Abstract
Background and Aims
Understanding the introduction history of invasive plant species is important for their management and identifying effective host-specific biological control agents. However, uncertain taxonomy, intra- and interspecific hybridization, and cryptic speciation may obscure introduction histories, making it difficult to identify native regions to explore for host-specific agents. The overall aim of this study was to identify the native source populations of Acacia pycnantha, a tree native to south-eastern Australia and invasive in South Africa, Western Australia and Portugal. Using a phylogeographical approach also allowed an exploration of the historical processes that have shaped the genetic structure of A. pycnantha in its native range.
Methods
Nuclear (nDNA) and plastid DNA sequence data were used in network and tree-building analyses to reconstruct phylogeographical relationships between native and invasive A. pycnantha populations. In addition, mismatch distributions, relative rates and Bayesian analyses were used to infer recent demographic processes and timing of events in Australia that led to population structure and diversification.
Key Results
The plastid network indicated that Australian populations of A. pycnantha are geographically structured into two informally recognized lineages, the wetland and dryland forms, whereas the nuclear phylogeny showed little geographical structure between these two forms. Moreover, the dryland form of A. pycnantha showed close genetic similarity to the wetland form based on nDNA sequence data. Hybrid zones may explain these findings, supported here by incongruent phylogenetic placement of some of these taxa between nuclear and plastid genealogies.
Conclusions
It is hypothesized that habitat fragmentation due to cycles of aridity inter-dispersed with periods of abundant rainfall during the Pleistocene (approx. 100 kya) probably gave rise to native dryland and wetland forms of A. pycnantha. Although the different lineages were confined to different ecological regions, we also found evidence for intraspecific hybridization in Victoria. The invasive populations in Portugal and South Africa represent wetland forms, whereas some South African populations resemble the Victorian dryland form. The success of the biological control programme for A. pycnantha in South Africa may therefore be attributed to the fact that the gall-forming wasp Trichilogaster signiventris was sourced from South Australian populations, which closely match most of the invasive populations in South Africa.
Keywords: Acacia pycnantha, biological control, biological invasions, genetic structure, hybridization, lineage divergence, native provenance, Pleistocene, systematics, taxonomy
INTRODUCTION
Understanding the processes that shape species distributions and their evolutionary trajectories has long interested biogeographers, ecologists and phylogeographers. From a phylogeographical perspective, delineating genealogical relationships among taxa informs us about the historical processes that shaped patterns of gene flow, local adaptation and, ultimately, speciation. There is growing interest in better understanding processes that underpin the evolutionary trajectories of contemporary species movements, especially those that involve the transfer of species that become established and invasive (Richardson et al., 2011). Historical and contemporary biogeographies are both important, with the former influencing the latter in several ways (e.g. Hui et al., 2011; Rosenmeier et al., 2013). Biological invasions are typically characterized by stochasticity, founder events and strong genetic drift, and introduced genotypes usually encompass only a small proportion of the total genetic diversity of the taxon (Bossdorf et al., 2005; Kliber and Eckert, 2005). It is therefore important to understand the structure of natal populations when building a framework for testing hypotheses about the processes driving biological invasions, such as introduction histories, hybridization, gene diversity and, ultimately, evolutionary potential (Stepien et al., 2002).
Relating historical biogeography to contemporary patterns of gene diversity associated with species introductions and invasions is, however, not always straightforward (Le Roux et al., 2011). Invasive plant populations can arise from individuals introduced from several previously allopatric parts of the native range. This creates opportunities for admixture, hybridization and, consequently, genetic novelty (Prentis et al., 2008). For example, Acacia cyclops sourced from different localities in Australia was introduced to a single location in South Africa that led to intraspecific hybridization (Le Roux et al., 2011). On the other hand, a congener, A. saligna, shows high intraspecific diversification in its native range (Millar et al., 2011), whereas invasive populations in South Africa comprise genetic entities not found in the native range (Thompson et al., 2012). Similarly, the most invasive Tamarix genotype in the United States is a hybrid of two species, which are allopatric in their native Eurasian range (Gaskin and Schaal, 2002). Such complex introduction scenarios are typical of species introduced for agroforestry in its widest sense, as species are often introduced from multiple sources on multiple occasions and are normally subjected to strong artificial selection following introduction.
Understanding introduction and invasion histories has important practical implications. The selection of effective host-specific biocontrol agents on invasive plants can depend on identifying which subspecific entities of the plant were introduced (Harris, 1998; Wardill et al., 2005; Goolsby et al., 2006). Identification of the native provenance of an invasive species can also improve the design of host-specificity lists and subsequent host testing under the assumption that historical biogeographical processes similarly influenced hosts and agents, and thus co-evolution (Wardill et al., 2005; McLeish et al., 2007). However, it should be noted that the identification of the native provenance of invasive species can be complicated by long histories of plantings and cultivation within the native range of the species (Thompson et al., 2012).
Here we aim to place populations of Acacia pycnantha that are invasive in South Africa, Western Australia and Portugal in the context of historical biogeographical patterns in the native range of the species in south-eastern Australia. Specifically, we use plastid and nuclear DNA (nDNA) markers to: (1) reconstruct phylogenetic relationships among invasive (South Africa, Portugal, western Australia) and native (eastern Australia) populations of A. pycnantha; (2) compare genetic diversities in these invasive and native populations; and (3) infer the historical processes that may have shaped genetic structure in the natal regions of the species.
MATERIALS AND METHODS
Study system
Australia's national flower, Acacia pycnantha (the golden wattle), is native to New South Wales, Victoria and South Australia and is introduced and invasive in Western Australia. It is probably also naturalizing in some areas of New South Wales and South Australia (Eyre Peninsula) from cultivated plantings in revegetation projects and along roadsides. The species, like many Australian acacias, has visible phenotypic variation across its native range (Maslin, 2001; Maslin and McDonald, 2004). In particular, informal morphological classification recognizes two distinct ecotypes, the dryland and wetland forms (Fig. 1). These have distinct phyllodes, flower colours and distributional ranges. Despite this variation, the species has not been formally separated into subspecific entities.
Fig. 1.

Herbarium specimens of the extreme ecotypes of Acacia pycnantha in Australia for the dryland form (A) characterized by thin phyllodes (C) and the wetland form (B) characterized by broad phyllodes (D).
Seeds of A. pycnantha were introduced to South Africa on at least two occasions (1865 and 1890) for tannin production and dune stabilization (Poynton, 2009), but the exact origin of the introduced seeds from the native range is unknown. Following introduction to South Africa, seeds of A. pycnantha were distributed extensively throughout the country (Poynton, 2009). Since then populations have become invasive in several locations with seven known localities of widespread invasions in the Eastern and Western Cape Provinces. A. pycnantha is now listed as a category-1 invasive plant meaning that all uses of the species are prohibited (Henderson, 2001; Nel et al., 2004).
