Abstract
Background and Aims
It has previously been shown that proanthocyanidins (PAs) in the seed coat of Arabidopsis thaliana have the ability to scavenge superoxide radicals (O2−). However, the physiological processess in PA-deficit seeds are not clear. It is hypothesized that there exist alternative ways in PA-deficient seeds to cope with oxidative stress.
Methods
The content of hydrogen peroxide (H2O2) and its relevance to the activities of superoxide dismutase (SOD), catalase (CAT) and peroxidases was investigated in both wild-type and PA-deficit mutant seeds. A biochemical staining approach was used to detect tissue localizations of peroxidase activities in PA-deficit mutant seeds.
Key Results
PA-deficient mutants possess significantly lower levels of H2O2 than the wild-type, despite their higher accumulation of superoxide radicals. Screening of the key antioxidant enzymes revealed that peroxidase activity was significantly over-activated in mutant seeds. This high peroxidase activity was mainly confined to the seed coat zone. Interestingly, neither ascorbate peroxidase nor glutathione peroxidase, just the guaiacol peroxidases (class III peroxidases), was specifically activated in the seed coat. However, no significant difference in peroxidase activity was observed in embryos of either mutants or the wild-type, although gene expressions of several candidate peroxidases were down-regulated in the embryos of PA-deficient seeds.
Conclusions
The results suggest that enhanced class III peroxidase activity in the seed coat of PA-deficient mutants is an adaptive strategy for seed development and survival.
Keywords: Proanthocyanidins, peroxidase, seed coat, seed development, seed germination, anti- oxidation, Arabidopsis thaliana
INTRODUCTION
The dual roles of reactive oxygen species (ROS) have been documented extensively in previous studies. At high concentrations they trigger genetically programmed cell suicide. However, they also have an important function as secondary messengers in signal transduction cascades, which regulate many physiological and developmental processes in plants (Apel and Hirt, 2004; Foyer and Noctor, 2005; Mittler et al., 2011). Several forms of ROS are present in plants, including the superoxide radical (O2−), singlet oxygen (1O2), hydroxyl radical (OH·) and hydrogen peroxide (H2O2) (Vranova et al., 2002). H2O2 has weak toxicity compared with other ROS species but it can trigger highly reactive hydroxyl radicals via the Haber–Weiss reaction. The relatively longer life span of H2O2 makes it harmful to organelles (Levine et al., 1994; Willekens et al., 1997; Apel and Hirt, 2004; Foyer and Noctor, 2005; Moller et al., 2007).
Plants have a complex antioxidant system for maintaining the homeostasis of ROS. In general, this system can be divided into enzymatic and non-enzymatic ROS-scavenging mechanisms. Three types of antioxidative enzymes, superoxide dismutase (SOD), catalase (CAT) and peroxidase, play a major role in keeping superoxide radicals and H2O2 at steady-state levels (Bowler et al., 1992; Willekens et al., 1997; Apel and Hirt, 2004). There are two classes of peroxidases in plants: class I and class III (Cosio and Dunand, 2009). Ascorbate peroxidase (APx) and glutathione peroxidase (GPx) belong to class I, which can detoxify H2O2 into H2O (Mittler, 2002). Class III peroxidases are encoded by a large multi-gene family. Altogether, 73 members have been identified in Arabidopsis thaliana (hereafter arabidopsis) (Hiraga et al., 2001; Almagro et al., 2009). Although they play a role in antioxidants by removing ROS, they also produce ROS. The bifunctional enzymes of class III peroxidases also participate in a broad range of physiological processes, such as defence against pathogens, the formation of lignin and suberin, cross-linking of cell-wall components, auxin catabolism, and seed germination and senescence (Passardi et al., 2004b; Almagro et al., 2009).
The main non-enzymatic antioxidants include ascorbate (vitamin C), α-tocopherol (vitamin E), glutathione (GSH), carotenoids and flavonoids, and these substances also play an important role in compromising oxidative damage as major cellular redox buffers (Noctor and Foyer, 1998; Sattler et al., 2004, 2006; Han et al., 2009; Foyer and Noctor, 2011). Most flavonoids outperform other antioxidants in in vitro antioxidant assays although relatively little has been reported compared with other non-enzymatic antioxidants such as ascorbate and α-tocopherol (Hernandez et al., 2009). Flavonoids are important secondary metabolites that originate from general phenopropanoid metabolism (Vogt, 2010). More than 9000 individual molecules have been identified, and it is believed that higher plants are the only natural source of flavonoids (Hernandez et al., 2009). Flavonoids are categorized into the following groups based on the oxidation levels of the central C-ring, stereochemistry, type and degree of polymerization, position and nature of the substitutions, and linkages between basic units: anthocyanidin, flavanone, proanthocyanidins (PAs) and flavone or flavonol (Winkel-Shirley, 2001; Lepiniec et al., 2006; Routaboul et al., 2006).
Several species of flavonoids have been investigated for their chemical identity and antioxidant capacity by in vitro experiments (Proteggente et al., 2003; Spencer et al., 2003). Flavonoids function as antioxidants mainly by scavenging peroxyl radicals but have minimal impact on OH radicals (Deng et al., 1997). Guaiacol peroxidase catalyses the H2O2-dependent oxidation of flavonols, which is evidence of an H2O2-scavenging mechanism mediated by a flavonoid–peroxidase reaction (Yamasaki et al., 1997). Although the antioxidant capacities of flavonoids are genuine and even greater than other extensively investigated antioxidants, many aspects of their putative functions as antioxidants remain unclear (Hernandez et al., 2009). The main obstacle to understanding their functions is the different subcellular localization of flavonoids from the site of ROS production. However, a recently discovered transport mechanism for anthocyanin from its synthesis site to the sites of storage and disposal, mediated by an endoplasmic reticulum-derived vesicle-like structure, provides the possibility of an interaction between flavonoids and ROS (Poustka et al., 2007; Gomez et al., 2011).
PAs are major end-products of the flavonoid biosynthesis pathway and mainly accumulate in the seed coat of arabidopsis (Debeaujon et al., 2003; Xie et al., 2003; Lepiniec et al., 2006). Among the many TRANSPARENT TESTA (TT) genes involved in PA biosynthesis, TT2, TT8 and TTG1 form a ternary complex to directly regulate BANYULS (BAN) expression (Baudry et al., 2004). The precise functions of the other three regulatory factors, including TT1, TT16 and TTG2, remain unclear and thus there is a need for further investigation (Lepiniec et al., 2006). TT18 encodes a leucocyanidin dioxygenase that is located downstream of the flavonoid pathway and is essential for the biosynthesis of PAs (Abrahams et al., 2003). PA-deficient mutants exhibit yellow or pale brown seed coat colour due to reduced flavonoid pigmentation and accumulation (Debeaujon et al., 2000; Lepiniec et al., 2006). Most PAs found in seeds are soluble procyanidin polymers, consisting of two stereoisomers: epicatechin (EC, 2-3-cis) and catechin (C, 2-3-trans) and their degree of polymerization can be up to 9 (Harborne and Williams, 2000; Routaboul et al., 2006). However, only EC with a mean degree of polymerization between 5 and 8 is detected in the seeds of arabidopsis (Abrahams et al., 2003; Routaboul et al., 2006).
Our most recent study also provided evidence of the antioxidation ability of PAs by in vivo experiments in arabidopsis seed. We found that the superoxide radical (O2−) accumulated more in PA-deficient mutant seeds than in wild-type seeds, which demonstrates that PAs function as antioxidants via the removal of O2− (Jia et al., 2012b). The PAs in the seed coat are likely to play an important role in buffering ROS homeostasis during maturation and germination (Marles et al., 2003; Routaboul et al., 2006). However, it would be difficult for PAs to exert their antioxidant functions following accumulation in the vacuole if they are unable to break the physical barrier created by the tonoplast (Gould et al., 2002; Hernandez et al., 2009). In the later stages of seed development, the seed coat experiences programmed cell death and releases PAs from the vacuole, making them available to exert their antioxidant properties (Debeaujon et al., 2003; Dixon et al., 2005; Haughn and Chaudhury, 2005). However, there are no reports of decreased seed quality of freshly harvested PA-deficient mutants. How the antioxidant system in these PA-deficient mutants operates is not clear. We investigated the hypothesis that these mutants have acquired an alternative antioxidant mechanism compared with the wild-type, which enables them to develop without the protection of PAs. Our results demonstrate that increased activity of class III peroxidases in the seed coat of PA-deficient mutant seeds may compensate for the loss of PAs.
