Abstract
Batten disease is a neurodegenerative disorder resulting from mutations in CLN3, a polytopic membrane protein, whose predominant intracellular destination in nonneuronal cells is the lysosome. The topology of CLN3 protein, its lysosomal targeting mechanism, and the development of Batten disease are poorly understood. We provide experimental evidence that both the N and C termini and one large loop domain of CLN3 face the cytoplasm. We have identified two lysosomal targeting motifs that mediate the sorting of CLN3 in transfected nonneuronal and neuronal cells: an unconventional motif in the long C-terminal cytosolic tail consisting of a methionine and a glycine separated by nine amino acids [M(X)9G], and a more conventional dileucine motif, located in the large cytosolic loop domain and preceded by an acidic patch. Each motif on its own was sufficient to mediate lysosomal targeting, but optimal efficiency required both. Interestingly, in primary neurons, CLN3 was prominently seen both in lysosomes in the cell body and in endosomes, containing early endosomal antigen-1 along neuronal processes. Because there are few lysosomes in axons and peripheral parts of dendrites, the presence of CLN3 in endosomes of neurons may be functionally important. Endosomal association of the protein was independent of the two lysosomal targeting motifs.
INTRODUCTION
Batten disease or juvenile-onset neuronal ceroid lipofuscinosis is the most common disorder in a group of severe neurodegenerative diseases, the NCLs, affecting people worldwide, and being as frequent as 1 in 12,500 births in some parts of northern Europe and the United States. The main manifestations of the NCLs are almost entirely restricted to the central nervous system with patients suffering from visual failures, epilepsy, phychomotor deterioration, and premature death (Santavuori, 1988). Various forms of NCLs have been shown to result from mutations in different genes, CLN1-CLN8, encoding proteins that are mostly localized in lysosomes but also in other organelles, including the endoplasmic reticulum (ER) (reviewed by Weimer et al., 2002). Recently, an interaction of three NCL proteins (CLN5 with CLN2 and CLN3, respectively) was described, linking for the first time different NCL proteins at the molecular level (Vesa et al., 2002).
The major Batten disease causing mutation is a 1.02-kb deletion in the CLN3 gene that is found in 85% of the disease chromosomes (The International Batten Disease Consortium, 1995). Additionally, 31 other Batten disease mutations have been described (http://www.ucl.ac.uk/ncl/CLN3.html). The CLN3 gene encodes a 438 amino acid protein, predicted to contain several transmembrane domains (TMDs), but no N-terminal signal sequence (The International Batten Disease Consortium, 1995; Janes et al., 1996; Mao et al., 2003). The low abundance of CLN3 protein in mammalian cells has contributed to difficulties in establishing its intracellular distribution. However, the consensus view based on various studies to localize the endogenous and exogenously expressed protein is that CLN3 is most likely a lysosomal/endosomal protein trafficking through the ER and Golgi (Järvelä et al., 1998; Kida et al., 1999; Haskell et al., 2000; reviewed in Pearce, 2000). A potentially important additional site of localization has been observed in neurons where CLN3 protein was found in the synaptic region (Järvelä et al., 1999; Haskell et al., 2000; Luiro et al., 2001). Consistent with a lysosomal location of CLN3 protein there is compelling evidence that it is functionally important in controlling the acidic pH in lysosomes. This evidence comes from studies of fibroblasts from Batten disease patients, which show elevated lysosomal pH (Holopainen et al., 2001), and of Btn1p, the yeast homologue of CLN3 protein, which localizes to the vacuole, the yeast equivalent of the lysosome. Yeast lacking Btn1p have abnormal vacuolar pH in the early phases of growth, a defect that can be reversed by complementation with either the yeast wild-type gene or human CLN3 (Pearce et al., 1999; Chattopadhyay et al., 2000). Alternative or additional functions of CLN3 protein have been suggested, including the maintenance of biophysical membrane properties (Das et al., 2001), control of apoptosis (Puranam et al., 1999; Persaud-Sawin et al., 2002) and control of protein trafficking (Chattopadhyay et al., 2003).
In this study, we have examined the intracellular trafficking of the polytopic CLN3 protein to lysosomes. Little is presently known about this except that it occurs independently of its glycosylation status (Kida et al., 1999). To locate targeting motifs in CLN3, we have experimentally determined the major cytosolic domains of the protein. Using this information, we have generated cDNA constructs encoding chimeric proteins with isolated cytosolic domains of CLN3 attached to a reporter molecule or encoding full-length CLN3 with mutations in putative targeting motifs. We have identified two targeting motifs that mediate trafficking of CLN3 to lysosomes both in transfected neuronal and nonneuronal cells. CLN3 protein expressed in neurons was found not only in lysosomes but also in early endosomes independently of the presence or absence of the lysosomal targeting motifs. This suggests that intracellular trafficking of CLN3 includes sorting in early endosomes and emphasizes the possible functional importance of CLN3 in endosomal compartments, especially in neurons.
MATERIALS AND METHODS
cDNA Constructs
Human CLN3 cDNA (Järvelä et al., 1998) was cloned into pCMV5 (Andersson et al., 1989) for transient expression experiments in HeLa cells and hippocampal neuronal cells or into Δ pMEP4 (Girotti and Banting, 1996) for stable transfection of normal rat kidney (NRK) cells. Chimeric CD8-CLN3 constructs were generated in CD8-pBluescript (Ihrke et al., 2000) by replacing the cytosolic tail of CD8 with either the cytosolic loop (amino acids 237-271) or the cytosolic tail (amino acids 398-438) of CLN3 produced by polymerase chain reaction, and subcloned into Δ pMEP4. Two constructs mimicking naturally occurring juvenile-onset neuronal ceroid lipofuscinosis mutations (Munroe et al., 1997) were created by introducing two stop codons (tgatga) into the CLN3-pCMV5 construct after the gag sequence encoding E399 in the CLN3 protein, or by deleting g1272 in a long CLN3p-CMV5 construct containing noncoding 3′ sequence of CLN3 (cloned from a human liver cDNA library; BD Biosciences Clontech, Palo Alto, CA). All point mutations, insertions, deletions, and addition of the FLAG-tag after S401, were generated in the constructs in pCMV5 or pBluescript, by using a QuikChange site-directed in vitro mutagenesis kit according to manufacturer's protocols (Stratagene, Amsterdam, The Netherlands). All constructs were verified by sequencing.
