs mapped specifically for WL and EOD, respectively. Furthermore, to identify specific QTLs associated with the SAS, we estimated the EOD response trait as the ratio of hypocotyl length in EOD/WL, a standardized shade response index used previously (Botto and Smith, 2002). We mapped five EODRATIO QTLs. EODRATIO1 was also found in WL, but the other four EODRATIO QTLs mapped in different loci to those identified in WL and EOD conditions (Fig. 2 and Supplementary Data Table S2). We suggest that candidate genes for EODRATIO1 might underlie variation in the shade-avoidance response primarily by controlling hypocotyl elongation in open areas, where the density of plants is low. Interestingly, using the same Ler × No-0 RIL population, a flowering time QTL co-localized with EODRATIO1 when Arabidopsis plants were grown in isolation at high R/FR ratios but not at high densities (Botto and Coluccio, 2007).
By using a candidate gene approach, we confirmed ERECTA as the gene responsible for EOD6. The leucine-rich repeat receptor-like Ser/Thr kinase gene ERECTA is involved in the plant developmental programme as well as in environmental and biotic responses controlling cell elongation (Torii et al., 1996; van Zanten et al., 2009; Fu et al., 2009). It has been described that ERECTA controls spatial and temporal patterns of epidermal cell number and size because the erecta allele leads to a low cell expansion rate (Tisné et al., 2011). It is therefore not surprising that ERECTA increased the plant growth elongation in WL and EOD light conditions. The novelty of out study is the demonstration that ERECTA is implicated in the SAS in a background-dependent manner and its effects are pleiotropic to other shade-avoidance responses. The erecta allele promoted the EOD response of hypocotyl growth in Col and Van-0, but not in Ler, Cvi-0, Hir-1 or Ws. Furthermore, the contribution of ERECTA in the EOD response was also found for leaf hyponastic growth, lamina and petiole elongation, and flowering time, suggesting that its role in shaded environments is relevant for certain populations in different phases of development. Other quantitative mapping studies also found a QTL mapped at the ERECTA marker for hypocotyl elongation in de-etiolated seedlings of a Ler × Cvi-0 RIL population grown in darkness, continuous blue light, or red and far-red pulses of light (Borevitz et al., 2002; Botto et al., 2003).
Here we propose that ERECTA is involved in canalization processes buffering the genetic variation of the SAS. Canalization is the ability of organisms to buffer their developmental processes against environmental fluctuations (Waddington, 1942; Meiklejohn and Hartl, 2002). Several kinases, such as ERECTA, are client proteins of Hsp90 that can act as a persuasive buffer against genetic variation (Jarosz and Linquist, 2010). In fact, Hsp90 protein modulated the expression of genetic variation and developmental stability in hypocotyl growth and flowering in A. thaliana (Sangster et al., 2008a, b). In one of these studies, an HSP90-responsive QTL affecting hypocotyl length was epistatic to the erecta mutation but not to wild-type ERECTA, suggesting that the pleiotropic erecta mutation may substitute Hsp90 inhibition to reveal a polymorphism at a second locus (Sangster et al., 2008a). In an independent study, Hall et al. (2007) found a QTL at ERECTA contributing to microenvironmental canalization for leaf number under long-day but not short-day photoperiods, the ERECTA effects being allele-specific and also dependent on the genetic background. The patterns of canalization could differ between pleiotropic traits because microenvironmental variation affects the degree of developmental buffering. In some circumstances, selection would favour reduced canalization and increased sensitivity to microenvironmental variations so that individuals maintain sufficient plasticity to respond to this variation. In summary, we suggest that the canalization penetrance of ERECTA in the SAS could be the result of the evolutionary trajectory of each genotype in different light environments.
Five EODRATIO QTLs were identified in this work. Interestingly, in all cases, the Ler alleles controlled hypocotyl elongation growth positively. This suggests a stronger function of Ler alleles than Col, Cvi-0 and No-0 alleles in the EOD response. In addition to the three EODRATIO QTLs co-localized on chromosome 2, we mapped EODRATIO3 to the middle of chromosome 5 in the Ler × Col RIL population (Supplementary Data Table S2). Interestingly, Coluccio et al. (2011) also detected a QTL co-localized with EODRATIO3 in the Bay × Sha RIL population, the Bay alleles having a positive contribution to hypocotyl elongation under shade. The candidate gene falling into the EODRATIO3 interval is TZP/PLUS3, a gene controlling cell-wall synthesis involved in hypocotyl length variation between Bay and Sha (Loudet et al., 2008). We also mapped EODRATIO4 to the top of chromosome 1 in the Ler × Cvi-0 RIL population. Several genes involved in the phyA responses, such as PHYA, PIF3 (Leivar and Quail, 2011) and CRY2 (Botto et al., 2003), mapped to the EODRATIO4 interval. Recently, it had been shown that genetic variants of phyA signalling loci are significantly associated with simulated shade in a genome-wide association study with 200 Arabidopsis accessions (Filiault and Maloof, 2012).
