Abstract
Background and Aims
Pantropical intercontinental disjunction is a common biogeographical pattern in flowering plants exhibiting a discontinuous distribution primarily in tropical Asia, Africa and the Americas. Only a few plant groups with this pattern have been investigated at the generic level with molecular phylogenetic and biogeographical methods. Paederia (Rubiaceae) is a pantropical genus of 31 species of woody lianas, with the greatest species diversity in continental Asia and Madagascar and only two species from tropical America. The aim of this study was to reconstruct the biogeographical history of Paederia based on phylogenetic analyses to explore how the genus attained its pantropical distribution.
Methods
Maximum parsimony and Bayesian inference were used for phylogenetic analyses using sequences of five plastid markers (the rbcL gene, rps16 intron, trnT-F region, atpB-rbcL spacer and psbA-trnH spacer). Biogeographical inferences were based on a Bayesian uncorrelated lognormal relaxed molecular clock together with both Bayesian and likelihood ancestral area reconstructions.
Key Results
The data suggest an early diverged Asian lineage sister to the clade of the remaining species consisting of a predominantly Asian sub-clade and a primarily Malagasy sub-clade. Paederia is inferred to have originated in the Oligocene in tropical continental Asia. It then reached Africa in the early to middle Miocene, most probably via long-distance dispersal across the Indian Ocean. The two Neotropical species are inferred to have derived independently in the late Miocene from ancestors of Asia and East Africa, respectively.
Conclusions
The results demonstrate the importance of post-Boreotropical long-distance dispersals (across three major oceans) in shaping the global pantropical disjunction in some plants, such as Paederia, with small, winged diaspores adapted to long-distance dispersal by various agents including wind, ocean currents or birds. Overland migration is less likely to explain its palaeotropical disjunction between Asia and Africa.
Keywords: Intercontinental disjunction, long-distance dispersal, Paederia, pantropical, post-Boreotropical, Rubiaceae
INTRODUCTION
Tropical forests are the most complex and richest terrestrial ecosystems on the planet (Morley, 2000, 2007). Studies of the origin and diversification of tropical intercontinental disjunctions in plants may provide important insights into the evolutionary assembly of tropical forests. Pantropical disjunctions usually involve tropical regions of the Americas, Africa, south-eastern Asia and Australia (Thorne, 1972). These disjunct patterns have sometimes been attributed to vicariance involving the break up of the Gondwanan supercontinent or continental rafting (Raven and Axelrod, 1974; Conti et al., 2002), but recent studies have shown that many tropical groups are of more recent origin (Givnish and Renner, 2004; Renner, 2004a). Molecular dating suggests that many disjunctions at the family or generic level are the result of degradation of the Boreotropical flora or other deep-time vicariance events with migrations across land bridges during times when the climate was favourable (Davis et al., 2002; Erkens et al., 2009). Many recent biogeographical analyses, especially at the generic level, have shown the importance of long-distance dispersal in the assembly of modern tropical floras (Renner, 2004a, b; Clayton et al., 2009; Christenhusz and Chase, 2013).
The number of studies on tropical plant groups has increased considerably in recent years, with improved phylogenetic reconstruction, molecular dating and ancestral area inferences (Lavin et al., 2000; Davis et al., 2002; Muellner et al., 2006; Clayton et al., 2009). However, most studies have focused on the family level, and only a few groups have been examined at the generic level with molecular phylogenetic and biogeographical methods, such as Anisophyllea (Zhang et al., 2007), Hernandia (Michalak et al., 2010), Peperomia and Piper (Smith et al., 2008), Sideroxylon (Smedmark and Anderberg, 2007) and Badula (Bone et al., 2012). In addition, among many genera of flowering plants showing intercontinental disjunctions, comparatively few of these genera are pantropical with taxa spanning all major tropical regions, and even fewer of these pantropical disjunct genera have well-resolved species-level molecular phylogenetic analyses, which are essential for testing alternative biogeographical hypotheses. The relatively few biogeographical analyses of this pattern may be partially due to the fact that many of the pantropical genera are species rich, making it difficult to obtain adequate taxon sampling, such as Begonia (approx. 1400 spp., Begoniaceae), Ficus (approx. 850 spp., Moraceae), Diospyros (approx. 550 spp., Ebenaceae), Terminalia (approx. 200 spp., Combretaceae) and Homalium (approx. 180 spp., Salicaceae) (Mabberley, 2008).
Paederia (Rubiaceae) is a small genus of only 31 species of woody climbers and shows a predominantly palaeotropical distribution, with most species in Asia and Madagascar–Africa and only two species in the Neotropics. The genus is an adequate group for testing hypotheses on the evolution of pantropical intercontinental disjunctions, although there is no species of the genus occurring in tropical Australia and the Pacific islands (Puff, 1991b; Razafimandimbison and Taylor, 2000). Moreover, Paederia exhibits a Malagasy–Asian disjunct pattern, with only two taxa in East Africa. This pattern is not common in Rubiaceae because relationships of taxa from Madagascar lie primarily with continental African relatives (Koechlin, 1972; Robbrecht, 1988).
Paederia was monographed by Puff (1991a), who recognized three subgenera: subgenus Paederia, subgenus Alatopaederia Puff and subgenus Lecontea (A. Rich.) Puff, based on corolla morphology and size, anther position, style length, presence of petaloid bracts and fruit morphology. Puff et al. (1991) hypothesized that the common ancestor of Paederia first diverged into subgenus Paederia (four Asian species with rounded fruits and unwinged diaspores) and subgenus Alatopaederia (12 Asian, eight Malagasy–African and two Neotropical species with laterally compressed fruits and winged diaspores). Subgenus Lecontea (five species endemic to Madagascar and Africa) was suggested to derive from subgenus Alatopaederia based on its similarly winged but larger diaspores (Puff, 1991a).
Backlund et al. (2007) conducted phylogenetic analyses of tribe Paederieae to which Paederia belongs, based on plastid DNA sequences. The results showed that Paederia consists of an Asian and an African clade, respectively. However, their study focused on tribal phylogeny and was limited in sampling of Paederia from Asia (16 species), one of the two species diversity centres. The recent biogeographical analysis by Wikström et al. (2010) suggested that Paederia originated in Asia with a single dispersal event to Madagascar and tropical East Africa from Asia, but similarly this biogeographical study included few samples from Asia and was not based on a dated inference. At present, a broader sampling from Asia in a temporal framework is needed to understand the biogeographical history of Paederia.
This study tests the systematic and biogeographical hypotheses of Puff (1991a) for Paederia. Phylogenetic relationships in Paederia are reconstructed with a broader sampling scheme from its entire distributional range, with extensive sampling from Asia based on sequences of five plastid DNA regions (the rbcL gene, rps16 intron, trnT-F region, atpB-rbcL spacer and psbA-trnH spacer). We employ a Bayesian uncorrelated lognormal relaxed molecular clock approach to estimate the ages of major diversification events in Paederia. Multiple calibration protocols including normal, uniform and exponential priors were applied to test the robustness of our dating estimation. The ancestral area of Paederia and subsequent range expansion are inferred and discussed in the context of the molecular dating results and palaeoclimatic evidence to explore the evolution of its pantropical distribution.
MATERIALS AND METHODS
Taxon sampling and sequencing
Sixty-six taxa were included in our phylogenetic data set (Appendix). We sampled 23 of the 31 Paederia spp. from continental Africa, Madagascar, Asia and the Neotropics. Although three species from Madagascar and five from Asia are unsampled, our sampling covered the entire extant geographical range of Paederia from the Palaeotropics and the Neotropics, and represented a wide range of morphological diversity of the three subgenera. Of the taxa analysed, sequences for five species were obtained from GenBank (Appendix). Widespread taxa were sampled with multiple accessions. Four representative species from the other three genera in the tribe (Spermadictyon, Leptodermis and Serissa) were selected as outgroups based on previous studies (Puff, 1982; Backlund et al., 2007).
Total DNA was extracted from silica gel-dried or herbarium materials using the DNeasy Plant Mini Kit (QIAGEN, Crawley, UK) or AutoGenPrep 965 (AutoGen, Holliston, USA) following the protocol specified by the manufacturers. Five different plastid regions were selected: the rbcL gene; the rps16 intron; the trnT-F region; and the atpB-rbcL and psbA-trnH intergenic spacers. Primers used for amplification and sequencing were Z1 and 3' for rbcL (Olmstead et al., 1992), and F and R2 for the rps16 intron (Oxelman et al., 1997). The trnT-F region was amplified and sequenced in two segments, with the first segment using the primer pair A (or A1) and D, and the second using the primer pair c and f (Taberlet et al., 1991; Bremer et al., 2002). The atpB-rbcL and psbA-trnH spacers were amplified and sequenced using the primers as described by Manen et al. (1994) and Sang et al. (1997), respectively. All sequences were amplified and sequenced based on the protocol in Nie et al. (2005). DNA sequences were assembled using Sequencher v4.1.4 (Gene Codes Corp., Ann Arbor, MI, USA). Sequence alignment was initially performed using with MUSCLE 3·8·31 (Edgar, 2004) in the multiple alignment routine followed by manual adjustment in Se-Al v2·0a11 (http://tree.bio.ed.ac.uk/software/seal/).
Phylogenetic analyses
Because all DNA sequences come from the plastid genome and preliminary analyses suggest no conflicts among each region (results not shown), we combined all the data (rbcL, rps16, trnT-F, atpB-rbcL and psbA-trnH) in our analysis using maximum parsimony (MP) and Bayesian inference (BI) as implemented in PAUP∗ 4·0b10 (Swofford, 2003) and MrBayes 3·2·1 (Huelsenbeck and Ronquist, 2001), respectively. The most parsimonious trees (MPTs) were estimated using heuristic searches with 1000 random addition-sequence replicates, tree bisection and reconnection (TBR) branch swapping, and no limiting MaxTrees. To assess clade support, non-parametric bootstrapping was conducted using 1000 bootstrap replicates, each with ten random-addition replicates, and only a single MPT was saved per replicate.