Cell Culture and Transfection
NRK and HeLa cells were grown on coverslips and transfected using Fu-GENE 6 transfection reagent as described previously (Ihrke et al., 2000). Transient expression of CLN3-pCMV5 constructs in HeLa cells was analyzed 48 h after transfection. Stable NRK cell lines expressing Δ pMEP4 constructs were generated as described previously (Ihrke et al., 2000). Expression of the Δ pMEP4 constructs was induced by adding 2-5 μM CdCl2 to culture media 16-24 h before experiments. Lysosomal degradation of the lumenal CD8 portion of chimeric proteins was inhibited by adding 21 μM leupeptin (Sigma Chemical, Poole, Dorset, United Kingdom) at the start of induction (Ihrke et al., 2000).
Hippocampal cultures were prepared from neonatal rats (age 6-24 h), as described previously (Schell et al., 2001). Briefly, ∼24 hippocampi were dissociated in papain, dispersed by trituration through pipets, plated onto 72 poly-d-lysinecoated coverslips (22 × 22 mm), and incubated overnight in Neurobasal medium (Invitrogen, Paisley, United Kingdom) supplemented with B27 (Invitrogen), and 20% horse serum. The next morning, medium was replaced with serum-free medium plus B27 and cells were maintained in this medium for up to 4 wk. In most experiments, cells were transfected after 7 d in culture by using a modified calcium phosphate method (Schell et al., 2001) and then fixed for immunocytochemistry 3-5 d later.
Immunofluorescence Labeling, Antibodies, and Microscopy
For immunofluorescence labeling, cells were fixed either with methanol at -20°C or 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS), pH 7.4. PFA-fixed cells were permeabilized by keeping 0.2% saponin (Sigma Chemical) in all solutions during labeling, or by 5-min incubation in 0.1% Triton X-100-PBS before adding antibodies [for hippocampal neurons and for labeling with antiprotein disulfide isomerase [PDI] and anti-early endosomal antigen-1 [EEA1] antibodies]. Recombinant CLN3 was detected either with rabbit anti-peptide antibody m385 against the amino acids 242-258 of mouse CLN3 (Luiro et al., 2001), h385 against amino acids 242-258 of human CLN3 (Järvelä et al., 1998), or a new affinity-purified rabbit antibody 33 aff against amino acids 1-33 of human CLN3 protein. Briefly, the sequence encoding amino acids 1-99 of the human CLN3 protein was cloned into pGEX4T-3 (Amersham Biosciences UK, Little Chalfont, Buckinghamshire, United Kingdom), expressed as a fusion protein with glutathione S-transferase (GST) in Escherichia coli, purified, and injected into two rabbits (3326 and 3327). Antiserum 3327 was affinity purified by removing the anti-GST antibodies on a GST-coupled cyanogen bromide column followed by binding to GST-CLN3 (aa 1-33) on a nitrocellulose membrane, and elution with 200 mM glycine, 0.1% gelatin, pH 2. Because antibody h385 was raised against a peptide containing LI(254-3), it did not recognize proteins carrying mutations in this motif and therefore, antibodies m385, 33aff, or 3326 were used for detection of these mutated proteins.
CD8-CLN3 chimeric proteins were detected with a rat monoclonal antibody (mAb) to human CD8α (a gift from Dr. G. Hale, University of Oxford, Oxford, United Kingdom). Lysosomes were detected with mouse monoclonal antibodies against rat lgp120 (GM10; Reaves et al., 1996) and human lamp-1 (H4A3; Developmental Studies Hybridoma Bank, Department of Biological Sciences, University of Iowa, Iowa City, IA) in NRK and HeLa cells, respectively. Mouse monoclonal anti-PDI antibody (ID3; a gift from Dr. David Vaux, Sir William Dunn School of Pathology, University of Oxford, Oxford, United Kingdom) and anti-EEA1 antibody (BD Transduction Laboratories, San Diego, CA) were used for detection of the ER and early endosomes, respectively. Mouse monoclonal anti-synapsin antibody (Chemicon, Hampshire, United Kingdom) was used as a presynaptic and neuronal marker in hippocampal cell cultures. Secondary antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA) or Molecular Probes Europe (Lerden, The Netherlands). The labeled coverslips were mounted using ProLong Antifade (Molecular Probes Europe) and visualized using either an Axioplan microscope equipped with a charge-coupled device camera (Figures 2, 7, and 9; Carl Zeiss, Jena, Germany) or an MRC 1024 confocal microscope (Bio-Rad, Hercules, CA). Adobe Photoshop software was used for image processing.
Figure 2.
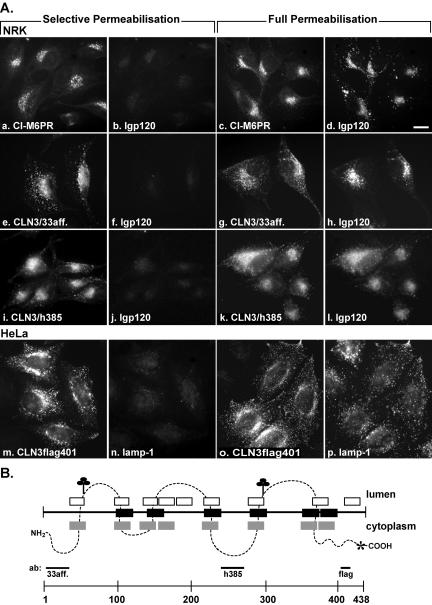
Topology of the CLN3 protein. (A) Stably transfected NRK cells stably expressing wt CLN3 were selectively permeabilized with 20 μg/ml digitonin and analyzed by epifluorescence microscopy. Selective permeabilization was monitored by staining the cytosolic tail of CI-M6PR with polyclonal 1001 antibody (a) and lack of staining of the lumenal domain of lgp120 with GM10 antibody (b, f, and j). Both CLN3 antibodies, 33 aff (e) and h385 (i) showed strong staining in selectively permeabilized cells. The lumenal epitopes of lgp120 were exposed only when the cells were fully permeabilized with 50 μg/ml digitonin (d, h, and l). The staining of CI-M6PR (c) and CLN3 protein with antibodies 33aff (g) and h385 (k) in fully permeabilized cells are shown. CLN3 protein containing the intramolecular FLAG-tag after S401 was expressed transiently in HeLa cells. A strong signal for the FLAG epitope was obtained both in selectively and fully permeabilized cells (m and o), whereas the lumenal epitope of lamp-1 was not recognized in selectively permeabilized cells (n) but only in completely permeabilized HeLa cells (p). Bar 10 μm. (B) Most probable topology model for the CLN3 protein (dotted line; also see text). TMDs suggested by Janes et al. (1996) or the SOSUI (Hirokawa et al., 1998) and TopPred2 (von Heijne, 1992) prediction programs are shown in black, gray, and white boxes, respectively. Two putative N-glycosylation sites are indicated by (♣). A prenylation site at C435 is indicated by asterisk. Antibodies used in selective permeabilization analysis and corresponding protein regions are shown.