MATERIALS AND METHODS
Plant material
The following mutants of arabidopsis (Arabidopsis thaliana) have been described previously: tt1-6 (SALK_107737C; Appelhagen et al., 2011), tt2-5 (SALK_005260; Nesi et al., 2001), tt8-4 (SALK_030966), tt8-5 (SALK_048673) (Nesi et al., 2000) and tt18-3 (SALK_028793; Shikazono et al., 2003; Jia et al., 2012a). Plants were kept in a growth chamber at 22 ± 2 °C with a 16-h photoperiod at a photon flux density of approx. 200 µmol m−2 s−1 and a relative humidity of 80 %. Seeds of the wild-type (Col0) and the mutants were grown in PlantMate compost (Huadu Co., Kong Kong) for all-purpose potting soil and harvested at the same time. The seed samples after harvest were stored in a dehumidifier cabinet for 2 months before using them in the experiments.
Seed coat isolation
Seed coats were isolated to investigate antioxidant enzyme activities. Gentle manipulation with fine forceps under a dissecting microscope allowed the release of embryos from the micropylar end of the seed coat. Isolated seed coats were washed with 5 % sucrose solution to remove the endosperm layer. Two hundred dissected seeds were collected in separate Eppendorf tubes for sample extraction (Dean et al., 2011).
Chemical treatments
Three oxidants or oxidative inducers were selected for H2O2 determination, antioxidant enzyme detection and gene expression analysis at the transcriptional level. All three chemicals [methyl viologen (MV), CuSO4 (Cu2+) and tert-butylhydroperoxide (t-BHP)] were obtained from Sigma-Aldrich (St Louis, MO, USA). A 200-μL solution including 0·5 µm MV, 0·4 mm Cu2+ or 0·5 mm t-BHP was added to approx. 50 mg seed sample for oxidative stress treatment, and the same volume of water was added to the control (Van Camp et al., 1996; Bae et al., 2010).
H2O2 measurements
An amplex red H2O2/peroxidase assay kit (Molecular Probes, Eugene, OR, USA) was used for the detection of endogenous concentrations of H2O2. Approximately 50 mg seed samples were ground in 20 mm sodium phosphate buffer (pH 7·4). The mixture was centrifuged at 10 000 g for 10 min at 4 °C, and the supernatant was used for subsequent assays. The detailed procedure followed the protocol of the assay kit and 50 µL supernatant was used for each reaction. The reaction proceeded for 30 min in darkness and the absorbance at 560 nm was read using FLUOStar Optima. The concentrations were calculated according to the standard curve (Shin and Schachtman, 2004; Lee et al., 2010).
Enzyme assays
Approximately 50 mg seed sample was crushed into a fine powder in a mortar and pestle under liquid nitrogen. One microlitre of 50 mm potassium phosphate buffer (pH 7·0) containing 1 mm EDTA and 1 % polyvinylpyrrolidone (PVP) was added and the sample was mixed. The homogenate was centrifuged at 15 000 g for 20 min at 4 °C and the supernatant was used for enzyme assays.
Total superoxide dismutase (SOD) (EC 1·15·1·1) activity was analysed using nitro blue tetrazolium (NBT) following the method of Giannopolitis and Ries (1977) with some modifications. The 1-mL reaction mixture contained 50 mm potassium phosphate buffer (pH 7·8), 13 mm methionine, 75 µm NBT, 2 µm riboflavin, 0·1 mm EDTA and 20 µL enzyme extract. The reaction mixtures were illuminated for 15 min at a light intensity of 5000 lx. One unit of SOD activity was defined as the amount of enzyme required to cause 50 % inhibition of the reduction of NBT as monitored at 560 nm (Giannopolitis and Ries, 1977).
Catalase (CAT) (EC 1·11·1·6) activity was determined by observing the consumption of H2O2 (extinction coefficient 39·4 mm−1 cm−1) at 240 nm for 3 min (Aebi, 1984). The reaction mixture contained 50 mm potassium phosphate buffer (pH 7·0), 10 mm H2O2 and 200 µL enzyme extract in a 3-mL volume (Jiang and Zhang, 2002).
Peroxidase was detected with an Amplex red H2O2/peroxidase assay kit (Molecular Probes). The extraction method was the same as that used for the H2O2 measurement but H2O2 was added to the seed sample solutions instead of horseradish peroxidase (HRP). A standard curve was produced using the diluted HRP solutions (0–2 mU mL−1) provided in the assay kit (Shin and Schachtman, 2004; Lee et al., 2010).
Guaiacol peroxidase activity was assayed in a reaction mixture consisting of 100 mm potassium phosphate buffer (pH 6·5), 15 mm guaiacol, 0·05 % (v/v) H2O2 and 60 µL enzyme extract diluted between 1 : 40 and 1 : 80 (v/v) with assay buffer. The reaction was initiated by adding H2O2, and the oxidation of guaiacol was determined based on the increase in A470 (ɛ = 26·6 mm−1 cm−1). One guaiacol peroxidase unit is defined as the amount of enzyme that produces 1 µmol min−1 oxidized guaiacol under the above assay conditions (Garcia-Limones et al., 2002). For the imaging of guaiacol peroxidase activity, 200 detached seed coats were ground in liquid nitrogen and reacted in the solution described above for 3 min before photos were taken.
APx activity was determined by observing the decrease in A290 (extinction coefficient 2·8 mm−1 cm−1) for 30 s in 1-mL reaction mixture containing 50 mm potassium phosphate buffer (pH 7·0), 0·5 mm ASC, 0·1 mm H2O2 and 20 µL enzyme extract. The reaction was initiated with an enzyme extract. A correction was made for the low, non-enzymatic oxidation of ascorbate by H2O2 (Nakano and Asada, 1981).
In-gel GPx activity was detected using 8 % native PAGE gels. Approximately 150 µg protein per sample was loaded onto a gel and run at 4 °C. The gel was removed from the glass plates, placed into a glass staining dish and washed for 3× 10 min in distilled water containing GSH using approx. 50 mL per wash. The stain was prepared in two 50-mL conical tubes by preparing 1 % ferric chloride in one tube and in a second tube preparing 1 % potassium ferricyanide. The gel was incubated in 100 mL ddH2O containing 0·008 % cumene hydroperoxide for 10 min. The gel was rinsed twice with ddH2O and incubated with the stains of a mixture of the two reagents prior to staining. They were poured directly on top of the gel. When achromatic bands began to form (5–15 min), the stain was poured off and the gel was rinsed with ddH2O. The formation of chromatic bands demonstrated the presence of GPx activity (Weydert and Cullen, 2010).
Histochemical detection of peroxidase activity
The imbibed seeds were spread between two glass slides and squeezed slightly to ensure that the cell inclusions were wholly or partially out of the testa. Then 3,3′, 5,5′-tetramethylbenzidine (TMB) Stabilized Substrate for Horseradish Peroxidase (Promega, Madison, WI, USA) and 1 mm H2O2 were added to squeezed seed samples. Peroxidase localization was achieved by short-term (10 min) staining for seed coats and long-term (1 h) staining for embryos (Linkies et al., 2010). For relative absorption value measurements of the reaction products spread from testa, 200 wild-type and/or PA-deficient mutant seeds were squeezed and the samples were collected into Eppendorf tubes. Following staining with 200 µL TMB solution, the supernatant was collected after being centrifuged at 10 000 g for 1 min. The reaction was stopped after 10 min with 50 µL H3PO4 and the absorbance at 450 nm was measured (Vanhercke et al., 2005).