For BI analyses, we partitioned our data set by sequence region and determined the evolutionary model and parameters that best fit each partition using the Akaike information criterion implemented in MrModelTest 2·3 (Nylander, 2004). Model parameters (statefreq, revmat and shape) were unlinked between partitions. We ran two independent analyses consisting of four Markov chains sampled every 1000 generations for 10 million generations. We used the online program AWTY (Nylander et al., 2008) to check for stationarity and estimate the burn-in parameter. After discarding the first 2 million generations as burn-in, the remaining trees from both analyses were pooled for a consensus tree. The proportions of bifurcations found in this consensus tree are given as Bayesian posterior probabilities (PPs).
Molecular dating
We analysed the Paederia clade in the broad phylogenetic framework of Rubiaceae to enable multiple fossil calibrations. In the dating data set, we sampled 80 taxa from Rubiaceae plus Gelsemium sempervirens (Gelsemiaceae) as outgroup, of which 60 were obtained from GenBank (Supplementary Data Table S1). For the molecular dating analyses, the strict molecular clock model was rejected (P < 0·01) for our data set based on a likelihood ratio test (Felsenstein, 1981). We thus estimated node ages of Paederia using a Bayesian relaxed clock model as implemented in BEAST v1.7.4 (Drummond and Rambaut, 2007).
The four DNA regions (rbcL, rps16, trnT-F and atpB-rbcL) were partitioned using BEAUti 1·7·4 (within BEAST) with the appropriate substitution model estimated by MrModeltest. The psbA-trnH sequences were not used in our estimation because there were relatively few sequences of Rubiaceae available in GenBank. A Yule speciation tree prior was specified; this prior assumes a constant rate of speciation per lineage and has been recommended for species-level phylogenetic analyses (Drummond and Rambaut, 2007). An uncorrelated lognormal distributed (UCLD) relaxed clock model was used which permits evolutionary rates to vary along branches according to lognormal distribution (Drummond and Rambaut, 2007). After optimal operator adjustment as suggested by the output diagnostics from several preliminary BEAST runs, two independent MCMC (Markov chain Monte Carlo) runs (each of 50 million generations, with sampling every 1000 generations) were performed on a cluster of Mac XServers at the Smithsonian Institution (http://topazweb.si.edu). Tracer version 1.5 was used to check for convergence between the runs (Drummond and Rambaut, 2007). Results were considered reliable once the effective sampling size (ESS) for all parameters exceeded 200 as suggested by the program manual. The sampled posterior trees were summarized to generate a maximum clade credibility tree using the program TreeAnnotator 1.7.4 (Drummond and Rambaut, 2007) with a PP limit of 0·5 and mean node heights.
Fossil calibration
The most reliable Rubiaceae fossils are fruits reported from Cephalanthus from the late Eocene to the Pliocene in almost 20 fossil sites (Dorofeev, 1960, 1963; Friis, 1985; Mai and Walther, 1985; Antonelli et al., 2009), with convincing morphological features that make them taxonomically recognizable by means of overall similarity (see more in Antonelli et al., 2009). The oldest fossil of Cephalanthus was found from Kireevski in western Siberia in the late Eocene (Dorofeev, 1960, 1963) and is used here to place a normal constraint of the stem age of Cephalanthus as 33·9 ± 1·0 million years ago (Mya). Another well-preserved fruit fossil of a head-shaped infructescence was described as a new species, Morinda chinensis Shi, Liu & Jin, from the Changchang Formation in Hainan of China (Shi et al., 2012). Because Morinda is paraphyletic in tribe Morideae (Razafimandimbison et al., 2009) and the phylogenetic position of this fossil species is unclear, we used this fossil to calibrate the crown age of the tribe with the prior set to 44 ± 1 Mya, which falls into the fossil age estimated from the late early Eocene to the early late Eocene (Shi et al., 2012).
Most of the reported Rubiaceae fossils are dispersed pollen grains of a common tricolporate type. However, we used only the two most reliable pollen fossils in our analyses. The oldest pollen fossils of Faramea from the late Eocene (34–40 Mya) in Panama to the Pliocene in Veracruz, Mexico (Graham, 2009), which are characterized by the orientation of the bacula at the apertures (two- to four-porate) and the size and the shape of the pollen (Erdtman, 1966; Bremer and Eriksson, 2009). Thus, the Faramea stem node was fixed at 37 ± 1 Mya. Two pollen fossils of Scyphiphora were reported at 16 Mya from Japan and at 23 Mya from the Marshall Islands in the northern Pacific Ocean (Leopold, 1969; Saenger, 1998). Scyphiphora is the only extant genus of Rubiaceae that inhabits mangrove vegetation, and its pollen character is unique in the family, with distinct pores having a protruding papilla-like rim (Bremer and Eriksson, 2009). We therefore used 23 ± 1 Mya as a normal prior for the Scyphiphora node.
Four fossils from Rubiaceae (two fruit and two pollen) were selected as calibration points in our analyses, which have been used to estimate divergence times in various groups in the family (Nie et al., 2005; Antonelli et al., 2009; Bremer and Eriksson, 2009; Smedmark et al., 2010; Huang et al., 2013). For rooting the tree, we followed Antonelli et al. (2009) to set the stem Rubiaceae age as 78 ± 1 Mya based on the crown age estimate of Gentianales (Bremer et al., 2004). To test the robustness of our dating results based on normal priors, we also performed the dating analyses using more conservative uniform priors, i.e. each calibrating point is extending from the age of the oldest reliable fossil to the age of the root. An exponential prior is a viable alternative to allow for some probability of an earlier divergence than the appearance of the fossils (Ho and Phillips, 2009). Here we also used exponential prior distributions with a mean of 1 for all the fossil calibrations.
Ancestral area reconstruction
We used Bayesian and likelihood methods for biogeographical inference that have been recently proposed that take into account genetic branch lengths and/or phylogenetic uncertainty (Ree and Smith, 2008; Lemey et al., 2009). A smaller data set including only Paederia and its relatives (derived from the dating data set) was used for the biogeographical analyses. Five areas of endemism were delimited based on the distributions of Paederia spp.: A, continental Asia; B, south-eastern Asia; C, Madagascar and Africa; D, Central America; and E, South America. For comparison, we repeated the biogeographical analyses using a six-area scheme with Madagascar (C) and Africa (F) split as independent range units.
The Bayesian method is a discrete phylogeographic analysis using a standard continuous-time Markov chain (CTMC) (Lemey et al., 2009) and integrates over phylogenetic uncertainty and Markov model parameter uncertainty (Sanmartín et al., 2008). Each taxon can be allocated to a geographical location, corresponding to the distribution of each species. For each node, CTMC reconstructs the probability distribution for the different states. The relative PP of each location state at any position along the phylogenetic tree is also estimated. The Bayesian CTMC method was implemented in BEAST using a 10 million MCMC chain length, saving trees every 10 000 steps.
The ancestral area of the Paederia was also reconstructed with the likelihood analysis using the program Lagrange version 20120508 (Ree et al., 2005; Ree and Smith, 2008). The likelihood approach incorporates an explicit dispersal–extinction–cladogenesis (DEC) model of dispersal routes available at historical intervals correlating stochastic events with lineage persistence (Ree and Smith, 2008).
In contrast to the CTMC method, DEC is prone to estimate unrealistically widespread ancestral ranges for early-branching lineages (Lamm and Redelings, 2009; Ree and Sanmartin, 2009). In our case, the maximum ancestral area size was constrained to 2 because there is no species in our studied taxa distributed in more than two areas.
RESULTS
Phylogenetic analyses
The combined five-marker data matrix consisted of 6078 nucleotides. In the combined MP analyses, 497 characters were variable, 246 of which were potentially parsimony-informative. The MP analyses resulted in >10 000 equally MPTs with a length of 635 steps, a consistency index of 0·83, a retention index of 0·92 and a rescaled consistency index of 0·76. For the Bayesian analysis, all partitions had a best-fit model of GTR + G + I, with the exception of rbcL, for which it was GTR + I.
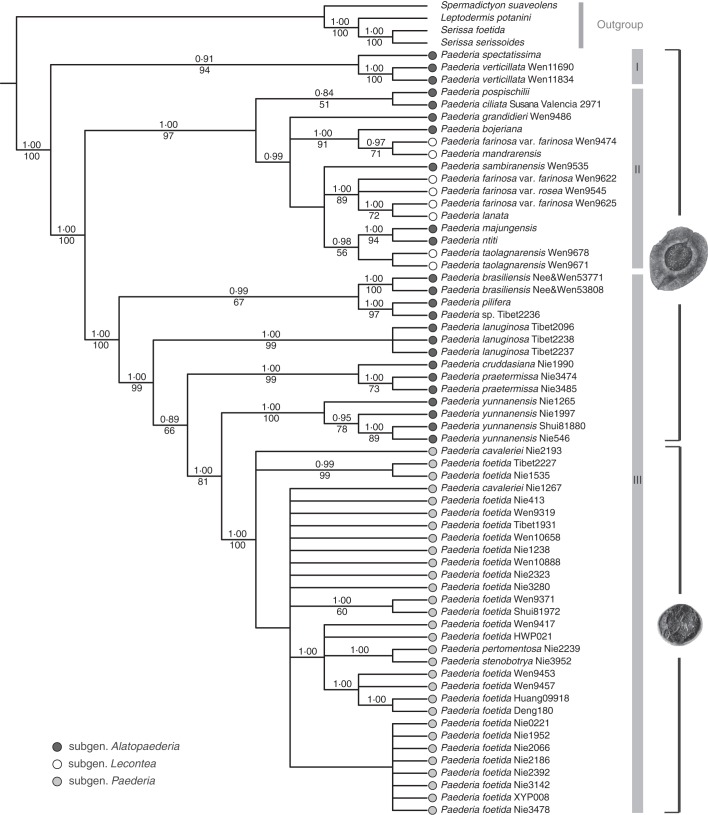
A strict consensus of the MPTs found in PAUP* indicates a backbone phylogenetic pattern for Paederia identical to the Bayesian result (Fig. 1), but low resolution for each major clade (results not shown). Paederia was strongly supported as monophyletic (MP bootstrap = 100 %; PP = 1·00). Three clades were recognizable in Paederia with robust support (clades I–III in Fig. 1). The first diverging group included two species (P. verticillata and P. spectatissima) endemic to tropical Asia (clade I, MP bootstrap = 94 %; PP = 0·91); this group was sister to clades II and III collectively. Clade II (MP bootstrap = 97 %; PP = 1·00) comprised all Malagasy and African species in subgenus Alatopaederia and all species of subgenus Lecontea plus the single species from Central America, P. ciliata (Fig. 1). Clade III (MP bootstrap = 100 %; PP = 1·00) consisted of almost all Asian members of subgenus Alatopaederia and all taxa of subgenus Paederia plus the single South American species, P. brasiliensis (Fig. 1).