Figure 7.
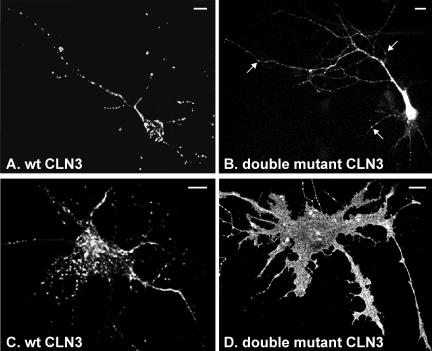
Localization of CLN3 protein in transfected neuronal cells. Postnatal rat hippocampal cells were transiently transfected with wt CLN3 or CLN3 carrying mutations in targeting motifs, fixed with PFA, permeabilized with Triton X-100, and analyzed by epifluorescence microscopy after labeling with anti-CLN3 antibody. (A and B) Neurons: the wt CLN3 was found in vesicular structures in the soma and along neuronal processes (A), whereas CLN3 carrying mutations in both lysosomal targeting motifs (double mutant LI253-4A+M409A+G419A) was mostly found at the plasma membrane in neurons (B), although some vesicular structures were still detectable along neuronal processes (see arrows). (C and D) Glial cells: localization of wt and double mutated CLN3 proteins in glial cells was similar to that in nonneuronal cells, i.e., with the wt protein in lysosomes (C; lysosomal marker not shown) and the double mutant at the plasma membrane (D). Bars, 10 μm.
Figure 9.
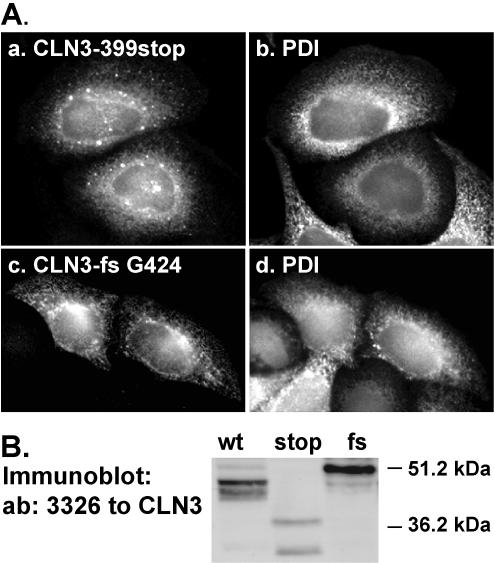
Batten disease mutations interfering with lysosomal targeting motifs. (A) CLN3 proteins carrying mutations that mimic two naturally occurring Batten disease-causing mutations, either introduction of a stop codon after E399 (stop) or a frameshift after G424 (fs), were transiently expressed in HeLa cells. Cells were fixed with PFA, permeabilized with Triton X-100, labeled with antibodies to CLN3 and PDI, and analyzed by epifluorescence microscopy. Both mutated proteins were retained in the ER (a and c, respectively), judged by colocalization with PDI (b and d). (B) Immunoblotting of cell lysates with a CLN3 specific antiserum 3326. Wt CLN3 occurs as a 43/46-kDa doublet, whereas the mutations resulted in proteins of 39 kDa (stop) and 50 kDa (fs), respectively.
Determination of Cytosolic Domains of CLN3 Protein by Selective Permeabilization
For selective permeabilization, we modified the protocol of Davies and Ioannou (2000). Cells were incubated at 4°C for 45 min in sucrose buffer (1% bovine serum albumin, 0.3 M sucrose, 0.1 M KCl, 2.5 mM MgCl2, 1 mM EDTA, 10 mM HEPES, pH 7.4) containing 0, 5, 10, 15, or 20 μg/ml freshly made digitonin (Sigma Chemical). NRK cells were then incubated with primary antibodies against CLN3 protein (h385or 33aff) and against the lumenal domain of lgp120 (GM10). As a control for selective permeabilization, cells were incubated with rabbit antibody 1001, raised against the cytosolic tail of the cation-independent mannose 6-phosphate receptor (CI-M6PR) (Reaves et al., 1996) and with GM10. The cells were washed once with cold PBS, fixed with methanol, blocked for 30 min in 1% bovine serum albumin-PBS, and labeled with secondary antibodies. For full permeabilization, the cells were treated as described above, but with 50-100 μg/ml digitonin in sucrose buffer. In selective permeabilization experiments with HeLa cells, FLAG-tagged CLN3 protein was detected by monoclonal mouse anti-FLAG antibody (clone M2; Sigma Chemical). The lumenal domain of lysosomal membrane protein lamp-1 was detected with fluorescein isothiocyanate-conjugated anti-CD107 antibody (BD Biosciences, Heidelberg, Germany).
Coimmunoprecipitation Assay
Coimmunoprecipitations of CLN3 and CLN5 proteins were performed as described previously (Vesa et al., 2002). Briefly, COS-1 cells were cotransfected with either wt or the SWE mutant of CLN5 (G253Stop) as well as with wild-type (wt), loop mutant (LI 253-4AA), tail mutant (M409A+G419A), or double mutant (LI253-4AA+M409A+G419A) of CLN3, all cloned in pCMV5. Two days posttransfection cells were lysed and supernatants were immunoprecipitated with CLN5-specific antibody CLN5/N (Vesa et al., 2002). Immunocomplexes were analyzed by Western blotting by using CLN3-specific antiserum 3326.