Following the success of other biological control agents against Australian acacias in South Africa, a gall-forming wasp, Trichilogaster signiventris, was introduced in 1987 (Hoffmann et al., 2002; Impson et al., 2011), and a seed-feeding weevil, Melanterius maculates, was introduced in 2003 (Impson et al., 2011). The wasp dramatically reduced seed production, but the impact of the weevil is still to be determined (Impson et al., 2011).
Elsewhere in the world, A. pycnantha is invasive in Portugal (Richardson and Rejmánek, 2011) and naturalized in the United States (California) (http://www.calflora.org). An initial assessment of the potential for biocontrol of Australian acacias based on the experience in South Africa is underway in Portugal (Marchante et al., 2011), with T. signiventris identified as a potential candidate for release.
Plant collection
Phyllodes of A. pycnantha were collected from throughout its native range (from what appear to be natural populations in south-eastern Australia) and invasive (Western Australia and South Africa) ranges during 2009 (Appendix). To prevent sampling individuals from reforestation projects, most native populations were sampled away from roadsides, e.g. in national parks. Between two and five trees were sampled from each population. A single population of A. pycnantha was also sampled in August 2010 in Portugal (see Appendix for locality data). Phyllode material was dried in silica gel and kept at room temperature until DNA extraction. Duplicate voucher specimens were collected from each sampled population and deposited at the State Herbarium in Adelaide, South Australia, and the Stellenbosch University Herbarium in Stellenbosch, South Africa.
DNA extraction, PCR amplification and sequencing
DNA was extracted from dried leaf samples using the CTAB method (Doyle and Doyle, 1987) modified by the addition of 5 m NaCl. DNA concentrations were measured using a Nanodrop spectrophotometer (Infinite 200 PRO NanoQuant, Tecan Group Ltd, Switzerland) and diluted to 100 ng μL−1 and stored at –80 °C until further use. The plastid rpl32-trnL region was amplified using the primers rpl32-F (5′-CAGTTCCAAAAAAACGTACTT-3′) and trnL(UAG) (5′-CTGCTTCCTAAGAGCAGCGT-3′) (Shaw et al., 2007). In addition, two nuclear loci, the external transcribed spacer region (ETS) and internal transcribed spacer region (ITS) were amplified using primers described in Murphy et al. (2010). All PCR reactions were carried out in 50-μL reactions consisting of 5 µL (5 µm) of each primer, 5 µL of DNA template, 1 µL dNTP mix (20 mm), 2·0 mm MgCl2, 5 µL of 10× buffer and 0·5 U Taq DNA polymerase (Super-Therm JMR-801; Southern Cross Biotechnologies, Cape Town, South Africa). For the plastid region the following thermocycler programme was used: an initial denaturation of 95 °C for 2 min followed by 30 cycles of 95 °C for 30 s, 60 °C for 30 s and 72 °C for 60 s, followed by a final extension at 72 °C for 10 min. For both nuclear loci the following programme was used: 95 °C for 3 min followed by 30 cycles of 94 °C for 1 min, 60 °C for 1 min and 72 °C for 2 min and a final extension of 72 °C for 10 min. Sequencing for all the three regions was carried out using Big dye Terminator cycle sequencing (Applied Biosystems, Foster City, CA, USA). Given the relative short lengths of all regions included here, sequencing was done in one direction only for all regions. All DNA sequences have been deposited in GenBank (http://www.ncbi.nlm.nih.gov/genbank/).
Phylogenetic analysis
DNA sequences were aligned using MAFFT (Katoh et al., 2002) with manual adjustments in BIOEDIT version 7·0·5·3 (Hall, 1999) for all regions separately. The nuclear ETS and ITS datasets were later combined into a single nDNA dataset. A haplotype network was constructed for the plastid dataset using statistical parsimony as implemented in TCS version 1·21 (Clement et al., 2000). Phylogeny reconstruction was conducted separately on the nuclear dataset. Bayesian inference of phylogenetic relationships was done for the nDNA datasets using Mr Bayes 3·1·2 (Ronquist and Huelsenbeck, 2003). The best fit models were first estimated using jModelTest (Guindon and Gascuel, 2003; Posada, 2008). The nDNA dataset was run for two million generations and trees sampled every 1000 generations. After discarding 25 % of the burn-in trees a consensus tree was generated. Posterior probabilities were calculated using the 50 % majority rule consensus method.
Genetic distances among A. pycnantha populations were estimated using the library seqinR in the R statistical environment (Charif and Lobry, 2007). To obtain an indication of the expected intraspecific variation for the rpl32-trnL(UAG) region, the genetic distances between subspecific entities of A. pycnantha and other species (A. nilotica, Carex elata, Linaria viscosa, Linaria multicaulis, Limnanthes floccosa and Centaurea aeolica) were calculated using data downloaded from GenBank.
To test for neutrality in the plastid and nuclear datasets we used Tajima's D test, Fu and Li's D* test and Fu and Li's F* test in dnaSP version 5·0 (Librado and Rozas, 2009). Mismatch distributions and Harpending's raggedness index for both datasets were also determined using dnaSP version 5·0 (Librado and Rozas, 2009).
An estimate of the divergence times of different native forms of A. pycnantha was based on previously published nucleotide substitution rates for acacias for the plastid genome of 0·1 % per million years (Byrne et al., 2002). The value obtained from the nucleotide diversity estimate is very broad (Byrne et al., 2002). Consequently, to validate this estimate, divergence times were also inferred using a relaxed molecular clock and a substitution rate of 0·1 per million years using a Bayesian Markov chain Monte Carlo (MCMC) procedure as implemented in BEAST 1·4·7 (Drummond and Rambaut, 2007). We used a relaxed molecular clock as our data did not meet the assumptions of a strict molecular clock, as inferred from a likelihood ratio test (Verbruggen et al., 2009). The MCMC model was run under a general time-reversible model of nucleotide substitution with rate variation among sites modelled using a gamma-distribution implemented in jModelTest (Guindon and Gascuel, 2003; Posada, 2008). The Yule speciation process was selected as the tree prior. Three independent MCMC analyses were each run for ten million steps and parameter values were sampled every ten thousand steps.