Fig. 1.
The Bayesian consensus tree of Paederia and closely related taxa based on five plastid sequences (rbcL, rps16, trnT-F, atpB-rbcL and psbA-trnH). The Bayesian posterior probabilities are shown above the branches and the bootstrap values below.
Biogeographical analyses
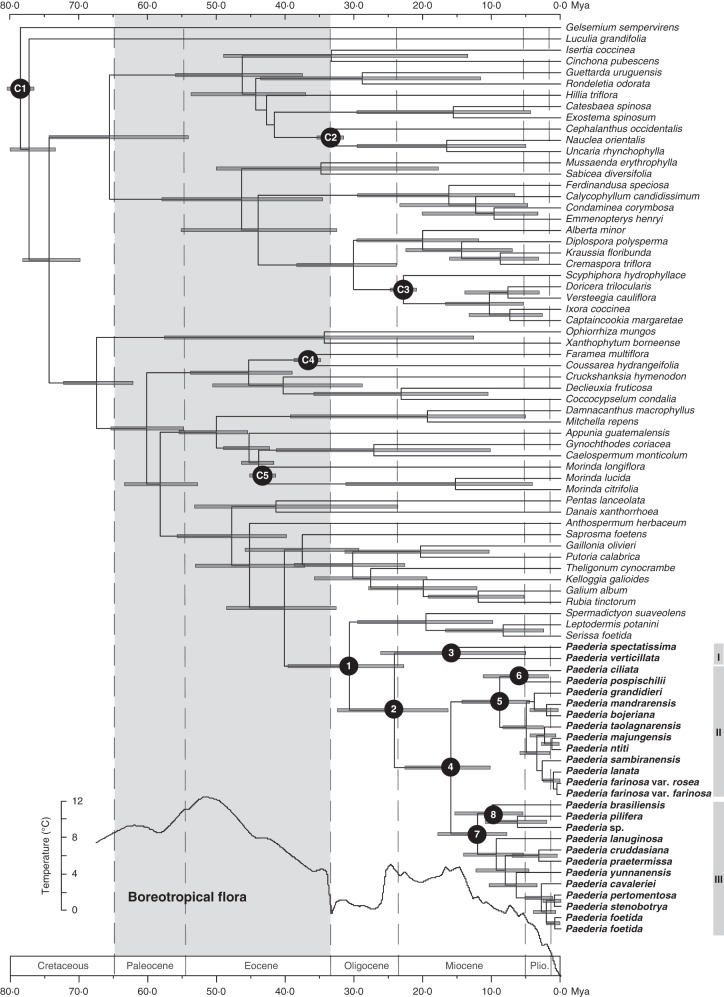
The BEAST analysis generated a well-resolved tree for Paederia, which was consistent with the topologies from the MP and Bayesian analyses (Fig. 1). Estimation from different calibration protocols (i.e. normal, uniform or exponential priors) produced similar results (Supplementary Data Table S2). Here we only report and discuss the mean ages and 95 % highest posterior density (HPD) intervals based on normal constraints as presented in Fig. 2 and Table 1. The uncorrelated-rates relaxed molecular clock suggested an origin of the Paederia stem lineage in the early Oligocene (30·73 Mya; 95 % HPD: 22·84–39·67 Mya; node 1 in Fig. 2) and the initial diversification of the Paederia crown group in the late Oligocene (24·21 Mya; 95 % HPD: 16·41–32·5 Mya; node 2 in Fig. 2). The Afro-Asian disjunction was dated to 15·99 (10·28–22·68) Mya (node 4 in Fig. 2). The two disjunctions between the Palaeotropics and the Neotropics were estimated at 6·08 (1·87–11·33) Mya for node 6 and 10·01 (5·57–15·47) Mya for node 8, respectively (Fig. 2, Table 1).
Fig. 2.
Chronogram of Paederia and relatives based on four plastid sequences (rbcL, rps16, trnT-F and atpB-rbcL) as inferred from BEAST. Grey bars represent the 95 % highest posterior density intervals for node ages. Global temperature means are shown by the curve adapted from Zachos et al. (2001). C1–C5 are calibration points; 1–8 indicate nodes with interests (see Table 1 for details).
Table Table 1.
Posterior age distributions of major nodes of Paederia in Rubiaceae, with results of ancestral reconstruction using the Bayesian CTMC and Lagrange
| Age estimates |
Bayesian CTMC |
Lagrange |
||||
|---|---|---|---|---|---|---|
| Node | Mean (Mya) | 95 % HPD (Mya) | Five-area | Six-area | Five-area | Six-area |
| C1: Rubiaceae stem | 78·57 | 76·64–80·54 | ||||
| C2: Cephalanthus stem | 33·48 | 31·59–35·47 | ||||
| C3: Scyphiphora stem | 22·88 | 20·98–24·82 | ||||
| C4: Faramea stem | 36·84 | 34·94–38·8 | ||||
| C5: Morinda crown | 43·36 | 41·46–45·27 | ||||
| 1: Paederia stem | 30·73 | 22·84–39·67 | A (0·83) | A (0·86) | A | A (0·43) | A | A (0·44) |
| 2: Paederia crown | 24·21 | 16·41–32·5 | A (0·83) | A (0·86) | A | A (0·40) | A | A (0·41) |
| 3: Paederia clade I crown | 15·92 | 5·11–26·21 | A (0·77) | A (0·81) | B | A (0·65) | B | A (0·49) |
| 4: Asian – African disjunction | 15·99 | 10·28–22·68 | A (0·80) | A (0·83) | C | A (0·68) | C | A (0·43) |
| 5: Paederia clade II crown | 8·68 | 4·59–14·43 | C (0·74) | C (0·23) | C | C (0·49) | D | C (0·39) |
| D (0·22) | ||||||
| F (0·23) | CD | C (0·49) | F | C (0·39) | ||||
| 6: African–Central American disjunction | 6·08 | 1·87–11·33 | C (0·74) | C (0·30) | C | D (0·86) | F | D (0·60) |
| D (0·30) | ||||||
| 7: Paederia clade III crown | 12·13 | 7·9–17·93 | A (0·95) | A (0·96) | A | A (0·78) | A | A (0·81) |
| 8: Asian–South American disjunction | 10·01 | 5·57–15·47 | A (0·95) | A (0·96) | E | A (0·65) | E | A (0·65) |
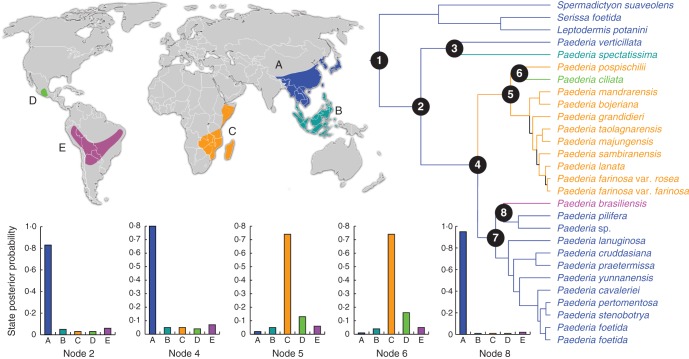
Bayesian CTMC analyses based on both the five-area and the six-area definitions produced similar results of ancestral areas with the probabilities of ancestral distributions (see Table 1). For example, both analyses suggested continental Asia as the ancestral area for Paederia (node 1 in Fig. 3). The clade dominant in Madagascar and Africa was inferred to have dispersed or migrated from continental Asia in the middle to late Miocene (node 4 in Figs 2 and 3). For node 6, dispersals were inferred from Africa–Madagascar to Mexico based on the five-area analysis, or each (Africa, Madagascar or Mexico) is equally possible as the ancestral area based on the six-area analysis. For the Asian–South American disjunction (node 8 in Fig. 3), all analyses suggested continental Asia as the ancestral area. As shown in Table 1, results estimated from Lagrange are similar to those from CTMC. Only CTMC results based on five-area analysis are shown in Fig. 3.
Fig. 3.
Bayesian CTMC ancestral area reconstruction from the BEAST tree (right) with branches coloured according to the most probable location state of their descendent nodes. The letter and colour coding for areas are indicated on the map (left upper). Left below are state posterior probability distributions for selected nodes as shown on the right-hand tree (see Fig. 2 and Table 1 for details).
DISCUSSION
Phylogenetics and morphology of Paederia
With a broader sampling from Asia, our analyses confirm the monophyly of Paederia as suggested by previous studies (Backlund et al., 2007; Rydin et al., 2009; Wikström et al., 2010). Paederia is a well-defined group of woody lianas, a growth form not particularly common in Rubiaceae. Paederia spp. all emit an unpleasant foetid odour with sulfur-containing iridoid glucosides when the tissue is bruised (Takeda et al., 1991; Zou et al., 2006). This chemical character is also known from a few other rubiaceous taxa, including Saprosma, Danais and Coprosma.