RESULTS
CLN3 Protein Is in Lysosomes of Transfected Nonneuronal Cells
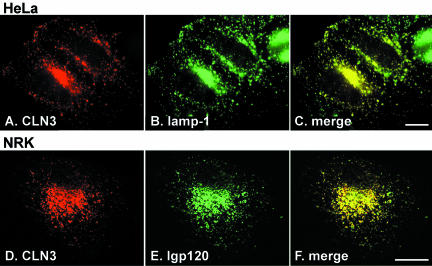
We first determined the steady-state localization of human CLN3 protein in transfected NRK and HeLa cells by immunofluorescence microscopy. In neither of these cell types were any of our antibodies to human CLN3 protein able to detect endogenous CLN3 protein by using this technique. In transfected cells transiently expressing CLN3, there was extensive overlap of the protein with endogenous lysosomal membrane glycoproteins (lgps; human lamp-1 or its rat homologue lgp120, respectively, in HeLa and NRK cells), consistent with the major site of localization being late endocytic organelles (HeLa cells shown in Figure 1, A-C; NRK cells not shown). There was little colocalization with the late endosome/trans-Golgi network marker CI-M6PR and minor overlap with the early endosomal marker EEA1 (our unpublished data), indicating that CLN3 protein was mainly lysosomal. To overcome the possibility that overexpressed protein may be mislocalized because of saturation of membrane traffic machinery, we prepared stably transfected NRK cell lines by using the inducible expression vector ΔpMEP4. This contains the metallothionein IIA promoter, allowing controlled, low levels of expression of membrane proteins after addition of CdCl2 (Reaves and Banting, 1994; Girotti and Banting, 1996). In previous studies using this promoter, sorting of membrane proteins to different destinations in post-Golgi membrane traffic pathways has not been compromised (Reaves et al., 1998; Ihrke et al., 2000; Rous et al., 2002). Even at the lowest levels of detectable expression in the stably transfected NRK cells, human CLN3 protein colocalized as well with lgp120 as at the higher, easily detectable level shown in Figure 1, D-F.
Figure 1.
Localization of CLN3 protein in transfected nonneuronal cells by confocal microscopy. (A-C) HeLa cells transiently expressing wt human CLN3 protein were fixed with 4% PFA, permeabilized with 0.2% saponin, and double labeled with rabbit polyclonal antibody h385 to human CLN3 protein (A) and a mouse mAb to lamp-1 (B) followed, respectively, by Texas Red- and fluorescein isothiocyanate-conjugated secondary antibodies. Merged confocal images shown in C. Bar, 10 μM. (D-F) Stably transfected NRK cells were induced to express wt human CLN3 protein (D) by incubation with 5 μM CdCl2 for 18 h. Cells were fixed, permeabilized, and labeled as described above except that a mouse mAb to lgp120 (E) was used; merged images shown in F. Bar, 10 μM.
Determination of the Cytoplasmic Domains of the CLN3 Protein
To obtain topological information for the human CLN3 protein, we used a selective cell permeabilization assay in stably transfected NRK cells. Such assays require the cells to be detergent treated before fixation. We could find no conditions where the extent of CLN3 protein colocalization with lgps was as complete, as observed after conventional fixation and permeabilization. Nevertheless, we were able to develop a protocol whereby the plasma membrane can be permeabilized by low concentrations of digitonin, leaving the late endosomal/lysosomal membranes intact. Under such conditions, cytosolic epitopes of lysosomal/endosomal membrane proteins are accessible to antibodies, whereas lumenal epitopes are hidden. We tested the extent of permeabilization by labeling cells with antibodies to the cytosolic tail of endogenous CI-M6PR, present in late endosomes, and to lumenal epitopes of endogenous lgp120. The signal for CI-M6PR was evident at low concentrations of digitonin, whereas lgp120 staining became visible only at high digitonin concentrations, i.e., with full permeabilization of the cells (Figure 2A, a-d). We next used two CLN3 antibodies under the same conditions: antibody h385, raised against amino acids 242-258 of the human CLN3 protein (Järvelä et al., 1998), and a new antibody, 33aff, directed against amino acids 1-33 of the N-terminus. Both antibodies gave a strong signal in selectively permeabilized cells (Figure 2A, e and f and i and j), whereas the lumenal epitope of lgp120 became detectable only in fully permeabilized cells (Figure 2A, g and h and k and l). These results show that epitopes in the sequences 1-33 and 242-258 of human CLN3 protein are exposed on the cytosolic side of intracellular membranes.
To obtain further information on the topology of CLN3, we examined the membrane orientation of a FLAG-tag inserted after S401, a site that still allowed some of the protein to be delivered to lysosomes in transiently transfected HeLa cells (all other sites at which we inserted a FLAG-tag resulted in protein that did not exit the ER; our unpublished data). The FLAG epitope was recognized by anti-FLAG antibodies in selectively permeabilized cells (Figure 2A, m and n), whereas detection of a lumenal epitope of lamp-1 required full permeabilization (Figure 2A, o and p). This suggests that, in contrast to many other lysosomal membrane proteins, CLN3 has a long cytosolic tail consisting of amino acids 401-438, if not more (see below). Overall, our experimental data suggest that CLN3 contains three large cytosolic domains, both termini and one large loop. Thus, CLN3 protein is a type III membrane protein containing at least four TMDs. Taking various topology predictions into account, it is more likely that there are six, or possibly eight, TMDs in CLN3, resulting in only one large cytosolic loop (at least 41 amino acids), confirmed in this study, and one or two additional small loops (see Figure 2B and DISCUSSION).
Expression of CD8-CLN3 Chimeras in Stably Transfected NRK Cells
Lysosomal membrane proteins with single or few TMDs are most often targeted to lysosomes by tyrosine-based GYXXØ (where X is any amino acid and Ø is a bulky hydrophophic residue) or dileucine-based sequence motifs present in short C-terminal cytosolic tails (Bonifacino and Traub, 2003). However, previous attempts to verify a sorting role for such motifs in CLN3 have failed (Kida et al., 1999; Haskell et al., 2000; our unpublished data), suggesting that trafficking of the CLN3 protein is controlled either by an unconventional sequence motif or by more than one targeting motif. Based on the new topological information, we made chimeric proteins consisting of the extracellular (lumenal) domain and TMD from the plasma membrane protein CD8, and individual cytosolic domains from CLN3. We chose the human T-cell surface marker CD8 as a reporter protein, because its usefulness as a “neutral” reporter is well established (Ponnambalam et al., 1994; Munro, 1995; Ihrke et al., 2000). The CD8-CLN3 chimeras contained either amino acids 237-271 of the large cytosolic loop domain of CLN3 or the most C-terminal 41 amino acids (398-438) of the predicted tail. Both chimeras were stably expressed in NRK cells and their intracellular distributions were analyzed by indirect immunofluorescence microscopy.