Total RNA extraction and transcript level analysis
A plant RNA Isolation Mini Kit (Agilent, Palo Alto, CA, USA) was used for total RNA extraction from seed samples according to the manufacturer's instructions. After being reverse transcribed using the SuperScript RT-PCR system (Invitrogen) the cDNA was used to perform the QRT-PCR analysis. IQ SYBR Green Supermix (Bio-Rad) and iCycle (Bio-Rad) were used. UBQ5 was used as an internal standard to normalize the data, and a second reference gene, ACT8, was used to re-confirm the results (Graeber et al., 2011). The primers used for gene expression analysis by QRT-PCR are shown in Table 1.
Table 1.
Sequence of primers used for QRT-PCR
| Primer name | Primer sequence |
|---|---|
| APX1-QRT-S | 5′ GGTCGTCTTCCTGATGCTAC 3′ |
| APX1-QRT-R | 5′ TGGCATCGTCCCAGAGTGT 3′ |
| Prx16-QRT-S | 5′ TTGCTTCGTCCGTGGATG 3′ |
| Prx16-QRT-R | 5′ CTATCAAGGGCCTGCTTTG 3′ |
| PrxIIE-QRT-S | 5′ AATGGCGAATTCACAGGG 3′ |
| PrxIIE-QRT-R | 5′ CACCTCCTTCTTCAAGATTCAG 3′ |
| UBQ5-QRT-S | 5′ GCATGCAAGCTTGGCGTAA 3′ |
| UBQ5-QRT-R | 5′ TGAGCGGATAACAATTTCACACA 3′ |
| ACT8-QRT-S | 5′ CTCAGGTATTGCAGACCGTATGAG 3′ |
| ACT8-QRT-R | 5′ CTGGACCTGCTTCATCATACTCTG 3′ |
Accession numbers
The sequence data described in this article can be found in the Arabidopsis Genome Initiative or TIGR databases under the following accession numbers: UBQ5 (AT3G62250); ACT8 (AT1G49240); APX1 (AT1G07890); Prx16 (AT2G18980); PrxIIE (AT3G52960).
RESULTS
Accumulation of endogenous H2O2 in PA-deficient mutant seeds
H2O2 was detected in both the non-dormant wild-type and several PA-deficient mutant seeds. Lower concentrations of H2O2 were observed in tt mutant seeds, including tt1-6, tt2-5, tt8-4 and tt18-3, than in the wild-type (Fig. 1A). Two PA-deficient mutants, tt2-5 and tt18-3, were selected to investigate H2O2 accumulation under normal and oxidative stress conditions in imbibed seeds. Three oxidants or oxidative inducers, MV, CuSO4 (Cu2+) and t-BHP, were chosen and two time points, short (3 h) and long (24 h), were set for detection. The mutants displayed more H2O2 accumulation over the short-term water imbibition period (Fig. 1B), which induced a rapid germination of tt mutants as reported earlier (Jia et al., 2012a). Under oxidative stress, the H2O2 content increased in wild-type seeds for both short- and long-term treatments. However, this did not occur in PA-deficient mutants. Not only was there no marked increase in the H2O2 content, but a decrease was detected under some oxidative treatments such as the Cu2+ and t-BHP in the short-time treatment (Fig. 1B).
Fig. 1.
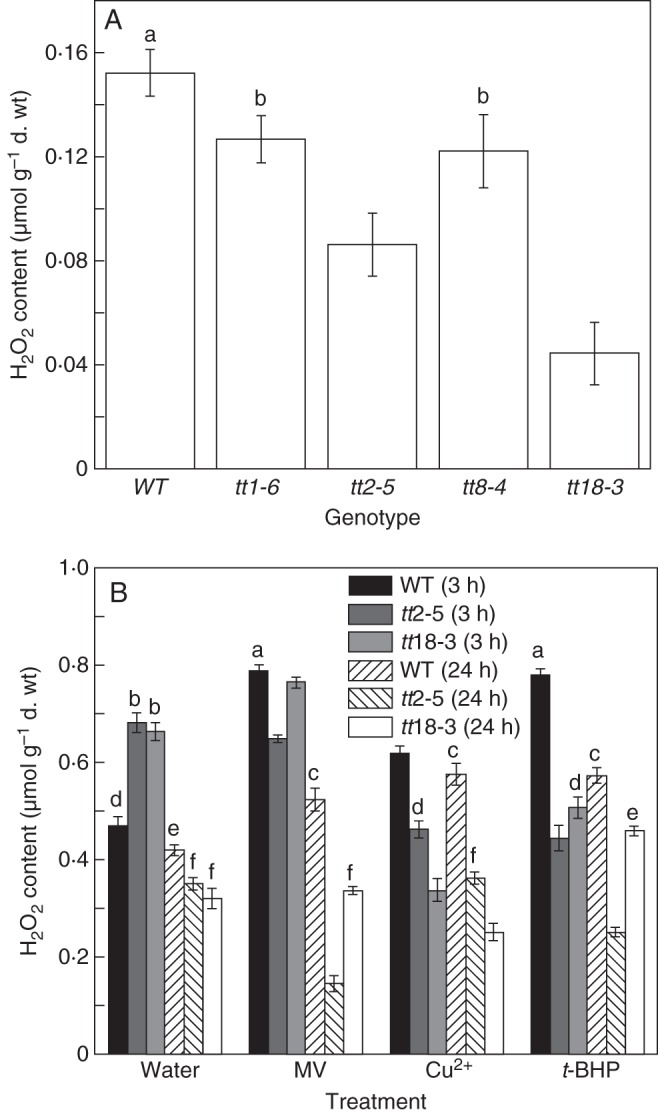
Comparison of hydrogen peroxide (H2O2) content between wild-type arabidopsis seeds and PA-deficient mutants under normal and oxidative stress conditions. (A) H2O2 content in non-dormant, dry, wild-type seeds and PA-deficient mutants including tt1-6, tt2-5, tt8-4 and tt18-3. (B) Three different inducers of oxidative stress [methyl viologen (MV), CuSO4 (Cu2+), t-butyl hydroperoxide (t-BHP)] and two PA-deficient mutants (tt2-5 and tt18-3) were selected for further detection of H2O2 concentrations over short-term (3 h) and long-term (24 h) imbibition periods. Water imbibition was used as a control. Values are means and s.e. (n = 3). Means denoted by different letters are significantly different at P < 0·05 according to Duncan's multiple range test.
Comparison of antioxidant enzymes between wild-type seeds and PA-deficient mutants
SOD, CAT and peroxidase activities were compared between the wild-type and mutant seeds under normal and oxidative conditions. Relatively lower CAT and SOD activities were detected in the mutants than in the wild-type seeds under not only normal conditions but also oxidative stress conditions induced by treatment with three oxidants (Fig. 2A, B). Increased SOD activity and decreased CAT activity under oxidative stress conditions might be the result of concentration effects of oxidative stress.
Fig. 2.