The first diverged lineage in Paederia includes only two Asian species: P. verticillata and P. spectatissima. Paederia verticillata is distributed in the Malay Peninsula, Sumatra, western Java, Borneo, Celebes and the Philippines, and P. spectatissima is confined to south-west China and adjacent parts of Vietnam (Puff, 1991c). The species share the synapomorphies of evergreen leaves, with a hypodermis and diaspores with by far the largest wings in the genus (Puff et al., 1991). The species pair was considered to be the most derived among the Asian taxa of subgenus Alatopaederia by Puff et al. (1991) based on these characters. Nevertheless, Puff et al. (1991) also pointed out that the two species remained ‘conservative’ with regard to chromosome number (diploid for P. verticillata, but unknown for P. spectatissima) and leaf arrangement (decussate for P. spectatissima and in whorls of three in P. verticillata; all other taxa have only opposite leaves), which is supported by our phylogenetic results (i.e. the first-branching position in the genus, Fig. 1).
Subgenus Alatopaederia is the largest subgenus recognized by Puff (1991b). Its monophyly is not supported by our results, with species from this subgenus placed into all three major clades of the genus (Fig.1); it was also considered to be paraphyletic by Puff et al. (1991) in their analyses based on morphological data. Our analyses also show that subgenus Lecontea is not monophyletic and is nested with the taxa of subgenus Alatopaederia from Madagascar (Fig. 1). Members of subgenus Lecontea have seeds resembling those of subgenus Alatopaederia, except that the former bears larger seeds, but subgenus Lecontea is distinguished from the others by its showy bracts, elongated and fused styles, and long and narrow corollas (Puff, 1991a). Subgenus Paederia is restricted to Asia with only four species (Puff, 1991c; Chen and Taylor, 2011), and is the only monophyletic subgenus recognized by Puff (1991b). However, phylogenetic resolution in this subgenus is extremely low when multiple collections were sampled, as shown by a large polytomy in Fig. 1.
Post-Boreotropical origin in continental Asia
Accurate calibration with multiple fossils has been suggested as a key factor in age estimation of lineages (Renner, 2005; Sauquet et al., 2009). Our age estimates employed four reliable fossils, and the results are largely close to those of Bremer and Eriksson (2009). The origin and early diversification of Paederia in the Oligocene (nodes 1 and 2; Fig. 2) are in a time frame well after the existence of the Boreotropical floras from the Palaeocene to the Eocene. Asia harbours the largest number of the Boreotropical genera, perhaps due to the continuous land connection between the northern latitudes and the equatorial zone in Asia (Tiffney, 1985a, b; Morley, 2003). The earliest diverging lineage of Paederia shows a disjunct distribution between the Malesian region (P. verticillata) and continental Asia (P. spectatissima) in the early to middle Miocene, which is consistent with this connection (node 3; Fig. 2).
Puff et al. (1991) generally hypothesized an Asian origin of Paederia according to its high species diversity in this region. Wikström et al. (2010) also suggested Asia as the ancestral area of Paederia based on molecular biogeographical analyses. Our results are largely congruent with the hypothesis of Asian origin of the genus (Fig. 3). However, our analyses further revealed that continental Asia (especially Indochina, the centre of highest diversity for the genus with 15 species) is the most likely ancestral range of Paederia, whereas there is only one endemic species (P. verticillata) found in the Malesian region of south-eastern Asia.
Palaeotropical intercontinental disjunction
The observed pattern of the African and Malagasy taxa (clade II in Figs 1 and 2) being nested in the continental Asian taxa is compatible with dispersal and/or migration from continental Asia to Africa and Madagascar. Both CTMC and Lagrange analyses further indicated a continental Asian origin of the Malagasy–African group in the middle to late Miocene (node 4 in Fig. 3 and Table 1). Since the estimated stem age for the Malagasy–African Paederia lineage at 15·99 Mya (95 % HPD: 10·28–22·68 Mya) in the early to mid Miocene is too recent (Fig. 2), several hypotheses seem to be unlikely to explain how such intercontinental disjunctions might have originated, such as ‘rafting’ on the Indian tectonic plate (Le Thomas and Doyle, 1996; Conti et al., 2002), migration through the Eocene Boreotropical flora (Wolfe, 1975) or Eocene–Oligocene dispersal from India to Madagascar through the ‘Lemurian stepping stones’ across the western Indian Ocean (Schatz, 1996). Long-distance dispersal across the Indian Ocean seems to be the most likely explanation for the disjunction of Paederia from continental Asia to Madagascar/Africa.
Many Afro-Asian plant disjunctions, especially those of very recent origin, have been explained by long-distance dispersal in both directions. It is of interest to note that fewer dispersal events have been reported from Asia to Africa. Nevertheless examples of the latter can be found in Bridelia (Phyllanthaceae) at approx. 10·00–1·85 Mya (Li et al., 2009) and Macaranga and Mallotus (Euphorbiaceae) at <27 Mya (Kulju et al., 2007). Dispersal from Africa and/or Madagascar to Asia has been suggested in Osbeckia (Melastomataceae) at approx. 16–7 Mya (Renner and Meyer, 2001; Renner, 2004b), Gaertnera (Rubiaceae) at approx. 6–5 Mya (Malcomber, 2002), Exacum (Gentianaceae) at less than approx. 35 Mya (Yuan et al., 2005), Cucumis (Cucurbitaceae) <10 Mya (Renner et al., 2007), Uvaria (Annonaceae) at 16·1–21·4 Mya (Zhou et al., 2012) and Eurycoma–Brucea–Soulamea (Simaroubaceae) during the Oligocene (Clayton et al., 2009).
As suggested by the biogeographical inferences based on our six-area analysis, either Madagascar or Africa is possible as the ancestral area of crown II (node 5 in Table 1). Wikström et al. (2010) also proposed that it is unclear whether the dispersal route is from Asia to Eastern Tropical Africa and finally to Madagascar (with further dispersal to the Comores because the Comorian species was nested in the Malagasy clade) or an opposite scenario (i.e. Asia → Madagascar → Africa). However, we prefer to adopt the former route, which seems to be consistent with the phylogenetic results (i.e. the single north-eastern African species of P. pospischilii plus the Mexican P. ciliata sister to a clade including all species from Madagascar, Fig. 2).
A recent comprehensive review on Malagasy biota suggested that dispersal of taxa from Africa was by far the most important contributing source to the Malagasy flora (Yoder and Nowak, 2006). For example, multiple dispersals from Africa to Madagascar have taken place in the Sapotaceae tribe Chrysophylloideae during the Tertiary (Bartish et al., 2011). Long-distance dispersals from Africa to Madagascar have also been reported from frogs (Vences et al., 2003, 2004), chameleons (Raxworthy et al., 2002), snakes (Nagy et al., 2003), lemurs (Yoder and Yang, 2004) and lorises (Masters et al., 2005). This hypothesis may be corroborated by our study on the Asian origin of Paederia with a recent dispersal from Africa to Madagascar in the late Miocene (node 5 in Figs 2 and 3, Table 1). Similarly, Wikström et al. (2010) found that the two other Rubiaceae tribes Knoxieae and Vanguerieae have their origins in eastern tropical and southern Africa and dispersed to Madagascar numerous times. Another taxon (P. bojeriana var. foetens, not sampled in this study) also occurs in Africa with its sister pair (P. bojeriana var. bojeriana) in Madagascar, which might indicate another dispersal event between Madagascar and Africa.
Another possibility is an overland migration from eastern Asia through south-west Asia to north-east Africa (Kappelman et al., 2003). This route of migration became available after the collision between the Arabian peninsula and the Anatolian Plate, the closing of the Tethys Sea with the formation of the ‘Gomphotherium land bridge’ (Rögl, 1998, 1999), and the global warming period between late Oligocene and middle Miocene (Zachos et al., 2001). The possibility of dispersal of tropical plants across northern Africa and Arabia is supported by fossil evidence in Meliaceae in Europe, Africa and Asia (Muellner et al., 2006) and the ‘out-of-Africa’ dispersal pattern in Uvaria of Annonaceae (Zhou et al., 2012). However, it seems less likely for the Paederia Asian–African disjunction, with a relatively young split age of 15·99 Mya at the boundary between the early and mid Miocene (Table 1). The rising of mountains (such as the Himalayas and the Tibetan Plateau) and increasingly cooler climates from the mid Miocene onwards (An et al., 2001; Zachos et al., 2001; Spicer et al., 2003; Sun et al., 2005) may have further constrained tropical and sub-tropical forests to lowlands and prevented floristic exchanges of tropical elements including Paederia between Asia and Africa.
Long-distance dispersals to the Neotropics
Paederia also shows a trans-Atlantic tropical disjunction between Central America and Africa at 6·08 (95 % HPD: 1·87–11·33) Mya in the late Miocene (node 6; Fig. 2). This divergence time is well after the last possible connection of Africa and South America at around 96–105 Mya and the last opening of the North Atlantic land bridge at 20–25 Mya (Tiffney, 1985a; McLoughlin, 2001; Morley, 2003). We thus argue that long-distance dispersal is also the most plausible mechanism for this trans-Atlantic disjunction in Paederia. Dispersal from the Caribbean or South America to São Tomé has been inferred for the primarily Neotropical genus Cayaponia (Cucurbitaceae), which has a single west African species estimated to have diverged at 2–5 Mya (Duchen and Renner, 2010). Trans-Atlantic tropical disjunctions are common in plants, and Renner (2004) enumerated a total of 109 plant lineages at the genus level showing this pattern, many apparently due to long-distance dispersal. However, most of them are disjunct between South America and west Africa, such as Vochysiaceae (Sytsma et al., 2004), Maschalocephalus in Rapateaceae (Givnish et al., 2004) and Pitcairnia in Bromeliaceae (Givnish et al., 2004). Paederia seems to be an unusual case exhibiting a disjunction between north-eastern Africa and Central America.