RESULTS
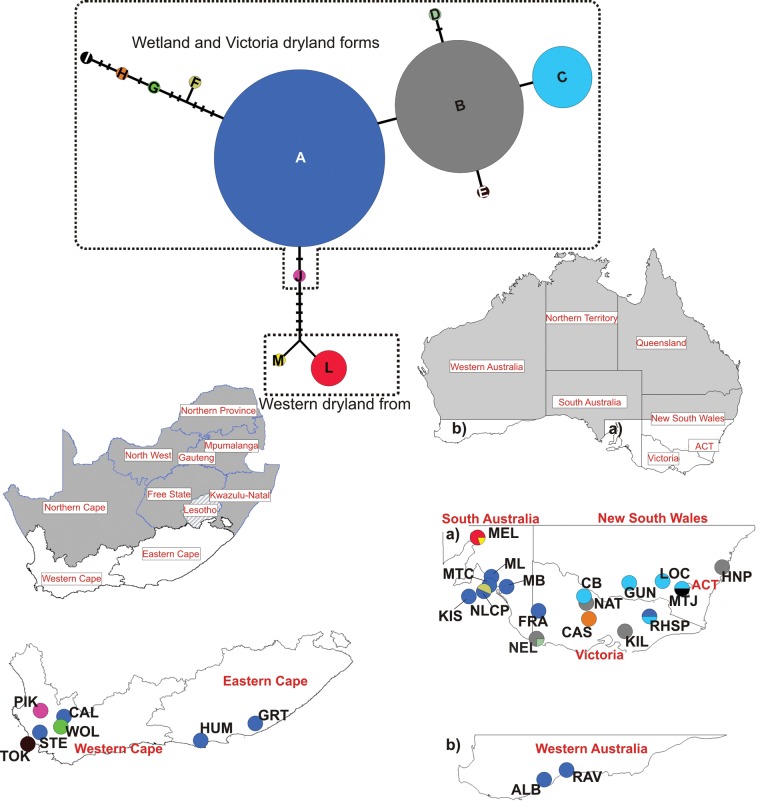
The aligned plastid DNA matrix was 608 bp long, requiring 12 gaps (indels) with an average length of 2 bp. Twelve haplotypes were identified in A. pycnantha (Fig. 2). Five (A, D, F, H and I) unique haplotypes occurred in the distributional ranges of the wetland form in Australia, two haplotypes (B and C) encompassed both Victorian dryland and wetland taxa, and two haplotypes (M and L) occurred in the drier Southern Flinders Ranges, where the dryland form occurs. Three haplotypes (E, G and J) were found in South Africa only (Fig. 2). The haplotype most commonly recorded in the natural range (A) also occurred in four invasive populations in South Africa (Fig. 2). Two of the invasive Australian populations (Western Australia) and the population from Portugal also had haplotype A. The three remaining invasive populations from South Africa were closely related to haplotypes A, B and G and haplotype H (Fig. 2). The Flinders Range haplotypes (L and M), corresponding to the dryland form of A. pycnantha, were not found in any of the invasive ranges.
Fig. 2.
Haplotype network of Acacia pycnantha (based on the rpl32-trnL(UAG) region) and geographical distribution of haplotypes in the native south-eastern Australia and invasive South Africa and western Australian ranges. Each unique haplotype is represented by a coloured circle and the size of the circle is proportional to the number of individuals possessing that haplotype. Pie charts represent the proportion and distribution of haplotypes across native (South Australia and New South Wales) and invasive (Western Australia and South Africa) ranges.
Strong spatial clustering occurred throughout the range of A. pycnantha in Australia, with most haplotypes having restricted distributions. Moreover, these groupings correspond to the morphologically recognized lineages in A. pycnantha: Flinders Range dryland (haplotypes M and L; slender trees with narrow phyllodes at the species western natural range); wetland and Victoria dryland (haplotypes A, B, C, D, F, H and I) (Fig. 2). There was geographical overlap of haplotypes in Reef Hills State Park, Victoria (haplotype A and C). A distinct haplotype (haplotype I) was found in Mount Jeramborerra close to the Australian Capital Territory (Fig. 2).
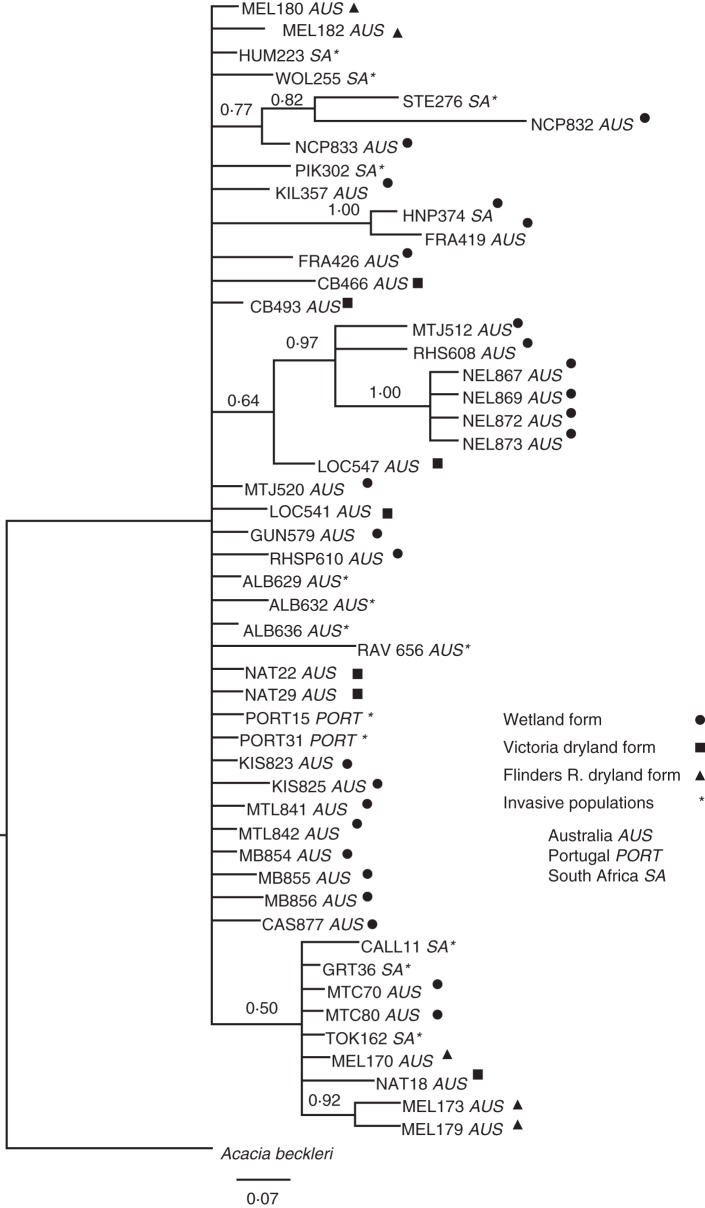
The best-fit model of evolution used in Bayesian inference, according to the Akaike information criterion (AIC) for the combined nuclear regions, was also the GTR model with a gamma-distribution. The nuclear gene tree retrieved from the Bayesian analysis was incongruent with the plastid DNA haplotype network in the placement of certain samples. Dryland samples from the Flinders Range clustered with the Natimuk samples from Victoria. In addition, South Australian populations from Mt Compass were shown to be closely related to the Flinders Range population. As in the plastid haplotype network, four South African populations clustered with South Australian populations. Two of the South African populations (Humansdorp and Wolseley) were closely related to the dryland form of A. pycnantha (Fig. 3). The Nelson population formed a distinct cluster which was closely related to nearby Victorian populations. Overall, there was no clear geographical structuring of A. pycnantha in the nuclear analysis.
Fig. 3.