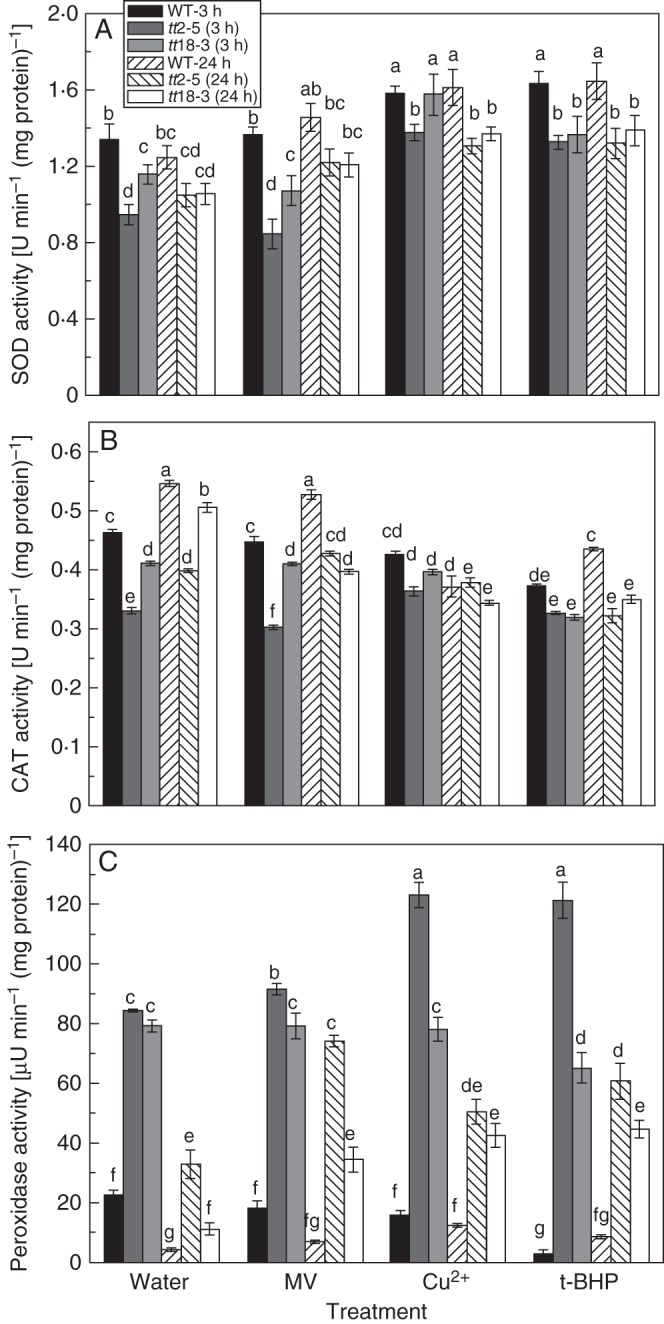
Changes in antioxidant enzyme activities under normal and oxidative stress conditions in wild-type and PA-deficient mutant arabidopsis seeds over short- (3 h) and long-term (24 h) imbibition periods. (A) Superoxide dismutase (SOD) activity, (B) catalase (CAT) activity, (C) peroxidase activity. Values are means and s.e. (n = 3). Means denoted by the same letter are not significantly different at P < 0·05 according to Duncan's multiple range test.
In contrast to the decreased activities of SOD and CAT in PA-deficient mutants, much higher activities of peroxidase were detected in the mutants than in the wild-type seeds (Fig. 2C). About a 4-fold (3·74- and 3·51-fold) increase in peroxidase activity was observed in the tt2-5 and tt18-3 seeds under normal conditions, which might have resulted from a strong response at an earlier stage before germination such as during seed development or the desiccation stage. Under oxidative stress conditions, peroxidase activity also increased substantially, for both short- and long-term treatments. We speculate that peroxidase may play a major role in the defence against oxidative stress, particularly by keeping H2O2 at low levels in PA-deficient seeds.
Tissue localization of peroxidase activity in seeds
Staining was used to localize and quantify the tissue distribution of the increased peroxidase activity in PA-deficient mutants, using TMB. A deeper colour was clearly exhibited in PA-deficient mutant seeds over short-term treatments but the staining was not observed in wild-type seeds (Fig. 3A). The result was further verified via quantitative analysis using a colorimetric method (Fig. 3B). The difference in peroxidase activity over short-term staining periods mainly occurred in the seed coat. After absorption of water, the peroxidase in the seed coat rapidly reacted with the staining substrate. We did not observe any staining on the surface of wild-type seeds but coloration was apparent in PA-deficient mutant seed coats, which is consistent with our hypothesis (Fig. 3B). There was also no staining in the cotyledon and radicle of mutants when embryos released from seeds were immersed into the same reaction solution as the seed coat (Fig. 3C).
Fig. 3.
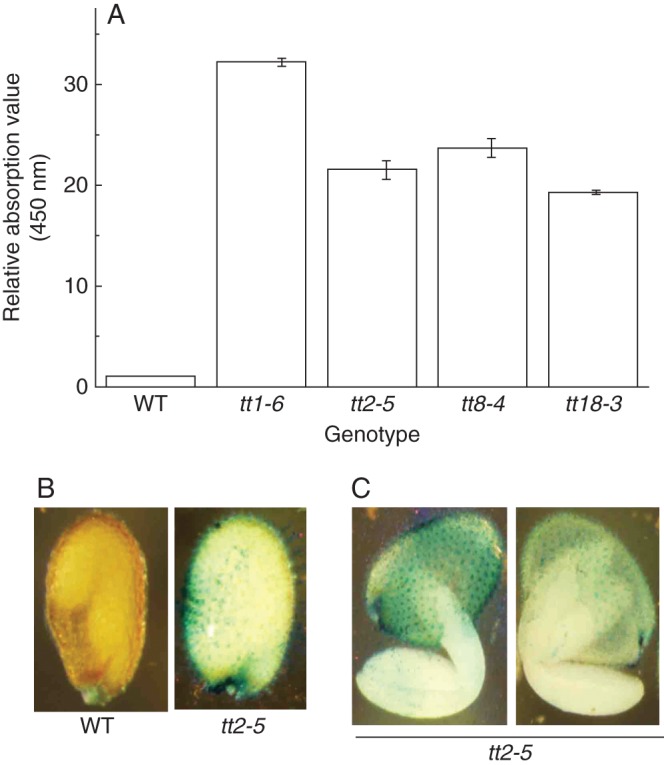
Activity and tissue localization of peroxidase in arabidopsis seeds. Comparison of peroxidase activity between wild-type and PA-deficient mutant seeds by 3,5,3′,5′-tetramethylbenzidine-HCl (TMB) staining and (A) relative quantitative analysis by absorptive value (values are means and s.e.; n = 3). (B) Tissue localization of peroxidase in seeds using the wild-type and PA-deficient mutants (tt2-5) as experimental material. (C) Staining effects of peroxidase over short-term periods (10 min) in arabidopsis seeds.
Distribution of peroxidase in the seed coat and embryo
We investigated the distribution of antioxidant activities, including peroxidase, in the seed coat and embryo (Fig. 4). Based on the activity per grain, we found almost equal distributions of SOD activity between the seed coat and embryo (49·62 and 50·38 %, respectively) in wild-type seeds. There was about a 10 % decrease in the seed coats of the mutants compared with the wild-type seeds (Fig. 4A). In contrast to the distribution of SOD activity, the activities of CAT and peroxidase in the seed coat of mutants were higher than those in the wild-type seeds. Although the activity of CAT was higher in mutant seeds than in wild-type seeds, it was still below 30 % of the total, accounting for 29·38 and 27·13 % in tt2-5 and tt18-3, respectively (Fig. 4B). Wild-type seeds showed more peroxidase activity than SOD or CAT activity. However, peroxidase activity was even higher in PA-deficient mutants, being 88·94 and 85·19 % for the tt2-5 and tt18-3 mutants, respectively, compared with 55·12 % in wild-type seeds (Fig. 4C). The high peroxidase activity in seeds and its widespread distribution in the coat of PA-deficient seeds indicate that it must have some essential function therein and may partially account for the antioxidant role of PAs.
Fig. 4.