The divergence time of 10·01 (95 % HPD: 5·57–15·47) Mya in the late Miocene was estimated for the Paederia disjunction between continental Asia and South America (node 8 in Fig. 2 and Table 1). Disjunctions between the tropical Pacific and the Neotropics are less common, with about 89 genera of flowering plants known with the amphi-Pacific tropical distribution (Thorne, 1972). The continental Asian–tropical American disjunction may invoke land bridge vicariance, such as the amphi-Pacific disjunction in Persea that may have resulted from the disruption of the Boreotropical flora by climatic cooling during the mid to late Eocene (Li et al., 2011). In the case of the tropical Asian–South American disjunction in Paederia, direct transoceanic dispersal across the Pacific seems to be the most plausible explanation due to its recent split in the late Miocene (node 8 in Figs 2 and 3). There are many examples of long-distance dispersal between the tropical Pacific islands and the Neotropics, such as in Hernandia (Hernandiaceae) exhibiting a trans-Pacific disjunction with transoceanic dispersal from tropical Australia to the Neotropics in the Miocene (Michalak et al., 2010). Paederia provides one of the few examples of disjunctions between continental Asia and tropical South America which are tentatively explained by transoceanic dispersal.
Dispersal mechanism in Paederia
A number of recent studies have attempted to explain the pantropical disjunctions in plants with relatively young divergence times (Li et al., 2009; Michalak et al., 2010; Zhou et al., 2012). The stepping-stone mechanism, birds capable of long-distance flight and monsoon trade winds coupled with oceanic currents may be important factors related to dispersal (Ali et al., 2012). The small subgenus Paederia has sub-globose fruits without obviously winged diaspores and without distinct carpophores, but they are restricted to Asia. Most taxa of the genus (subgenus Alatopaederia and subgenus Lecontea) possess small, winged diaspores, showing a remarkable adaptation to wind dispersal (Puff, 1991a; Puff et al., 1991). It is not only the conspicuous diaspore wings but also the presentation and orientation of the diaspores which have been interpreted as a functional adaptation (Igersheim and Puff, 1991). The winged diaspores are held in a vertical position by the stout carpophores in order to be exposed effectively to the wind (Igersheim and Puff, 1991).
For many tropical tree diaspores, on the other hand, transport via ocean currents or in rafting mats of vegetation is a much more feasible scenario than dispersal by wind. Houle's study demonstrated that during the Miocene, intercontinental rafting could have occurred in <2 weeks on the North and South Equatorial counter currents between Africa and the Neotropics and on the North and South Equatorial counter current between East Africa and South India/south-east Asia (Houle, 1998). With small and winged seeds, it is also likely that seeds of Paederia spp. could travel by attaching to birds (while wet) or that propagules could have rafted across water barriers in large mats of vegetation. Therefore, the winged diaspores in Paederia may have facilitated long-distance dispersal by various agents including wind, ocean currents or even birds. The noni plant (Morinda citrifolia) is another example in Rubiaceae with a pantropical disjunction, and buoyant fruits and seeds that are transported by oceanic current drifting (Razafimandimbison et al., 2010).
Conclusions
Long-distance dispersal may be much more common than previously thought and may represent an important mechanism in the assembly of modern tropical floras (Givnish and Renner, 2004; Renner, 2004a; de Queiroz, 2005; Michalak et al., 2010). Our results from Paederia with robust molecular dating provide strong evidence for the divergence of Paederia from its sister group in tropical continental Asia in the Oligocene, with three dispersal events inferred into the other tropical areas. The trans-Indian Ocean dispersal into Madagascar–Africa is inferred to have occurred between the early and mid Miocene. The two Neotropical species are suggested to have dispersed from south-eastern Asia (trans-Pacific) and east Africa (trans-Atlantic) in the late Miocene via long-distance dispersal, respectively. The data presented here suggest the potential of post-Boreotropical divergence and long-distance dispersals across the three major oceans in shaping the global pantropical disjunction in Paederia.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
This study was supported by grants from Natural Sciences Foundation of China (NSFC 30970193 to Z.-L.N.), the One Hundred Talents Program of the Chinese Academy of Sciences (2011312D11022 to H.S.), United Fund of the NSFC and Yunnan Natural Science Foundation (U1136601 to H.S.), NSFC–International (Region) Cooperation Project (31061160184 to H.S.), the “Strategic Priority Research Program (B)” of the Chinese Academy of Sciences, XDB03030112 to H.S.) and the John D. and Catherine T. MacArthur Foundation to J. Wen, R. Ree and G. Mueller. Laboratory work was done at and partially supported by the Laboratory of Analytical Biology of the National Museum of Natural History, Smithsonian Institution. Fieldwork in North America was supported by the Small Grants Program of the National Museum of Natural History, the Smithsonian Institution. We thank Michael Nee, Fernando Chang, Ruth Kiew, Tze Leong Yao, Richard Chung, Leng Guan Saw, Elizabeth Widjaja, Zhiduan Chen, Tieyao Tu, Yumin Shui and Wenhong Chen for field assistance and sample collection.
APPENDIX
Taxa sequenced with voucher information and GenBank accessions
| Taxa | Voucher* | Location | rbcL | trnT-F | rps16 | atpB-rbcL | psbA-trnH |
|---|---|---|---|---|---|---|---|
| Paederia brasiliensis (Hook.f.) Puff | Nee & Wen 53771 (US) | Bolivia: Santa Cruz | KC305897 | KC306110 | KC306055 | KC306006 | KC305951 |
| Nee & Wen 53808 (US) | Bolivia: Santa Cruz | KC305898 | KC306111 | KC306056 | – | KC305952 | |
| Paederia cavaleriei H.Lév. | Nie 1267 (KUN) | China: Sichuan | KC305899 | KC306112 | KC306057 | KC306007 | KC305953 |
| Nie 2193 (KUN) | China: Guizhou | KC305900 | KC306113 | KC306058 | KC306008 | KC305954 | |
| Paederia ciliata (Bartl. ex DC.) Standl. | Susana Valencia 2971 (FCME) | Mexico: Guerrero | KC305901 | KC306114 | – | – | KC305955 |
| Paederia cruddasiana Prain | Nie 1990 (KUN) | China: Yunnan | KC305902 | KC306115 | KC306059 | KC306009 | KC305956 |
| Paederia farinosa var. farinosa (Baker) Puff | Wen 9474 (US) | Madagascar: Fianarantsoa | KC305903 | KC306116 | KC306060 | KC306010 | KC305957 |
| Wen 9622 (US) | Madagascar: Antsiranana | KC305904 | KC306117 | KC306061 | KC306011 | KC305958 | |
| Wen 9625 (US) | Madagascar: Antsiranana | KC305905 | KC306118 | KC306062 | KC306012 | KC305959 | |
| Paederia farinosa var. rosea Puff | Wen 9545 (US) | Madagascar: Antsiranana | KC305906 | KC306119 | KC306063 | KC306013 | KC305960 |
| Paederia foetida L. | Nie 413 (KUN) | China: Yunnan | KC305907 | KC306120 | KC306064 | KC306014 | KC305961 |
| Wen 9319 (US) | China: Hunan | KC305908 | KC306121 | KC306065 | KC306015 | KC305962 | |
| Wen 9371 (US) | China: Hunan | KC305909 | KC306122 | KC306066 | KC306016 | KC305963 | |
| Wen 9417 (US) | China: Taiwan | KC305910 | KC306123 | KC306067 | KC306017 | KC305964 | |
| Wen 9453 (US) | China: Taiwan | KC305911 | KC306124 | KC306068 | KC306018 | KC305965 | |
| Wen 9457 (US) | China: Taiwan | KC305912 | KC306125 | KC306069 | KC306019 | KC305966 | |
| Tibet-MacArthur 1931 (US, KUN) | China: Sichuan | KC305913 | KC306126 | KC306070 | KC306020 | KC305967 | |
| Shui et al. 81972 (KUN) | China: Yunnan | KC305914 | KC306127 | KC306071 | KC306021 | KC305968 | |
| Tibet-MacArthur 2227 (US, KUN) | China: Yunnan | KC305915 | KC306128 | KC306072 | KC306022 | KC305969 | |
| Wen 10658 (US) | China: Beijing | KC305916 | KC306129 | KC306073 | KC306023 | KC305970 | |
| Nie 1238 (KUN) | China: Sichuan | KC305917 | KC306130 | KC306074 | KC306024 | KC305971 | |
| Nie 1535 (KUN) | China: Yunnan | KC305918 | KC306131 | KC306075 | KC306025 | KC305972 | |
| Wen 10888 (US) | Vietnam: Lao Cai | KC305919 | KC306132 | KC306076 | – | KC305973 | |
| Huang 09918 (IBSC, KUN) | China: Guangxi | KC305920 | KC306133 | KC306077 | KC306026 | KC305974 | |
| Nie 0221 (KUN) | China: Chongqin | KC305921 | KC306134 | KC306078 | KC306027 | KC305975 | |
| Nie 1952 (KUN) | China: Yunnan | KC305922 | KC306135 | KC306079 | KC306028 | KC305976 | |
| Nie 2066 (KUN) | China: Sichuan | KC305923 | KC306136 | KC306080 | KC306029 | KC305977 | |
| Nie 2186 (KUN) | China: Guizhou | KC305924 | KC306137 | KC306081 | KC306030 | KC305978 | |