Bayesian tree for Acacia pycnantha using combined ITS and ETS regions. Numbers above the branches represent posterior probabilities derived from the analysis. Invasive taxa are indicated by asterisks for Australian, Portuguese and South African collections, as indicated in the key; also indicated are wetland, Victoria dryland and Flinders Range dryland forms of A. pycnantha.
Genetic distances ranged from 0 to 0·018 % between plastid DNA haplotypes (Table 1). Haplotypes L and M, which were collected from Melrose (MEL, Flinders Range), had larger distances of up to 0·018 % from the South Australia, Victoria and New South Wales haplotypes (mostly wetland haplotypes; Table S1).
Table 1.
Genetic distances between rpl32-trnL(UAG) plastid haplotypes of Acacia pycnantha; A–H correspond to haplotypes as indicated in Fig. 1
| A | B | C | D | E | F | G | H | I | J | L | M | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | 0 | |||||||||||
| C | 0 | 0 | ||||||||||
| D | 0 | 0 | 0 | |||||||||
| E | 0 | 0 | 0·002 | 0 | ||||||||
| F | 0 | 0·004 | 0·007 | 0·004 | 0·004 | |||||||
| G | 0·002 | 0·002 | 0·002 | 0·002 | 0·002 | 0·006 | ||||||
| H | 0·004 | 0·004 | 0·002 | 0·004 | 0·004 | 0·004 | 0·002 | |||||
| I | 0·008 | 0·009 | 0·009 | 0·009 | 0·009 | 0·012 | 0·005 | 0·009 | ||||
| J | 0·002 | 0·002 | 0·002 | 0·002 | 0·002 | 0·007 | 0 | 0 | 0·005 | |||
| L | 0·01 | 0·009 | 0·009 | 0·009 | 0·009 | 0·012 | 0·011 | 0·012 | 0·018 | 0·002 | ||
| M | 0·01 | 0·011 | 0·012 | 0·011 | 0·011 | 0·015 | 0·011 | 0·015 | 0·016 | 0·01 | 0·002 |
Tests of neutrality for the plastid region using Tajima's D (–2·10088, P < 0·05), Fu and Li's D* (–3·54960, P < 0·02) and Fu and Li's F* (–3·62302, P < 0·02) all yielded negative values that were significantly different from zero. The population expansion hypothesis was investigated by computing the distribution of pairwise differences using dnaSP version 5. The mismatch distribution showed slight bimodality (Fig. 4A) with a Harpending's raggedness value resembling constant size population (r = 0·3667, see Zink et al., 2000). Using the rate of nucleotide divergence previously published for the same region (rpl32-trnL) revealed that wetland and Flinders Range dryland forms of A. pycnantha diverged approx. 110 kya. The upper limit of the Bayesian estimate of the age of the most recent common ancestor (TMRCA) was in agreement with the nucleotide diversity divergence times, indicating that these lineages split approx. 66 kya (95 % confidence interval 58–87 kya).
Fig. 4.
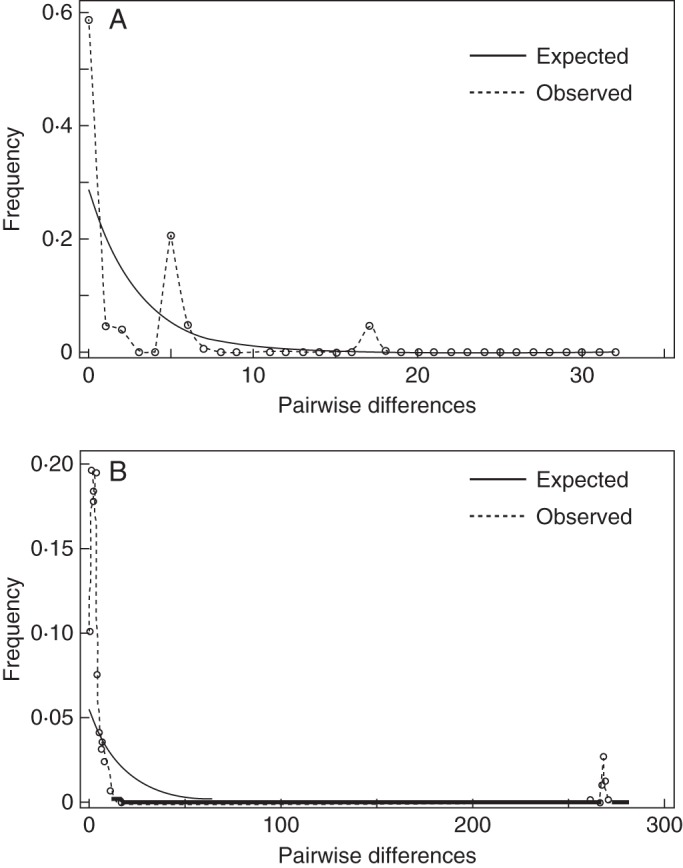
Mismatch distributions for DNA sequence data of the plastid (A) and nuclear (B) genomes for Acacia pycnantha in Australia (native range). The solid line represents the expected mismatch distribution of a constant-size population and the dotted line represents the observed mismatch distribution based on A. pycnantha DNA sequence data.
Similar to plastid DNA, neutrality tests on the nDNA of Tajima's D (–2·84, P < 0·001), Fu and Li's D* (–5·88, P < 0·02) and Fu and Li's F* (–5·71, P < 0·02) yielded negatively significant values. However, pairwise mismatch distribution of the nuclear data yielded was unimodal with a raggedness index of 0·0265, indicative of recent population expansion (Fig. 4B).
DISCUSSION
Linking contemporary and historical biogeography remains a central theme in invasion biology (Stepien et al., 2002). We were able to identify that invasive South African populations of A. pycnantha originated from the wetland form and Victorian dryland form of the species, whereas Portuguese and Western Australian invasive populations represent the wetland form only. Most invasive populations originated from the areas around Adelaide and the Mt Lofty ranges. This has important implications for the management of A. pycnantha in South Africa and Portugal as discussed below. We found evidence to support previous informal morphological classifications of A. pycnantha as two distinct ecotypes (wetland and dryland forms) based on plastid genealogies, providing a platform for guiding conservation efforts in the native range. However, the link between phylogenetic relatedness and morphological classification is not clear cut. The dryland forms of A. pycnantha found in Victoria, which had similar phyllode morphologies to the Flinders Range dryland variant, shared a close phylogenetic relationship with the wetland variants. However, the dryland variants found in the Flinders Range showed distinct genetic variation from the wetland/dryland Victoria group (Fig. 2).