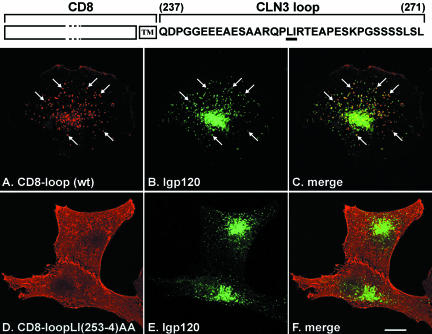
The CD8-CLN3loop chimera colocalized extensively with lgp120, indicating that a significant proportion was targeted to lysosomes, although a minor fraction was seen at the plasma membrane (Figure 3, A-C). The CLN3loop contains a putative dileucine motif, LI (253-254), and indeed, simultaneous alanine substitution of these hydrophobic amino acids completely blocked lysosomal targeting, such that the mutated chimera was observed solely at the plasma membrane (Figure 3, D-F), similar to wt CD8 (Ihrke et al., 2000). The separate mutations L253A or I254A also resulted in loss of lysosomal targeting of the corresponding CD8-CLN3loop chimeras (our unpublished data). Because functional dileucine motifs often include upstream acidic residues (D/E) (Pond et al., 1995; Bonifacino and Traub, 2003), we examined whether the patch of acidic residues at positions -9 to -11 N-terminal to the LI sequence was part of the motif. All three glutamic acid residues in positions -9 to -11 were simultaneously substituted with alanines, without changing the LI sequence. When localization of the resultant CD8-CLN3loop chimera was examined, we found that the majority of chimeric protein was still present in lysosomes and only a moderate proportion was mistargeted to the plasma membrane (our unpublished data).
Figure 3.
CD8-CLN3loop chimeras in NRK cells. The CD8-CLN3loop chimeras were expressed in stably transfected NRK cells and localized by confocal microscopy after fixation and permeabilization with methanol and double labeling with antibodies to CD8α (A and D) and lgp120 (B and E). The chimera containing the wt CLN3 loop (A) showed extensive overlap with with lgp120 (B). Arrows point to some of many structures that are positive for both proteins; merged images shown in C. The CD8-CLN3loopLI(253-4)AA chimera was detected at the plasma membrane (D) and showed no colocalization with lgp120 (E); merged images shown in F. Bar, 10 μm.
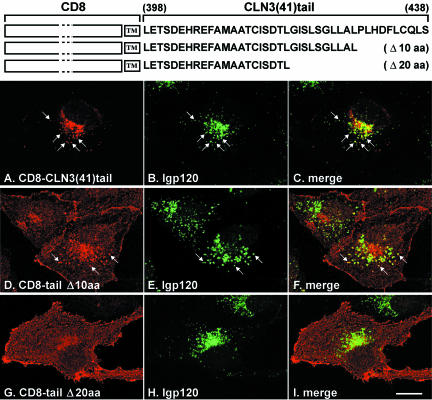
A significant fraction of the CD8-CLN3tail(41) chimera was also targeted to lysosomes, suggesting that there was a second signal in the C-terminus of CLN3 (Figure 4, A-C). However, the expression level of this protein was low and a fraction was retained in the ER, suggesting problems in folding and/or stability. To test whether these problems were due to incorrect length of the tail domain, we also made chimeras with longer and shorter CLN3 tails of 59 and 28 amino acids, respectively, but always observed dual ER/ lysosomal localization (our unpublished data). To narrow down the location of lysosomal targeting information within the 41 amino acid long tail, we made C-terminal deletions of 10 or 20 amino acids. Whereas deletion of 10 amino acids resulted in an intermediate phenotype with increased plasma membrane localization of the CD8-CLN3 tail chimera (Figure 4D), targeting to lysosomes was completely abolished by deletion of 20 amino acids (Figure 4G). Thus, it was likely that targeting information would be located within the carboxy-terminal 20 amino acids.
Figure 4.
CD8-CLN3tail chimeras in NRK cells. CD8-CLN3(41)tail chimeras were expressed in stably transfected NRK cells and localized confocal microscopy after fixation and permeabilization with methanol and double labeling with antibodies to CD8α (A, D, and G) and lgp120 (B, E, and H). The CD8-CLN3(41)tail chimera (A) colocalized significantly (see arrows and merged images in C) with lpg120 in lysosomes (B). Removal of 10 amino acids from the C-terminus of the chimera (D) resulted in partial colocalization with lgp120 (E), but a significant fraction was seen at the cell surface. Lysosomal targeting of the chimera was completely lost by deletion of 20 amino acids (G) resulting in most of the protein being on the cell surface and essentially no overlap with lgp120 (H). Merged images of D/E and G/H are shown in F and I, respectively. Bar, 10 μm.
Determination of Lysosomal Targeting Motifs in the Full-Length CLN3 Protein
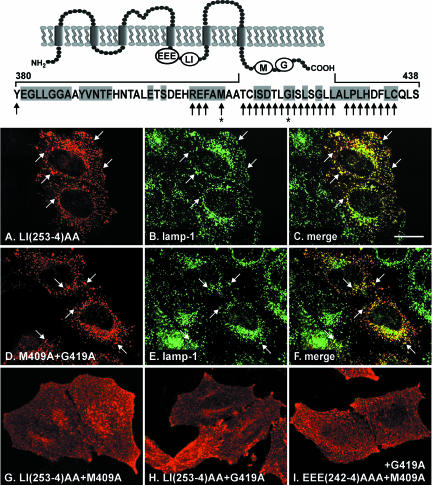
Expression of the CD8-CLN3 chimeras provided evidence for the existence of lysosomal targeting information in two different cytosolic domains of the CLN3 protein. To establish whether this held true in the native CLN3 protein, we investigated the steady-state localization of full-length CLN3 carrying different mutations in transiently transfected HeLa cells. The substitution LI(253-4)AA in the CLN3 loop resulted in a protein that colocalized well with lamp-1, but additionally showed minor plasma membrane localization (Figure 5, A-C). To determine the amino acids in the C-terminal tail responsible for targeting, we investigated the localization of mutated CLN3 proteins containing the LI(253-4)AA together with single alanine substitutions throughout the predicted tail domain (Figure 5). By using this approach, we identified two amino acid mutations, M409A (Figure 5G) and G419A (Figure 5H), either of which, in combination with the loop mutation, resulted in localization of CLN3 at the plasma membrane. To assess whether the two amino acids M409 and G419 were part of the same targeting motif, we deleted a single amino acid residue, A410, between these amino acids, whereby M409 and G419 themselves were left intact, but the LI(253-4) motif was mutated. The resultant CLN3 protein was again relocated to the plasma membrane, indicating that the distance between the M409 and G419 was critical for targeting (our unpublished data). When all nine residues between M409 and G419 were simultaneously substituted by alanines (together with mutation of LI), targeting information to lysosomes was also lost (our unpublished data). However, single alanine substitutions of these residues did not have detectable effect on targeting, except in two cases, C413A and L418A, where an intermediate phenotype with increased cell surface expression was observed (our unpublished data). Having identified specific amino acids in the cytosolic tail of CLN3 mediating lysosomal targeting, we next investigated whether the cytosolic loop alone was sufficient to mediate lysosomal delivery. A mutant protein with an intact LI motif, but containing both M409A and G419A substitutions was mostly found in lysosomes (Figure 5, D-F), indicating that both the loop domain and the cytosolic tail could independently facilitate lysosomal targeting of CLN3. We also reexamined the putative role of the acidic sequence N-terminal to the LI-motif by using CLN3 constructs with a mutated tail motif (M409A+G419A). In this context, replacement of glutamic acids -9 to -11 with alanines [EEE(242-4)AAA] in the presence of an intact LI(253-4) motif led to accumulation of CLN3 at the plasma membrane with little lysosomal localization (Figure 5I). This finding suggests that the dileucine targeting motif in the loop domain of CLN3 includes the upstream acid patch.