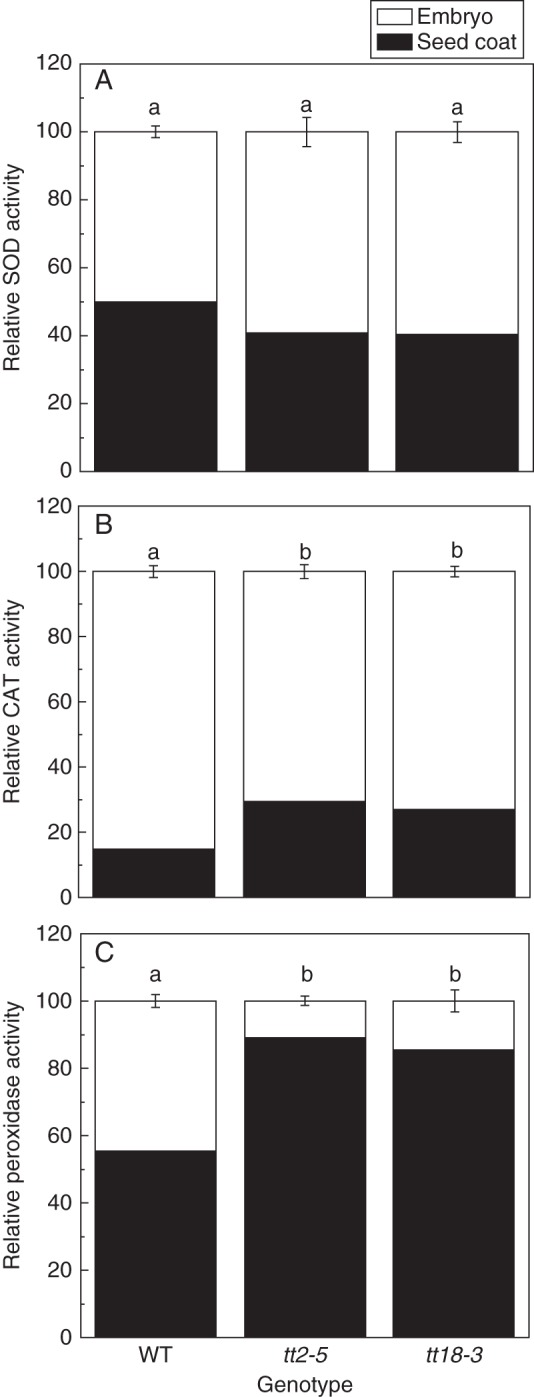
Comparison of antioxidant enzyme activities in the testa and embryo between wild-type and PA-deficient mutant seeds (tt2-5 and tt18-3). (A) Relative SOD activity, (B) relative CAT activity, (C) relative peroxidase activity. Values are means and s.e. (n = 3). Means denoted by different letters are significantly different at P < 0·05 according to Duncan's multiple range test.
Guaiacol peroxidase is specifically activated in the seed coat of tt mutants
To investigate which type of peroxidase contributes most when PAs are deficient, we determined the activities of APx, GPx and guaiacol peroxidase (class III peroxidase). Using a leaf as a control, we found that there was very low APx activity and no GPx activity in the seed. There was also no significant difference in the APx activities of the wild-type and PA-deficient (tt2-5 and tt18-3) seeds (Fig. 5B, C). We also did not detect guaiacol peroxidase activity in non-dormant wild-type seeds. However, guaiacol peroxidase activity was dramatically activated in the tt2-5 and tt18-3 seeds (Fig. 5A). Further investigation indicated that the guaiacol peroxidase activity was confined to the mutant seed coats (data not shown).
Fig. 5.
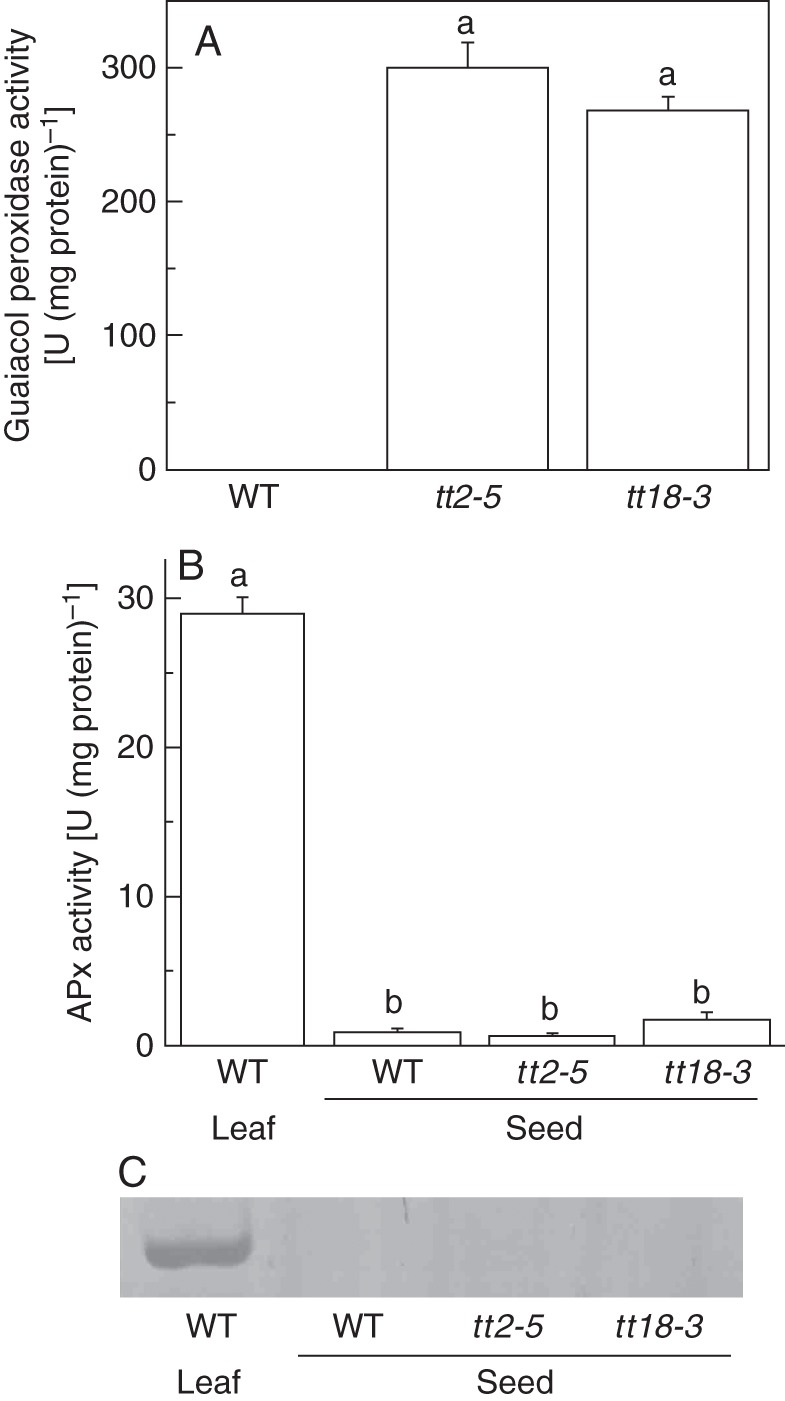
Peroxidase activities in the seed coat of arabidopsis. (A) Comparison of guaiacol peroxidase activity between wild-type seeds and PA-deficient mutants. (B) APx activities in wild-type and PA-deficient mutant seeds. The leaf of the wild-type was used as a control. (C) In-gel GPx activities of wild-type and PA-deficient mutant seeds. The leaf of the wild-type was used as a control. Values are means and s.e. (n = 3 for SA). Means denoted by the same letter are not significantly different at P < 0·05 according to Duncan's multiple range test.
Peroxidase in the embryo at transcriptional and post-transcriptional levels
Despite the high peroxidase activity in the mutant testa, there was no significant difference in its activity in the embryo between the wild-type and the mutants (Figs 2 and 4). This was consistent with the results of histochemical analysis by immersing the embryo in staining solution for a longer time. Peroxidase activity was present at the radicle tip for both the wild-type and the mutants (Fig. 6A). However, the transcriptional levels of peroxidase-encoding genes, including APX1, Prx16 and PrxIIE, were different between the wild-type and PA-deficient mutant seeds. All three genes displayed lower expressions in the mutants compared with non-dormant wild-type seeds (Fig. 6B). Under oxidative treatment, the expression patterns became complex with APX1 being inhibited and PrxIIE being promoted at the transcriptional level.