We have shown here that PHYB is the candidate gene for EODRATIO5 mapped in the Ler × Cvi-0 RIL population, but not for two co-localized EODRATIO QTLs detected in Ler × Col and Ler × No-0 RIL populations. We also found a lower sensitivity response of Cvi-PHYB than the Ler-PHYB allele under WL and EOD responses. Seedlings of both genotypes exposed to continuous light also showed a low red sensitivity response for the Cvi-PHYB allele (Filiault et al., 2008). Interestingly, in a sequence diversity analysis of PHYB, Ler, Col and No-0 PHYB variants segregated together in a Central European clade and the Cvi-0 variant grouped with four Spanish accessions in an isolated clade (Filiault et al., 2008). Furthermore, we showed a mild positive tendency to display a higher EOD response for hypocotyl elongation in a north–south latitudinal cline, probably as a consequence of a lower light sensitivity in WL of the south-located genotypes (Supplementary Data Fig. S1). However, in certain cases, the evolution of shade response would be the consequence of local adaptations to light variations. For example, we found two accessions from the same geographical location in Germany, Got-7 and Got-22, with contrasting EOD response (1·35 vs. 2·74, respectively, Fig. 1). In fact, the distribution of each population could be the result of individual adaptation to a particular habitat integrating complex networks of light signals and other environmental signals to increase its fitness.
While this manuscript was under final revision, Patel et al. (2013) reported similar findings in the Plant Journal. These authors found that ERECTA function in shade environments is dependent on growth temperature and genetic background.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank Joost Keurentjes, Keiko Torii, Julin Maloof, Ton Peeters, Martijn van Zanten and ABRC for providing seed materials. We also thank Paula Coluccio for helping in the Ler × Col RIL measurements. Financial support to J.F.B. came from the University of Buenos Aires (UBACyT20020100100774) and the Agencia Nacional de Promoción Científica y Tecnológica (PICT08-1061).
LITERATURE CITED
- Alonso-Blanco C, Peeters AJM, Koornneef M, et al. Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant Journal. 1998;14:259–271. doi: 10.1046/j.1365-313x.1998.00115.x. [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Hirschfeld M, Wester L, et al. A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. Plant Cell. 1997;9:1317–1326. doi: 10.1105/tpc.9.8.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Maloof J, Lutes J, et al. Quantitative trait loci controlling light and hormone response in two accessions of Arabidopsis thaliana. Genetics. 2002;160:683–696. doi: 10.1093/genetics/160.2.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto JF, Coluccio MP. Seasonal and plant-density dependency for quantitative trait loci affecting flowering time in multiple populations of Arabidopsis thaliana. Plant, Cell and Environment. 2007;30:1465–1479. doi: 10.1111/j.1365-3040.2007.01722.x. [DOI] [PubMed] [Google Scholar]
- Botto JF, Smith H. Differential genetic variation in adaptive strategies to a common environmental signal in Arabidopsis accessions: phytochrome-mediated shade avoidance. Plant, Cell and Environment. 2002;25:53–63. [Google Scholar]
- Botto JF, Alonso-Blanco C, Garzaron I, Sanchez RA, Casal JJ. The Cape Verde Islands allele of cryptochrome 2 enhances cotyledon unfolding in the absence of blue light in Arabidopsis. Plant Physiology. 2003;133:1547–1556. doi: 10.1104/pp.103.029546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock MT, Maloof JN, Weinig C. Genes underlying quantitative variation in ecologically important traits: PIF4 (phytochrome interacting factor 4) is associated with variation in internode length, owering time, and fruit set in Arabidopsis thaliana. Molecular Ecology. 2010;19:1187–1199. doi: 10.1111/j.1365-294X.2010.04538.x. [DOI] [PubMed] [Google Scholar]