| Nie 2323 (KUN) | China: Guangxi | KC305925 | KC306138 | KC306082 | KC306031 | KC305979 | |
| Nie 2392 (KUN) | China: Guangxi | KC305926 | KC306139 | KC306083 | KC306032 | KC305980 | |
| Nie 3142 (KUN) | China: Sichuan | KC305927 | KC306140 | KC306084 | KC306033 | KC305981 | |
| Huang WP 021 (KUN) | China: Jiangxi | KC305928 | KC306141 | KC306085 | KC306034 | KC305982 | |
| Xie YP 008 (KUN) | China: Shanxi | KC305929 | KC306142 | KC306086 | KC306035 | KC305983 | |
| Deng T 180 (KUN) | China: Anhui | KC305930 | KC306143 | KC306087 | KC306036 | KC305984 | |
| Nie 3280 (KUN) | China: Yunnan | KC305931 | KC306144 | KC306088 | KC306037 | KC305985 | |
| Nie 3478 (KUN) | China: Yunnan | KC305932 | KC306145 | KC306089 | KC306038 | KC305986 | |
| Paederia grandidieri Drake | Wen 9486 (US) | Madagascar: Fianarantsoa | KC305933 | KC306146 | KC306090 | KC306039 | KC305987 |
| Paederia lanuginosa Wall. | Tibet-MacArthur 2096 (US, KUN) | China: Yunnan | KC305934 | KC306147 | KC306091 | KC306040 | KC305988 |
| Tibet-MacArthur 2238 (US, KUN) | China: Yunnan | KC305935 | KC306148 | KC306092 | KC306041 | KC305989 | |
| Tibet-MacArthur 2237 (US, KUN) | China: Yunnan | KC305936 | – | KC306093 | KC306042 | KC305990 | |
| Paederia pertomentosa Merr. ex H.L.Li | Nie 2239 (KUN) | China: Guizhou | KC305937 | KC306149 | KC306094 | KC306043 | KC305991 |
| Paederia praetermissa Puff | Nie 3474 (KUN) | China: Yunnan | KC305938 | KC306150 | KC306095 | KC306044 | KC305992 |
| Nie 3485 (KUN) | China: Yunnan | – | KC306164 | KC306109 | – | KC306005 | |
| Paederia sambiranensis Homolle ex Puff | Wen 9535 (US) | Madagascar: Antsiranana | KC305939 | KC306151 | KC306096 | KC306045 | KC305993 |
| Paederia sp. | Tibet-MacArthur 2236 (US, KUN) | China: Yunnan | KC305940 | KC306152 | KC306097 | KC306046 | KC305994 |
| Paederia spectatissima H.Li | Nie 4375 (KUN) | China: Yunnan | KC305941 | KC306153 | KC306098 | – | KC305995 |
| Paederia stenobotrya Merr. | Nie 3952 (KUN) | China: Hainan | KC305942 | KC306154 | KC306099 | – | KC305996 |
| Paederia taolagnarensis Razafim. & C.M.Taylor | Wen 9678 (US) | Madagascar: Toliara | KC305943 | KC306155 | KC306100 | KC306047 | KC305997 |
| Wen 9671 (US) | Madagascar: Toliara | KC305944 | KC306156 | KC306101 | KC306048 | KC305998 | |
| Paederia verticillata Blume | Wen 11690 (US) | Malaysia: Malay | KC305945 | KC306157 | KC306102 | KC306049 | KC305999 |
| Wen 11834 (US) | Malaysia: Borneo | KC305946 | KC306158 | KC306103 | KC306050 | KC306000 | |
| Paederia yunnanensis (H.Lév.) Rehder | Shui et al. 81880 (KUN) | China: Yunnan | KC305947 | KC306159 | KC306104 | KC306051 | KC306001 |
| Nie 546 (KUN) | China: Yunnan | KC305948 | KC306160 | KC306105 | KC306052 | – | |
| Nie 1265 (KUN) | China: Sichuan | KC305949 | KC306161 | KC306106 | KC306053 | KC306002 | |
| Nie 1997 (KUN) | China: Yunnan | KC305950 | KC306162 | KC306107 | KC306054 | KC306003 | |
| Serissa serissoides (DC.) Druce | Chen et al. 20110431 (PE) | China: Taiwan | – | KC306163 | KC306108 | – | KC306004 |
* Herbarium acronyms are as follows: IBSC, South China Botanical Garden, Chinese Academy of Sciences; KUN, Kunming Institute of Botany, Chinese Academy of Sciences; FCME, Universidad Nacional Autónoma de México, Ciudad Universitaria; PE, Institute of Botany, Chinese Academy of Sciences; US, US National Herbarium of the Smithsonian Institution.
LITERATURE CITED
- Ali SS, Yu Y, Pfosser M, Wetschnig W. Inferences of biogeographical histories within subfamily Hyacinthoideae using S-DIVA and Bayesian binary MCMC analysis implemented in RASP (Reconstruct Ancestral State in Phylogenies) Annals of Botany. 2012;109:95–107. doi: 10.1093/aob/mcr274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Z, Kutzbach JE, Prell WL, Porter SC. Evolution of Asian monsoons and phased uplift of the Himalaya–Tibetan plateau since Late Miocene times. Nature. 2001;411:62–66. doi: 10.1038/35075035. [DOI] [PubMed] [Google Scholar]
- Antonelli A, Nylander JAA, Persson C, Sanmartín I. Tracing the impact of the Andean uplift on Neotropical plant evolution. Proceedings of the National Academy of Sciences, USA. 2009;106:9749–9754. doi: 10.1073/pnas.0811421106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backlund M, Bremer B, Thulin M. Paraphyly of Paederieae, recognition of Putorieae and expansion of Plocama (Rubiaceae–Rubioideae) Taxon. 2007;56:315–328. [Google Scholar]
- Bartish IV, Antonelli A, Richardson JE, Swenson U. Vicariance or long-distance dispersal: historical biogeography of the pantropical subfamily Chrysophylloideae (Sapotaceae) Journal of Biogeography. 2011;38:177–190. [Google Scholar]
- Bone RE, Strijk JS, Fritsch PW, et al. Phylogenetic inference of Badula (Primulaceae), a rare and threatened genus endemic to the Mascarene Archipelago. Botanical Journal of the Linnean Society. 2012;169:284–296. [Google Scholar]
- Bremer B, Bremer K, Heidari N, et al. Phylogenetics of asterids based on 3 coding and 3 non-coding chloroplast DNA markers and the utility of non-coding DNA at higher taxonomic levels. Molecular Phylogenetics and Evolution. 2002;24:274–301. doi: 10.1016/s1055-7903(02)00240-3. [DOI] [PubMed] [Google Scholar]
- Bremer B, Eriksson T. Time tree of Rubiaceae: phylogeny and dating the family, subfamilies, and tribes. International Journal of Plant Sciences. 2009;170:766–793. [Google Scholar]
- Bremer K, Friis EM, Bremer B. Molecular phylogenetic dating of asterid flowering plants shows early Cretaceous diversification. Systematic Biology. 2004;53:496–505. doi: 10.1080/10635150490445913. [DOI] [PubMed] [Google Scholar]
- Chen T, Taylor CM. In: Paederia Flora of China Volume 19 (Rubiaceae). Wu ZY, Raven PH, Hong DY, editors. Beijing: Science Press and St; 2011. pp. 282–287. [Google Scholar]
- Christenhusz MJM, Chase MW. Biogeographical patterns of plants in the Neotropics – dispersal rather than plate tectonics is most explanatory. Botanical Journal of the Linnean Society. 2013;171:277–286. [Google Scholar]
- Clayton JW, Soltis PS, Soltis DE. Recent long-distance dispersal overshadows ancient biogeographical patterns in a pantropical angiosperm family (Simaroubaceae, Sapindales) Systematic Biology. 2009;58:395–410. doi: 10.1093/sysbio/syp041. [DOI] [PubMed] [Google Scholar]
- Conti E, Eriksson T, Schonenberger J, Sytsma KJ, Baum DA. Early tertiary out-of-India dispersal of Crypteroniaceae: evidence from phylogeny and molecular dating. Evolution. 2002;56:1931–1942. doi: 10.1554/0014-3820(2002)056[1931:ETOOID]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Davis CC, Bell CD, Mathews S, Donoghue MJ. Laurasian migration explains Gondwanan disjunctions: evidence from Malpighiaceae. Proceedings of the National Academy of Sciences, USA. 2002;99:6833–6837. doi: 10.1073/pnas.102175899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorofeev PI. New data about Tertiary floras of Kirrenskiy ravine on the Ob River. Doklady Akademii Nauk SSSR. 1960;133:211–213. [Google Scholar]
- Dorofeev PI. Moscow: Izd-vo Akademii nauk SSSR; 1963. Tertiary floras of western Siberia. [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7(214) doi: 10.1186/1471-2148-7-214. http://dx.doi.org/10.1186/1471-2148-7-214 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchen P, Renner SS. The evolution of Cayaponia (Cucurbitaceae): repeated shifts from bat to bee pollination and long-distance dispersal to Africa 2–5 million years ago. American Journal of Botany. 2010;97:1129–1141. doi: 10.3732/ajb.0900385. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdtman G. Pollen morphology and plant taxonomy. New York: Hafner; 1966. [Google Scholar]
- Erkens RHJ, Maas JW, Couvreur TLP. From Africa via Europe to South America: migrational route of a species-rich genus of Neotropical lowland rain forest trees (Guatteria, Annonaceae) Journal of Biogeography. 2009;36:2338–2352. [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. Journal of Molecular Evolution. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Friis EM. Angiosperm fruits and seeds from the Middle Miocene of Jutland, Denmark. Biologiske Skrifter. 1985;24:1–165. [Google Scholar]
- Givnish TJ, Renner SS. Tropical intercontinental disjunctions: Gondwana breakup, immigration from the Boreotropics, and transoceanic dispersal. International Journal of Plant Sciences. 2004;165:S1–S6. [Google Scholar]
- Givnish TJ, Millam KC, Evans TM, et al. Ancient vicariance or recent long-distance dispersal? Inferences about phylogeny and South American–African disjunctions in Rapateaceae and Bromeliaceae based on ndhF sequence data. International Journal of Plant Sciences. 2004;165:s35–s54. [Google Scholar]
- Graham A. Fossil record of the Rubiaceae. Annals of the Missouri Botanical Garden. 2009;96:90–108. [Google Scholar]