Figure 5.
The lysosomal targeting motifs of CLN3 protein. Full-length CLN3 proteins carrying different alanine substitutions were transiently expressed in HeLa cells, and localization of the mutated proteins was investigated by double labeling with CLN3 specific antibody (33aff) and antibody to lamp-1 (H4A3) followed by confocal microscopy. CLN3 carrying the LI(253-4)AA substitution alone (A) was mostly targeted to lysosomes (A), colocalizing well with lamp-1 (B); merged images shown in C. Single alanine scanning mutagenesis was carried out along the tail domain of CLN3 in the presence or absence of the LI(253-4)AA substitution in the loop domain (mutated amino acids are indicated by black arrows along the sequence given above confocal images). CLN3 carrying both M409A+G419A alone (intact LI) was localized to lysosomes (D, CLN3; E, lgp120; and F, merged images). When expressed in conjunction with the LI(253-4)AA mutation the two separate amino acid substitutions (asterisks), M409A (G) and G419A (H), resulted in mistargeting of CLN3 to the plasma membrane. When M409A and G419A mutations were both present together with intact LI(253-4), additional alanine substitutions in the acidic patch EEE (242-4)AAA upstream of the LI-motif resulted in a protein that was again mostly mistargeted to plasma membrane (I). Examples of lysosomal colocalization of CLN3 and lgp120 are indicated by white arrows in A-F. Bar, 10 μM. Topology diagram: positions of important amino acid motifs, EEE(242-4), LI(253-4), M409, and G419 are indicated. The amino acid sequence shows the most C-terminal 59 amino acids of CLN3, highly conserved residues are indicated by gray boxes.
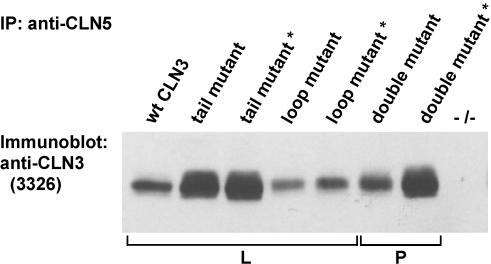
The recently described interaction of CLN5, another NCL protein of lysosomal membranes, with CLN3 (Vesa et al., 2002) raised the question of whether this interaction is important for correct lysosomal targeting of CLN3. We therefore coexpressed CLN5, either wild-type or a C-terminally truncated protein (CLN5 SWE mutation; Vesa et al., 2002), transiently in COS-1 cells together with CLN3 carrying mutations in the targeting motifs. Immunoprecipitation with a CLN5 specific antibody, followed by immunoblotting with CLN3-specific antibody 3326 revealed that the two NCL proteins were able to retain their physical interaction even when both lysosomal targeting motifs in CLN3 were mutated simultaneously (Figure 6). This result indicates that the targeting motifs of CLN3 act independently of an interaction with CLN5 protein, presumably by direct binding to the lysosomal sorting machinery.
Figure 6.
Physical interaction between CLN5 and CLN3 in COS-1 cells. COS-1 cells were transiently transfected with either the full-length or C-terminally truncated CLN5 carrying SWE mutation (asterisk) and wt CLN3 or CLN3 carrying mutations either in the loop [LI(253-4)AA] or tail (M409A+G419A) targeting motifs or both (double mutant). Cell lysates were immunoprecipitated with CLN5 specific antibody, followed by immunoblotting with CLN3 specific antiserum (3326). -/- lane corresponds to nontransfected control cells. The observed differences in the strength of signal were directly proportional to expression levels of the different CLN3 constructs (our unpublished data). L and P indicate primary localization of CLN3 proteins in lysosomes and at the plasma membrane, respectively, in transfected HeLa cells (see Figures 1 and 5).