Fig. 6.
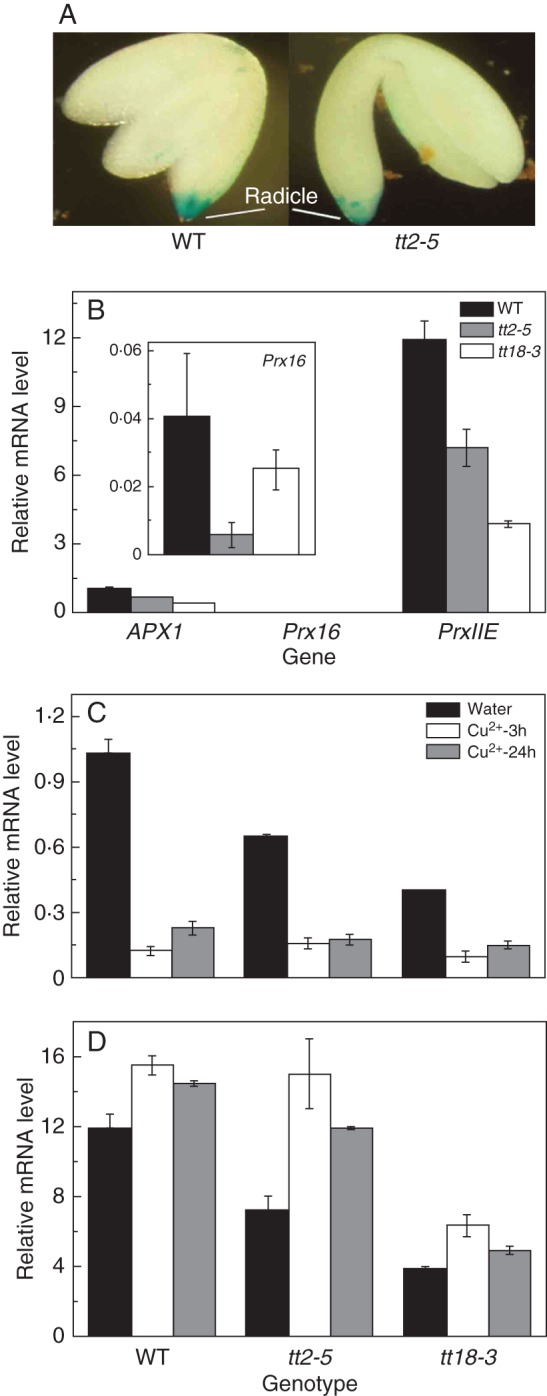
Peroxidase localization and its encoded gene expression in the embryo of arabidopsis. (A) TMB staining over long-term periods (2 h). (B) Expression of peroxidase-encoded genes (APX1, Prx16 and PrxIIE) in wild-type seeds and PA-deficient mutants. (C,D) Comparison of (C) APX1 and (D) PrxIIE between wild-type seeds and PA-deficient mutants at transcriptional levels under oxidative stress conditions. In (B–D) values are means and s.e. (n = 3).
DISCUSSION
Seeds experience substantial water content changes from maturation to germination, during which large amounts of ROS can accumulate and generate oxidative stress. In arabidopsis, PAs are mainly produced in the endothelium and have functions as antioxidants to cope with such oxidative stress (Debeaujon et al., 2000, 2003; Jia et al., 2012a). However, PA-deficient mutant seeds can still germinate and seedlings can establish under normal conditions (Bailly, 2004; El-Maarouf-Bouteau and Bailly, 2008). In a previous study, we found that more O2− was produced in PA-deficient mutants under oxidative stress and that these ROS can be removed by PAs in wild-type seeds in the germination stage (Jia et al., 2012b). However, H2O2 is reduced in the mutants under normal and oxidative stress conditions (Fig. 1). This finding is inconsistent with the results of a previous study regarding the function of flavonoids as ROS scavengers (Yamasaki et al., 1997). We hypothesize that there may be another antioxidant system that is activated or initiated in the mutants in response to the lack of PAs in seeds.
The enzymatic ROS-scavenging mechanism plays a pivotal role in multiple developmental stages and defends against biotic and abiotic stresses. Such a mechanism mainly includes SOD, CAT and peroxidase (Bowler et al., 1992; Willekens et al., 1997; Apel and Hirt, 2004). In the present study, all three antioxidant enzymes were investigated under different time periods and with different ROS-generated xenobiotics (Fig. 2). Among the enzymes, only peroxidase was substantially more activated in mutants compared with wild-type seeds (Fig. 2C).
Based on these results, we hypothesize that peroxidase is a candidate antioxidant that compensates for the absence of PAs in tt mutants. If so, peroxidase expression should be mainly distributed in the seed coat because PAs are biosynthesized and accumulate in the endothelium (Debeaujon et al., 2003; Xie et al., 2003; Lepiniec et al., 2006). To detail the tissue localization of peroxidase inside seeds, a histochemical method was used. We found that peroxidase activity is focused on the seed coat (Fig. 3). This is because the cells in all layers of the seed coat experience programmed cell death during the later stages of seed development. Cell contents, including some metabolites and proteins, are readily released (Haughn and Chaudhury, 2005). In the short-term staining reactions, colour appeared only in PA-deficient mutants (Fig. 3A, C). When we immersed mutant seeds with radicles or embryos extruded from the testa in the same reaction solution, we only observed colour on the seed coat in the short-term reaction, which further confirms the different peroxidase activities between wild-type and PA-deficient mutant seed coats (Fig. 3D).
The distribution of peroxidase in seed coats and embryos provides further evidence for this. A lower percentage of SOD activity and higher CAT and peroxidase activities occurred in the mutant seed coats compared with the wild-type (Fig. 4). However, CAT in the seed coat contributed less activity to the whole seed, not only in the wild-type but also in the mutants (Fig. 4B). Compared with other antioxidant enzymes, peroxidase seems to play a more important role in testa, because it accounted for almost 90 % of activity in the mutants (Fig. 4C). Due to the large distribution of peroxidase activities in the seed coat, there was no significant difference in the embryos between the wild-type and PA-deficient mutants, although the total peroxidase activity was higher in the mutants.
Class III peroxidases are bifunctional enzymes, participating in two possible catalytic cycles, peroxidative and hydroxylic, to detoxify or generate ROS and regulate H2O2 levels (Passardi et al., 2004a). They can also cross-link cell-wall macromolecules to make cell walls rigid, particularly the class III peroxidases when secreted into a cell wall or surrounding medium (Chen and Schopfer, 1999; Passardi et al., 2004b). Such functions of class III peroxidases make them good candidates to substitute for PAs in a PA-deficient seed coat.
Three major plant peroxidases, ascorbate peroxidase, glutathione peroxidase and class III peroxidase, were considered in our study. Guaiacol, as an artificial phenolic substrate, can be used to detect all class III peroxidases present independent of their different in vivo substrate specificities (Mika et al., 2010). Only guaiacol peroxidase activity was increased in the testa of PA-deficient mutant seeds.
We also compared peroxidase activities in embryos between the wild-type and PA-deficient mutants due to their different germination behaviour (Fig. 6). A recent study has shown that peroxidases play an important role during seed germination and are specifically expressed in the micropylar endosperm and radicle, which are the key sites determining the seed germination of Lepidium sativum. The corresponding SALK lines of orthologues in arabidopsis display germination phenotypes (Linkies et al., 2010). We investigated whether there is any difference in peroxidase activity between wild-type and PA-deficient mutant embryos in arabidopsis. We found that peroxidase is specifically localized at the radicle tip in arabidopsis seeds although we could not exclude its distribution in endosperm (Fig. 6A), which is consistent with the results from L. sativum (Linkies et al., 2010). However, three candidate peroxidase-encoding genes are down-regulated at the transcriptional level in PA-deficient mutant seeds. This might be negative feedback regulation due to high peroxidase activity in the seed coat of PA-deficient mutants (Fig. 6b). Under oxidative stresses, PrxIIE is upregulated in the mutants but APX1 expression is still lower compared with the wild-type, which demonstrates a complex regulatory network of peroxidase at the transcriptional level (Fig. 6C, D). This might be because many genes encoding peroxidase are universal with gene redundancy and result in an inconsistency between enzyme activity and gene expression (Apel and Hirt, 2004; Cosio and Dunand, 2009).