Native range phylogeography
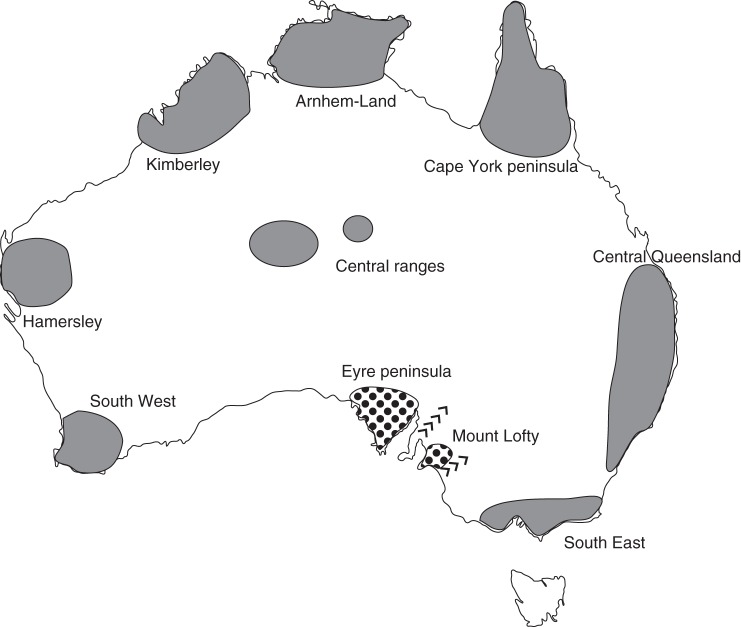
We suspect that the phylogeographic structure of A. pycnantha in eastern Australia is a result of relatively recent geological climatic shifts during the Pleistocene (Worth et al., 2009, 2010). During the Pleistocene (approx. 16 kya – 1·8 Mya) eastern Australia experienced a series of cycles of pluviality and aridity (Toon et al., 2007). Numerous and isolated Pleistocene refugia during the extreme dry periods have been previously identified (Fig. 5; Ford, 1987), separated by arid barriers, and climatic and edaphic factors (Ladiges et al., 2011). This fragmented landscape resulted in allopatric speciation in many Australian plant groups (Byrne et al., 2002, 2003; Millar et al., 2008). Using an estimate of 0·1 % DNA sequence divergence per million years for plastid DNA as reported for other acacias (Byrne et al., 2003), the divergence between the Flinders dryland and wetland/Victoria dryland form of A. pycnantha is estimated to have taken place around 100 kya. This coincides with the mid Pleistocene, a period with high sedimentation rates and aridity in eastern Australia (Prideaux et al., 2007). This is supported by the A. pycnantha plastid structure, which indicated the presence of two main haplotype groups that is indicative of the two distinct ecological zones where the different A. pycnantha variants occur. Kangaroo Island was connected to mainland Australia during the late Pleistocene and the cut-off time of the island from mainland Australia is estimated at about 8800–13 500 years BP (Hope et al., 1977). This timeframe is in agreement with the A. pycnantha collections from Kangaroo Island, which are genetically closely related to the wetland form (KIS 823 and KIS 825, Fig. 2).
Fig. 5.
Refuge areas and geographical barriers in Australia during the Pleistocene. Shaded areas represent refugia in Australia and those with black dots represent refugia for Acacia pycnantha (Eyre Peninsula and Mt Lofty). The arrows show the Eyrean and Mallee geographical barriers (Ford et al., 1987).
For the dryland and wetland lineages, three putative refugia existed in the past: the Eyre Peninsula (Flinders Range dryland form), South East and Mt Lofty refugia (for the wetland and dryland Victorian lineage) (Fig. 5; Ford, 1987). These refugia were separated by the Eyrean and Mallee geographical barriers (Ford, 1987; Toon et al., 2007) and represent areas that had low rainfall (Ford, 1987). Since this time, A. pycnantha has expanded its range eastwards and northwards. These expansions are supported by the presence of putative intraspecific hybrids identified by incongruent nDNA and plastid DNA phylogenetic placements (e.g. NAT18 Victoria, Natimuk) and by the patterns of nDNA mismatch distribution, which was unimodal, indicative of recent population expansion (Hwang et al., 2003).
Morphologically, the wetland form of A. pycnantha has broader phyllodes and darker golden inflorescences than the dryland form (with narrower phyllodes and often pale yellow flowers, Fig. 1). However, slight morphological variants of A. pycnantha also exist in the wetland distributional range in South Australia, Victoria and New South Wales (Maslin and McDonald, 2004). The putative hybrid from Natimuk, Victoria, showed intermediate morphological characters between the wetland haplotypes and the Flinders Range haplotype (J. Ndlovu and J. J. Le Roux, pers. obs.). The presence of these intermediates in Victoria is probably the result of hybridization through pollen exchange between the wetland and the dryland forms.
To determine whether the genetic divergences between the extreme forms of A. pycnantha potentially represent different subspecies, we calculated genetic differences for known subspecies complexes and closely related species. At the interspecific level genetic divergence ranged from 0·04 % (Acacia pulchella and A. koa) to 0·15 % (A. pulchella and A. longifolia) and at the intraspecific level between 0 % (recognized subspecies of A. nilotica) and 0·005 % (recognized subspecies of Linaria multicaulis); the level of genetic divergence at the intraspecific level for A. pycnantha ranged between 0 and 0·018 %.
Overall, informal morphological classifications and our genetic results suggest that the A. pycnantha species complex requires taxonomic revision. The genetic divergence found between different forms, at least for plastid DNA, exceeds known divergences between some other taxa consisting of subspecies (Table 1). However, the combination of our genetic and morphological data is not sufficient to separate the species into a subspecies complex. These attempts are hampered by many (other) characters showing geographical variation between the distributional ranges of the dry and wetland forms. For example, the Victorian dryland form, despite its geographical disparity, resembles the variant from the Flinders Range. These problems are also exacerbated by the history of cultivation of A. pycnantha in its natural range, possibly obscuring the natural distribution of subspecific taxa in the species. We recommend a finer-scale population genetic study with a more extensive sampling scheme to resolve some of these issues.
Invasive range phylogeography and consequences for management
High-frequency genotypes in the invasive range in South Africa, Western Australia and Portugal occur predominantly in South Australia, with some evidence of wetland variants from Victoria and New South Wales having also been introduced. Notably, however, A. pycnantha in South Africa harbours only a fraction of the genetic diversity found in its Australian range (Fig. 2). Clearly there has been a genetic bottleneck during introduction and/or invasion (Fig. 2).
Assuming that historical genetic structure possibly determines co-evolutionary relationships between host plants and their antagonists and mutualists, our findings may explain why Trichilogaster signiventris, a gall-forming wasp initially released from Lake Natimuk in Victoria for biological control, did not establish successfully and did not achieve significant control of A. pycnantha in South Africa (Hoffmann et al., 2002), as this region does not appear to be the source of invasive populations. However, the reintroduction of the same wasp species collected from Mt Compass in South Australia, a region identified here as a potential source of the invasive populations in South Africa, has led to successful establishment and substantial impacts and control of invasive populations in South Africa (Hoffmann et al., 2002). We therefore have reason to conclude that host-specificity and compatibility may be linked to natal phylogeographical structure in this system.