Localization of Wild-Type and Mutated CLN3 Protein in Neurons
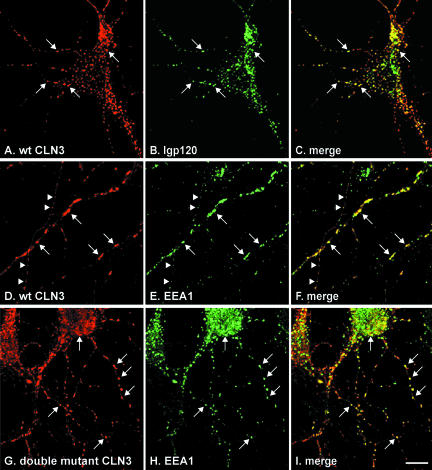
Because the main manifestations of Batten disease affect the CNS, we investigated the role of the two lysosomal targeting motifs on the localization of CLN3 in neuronal cells. Postnatal rat hippocampal primary cultures were transiently transfected with wt CLN3 or CLN3 containing mutations in either one or both motifs (LI253-4AA and/or M409A+G419A) and analyzed by indirect immunofluorescence. In the neuronal cell soma, wt CLN3 was found in vesicular structures, most of which were identified as lysosomes by positive staining for lgp120 (Figures 7A and 8, A-C), and some as EEA1-positive endosomes (our unpublished data). CLN3 staining was also detected along the neuronal processes, where it was mainly associated with EEA1-positive structures (Figure 8, D-F). Punctate structures containing CLN3 were not only visible in dendrites but also along the entire length of axons identified by their different morphology (thinner process with constant diameter; Figure 8, D-F). No significant colocalization was seen between CLN3 and the presynaptic marker synapsin I, although occasionally CLN3-positive structures were detected in proximity to synapsin I staining (our unpublished data). CLN3 proteins carrying mutations in either of the lysosomal targeting motifs were sorted similarly to the wt protein (our unpublished data). When both motifs were mutated (double mutant, LI253-4A+M409A+G419A) the resultant CLN3 protein no longer colocalized with lgp120, but was mostly on the plasma membrane of both soma and neuronal processes (Figure 7B). A similar striking difference in localization between the wt and double mutated CLN3 was seen in glial cells (Figure 7, C and D, respectively). In neurons, however, despite silencing of the two lysosomal targeting motifs, the double mutant was still associated with EEA1-positive structures, which was particularly evident in the neuronal processes (Figure 8, G-I). This indicates that CLN3 can reach early endosomes even in the absence of these motifs.
Figure 8.
Colocalization of CLN3 protein with lysosomal and early endosomal markers in transfected neurons. Postnatal rat hippocampal cells were transiently transfected with wt CLN3 or CLN3 carrying mutations in targeting motifs, fixed with PFA, permeabilized with Triton X-100 and analyzed by confocal microscopy after antibody labeling. The wt CLN3 protein (A and D) was found in vesicular structures in the soma and along neuronal processes. In the soma and proximal processes wt CLN3 (A) colocalized well with lgp120 (B) and to a lesser degree with EEA1 (our unpublished data); C, merge of A and B. The wt CLN3 found in vesicular structures in peripheral neuronal processes (D) was associated with EEA1-positive structures (E), indicating its presence in early endosomes; merge shown in F. A likely axon is marked by arrowheads, D-F. CLN3 carrying mutations in both targeting motifs (double mutant LI253-4A+M409A+G419A) was not found in lysosomes (our unpublished data), but at the plasma membrane (see cell bodies in G and Figure 6B). And also in vesicular structures in neuronal processes (G), which were positive for EEA1 (H); I, merge of G and H. Examples of colocalization are indicated by white arrows and arrowheads. Bar, 10 μm.
Analysis of Batten Disease Mutations Affecting Lysosomal Targeting Motifs of CLN3
We next reviewed all known disease causing mutations in CLN3 (http://www.ucl.ac.uk/ncl/CLN3.html) to identify those that might result in a defect in intracellular trafficking of CLN3 by interfering with its lysosomal sorting. Although most of the mutations result in dramatic truncations, we found two mutations that could affect the C-terminal targeting motif more specifically. The first mutation leads to aberrant splicing of exon 15 and the introduction of two early stop codons after E399 (Munroe et al., 1997). When a CLN3 protein mimicking this mutation (CLN3-399stop) was transiently expressed in HeLa cells, the protein was found to have a molecular mass of 39 kDa, differing from the 43/46-kDa doublet of wt CLN3 (Figure 9B). Immunolocalization of the truncated protein revealed that it was retained in the ER, judged by its colocalization with PDI (Figure 9A, a and b). The second mutation, a deletion of a single nucleotide (g1272), causes a frameshift after G424, and due to this mutation is predicted to result in a protein extended for 75 amino acids compared with wt CLN3 (Munroe et al., 1997). The abnormally long tail still contains both amino acids, M409 and G419, shown to be important for targeting of CLN3. Expression of this large CLN3 variant (50 kDa; Figure 9B) in HeLa cells, showed that this protein was also retained in the ER (Figure 9A, c and d). Together, these findings suggest that in addition to containing lysosomal targeting information, the C-terminal tail domain of CLN3 protein has an important role in folding and exit from the ER.
DISCUSSION
Since the discovery of the Batten disease protein CLN3 in 1995, its topology has been based mainly on predictions. Lack of topological knowledge has complicated the identification of trafficking motifs and pathways, as well as putative domains responsible for interactions with other proteins. In this study, we experimentally determined that both the N and C termini and one large loop domain of CLN3 protrude into the cytoplasm and that there are consequently at least four TMDs and two loop domains facing the lumen of the lysosomes (Figure 2B). This is consistent with previous data suggesting that at least two putative N-glycosylation sites in the CLN3 protein (N71 and N310) are indeed glycosylated and therefore must be lumenal (Persaud-Sawin et al., 2002). However, different topology predictions all suggest that there are three or five TMDs between the first lumenal loop (containing N71) and the large cytosolic loop domain confirmed in this study. Because we showed that amino acid S401 faces the cytoplasm, and data by others suggest that the C terminus is prenylated at C435 (Kaczmarski et al., 1999), it is unlikely that there are additional TMDs at the C-terminus. Recently, in vitro translation experiments, combined with immunoprecipitation, have provided data consistent with CLN3 protein having two cytosolic loop domains and a cytosolic C terminus as shown in Figure 2B (Mao et al., 2003). However, these authors suggest that CLN3 has five TMDs with a long lumenal N terminus because they were unable to immunoprecipitate in vitro translated protein by using an antibody to the N terminus in the presence of microsomes. In contrast, our N-terminal antibody (33aff) gives a positive signal for CLN3 in selectively permeabilized cells, indicating that, in the in vivo situation, at least the first 33 N-terminal residues face the cytosol. It is likely that the following stretch of hydrophobic amino acids (38-60) constitutes the first TMD of human CLN3, as predicted by various topology programs. In this model, N71 would be available for glycosylation at the lumenal side, consistent with data published by Persaud-Sawin et al. (2002) and Mao et al. (2003). Therefore, we propose that CLN3 is a type III membrane protein with six (or possibly eight) TMDs.
Identification of the major cytosolic domains of CLN3 was a key to localize motifs, which mediate lysosomal targeting of the protein. In contrast to many other lysosomal membrane proteins targeted by single motifs in their short C-terminal cytosolic tails (Bonifacino and Traub, 2003), we found that efficient lysosomal targeting of polytopic CLN3 protein requires two sequence motifs in separate protein domains. One of the motifs is a conventional dileucine motif (LI), but it is located in an unconventional position, i.e., in a cytosolic loop domain. In addition, a novel sequence motif M(X)9G in the long C-terminal tail of the protein is needed for optimal lysosomal sorting of CLN3.