In conclusion, class III peroxidase activity is significantly enhanced in the seed coats of PA-deficient mutants as a consequence of the suppression of PA biosynthesis. Our results suggest that enhanced peroxidase activity may be an adaptive mechanism for substituting the antioxidant function of PAs that is required during seed development and later in seed germination. Further study is required to investigate the role and mechanism of peroxidase activity during seed development and how the regulatory mechanism operates to coordinate peroxidase activity and PA production.
ACKNOWLEDGEMENTS
We are grateful for grant support from the National Basic Research Program of China (973 project, 2012CB114300), Hong Kong Research Grants Council (HKBU1/CRF/10), Shenzhen Overseas Talents Innovation & Entrepreneurship Funding Scheme (The Peacock Scheme) and the Chinese University of Hong Kong.
LITERATURE CITED
- Abrahams S, Lee E, Walker AR, Tanner GJ, Larkin PJ, Ashton AR. The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. Plant Journal. 2003;35:624–636. doi: 10.1046/j.1365-313x.2003.01834.x. [DOI] [PubMed] [Google Scholar]
- Aebi H. Catalase in vitro. Methods in Enzymology. 1984;105:121–126. doi: 10.1016/s0076-6879(84)05016-3. [DOI] [PubMed] [Google Scholar]
- Almagro L, Gomez Ros LV, Belchi-Navarro S, Bru R, Ros Barcelo A, Pedreno MA. Class III peroxidases in plant defence reactions. Journal of Experimental Botany. 2009;60:377–390. doi: 10.1093/jxb/ern277. [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annual Review of Plant Biology. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Appelhagen I, Lu G, Huep G, Schmelzer E, Weisshaar B, Sagasser M. TRANSPARENT TESTA1 interacts with R2R3-MYB factors and affects early and late steps of flavonoid biosynthesis in the endothelium of Arabidopsis thaliana seeds. The Plant Journal. 2011;67:406–419. doi: 10.1111/j.1365-313X.2011.04603.x. [DOI] [PubMed] [Google Scholar]
- Bae SJ, Lee JS, Kim JM, et al. 5-Hydroxytrytophan inhibits tert-butylhydroperoxide (t-BHP)-induced oxidative damage via the suppression of reactive species (RS) and nuclear factor-kappaB (NF-kappaB) activation on human fibroblast. Journal of Agricultural and Food Chemistry. 2010;58:6387–6394. doi: 10.1021/jf904201h. [DOI] [PubMed] [Google Scholar]
- Bailly C. Active oxygen species and antioxidants in seed biology. Seed Science Research. 2004;14:93–107. [Google Scholar]
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant Journal. 2004;39:366–380. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- Bowler C, Van Montagu M, Inze D. Superoxide dismutase and stress tolerance. Annual Review of Plant Biology. 1992;43:83–116. [Google Scholar]
- Chen SX, Schopfer P. Hydroxyl-radical production in physiological reactions A novel function of peroxidase. European Journal of Biochemistry. 1999;260:726–735. doi: 10.1046/j.1432-1327.1999.00199.x. [DOI] [PubMed] [Google Scholar]
- Cosio C, Dunand C. Specific functions of individual class III peroxidase genes. Journal of Experimental Botany. 2009;60:391–408. doi: 10.1093/jxb/ern318. [DOI] [PubMed] [Google Scholar]
- Dean G, Cao Y, Xiang D, et al. Analysis of gene expression patterns during seed coat development in Arabidopsis. Molecular Plant. 2011;4:1074–1091. doi: 10.1093/mp/ssr040. [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Leon-Kloosterziel KM, Koornneef M. Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiology. 2000;122:403–414. doi: 10.1104/pp.122.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Nesi N, Perez P, et al. Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell. 2003;15:2514–2531. doi: 10.1105/tpc.014043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Fang X, Wu J. Flavonoids function as antioxidants: by scavenging reactive oxygen species or by chelating iron? Radiation Physics and Chemistry. 1997;50:271–276. [Google Scholar]
- Dixon RA, Xie DY, Sharma SB. Proanthocyanidins–a final frontier in flavonoid research? New Phytologist. 2005;165:9–28. doi: 10.1111/j.1469-8137.2004.01217.x. [DOI] [PubMed] [Google Scholar]
- El-Maarouf-Bouteau H, Bailly C. Oxidative signaling in seed germination and dormancy. Plant Signaling & Behavior. 2008;3:175–182. doi: 10.4161/psb.3.3.5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. Oxidant and antioxidant signalling in plants: a re-evaluation of the concept of oxidative stress in a physiological context. Plant Cell & Environment. 2005;28:1056–1071. [Google Scholar]
- Foyer CH, Noctor G. Ascorbate and glutathione: the heart of the redox hub. Plant Physiology. 2011;155:2–18. doi: 10.1104/pp.110.167569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Limones C, Hervas A, Navas-Cortes JA, Jimenez-Diaz RM, Tena M. Induction of an antioxidant enzyme system and other oxidative stress markers associated with compatible and incompatible interactions between chickpea (Cicer arietinum L.) and Fusarium oxysporum f.sp. ciceris. Physiological and Molecular Plant Pathology. 2002;61:325–337. [Google Scholar]
- Giannopolitis C, Ries S. Superoxide dismutase. I. Occurrence in higher plants. Plant Physiology. 1977;59:309–314. doi: 10.1104/pp.59.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez C, Conejero G, Torregrosa L, Cheynier V, Terrier N, Ageorges A. In vivo grapevine anthocyanin transport involves vesicle-mediated trafficking and the contribution of anthoMATE transporters and GST. Plant Journal. 2011;67:960–970. doi: 10.1111/j.1365-313X.2011.04648.x. [DOI] [PubMed] [Google Scholar]
- Gould KS, McKelvie J, Markham KR. Do anthocyanins function as antioxidants in leaves? Imaging of H2O2 in red and green leaves after mechanical injury. Plant Cell & Environment. 2002;25:1261–1269. [Google Scholar]
- Graeber K, Linkies A, Wood AT, Leubner-Metzger G. A guideline to family wide comparative state-of-the-art quantitative RT-PCR analysis exemplified with a brassicaceae cross-species seed germination case study. Plant Cell. 2011;23:2045–2063. doi: 10.1105/tpc.111.084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han RM, Tian YX, Liu Y, et al. Comparison of flavonoids and isoflavonoids as antioxidants. Journal of Agricultural and Food Chemistry. 2009;57:3780–3785. doi: 10.1021/jf803850p. [DOI] [PubMed] [Google Scholar]
- Harborne JB, Williams CA. Advances in flavonoid research since 1992. Phytochemistry. 2000;55:481–504. doi: 10.1016/s0031-9422(00)00235-1. [DOI] [PubMed] [Google Scholar]
- Haughn G, Chaudhury A. Genetic analysis of seed coat development in Arabidopsis. Trends in Plant Science. 2005;10:472–477. doi: 10.1016/j.tplants.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Hernandez I, Alegre L, Van Breusegem F, Munne-Bosch S. How relevant are flavonoids as antioxidants in plants? Trends in Plant Science. 2009;14:125–132. doi: 10.1016/j.tplants.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Hiraga S, Sasaki K, Ito H, Ohashi Y, Matsui H. A large family of class III plant peroxidases. Plant and Cell Physiology. 2001;42:462–468. doi: 10.1093/pcp/pce061. [DOI] [PubMed] [Google Scholar]
- Jia LG Wu QY, Ye NH, et al. Proanthocyanidins inhibit seed germination by maintaining high level of abscisic acid in Arabidopsis. Journal of Integrative Plant Biology. 2012a;54:663–673. doi: 10.1111/j.1744-7909.2012.01142.x. [DOI] [PubMed] [Google Scholar]
- Jia LG, Sheng ZW, Xu WF, et al. Modulation of anti-oxidation ability by proanthocyanidins during germination of Arabidopsis thaliana seeds. Molecular Plant. 2012b;5:472–481. doi: 10.1093/mp/ssr089. [DOI] [PubMed] [Google Scholar]