These results have important implications. First, we would expect populations in the native range to be able to survive drier conditions. If the dryland form was introduced, there could potentially be a large expansion in the invasive range in South Africa. Second, the recommendations that biocontrol agents that have proved successful in South Africa should be considered for introduction to other countries where the species is invasive (e.g. Wilson et al., 2011) might need to take subspecific identity and phylogenetic affinity into account (e.g. St. Quinton et al., 2011). Initial results, however, are promising. A. pycnantha in Portugal appears to have a similar genetic origin to A. pycnantha in South Africa, suggesting the substantial reductions in seed production caused by T. signiventris in South Africa (Hoffmann et al., 2002; Impson et al., 2011) might be replicated in Portugal.
Concluding remarks
Identifying where invasive Acacia spp. originate from in their natal range is important for determining priorities for biological control. A comprehensive genetic diversity and phylogeographical study in the native range for all the known invasive acacias could potentially form a basis for recommendations of host-specific biological control organisms. In the case of A. pycnantha, the invasive genotype found in South Africa is similar to the invasive genotypes in Portugal and Western Australia and thus introduction of the same variant of T. signiventris for biological control released in South Africa is recommended. Such studies for all the Acacia spp. should be based on plastid genetic diversity as the plastid region has been shown to be more reliable for identifying source populations in this study and in other species in Australia. We therefore recommend that biological control efforts for Australian acacias must recognize the importance of genetic diversity by verifying the source of the invader before releasing biocontrol organisms.
ACKNOWLEDGEMENTS
This work was funded by the DST-NRF Centre of Excellence for Invasion Biology (CIB) and the Working for Water Programme through their collaborative research project on ‘Research for Integrated Management of Invasive Alien Species’ and grants from South Africa's National Research Foundation and the Hans Sigrist Foundation. We thank Kate Le Roux for assistance during field collections.
Appendix
APPENDIX Localities in South Africa, Australia and Portugal where Acacia pycnantha was collected.
| Sample ID | Latitude | Longitude | Country | Locality and abbreviation | Status | rpl32-trnL | ETS | ITS |
|---|---|---|---|---|---|---|---|---|
| CAL11 | −33·10701 | 19·29755 | RSA | Caledon (CAL), WC | invasive | JF276987 | KC261682 | KC261745 |
| GRT36 | −33·46032 | 26·15991 | RSA | Grahamstown (GRT), EC | invasive | JF276999 | KC261683 | KC261746 |
| MTC70 | −35·40585 | 145·95586 | AUS | Mt Compass (MTC), SA | native | JF276989 | KC261684 | KC261747 |
| MTC80 | −35·40585 | 145·95586 | AUS | Mt Compass (MTC), SA | native | JF276990 | KC261685 | KC261748 |
| TOK162 | −33·84179 | 18·66602 | RSA | Tokai (TOK), WC | invasive | JF276991 | KC261686 | KC261749 |
| MEL170 | −32·78187 | 138·1973 | AUS | Melrose (MEL), SEA | native | KC261785 | KC261687 | KC261750 |
| MEL173 | −32·78187 | 138·1973 | AUS | Melrose (MEL), SEA | native | KC261786 | KC261688 | KC261751 |
| MEL179 | −32·78187 | 138·1973 | AUS | Melrose (MEL), SEA | native | KC261787 | KC261689 | KC261752 |
| MEL180 | −32·78187 | 138·1973 | AUS | Melrose (MEL), SEA | native | JF276992 | KC261690 | KC261753 |
| MEL182 | −32·78187 | 138·1973 | AUS | Melrose (MEL), SEA | native | KC261788 | KC261691 | KC261754 |
| HUM223 | −34·03989 | 24·78687 | RSA | Humansdorp (HUM), EC | invasive | JF276993 | KC261692 | KC261755 |
| WOL255 | −33·34012 | 19·16109 | RSA | Wolseley (HUM), WC | invasive | JF276994 | KC261693 | KC261756 |
| STE276 | −34·06024 | 18·41480 | RSA | Stellenrust (HUM), WC | invasive | JF276995 | KC261694 | KC261757 |
| PIK302 | −32·80084 | 18·71501FC | RSA | Piketberg (PIK), WC | invasive | JF276996 | KC261695 | KC261758 |
| KIL357 | −37·22176 | 145·021 | AUS | Kilmore (PIK), VIC | native | JF276997 | KC261696 | KC261759 |
| HNP374 | −34·08589 | 150·989 | AUS | Heathcote NP (HNP), NSW | native | JF276988 | KC261697 | KC261760 |
| FRA419 | −36·77054 | 141·18135 | AUS | Frances (FRA), VIC | native | JF277005 | KC261698 | KC261762 |
| FRA426 | −36·77054 | 141·18135 | AUS | Frances (FRA), VIC | native | JF277008 | KC261699 | KC261763 |
| CB466 | −35·99273 | 143·76538 | AUS | between Charlton & Boorte (CB) | native | JF277011 | KC261700 | KC261764 |
| CB493 | −35·99273 | 143·76538 | AUS | between Charlton & Boorte (CB) | native | JF277022 | KC261701 | KC261765 |
| MTJ512 | −35·36866 | 149·20332 | AUS | Mt Jeramborera (MTJ), ACT | native | JF276998 | KC261702 | KC261767 |
| MTJ520 | −35·36866 | 149·20332 | AUS | Mt Jeramborera (MTJ), ACT | native | JF277009 | KC261703 | KC261768 |
| LOC541 | −35·36866 | 146·64549 | AUS | Lockheart (LOC), NSW | native | JF277012 | KC261704 | KC261769 |
| LOC547 | −35·36866 | 146·64549 | AUS | Lockheart (LOC), NSW | native | JF277021 | KC261705 | KC261771 |
| GUN579 | −35·21065 | 147·76425 | AUS | Gundagai (GUN), NSW | native | JF277023 | KC261706 | KC261772 |
| RHSP608 | −36·59888 | 145·95586 | AUS | Reef Hills SP (RHSP), VIC | native | JF277004 | KC261707 | KC261773 |
| RHSP610 | −36·59888 | 145·95586 | AUS | Reef Hills SP (RHSP), VIC | native | JF277007 | KC261708 | KC261774 |
| ALB629 | −34·31586 | 118·79919 | AUS | Albany (ALB), WA | invasive | JF277016 | KC261709 | KC261775 |
| ALB632 | −34·31586 | 118·79919 | AUS | Albany (ALB), WA | invasive | JF277013 | KC261710 | KC261776 |
| ALB636 | −34·31586 | 118·79919 | AUS | Albany (ALB), WA | invasive | JF277019 | KC261711 | KC261777 |
| RAV656 | −33·59650 | 120·17688 | AUS | Ravernsthorpe (RAV), WA | invasive | JF277016 | KC261712 | KC261778 |
| NAT18 | −36·00409 | 143·76041 | AUS | Natimuk (NAT), VIC | native | KC261791 | KC261814 | KC261779 |
| NAT22 | −36·00409 | 143·76041 | AUS | Natimuk (NAT), VIC | native | KC261792 | KC261815 | KC261780 |