- Ho SYW, Phillips MJ. Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Systematic Biology. 2009;58:367–380. doi: 10.1093/sysbio/syp035. [DOI] [PubMed] [Google Scholar]
- Houle A. Floating islands: a mode of long-distance dispersal for small and medium-sized terrestrial vertebrates. Diversity and Distributions. 1998;4:201–216. [Google Scholar]
- Huang W-P, Wen J, Sun H, Nie Z-L. Molecular phylogeny of the eastern Asian – eastern North American disjunct Mitchella and its close relative Damnacanthus (Rubiaceae, Mitchelleae) Botanical Journal of the Linnean Society. 2013;171:395–412. [Google Scholar]
- Huelsenbeck JP, Ronquist R. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Igersheim A, Puff C. The fruits, diaspores and seeds of Paederia L. (Rubiaceae-Paederieae) Opera Botanica Belgica. 1991;3:89–102. [Google Scholar]
- Kappelman J, Rasmussen DT, Sanders WJ, et al. Oligocene mammals from Ethiopia and faunal exchange between Afro-Arabia and Eurasia. Nature. 2003;426:549–552. doi: 10.1038/nature02102. [DOI] [PubMed] [Google Scholar]
- Koechlin J. Flora and vegetation of Madagascar. Biogeography and ecology in Madagascar. 1972:145–190. The Hague: Junk. [Google Scholar]
- Kulju KKM, Sierra SEC, Draisma SGA, Samuel R, van Welzen PC. Molecular phylogeny of Macaranga, Mallotus, and related genera (Euphorbiaceae s.s.): insights from plastid and nuclear DNA sequence data. American Journal of Botany. 2007;94:1726–1743. doi: 10.3732/ajb.94.10.1726. [DOI] [PubMed] [Google Scholar]
- Lamm KS, Redelings BD. Reconstructing ancestral ranges in historical biogeography: properties and prospects. Journal of Systematics and Evolution. 2009;47:369–382. [Google Scholar]
- Lavin M, Thulin M, Labat JN, Pennington RT. Africa, the odd man out: molecular biogeography of dalbergioid legumes (Fabaceae) suggests otherwise. Systematic Botany. 2000;25:449–467. [Google Scholar]
- Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLoS Computational Biology. 2009;5 doi: 10.1371/journal.pcbi.1000520. e1000520. http://dx.doi.org/10.1371/journal.pcbi.1000520 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopold E. Miocene pollen and spore flora of Eniwetok Atoll, Marshall Island. Geological Survey Professional Paper260-II. 1969:1133–1185. [Google Scholar]
- Le Thomas A, Doyle JA. Geographic relationships of Malagasy Annonaceae. In: Lourenco WR, editor. Biogéographie de Madagascar. 1996. pp. 85–94. Paris: Institut Français de Recherche Scientifique pour le Développement en Coopération. [Google Scholar]
- Li L, Li J, Rohwer JG, van der Werff H, Wang ZH, Li HW. Molecular phylogenetic analysis of the Persea group (Lauraceae) and its biogeographic implications on the evolution of tropical and subtropical amphi-Pacific disjunctions. American Journal of Botany. 2011;98:1520–1536. doi: 10.3732/ajb.1100006. [DOI] [PubMed] [Google Scholar]
- Li YQ, Dressler S, Zhang DX, Renner SS. More Miocene dispersal between Africa and Asia – the case of Bridelia (Phyllanthaceae) Systematic Botany. 2009;34:521–529. [Google Scholar]
- Mabberley DJ. Mabberley's Plant-book: a portable dictionary of plants, their classifications, and uses. Cambridge: Cambridge University Press; 2008. [Google Scholar]
- Mai DH, Walther H. The upper Eocene floras of the Weisselster basin and adjacent areas. Abhandlungen des Staatlichen Museums fu"r Mineralogie und Geologie zu Dresden. 1985;33:1–260. [Google Scholar]
- Malcomber ST. Phylogeny of Gaertnera Lam. (Rubiaceae) based on multiple DNA markers: evidence of a rapid radiation in a widespread, morphologically diverse genus. Evolution. 2002;56:42–57. doi: 10.1111/j.0014-3820.2002.tb00848.x. [DOI] [PubMed] [Google Scholar]
- Manen J-F, Natali A, Ehrendorfer F. Phylogeny of Rubiaceae-Rubieae inferred from the sequence of a cpDNA intergene region. Plant Systematics and Evolution. 1994;190:195–211. [Google Scholar]
- Masters JC, Anthony NM, de Wit MJ, Mitchell A. Reconstructing the evolutionary history of the Lorisidae using morphological, molecular, and geological data. American Journal of Physical Anthropology. 2005;127:465–480. doi: 10.1002/ajpa.20149. [DOI] [PubMed] [Google Scholar]
- McLoughlin S. The breakup history of Gondwana and its impact on pre-Cenozoic floristic provincialism. Australian Journal of Botany. 2001;49:271–300. [Google Scholar]
- Michalak I, Zhang LB, Renner SS. Trans-Atlantic, trans-Pacific and trans-Indian Ocean dispersal in the small Gondwanan Laurales family Hernandiaceae. Journal of Biogeography. 2010;37:1214–1226. [Google Scholar]
- Morley RJ. Chichester: John Wiley & Sons, Ltd. Origin and evolution of tropical rain forests. 2000 [Google Scholar]
- Morley RJ. Interplate dispersal paths for megathermal angiosperms. Perspectives in Plant Ecology, Evolution and Systematics. 2003;6:5–20. [Google Scholar]
- Morley RJ. Tropical rainforest responses to climatic change. Springer: Berlin; 2007. Cretaceous and Tertiary climate change and the past distribution of megathermal rainforests; pp. 1–31. [Google Scholar]
- Muellner AN, Savolainen V, Samuel R, Chase MW. The mahogany family ‘out-of-Africa’: divergence time estimation, global biogeographic patterns inferred from plastid rbcL DNA sequences, extant, and fossil distribution of diversity. Molecular Phylogenetics and Evolution. 2006;40:236–250. doi: 10.1016/j.ympev.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Nagy ZT, Joger U, Wink M, Glaw F, Vences M. Multiple colonization of Madagascar and Socotra by colubrid snakes: evidence from nuclear and mitochondrial gene phylogenies. Proceedings of the Royal Society B: Biological Sciences. 2003;270:2613–2621. doi: 10.1098/rspb.2003.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z-L, Wen J, Sun H, Bartholomew B. Monophyly of Kelloggia Torrey ex Benth. (Rubiaceae) and evolution of its intercontinental disjunction between western North America and eastern Asia. American Journal of Botany. 2005;92:642–652. doi: 10.3732/ajb.92.4.642. [DOI] [PubMed] [Google Scholar]
- Nylander JAA. Program distributed by the author. MrModeltest v2. 2004 Evolutionary Biology Centre, Uppsala University. http://www.abc.se/~nylander/ [Google Scholar]
- Nylander JAA, Wilgenbusch JC, Warren DL, Swofford DL. AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2008;24:581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
- Olmstead RG, Michaels HJ, Scott KM, Palmer JD. Monophyly of the Asteridae and identification of their major lineages inferred from DNA sequences of rbcL. Annals of the Missouri Botanical Garden. 1992;79:249–265. [Google Scholar]
- Oxelman B, Liden M, Berglund D. Chloroplast rps16 intron phylogeny of the tribe Sileneae (Caryophyllaceae) Plant Systematics and Evolution. 1997;206:393–410. [Google Scholar]
- Puff C. The delimitation of the tribe Anthospermeae and its affinities to the Paederieae (Rubiaceae) Botanical Journal of the Linnean Society. 1982;84:355–377. [Google Scholar]
- Puff C. The genus Paederia L. (Rubiaceae-Paederieae): taxonomic history, revised generic description, and subgeneric division. Opera Botanica Belgica. 1991;3:195–204. [Google Scholar]
- Puff C. Revision of the genus Paederia L. (Rubiaceae-Paederieae) in America. Opera Botanica Belgica. 1991;3:325–333. [Google Scholar]
- Puff C. Revision of the genus Paederia L. (Rubiaceae-Paederieae) in Asia. Opera Botanica Belgica. 1991;3:207–289. [Google Scholar]
- Puff C, Robbrecht E, Andersson L. Relationships and evolution in Paederia L. (Rubiaceae-Paederieae): a comparison of classical approach, numerical evaluation and cladistic analysis. Opera Botanica Belgica. 1991;3:337–358. [Google Scholar]
- de Queiroz A. The resurrection of oceanic dispersal in historical biogeography. Trends in Ecology and Evolution. 2005;20:68–73. doi: 10.1016/j.tree.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Raven PH, Axelrod DI. Angiosperm biogeography and past continental movements. Annals of the Missouri Botanical Garden. 1974;61:539–673. [Google Scholar]
- Raxworthy CJ, Forstner MRJ, Nussbaum RA. Chameleon radiation by oceanic dispersal. Nature. 2002;415:784–787. doi: 10.1038/415784a. [DOI] [PubMed] [Google Scholar]
- Razafimandimbison SG, McDowell TD, Halford DA, Bremer B. Molecular phylogenetics and generic assessment in the tribe Morindeae (Rubiaceae-Rubioideae): how to circumscribe Morinda L. to be monophyletic? Molecular Phylogenetics and Evolution. 2009;52:879–886. doi: 10.1016/j.ympev.2009.04.007. [DOI] [PubMed] [Google Scholar]
- Razafimandimbison SG, McDowell TD, Halford DA, Bremer B. Origin of the pantropical and nutriceutical Morinda citrifolia L. (Rubiaceae): comments on its distribution range and circumscription. Journal of Biogeography. 2010;37:520–529. [Google Scholar]