- Jiang M, Zhang J. Water stress-induced abscisic acid accumulation triggers the increased generation of reactive oxygen species and upregulates the activities of antioxidant enzymes in maize leaves. Journal of Experimental Botany. 2002;53:2401–2410. doi: 10.1093/jxb/erf090. [DOI] [PubMed] [Google Scholar]
- Lee S, Kim SG, Park CM. Salicylic acid promotes seed germination under high salinity by modulating antioxidant activity in Arabidopsis. New Phytologist. 2010;188:626–637. doi: 10.1111/j.1469-8137.2010.03378.x. [DOI] [PubMed] [Google Scholar]
- Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M. Genetics and biochemistry of seed flavonoids. Annual Review of Plant Biology. 2006;57:405–430. doi: 10.1146/annurev.arplant.57.032905.105252. [DOI] [PubMed] [Google Scholar]
- Levine A, Tenhaken R, Dixon R, Lamb C. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- Linkies A, Schuster-Sherpa U, Tintelnot S, Leubner-Metzger G, Muller K. Peroxidases identified in a subtractive cDNA library approach show tissue-specific transcript abundance and enzyme activity during seed germination of Lepidium sativum. Journal of Experimental Botany. 2010;61:491–502. doi: 10.1093/jxb/erp318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marles MA, Ray H, Gruber MY. New perspectives on proanthocyanidin biochemistry and molecular regulation. Phytochemistry. 2003;64:367–383. doi: 10.1016/s0031-9422(03)00377-7. [DOI] [PubMed] [Google Scholar]
- Mika A, Boenisch MJ, Hopff D, Luthje S. Membrane-bound guaiacol peroxidases from maize (Zea mays L.) roots are regulated by methyl jasmonate, salicylic acid, and pathogen elicitors. Journal of Experimental Botany. 2010;61:831–841. doi: 10.1093/jxb/erp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends in Plant Science. 2002;7:405–410. doi: 10.1016/s1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- Mittler R, Verauwera S, Suzuki N, et al. ROS signaling: the new wave? Trends in Plant Science. 2011;16:300–309. doi: 10.1016/j.tplants.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Moller IM, Jensen PE, Hansson A. Oxidative modifications to cellular components in plants. Annual Review of Plant Biology. 2007;58:459–481. doi: 10.1146/annurev.arplant.58.032806.103946. [DOI] [PubMed] [Google Scholar]
- Nakano Y, Asada K. Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant and Cell Physiology. 1981;22:867–880. [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L. The TT8 gene encodes a basic helix–loop–helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell. 2000;12:1863–1878. doi: 10.1105/tpc.12.10.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell. 2001;13:2099–2114. doi: 10.1105/TPC.010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Foyer CH. Ascorbate and glutathione: keeping active oxygen under control. Annual Review of Plant Physiology and Plant Molecular Biology. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- Passardi F, Longet D, Penel C, Dun C. The class III peroxidase multigenic family in rice and its evolution in land plants. Phytochemistry. 2004a;65:1879–1893. doi: 10.1016/j.phytochem.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Passardi F, Penel C, Dun C. Performing the paradoxical: how plant peroxidases modify the cell wall. Trends in Plant Science. 2004b;9:534–540. doi: 10.1016/j.tplants.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Poustka F, Irani NG, Feller A, et al. A trafficking pathway for anthocyanins overlaps with the endoplasmic reticulum-to-vacuole protein-sorting route in Arabidopsis and contributes to the formation of vacuolar inclusions. Plant Physiology. 2007;145:1323–1335. doi: 10.1104/pp.107.105064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proteggente AR, Saija A, De Pasquale A, Rice-Evans CA. The compositional characterisation and antioxidant activity of fresh juices from sicilian sweet orange (Citrus sinensis L. Osbeck) varieties. Free Radical Research. 2003;37:681–687. doi: 10.1080/1071576031000083198. [DOI] [PubMed] [Google Scholar]
- Routaboul JM, Kerhoas L, Debeaujon I, Pourcel L, Caboche M, Einhorn J, Lepiniec L. Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta. 2006;224:96–107. doi: 10.1007/s00425-005-0197-5. [DOI] [PubMed] [Google Scholar]
- Sattler SE, Gillil LU, Magallanes-Lundback M, Pollard M, DellaPenna D. Vitamin E is essential for seed longevity and for preventing lipid peroxidation during germination. Plant Cell. 2004;16:1419–1432. doi: 10.1105/tpc.021360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler SE, Mene-Saffrane L, Farmer EE, Krischke M, Mueller MJ, DellaPenna D. Nonenzymatic lipid peroxidation reprograms gene expression and activates defense markers in Arabidopsis tocopherol-deficient mutants. Plant Cell. 2006;18:3706–3720. doi: 10.1105/tpc.106.044065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikazono N, Yokota Y, Kitamura S, Suzuki C, Watanabe H, Tano S, Tanaka A. Mutation rate and novel tt mutants of Arabidopsis thaliana induced by carbon. Genetics. 2003;163:1449–1455. doi: 10.1093/genetics/163.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R, Schachtman DP. Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proceedings of the National Academy Sciences USA. 2004;101:8827–8832. doi: 10.1073/pnas.0401707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer JP, Kuhnle GG, Williams RJ, Rice-Evans C. Intracellular metabolism and bioactivity of quercetin and its in vivo metabolites. Biochemical Journal. 2003;372:173–181. doi: 10.1042/BJ20021972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Camp W, Capiau K, Van Montagu M, Inze D, Slooten L. Enhancement of oxidative stress tolerance in transgenic tobacco plants overproducing Fe-superoxide dismutase in chloroplasts. Plant Physiology. 1996;112:1703–1714. doi: 10.1104/pp.112.4.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhercke T, Ampe C, Tirry L, Denolf P. Rescue and in situ selection and evaluation (RISE): a method for high-throughput panning of phage display libraries. Journal of Biomolecular Screening. 2005;10:108–117. doi: 10.1177/1087057104271956. [DOI] [PubMed] [Google Scholar]
- Vogt T. Phenylpropanoid biosynthesis. Molecular Plant. 2010;3:2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- Vranova E, Inze D, Van Breusegem F. Signal transduction during oxidative stress. Journal of Experimental Botany. 2002;53:1227–1236. [PubMed] [Google Scholar]
- Weydert CJ, Cullen JJ. Measurement of superoxide dismutase, catalase and glutathione peroxidase in cultured cells and tissue. Nature Protocols. 2010;5:51–66. doi: 10.1038/nprot.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willekens H, Chamnongpol S, Davey M, et al. Catalase is a sink for H2O2 and is indispensable for stress defence in C3 plants. The EMBO Journal. 1997;16:4806–4816. doi: 10.1093/emboj/16.16.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiology. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DY, Sharma SB, Paiva NL, Ferreira D, Dixon RA. Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science. 2003;299:396–399. doi: 10.1126/science.1078540. [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Sakihama Y, Ikehara N. Flavonoid–peroxidase reaction as a detoxification mechanism of plant cells against H2O2. Plant Physiology. 1997;115:1405–1412. doi: 10.1104/pp.115.4.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]