| NAT29 | −36·00409 | 143·76041 | AUS | Natimuk (NAT), VIC | native | KC261793 | KC261816 | KC261781 |
| PORT15 | NA | NA | PORT | invasive | KC261794 | KC261817 | KC261783 | |
| PORT31 | NA | NA | PORT | invasive | KC261795 | KC261818 | KC261784 | |
| KIS823 | −35·75669 | 137·89486 | AUS | Kangaroo Island (KIS), SA | native | KC261796 | KC261713 | KC261727 |
| KIS825 | −35·75669 | 137·89486 | AUS | Kangaroo Island (KIS), SA | native | KC261797 | KC261714 | KC261728 |
| NCP832 | −35·61298 | 138·47950 | AUS | Newlands C. Park (NLCP), SA | native | KC261798 | KC261715 | KC261729 |
| NCP833 | −35·61298 | 138·47950 | AUS | Newlands C. Park (NLCP), SA | native | KC261799 | KC261716 | KC261730 |
| MTL841 | −34·97175 | 138·6653 | AUS | Mt Lofty (ML), SA | native | KC261800 | KC261717 | KC261731 |
| MTL842 | −34·97175 | 138·6653 | AUS | Mt Lofty (ML), SA | native | KC261801 | KC261718 | KC261732 |
| MB854 | −35·31895 | 139·51193 | AUS | Murray Bridge (ML), SA | native | KC261803 | KC261719 | KC261734 |
| MB855 | −35·31895 | 139·51193 | AUS | Murray Bridge (ML), SA | native | KC261804 | KC261720 | KC261735 |
| MB856 | −35·31895 | 139·51193 | AUS | Murray Bridge (ML), SA | native | KC261805 | KC261721 | KC261736 |
| NEL867 | −38·05003 | 141·01510 | AUS | Nelson (NEL), VIC | native | KC261807 | KC261722 | KC261738 |
| NEL869 | −38·05003 | 141·01510 | AUS | Nelson (NEL), VIC | native | KC261808 | KC261723 | KC261739 |
| NEL872 | −38·05003 | 141·01510 | AUS | Nelson (NEL), VIC | native | KC261810 | KC261724 | KC261741 |
| NEL873 | −38·05003 | 141·01510 | AUS | Nelson (NEL), VIC | native | KC261811 | KC261725 | KC261742 |
| CAS877 | −37·10758 | 144·09283 | AUS | Castlemaine (CAS), VIC | native | KC261813 | KC261726 | KC261744 |
AUS, Australia; RSA, Republic of South Africa; Port, Portugal. WC, Western Cape; EC, Eastern Cape; VIC, Victoria; WA, Western Australia; NSW, New South Wales; SA, South Australia; SEA, south-east Australia; ACT, Australian Capital Territory; NA, not available.
LITERATURE CITED
- Bossdorf O, Auge H, Lafuma L, Rogers WE, Siemann E, Pratti D. Phenotypic and genetic differentiation between native and introduced plant populations. Oecologia. 2005;144:1–11. doi: 10.1007/s00442-005-0070-z. [DOI] [PubMed] [Google Scholar]
- Byrne M, Macdonald B, Coates D. Phylogeographical patterns in chloroplast DNA variation within Acacia acuminata (Leguminosae: Mimosoideae) complex in Western Australia. Journal of Evolutionary Biology. 2002;15:576–587. [Google Scholar]
- Byrne M, Macdonald B, Brand J. Phylogeography and divergence in the chloroplast genome of Western Australian sandalwood (Santalum spicatum) Heredity. 2003;91:389–395. doi: 10.1038/sj.hdy.6800346. [DOI] [PubMed] [Google Scholar]
- Charif D, Lobry JR. New York: Springer; 2007. SeqinR 1·0-2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis. Structural approaches to sequence evolution: molecules, networks, populations; pp. 207–232. [Google Scholar]
- Clement M, Posada D, Crandall K. TCS: a computer program to estimate gene genealogies. Molecular Ecology. 2000;9:1657–1660. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochemical Bulletin, Botanical Society of America. 1987;19:11–15. [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. http://dx.doi.org/10.1186/1471-2148-7-214 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford J. Hybrid zones in Australian birds. EMU. 1987;87:158–178. [Google Scholar]
- Gaskin JF, Schaal BA. Hybrid Tamarix widespread in U.S. invasion and undetected in native Asian range. Proceedings of the National Academy of Sciences USA. 2002;99:11256–11259. doi: 10.1073/pnas.132403299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goolsby JA, De Barro PJ, Markinson JR, Pemberton RW, Hurtley DM, Frohlich DR. Matching the origin of an invasive weed for selection of a herbivore haplotype for a biological control programme. Molecular Ecology. 2006;15:287–297. doi: 10.1111/j.1365-294X.2005.02788.x. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Harris P. Evolution of classical weed biocontrol: meeting survival challenges. Bulletin of the Entomological Society of Canada. 1998;30:134–143. [Google Scholar]
- Henderson L. Alien weeds and invasive plants. A complete guide to weeds and invaders in South Africa. PPRI handbooks No. 12, 260. Pretoria: Plant Protection Research Institute; 2001. [Google Scholar]
- Hoffmann JH, Impson FAC, Moran VC, Donnelly D. Biological control of golden wattle trees (Acacia pycnantha) by a gall wasp Trichilogaster species (Hymenoptera: Pteromalidae) in South Africa. Biological Control. 2002;25:64–73. [Google Scholar]
- Hope JH, Lampert RJ, Edmondson E, Smith MJ, van Tets GF. Late Pleistocene faunal remains from Seton Rock Shelter, Kangaroo Island, South Australia. Journal of Biogeography. 1977;4:363–385. [Google Scholar]
- Hui C, Richardson DM, Robertson MP, Wilson JRU, Yates CJ. Macroecology meets invasion ecology: linking native distribution of Australian acacias to invasiveness. Diversity and Distributions. 2011;17:872–883. [Google Scholar]
- Hwang SY, Lin TP, Ma CS, Lin CL, Chung JD, Yang JC. Postglacial population growth of Cunninghammia konishii (Cupressaceae) inferred from phylogeographical analysis of chloroplast DNA variation. Molecular Ecology. 2003;12:2689–2695. doi: 10.1046/j.1365-294x.2003.01935.x. [DOI] [PubMed] [Google Scholar]
- Impson FAC, Kleinjan CA, Hoffmann JH, Post JA, Wood AR. Biological control of Australian Acacia species and Paraserianthes lophantha (Willd.) Nielsen (Mimosaceae) in South Africa. African Entomology. 2011;19:186–207. [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliber A, Eckert CG. Interaction between founder effect and selection during biological invasion in an aquatic plant. Evolution. 2005;59:1900–1913. [PubMed] [Google Scholar]
- Ladiges P, ParraOC Gibbs A, Udovicic F, Nelson G, Bayly M. Historical biogeographical patterns in continental Australia: congruence among areas of endemism of two major clades of eucalypts. Cladistics. 2011;27:29–41. doi: 10.1111/j.1096-0031.2010.00315.x. [DOI] [PubMed] [Google Scholar]