- Razafimandimbison SG, Taylor CM. A new species of the genus Paederia (Rubiaceae) from the Petriky Forest, Taolagnaro, Madagascar. Novon. 2000;10:71–73. [Google Scholar]
- Ree RH, Moore BR, Webb CO, Donoghue MJ. A likelihood framework for inferring the evolution of geographic range on phylogenetic trees. Evolution. 2005;59:2299–2311. [PubMed] [Google Scholar]
- Ree RH, Sanmartin I. Prospects and challenges for parametric models in historical biogeographical inference. Journal of Biogeography. 2009;36:1211–1220. [Google Scholar]
- Ree RH, Smith SA. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Systematic Biology. 2008;57:4–14. doi: 10.1080/10635150701883881. [DOI] [PubMed] [Google Scholar]
- Renner S. Plant dispersal across the tropical Atlantic by wind and sea currents. International Journal of Plant Sciences. 2004;165:S23–S33. [Google Scholar]
- Renner SS. Multiple Miocene Melastomataceae dispersal between Madagascar, Africa and India. Philosophical Transactions of the Royal Society B: Biological Sciences. 2004;359:1485–1494. doi: 10.1098/rstb.2004.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner SS. Relaxed molecular clocks for dating historical plant dispersal events. Trends in Plant Science. 2005;10:550–558. doi: 10.1016/j.tplants.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Renner SS, Meyer K. Melastomeae come full circle: biogeographic reconstruction and molecular clock dating. Evolution. 2001;55:1315–1324. doi: 10.1111/j.0014-3820.2001.tb00654.x. [DOI] [PubMed] [Google Scholar]
- Renner SS, Schaefer H, Kocyan A. Phylogenetics of Cucumis (Cucurbitaceae): cucumber (C. sativus) belongs in an Asian/Australian clade far from melon (C. melo) BMC Evolutionary Biology. 2007;7(58) doi: 10.1186/1471-2148-7-58. http://dx.doi.org/10.1186/1471-2148-7-58 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbrecht E. Tropical woody Rubiaceae. Opera Botanica Belgica. 1988;1:1–271. [Google Scholar]
- Rögl F. Palaeogeographic considerations for Mediterranean and Paratethys seaways (Oligocene to Miocene) Annalen des Naturhistorischen Museums in Wien. 1998;99:279–310. [Google Scholar]
- Rögl F. Mediterranean and Paratethys. Geologica Carpathica. 1999;50:339–349. [Google Scholar]
- Rydin C, Razafimandimbison SG, Khodabandeh A, Bremer B. Evolutionary relationships in the Spermacoceae alliance (Rubiaceae) using information from six molecular loci: insights into systematic affinities of Neohymenopogon and Mouretia. Taxon. 2009;58:793–810. [Google Scholar]
- Saenger P. Mangrove vegetation: an evolutionary perspective. Marine Freshwater Research. 1998;49:277–286. [Google Scholar]
- Sang T, Crawford DJ, Stuessy TF. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae) American Journal of Botany. 1997;84:1120–1136. [PubMed] [Google Scholar]
- Sanmartín I, Van der Mark P, Ronquist F. Inferring dispersal: a Bayesian approach to phylogeny-based island biogeography, with special reference to the Canary Islands. Journal of Biogeography. 2008;35:428–449. [Google Scholar]
- Sauquet H, Weston PH, Anderson CL, et al. Contrasted patterns of hyperdiversification in Mediterranean hotspots. Proceedings of the National Academy of Sciences, USA. 2009;106:221–225. doi: 10.1073/pnas.0805607106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz GE. Malagasy/Indo-Australo-Malesian phytogeographic connections. In: Lourenco WR, editor. Biogeography of Madagascar. Paris: 1996. pp. 73–83. Editions de l'ORSTOM. [Google Scholar]
- Shi X, Jin J, Ye C, Liu W. First fruit fossil record of Morinda (Rubiaceae) from China. Review of Palaeobotany and Palynology. 2012;179:13–16. [Google Scholar]
- Smedmark JEE, Anderberg AA. Boreotropical migration explains hybridization between geographically distant lineages in the pantropical clade Sideroxyleae (Sapotaceae) American Journal of Botany. 2007;94:1491–1505. doi: 10.3732/ajb.94.9.1491. [DOI] [PubMed] [Google Scholar]
- Smedmark JEE, Eriksson T, Bremer B. Divergence time uncertainty and historical biogeography reconstruction – an example from Urophylleae (Rubiaceae) Journal of Biogeography. 2010;37:2260–2274. [Google Scholar]
- Smith JF, Stevens AC, Tepe EJ, Davidson C. Placing the origin of two species-rich genera in the late Cretaceous with later species divergence in the tertiary: a phylogenetic, biogeographic and molecular dating analysis of Piper and Peperomia (Piperaceae) Plant Systematics and Evolution. 2008;275:9–30. [Google Scholar]
- Spicer RA, Harris NBW, Widdowson M, et al. Constant elevation of southern Tibet over the past 15 million years. Nature. 2003;421:622–624. doi: 10.1038/nature01356. [DOI] [PubMed] [Google Scholar]
- Sun J, Zhu R, An Z. Tectonic uplift in the northern Tibetan Plateau since 13·7 Ma ago inferred from molasse deposits along the Altyn Tagh Fault. Earth and Planetary Science Letters. 2005;235:641–653. [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sunderland, MA: Sinauer Associates; 2003. [Google Scholar]
- Sytsma KJ, Litt A, Zjhra ML, et al. Clades, clocks, and continents: historical and biogeographical analysis of Myrtaceae, Vochysiaceae, and relatives in the Southern Hemisphere. International Journal of Plant Sciences. 2004;165:S85–S105. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Takeda Y, Puff C, Kondoh Y, Yoshioka Y. Analysis of iridoid glycosides of Paederia L. species (Rubiaceae-Paederieae) by gas-chromatography and gas chromatography-mass spectrometry. Opera Botanica Belgica. 1991;3:153–158. [Google Scholar]
- Thorne RF. Major disjunctions in the geographic ranges of seed plants. Quarterly Review of Biology. 1972;47:365–411. [Google Scholar]
- Tiffney BH. The Eocene North Atlantic land bridge: its importance in Tertiary and modern phytogeography of the Northern Hemisphere. Journal of the Arnold Arboretum. 1985;66:243–273. [Google Scholar]
- Tiffney BH. Perspectives on the origin of the floristic similarity between eastern Asia and eastern North America. Journal of the Arnold Arboretum. 1985;66:73–94. [Google Scholar]
- Vences M, Kosuch J, Glaw F, Böhme W, Veith M. Molecular phylogeny of hyperoliid treefrogs: biogeographic origin of Malagasy and Seychellean taxa and re-analysis of familial paraphyly. Journal of Zoological Systematics and Evolutionary Research. 2003;41:205–215. [Google Scholar]
- Vences M, Kosuch J, Rödel M-O, Lötters S, Channing A, Glaw F, Böhme W. Phylogeography of Ptychadena mascareniensis suggests transoceanic dispersal in a widespread African-Malagasy frog lineage. Journal of Biogeography. 2004;31:593–601. [Google Scholar]
- Wikström N, Avino M, Razafimandimbison SG, Bremer B. Historical biogeography of the coffee family (Rubiaceae, Gentianales) in Madagascar: case studies from the tribes Knoxieae, Naucleeae, Paederieae and Vanguerieae. Journal of Biogeography. 2010;37:1094–1113. [Google Scholar]
- Wolfe JA. Some aspects of plant geography of the Northern Hemisphere during the late Cretaceous and Tertiary. Annals of the Missouri Botanical Garden. 1975;62:264–279. [Google Scholar]
- Yoder AD, Nowak MD. Has vicariance or dispersal been the predominant biogeographic force in Madagascar? Only time will tell. Annual Review of Ecology Evolution and Systematics. 2006;37:405–431. [Google Scholar]
- Yoder AD, Yang ZH. Divergence dates for Malagasy lemurs estimated from multiple gene loci: geological and evolutionary context. Molecular Ecology. 2004;13:757–773. doi: 10.1046/j.1365-294x.2004.02106.x. [DOI] [PubMed] [Google Scholar]
- Yuan YM, Wohlhauser S, Moller M, Klackenberg J, Callmander MW, Kupfer P. Phylogeny and biogeography of Exacum (Gentianaceae): a disjunctive distribution in the Indian Ocean Basin resulted from long distance dispersal and extensive radiation. Systematic Biology. 2005;54:21–34. doi: 10.1080/10635150590905867. [DOI] [PubMed] [Google Scholar]
- Zachos J, Pagani M, Sloan L, Thomas E, Billups K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science. 2001;292:686–693. doi: 10.1126/science.1059412. [DOI] [PubMed] [Google Scholar]
- Zhang LB, Simmons MP, Renner SS. A phylogeny of Anisophylleaceae based on six nuclear and plastid loci: ancient disjunctions and recent dispersal between South America, Africa, and Asia. Molecular Phylogenetics and Evolution. 2007;44:1057–1067. doi: 10.1016/j.ympev.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Zhou L, Su YCF, Thomas DC, Saunders RMK. ‘Out-of-Africa’ dispersal of tropical floras during the Miocene climatic optimum: evidence from Uvaria (Annonaceae) Journal of Biogeography. 2012;39:322–335. [Google Scholar]
- Zou X, Peng SL, Liu X, Bai BR, Ding LS. Sulfur-containing iridoid glucosides from Paederia scandens. Fitoterapia. 2006;77:374–377. doi: 10.1016/j.fitote.2006.05.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.