- Casal JJ. Shade avoidance. The Arabidopsis Book. 2012;10 doi: 10.1199/tab.0157. e0157. http://dx.doi.org/10.1199/tab.0157 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluccio MP, Sanchez SE, Kasulin L, Yanovsky MJ, Botto JF. Genetic mapping of natural variation in a shade avoidance response: ELF3 is the candidate gene for a QTL in hypocotyl growth regulation. Journal of Experimental Botany. 2011;62:167–176. doi: 10.1093/jxb/erq253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocco C, Holm M, Yanovsky MJ, Botto JF. AtBBX21 and COP1 genetically interact in the regulation of shade avoidance. Plant Journal. 2010;64:551–562. doi: 10.1111/j.1365-313X.2010.04360.x. [DOI] [PubMed] [Google Scholar]
- Crocco CD, Holm M, Yanovsky MJ, Botto JF. Function of B-BOX under shade. Plant Signaling Behavior. 2011;6:101–104. doi: 10.4161/psb.6.1.14185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djakovic-Petrovic T, de Wit M, Voesenek LACJ, Pierik R. DELLA protein function in growth responses to canopy signals. Plant Journal. 2007;51:117–126. doi: 10.1111/j.1365-313X.2007.03122.x. [DOI] [PubMed] [Google Scholar]
- Doerge RW, Churchill GA. Permutation tests for multiple loci affecting a quantitative character. Genetics. 1996;142:285–294. doi: 10.1093/genetics/142.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas SJ, Chuck G, Dengler RE, Pelecanda L, Riggs CD. KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. Plant Cell. 2002;14:547–558. doi: 10.1105/tpc.010391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiault DL, Maloof JN. A genome-wide association study identifies variants underlying the Arabidopsis thaliana shade avoidance response. PLoS Genetics. 2012;8 doi: 10.1371/journal.pgen.1002589. e1002589. http://dx.doi.org/10.1371/journal.pgen.1002589 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiault DL, Wessinger CA, Dinneny JR, et al. Amino acid polymorphisms in Arabidopsis phytochrome B cause differential responses to light. Proceedings of the National Academy of Sciences, USA. 2008;105:3157–3162. doi: 10.1073/pnas.0712174105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Keurentjes JJB, Bouwmeester H, et al. System-wide molecular evidence for phenotypic buffering in Arabidopsis. Nature Genetics. 2009;41:166–167. doi: 10.1038/ng.308. [DOI] [PubMed] [Google Scholar]
- Galstyan A, Cifuentes-Esquivel N, Bou-Torrent J, MartinezGarcia JF. The shade avoidance syndrome in Arabidopsis: a fundamental role for atypical basic helix–loop–helix proteins as transcriptional cofactors. Plant Journal. 2011;66:258–267. doi: 10.1111/j.1365-313X.2011.04485.x. [DOI] [PubMed] [Google Scholar]
- Godiard L, Sauviac L, Torii KU, et al. ERECTA, an LRR receptor-like kinase protein controlling development pleiotropically affects resistance to bacterial wilt. Plant Journal. 2003;36:353–65. doi: 10.1046/j.1365-313x.2003.01877.x. [DOI] [PubMed] [Google Scholar]
- Hall MC, Dworkin I, Ungerer MC, Purugganan M. Genetics of microenvironmental canalization in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 2007;104:13717–13722. doi: 10.1073/pnas.0701936104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO Journal. 2009;28:3893–3902. doi: 10.1038/emboj.2009.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz DF, Linquist S. Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science. 2010;330:1820–1824. doi: 10.1126/science.1195487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Gómez JM, Wallace AD, Maloof JN. Network analysis identifies ELF3 as a QTL for the shade avoidance response in Arabidopsis. PLoS Genetics. 2010;6 doi: 10.1371/journal.pgen.1001100. e1001100. http://dx.doi.org/10.1371/journal.pgen.1001100 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes JJB, Fu J, Terpstra IR, et al. Regulatory network construction in Arabidopsis by using genome-wide gene expression quantitative trait loci. Proceedings of the National Academy of Sciences, USA. 2007;104:1708–1713. doi: 10.1073/pnas.0610429104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH. PIFs: pivotal components in a cellular signaling hub. Trends in Plant Science. 2011;16:19–28. doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, et al. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Lister C, Dean C. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant Journal. 1993;4:745–750. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- Llorente F, Alonso-Blanco C, Sanchez-Rodriguez C, Jorda L, Molina A. ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant Journal. 2005;43:165–180. doi: 10.1111/j.1365-313X.2005.02440.x. [DOI] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant Journal. 2008;53:312–323. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- Loudet O, Michael TP, Burger BT, et al. A zinc knuckle protein that negatively controls morning-specific growth in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 2008;105:17193–17198. doi: 10.1073/pnas.0807264105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Walsh J. Genetics and analysis of quantitative traits. Sunderland, MA: Sinauer Associates; 1998. [Google Scholar]
- Mackay TFC. The genetic architecture of quantitative traits. Annual Review of Genetics. 2001;35:303–339. doi: 10.1146/annurev.genet.35.102401.090633. [DOI] [PubMed] [Google Scholar]
- Magliano TM, Botto JF, Godoy AV, et al. New Arabidopsis recombinant inbred lines (Landsberg erecta × Nossen) reveal natural variation in phytochrome-mediated responses. Plant Physiology. 2005;138:1126–1135. doi: 10.1104/pp.104.059071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masle J, Gilmore SR, Farquhar GD. The ERECTA gene regulates plant transpiration efficiency in Arabidopsis. Nature. 2005;436:866–870. doi: 10.1038/nature03835. [DOI] [PubMed] [Google Scholar]
- Meiklejohn CD, Hartl DL. A single mode of canalization. Trends in Ecology & Evolution. 2002;17:468–473. [Google Scholar]
- Patel D, Basu M, Hayes S, et al. Temperature-dependent shade avoidance involves the receptor-like kinase ERECTA. Plant Journal. 2013 doi: 10.1111/tpj.12088. in press. http://dx.doi.org/10.1111/tpj.12088 . [DOI] [PubMed] [Google Scholar]
- Salathia N, Lee HN, Sangster TA, et al. Indel arrays: an affordable alternative for genotyping. Plant Journal. 2007;51:727–737. doi: 10.1111/j.1365-313X.2007.03194.x. [DOI] [PubMed] [Google Scholar]
- Sangster TA, Salathia N, Undurraga S. HSP90 affects the expression of genetic variation and developmental stability in quantitative traits. Proceedings of the National Academy of Sciences, USA. 2008a;105:2963–2968. doi: 10.1073/pnas.0712200105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangster TA, Salathia N, Lee HN, et al. HSP90-buffered genetic variation is common in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 2008b;105:2969–2974. doi: 10.1073/pnas.0712210105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt J. Is photomorphogenic shade avoidance adaptative? Perspectives from population biology. Plant, Cell and Environment. 1997;20:826–830. [Google Scholar]
- Smith H. Light quality, photoperception and plant strategy. Annual Review of Plant Physiology and Plant Molecular Biology. 1982;33:481–518. [Google Scholar]
- Tisné S, Barbier F, Granier C. The ERECTA gene controls spatial and temporal patterns of epidermal cell number and size in successive developing leaves of Arabidopsis thaliana. Annals of Botany. 2011;108:159–168. doi: 10.1093/aob/mcr091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, et al. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell. 1996;8:735–746. doi: 10.1105/tpc.8.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuinstra MR, Ejeta G, Goldsbrough PB. Heterogeneous inbred family (HIF) analysis: a method for developing near-isogenic lines that differ at quantitative trait loci. Theoretical and Applied Genetics. 1997;95:1005–1011. [Google Scholar]
- Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- Wang X, Korstanje R, Higgins D, Paigen B. Haplotype analysis in multiple crosses to identify a QTL gene. Genome Research. 2004;14:1767–1772. doi: 10.1101/gr.2668204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zanten M, Snoek LB, Proveniers MC, Peeters AJ. The many functions of ERECTA. Trends Plant Science. 2009;14:214–218. doi: 10.1016/j.tplants.2009.01.010. [DOI] [PubMed] [Google Scholar]
- van Zanten M, Snoek LB, van Eck-Stouten E. Ethylene-induced hyponastic growth in Arabidopsis thaliana is controlled by ERECTA. Plant Journal. 2010;61:83–95. doi: 10.1111/j.1365-313X.2009.04035.x. [DOI] [PubMed] [Google Scholar]
- Zeng ZB. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–1468. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.