- Le Roux JJ, Brown GK, Byrne M, et al. Phylogeographic consequences of different introduction histories of invasive Australian Acacia species and Paraserianthes lophantha (Fabaceae) Diversity and Distributions. 2011;17:861–871. [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphisms data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Marchante H, Freitas H, Hoffmann JH. Assessing the suitability and safety of a well-known bud-galling wasp, Trichilogaster acaciaelongifoliae for biological control of Acacia longifolia in Portugal. Biological Control. 2011;56:193–201. [Google Scholar]
- Maslin BR. Canberra: Australian Biological Resources Study & Melbourne; CSIRO Publishing; 2001. Acacia pycnantha. Flora of Australia 11A: 297–299. [Google Scholar]
- Maslin BR, McDonald MW. Report to the Rural Industries Research and Development Corporation, Publication No. 03/017. Joint Venture Agroforestry Program. Acacia search: evaluation of Acacia as a woody crop option for Southern Australia. 2004 [Google Scholar]
- McLeish MJ, Chapman TW, Schwarz MP. Host-driven diversification of gall-inducing Acacia thrips and the aridification of Australia. BMC Biology. 2007;5:1–13. doi: 10.1186/1741-7007-5-3. http://dx.doi.org/10.1186/1741-7007-5-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar MA, Byrne M, Nuberg IK, Sedgley M. A rapid PCR based diagnostic test for the identification of subspecies of Acacia saligna. Tree Genetics and Genomes. 2008;4:625–635. [Google Scholar]
- Millar MA, Byrne M, Nuberg IK, Sedgley M. High levels of genetic contamination in remnant populations of Acacia saligna from a genetically divergent planted stand. Restoration Ecology. 2011;20:260–267. [Google Scholar]
- Murphy DJ, Brown GK, Miller JT, Ladiges PY. Molecular phylogeny of acacias (Mimosoideae: Leguminosae) – evidence for major clades and informal classifications. Taxon. 2010;59:7–19. [Google Scholar]
- Nel JL, Richardson DM, Rouget M, et al. A proposed classification of invasive alien plant species in South Africa: towards prioritising species and areas for management action. South African Journal of Science. 2004;100:53–64. [Google Scholar]
- Posada D. ModelTest: phylogenetic model averaging. Molecular Biology and Evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Poynton RJ. Tree planting in southern Africa, Vol. 3. Other genera. Pretoria: Department of Agriculture, Forestry and Fisheries. 2009 [Google Scholar]
- Prentis PJ, Wilson JRU, Dormontt EE, Richardson DM, Lowe AJ. Adaptive evolution in invasive species. Trends in Plant Science. 2008;13:288–294. doi: 10.1016/j.tplants.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Prideaux GJ, Long JA, Ayliffe LK, et al. An arid-adapted middle Pleistocene vertebrate fauna from South Central Australia. Nature. 2007;445:422–425. doi: 10.1038/nature05471. [DOI] [PubMed] [Google Scholar]
- Richardson DM, Rejmánek M. Trees and shrubs as invasive alien species – a global review. Diversity and Distributions. 2011;17:788–809. [Google Scholar]
- Richardson DM, Caruthers J, Hui C, et al. Human-mediated introductions of Australian acacias – a global experiment in biogeography. Diversity and Distributions. 2011;17:771–787. [Google Scholar]
- Ronquist F, Huelsenbeck JP. Mr Bayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rosenmeier L, Kjær ED, Nielsen LR. The Scotch broom, Cytisus scoparius (Fabaceae), a paradox in Denmark – an invasive plant or endangered native species? Botanical Journal of the Linnean Society. 2013;171:429–440. [Google Scholar]
- Shaw J, Lickey EB, Schilling EE, Small RL. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. American Journal of Botany. 2007;94:275–288. doi: 10.3732/ajb.94.3.275. [DOI] [PubMed] [Google Scholar]
- St. Quinton JM, Fay MF, Ingrouille M, Faull J. Characterisation of Rubus niveus: a prerequisite to its biological control in oceanic islands. Biocontrol Science and Technology. 2011;21:733–752. [Google Scholar]
- Stepien CA, Taylor CD, Dabrowska KA. Genetic variability and phylogeographic patterns of a non-indigenous species invasion: a comparison of exotic vs. native zebra and quagga mussel populations. Journal of Evolutionary Biology. 2002;15:314–328. [Google Scholar]
- Thompson GD, Bellstedt DU, Byrne M, et al. Cultivation shapes genetic novelty in a globally important invader. Molecular Ecology. 2012;21:3187–3199. doi: 10.1111/j.1365-294X.2012.05601.x. [DOI] [PubMed] [Google Scholar]
- Toon A, Mather PB, Baker AM, Durrant KL. Pleistocene refugia in an arid landscape: analysis of a widely distributed Australian passerine. Molecular Ecology. 2007;16:2525–2541. doi: 10.1111/j.1365-294X.2007.03289.x. [DOI] [PubMed] [Google Scholar]
- Verbruggen H, Ashworth M, LoDuca ST, et al. A multi-locus time-calibrated phylogeny of siphonous green algae. Molecular Phylogenetics and Evolution. 2009;50:642–653. doi: 10.1016/j.ympev.2008.12.018. [DOI] [PubMed] [Google Scholar]
- Wardill TJ, Graham GC, Zalucki M, Palmer WA, Playford J, Scott KD. The importance of species identity in the biocontrol process: identifying the subspecies of Acacia nilotica (Leguminosae: Mimosoideae) by genetic distance and the implications for biological control. Journal of Biogeography. 2005;32:2145–2159. [Google Scholar]
- Wilson JRU, Gaifiro C, Gibson MR, et al. Risk assessment, eradication, and biological control: global efforts to limit Australian Acacia invasions. Diversity and Distributions. 2011;17:1030–1046. [Google Scholar]
- Worth J, Jordan G, Mackinnon G, Vaillancourt R. The major Australian cool temperate rainforest tree Nothofagus cunninghamii withstood Pleistocene glacial aridity within multiple regions: evidence from the chloroplast. New Phytologist. 2009;182:519–532. doi: 10.1111/j.1469-8137.2008.02761.x. [DOI] [PubMed] [Google Scholar]
- Worth JRP, Jordan GJ, Marthick JR, McKinnon GE, Vaillancourt RE. Chloroplast evidence forgeographic stasis of the Australian bird-dispersed shrub Tasmannia lanceolata (Winteraceae) Molecular Ecology. 2010;19:2949–2963. doi: 10.1111/j.1365-294X.2010.04725.x. [DOI] [PubMed] [Google Scholar]
- Zink RM, Barrowclough GF, Atwood JL, Blackwell-Rago RC. Genetics, taxonomy and conservation of the threatened California gnatcatcher. Conservation Biology. 2000;14:1394–1405. [Google Scholar]