Dileucine motifs have a well established role in mediating lysosomal trafficking, endocytosis, and/or basolateral sorting in polarized epithelial cells. Amino acids surrounding the hydrophobic residues have previously been shown to be critical for recognition of these motifs by specific adaptor proteins that mediate different sorting events (Pond et al., 1995; Rodionov et al., 2002). Acidic amino acids (D/E) located N-terminally to dileucine sequences have often been shown to be important functional parts of such motifs (Pond et al., 1995; Sandoval et al., 2000; Misra et al., 2002; Shiba et al., 2002; Bonifacino and Traub, 2003). Acidic patches may function independently as internalization signals (Voorhees et al., 1995; Johnson et al., 2001), but in most reported examples, the vicinity of acidic residues to a dileucine is a specifying factor for adaptor recognition. In the present study, we show that acidic amino acids located nine to 11 residues N-terminal of LI (253-4) are required for a functional dileucine motif in CLN3. Interestingly, this was only fully evident in the context of full-length CLN3, indicating that the presentation of the complete motif (acidic patch + LI) in a peptide loop rather than in a free tail (as in CD8-chimeras) is important.
Which adaptor molecules could be involved in recognition of this unusually presented dileucine motif? Heterotetrameric adaptor proteins AP1 and AP3, as well as monomeric Golgi localized γ-ear-containing ARF binding proteins (GGAs) all are believed to recognize dileucine motifs and are involved in lysosomal sorting in the trans-Golgi network and/or endosomes (Robinson and Bonifacino, 2001), whereas AP2 binds these motifs at the plasma membrane and mediates endocytosis. Location of the acidic patch 9-11 residues upstream of LI in the loop of CLN3 does not fulfill the positional criteria described for optimal recognition of dileucine motifs by AP1, AP3, and GGAs, which prefer acidic residues in positions -4, -4/-5, and -3/-4, respectively (Misra et al., 2002; Reusch et al., 2002; Shiba et al., 2002). However, the sequence directly adjacent to LI in CLN3 is very similar to that surrounding the LI motif of another lysosomal membrane protein, limp II, which has been shown to be recognized by AP3 (Höning et al., 1998). Both proteins contain an arginine and a proline in positions -3 and -1, respectively, the exact amino acids that have been recently shown to determine specific binding to AP3 (Rodionov et al., 2002).
The second lysosomal targeting signal in the C-terminal cytosolic tail of CLN3 is a novel sequence motif M(X)9G. It differs from all previously determined lysosomal targeting motifs, which do not conform with the typical GYXXØ or dileucine type sequences, including the KCPL sequence in P-selectin (Blagoveshchenskaya et al., 1998) and YFPQA in cystinosin (Cherqui et al., 2001). In the M(X)9G motif, methionine and glycine were found to be the most important residues. Although glycines, such as in the GYXXØ-motif, usually do not interact with adaptor binding pockets per se, they are involved in presentation of the signal (Owen et al., 2001). Therefore, the glycine at position 419 in the M(X)9G motif of CLN3 may fulfill a similar role. Because changes both in the number or the composition of residues between M409 and G419 also altered localization of CLN3, it is possible that this motif is part of a larger three-dimensional determinant, rather than being a linear sequence motif. The C-terminal tail structure of CLN3 could be stabilized by interaction of hydrophobic amino acids and/or lipid anchors with the cytosolic leaflet of the membrane and thereby be important for the presentation of the targeting motif. One possible attachment site is C435, which has been shown to be prenylated in vitro (Kaczmarski et al., 1999). However, our present data are consistent with a previous report indicating that substitution of C435 with alanine had no detectable effect on steady state localization of CLN3 (Haskell et al., 2000). Thus, additional factors may contribute to the presentation of the M(X)9G motif in CLN3.
There is currently no information about the trafficking machinery that might interact with unusual targeting motifs. It was recently reported that the sequence DVPM, found in the sorting receptor sorLA, was the minimal sequence requirement of the form Ψ XXΦ (where Ψ is an acidic amino acid) for binding to GGA1 (Jacobsen et al., 2002). A similar sequence EFAM409 in CLN3 overlaps with the M(X)9G motif. Because substitution of E406 by alanine had no detectable effect on lysosomal targeting of CLN3, it is unlikely that M(X)9G motif is a Ψ XXΦ-type signal recognized by GGA1. Our experiments also showed that interaction with another NCL protein, CLN5, did not depend on the presence of the lysosomal targeting motifs of CLN3, suggesting that CLN5 binding is not a prerequisite for lysosomal targeting of CLN3.
An unresolved question is why the most severe symptoms of Batten disease manifest in the brain. Thus, expression of CLN3 in neurons has been investigated extensively with the hope of uncovering any putative differences with nonneuronal cells. Consistent with previous studies, we found that CLN3 was present in vesicular structures both in soma and in neuronal processes (including axons) of cultured rat hippocampal neurons (Järvelä et al., 1999; Haskell et al., 2000; Luiro et al., 2001). Because lysosomes and late endosomes are predominantly located in the cell soma and proximal regions of dendrites (Parton et al., 1992), it is likely that the presence of CLN3 in distal parts of neuronal processes represents localizations other than lysosomes/late endosomes. Previously, CLN3 expressed in neuronal processes has been colocalized with presynaptic markers and therefore suggested to have a role in synaptic transmission (Järvelä et al., 1999; Haskell et al., 2000). However, fractionation analysis of mouse brain later revealed that CLN3 resides in a compartment different from synaptic vesicles (Luiro et al., 2001). We also frequently observed CLN3 close to a presynaptic marker (synapsin I), but saw very little precise overlap. In contrast, we found that CLN3 was associated prominently with EEA1 along neuronal processes demonstrating a novel localization of CLN3 in early endosomes. It has been previously shown that early endosomes are concentrated at presynaptic terminals and varicosities in axons of hippocampal neurons (Parton et al., 1992). The presence of CLN3 in early endosomes was much less evident in nonneuronal cells (HeLa), suggesting this localization could be especially important in neurons. The same lysosomal targeting motifs of CLN3 operated both in neuronal and nonneuronal cells; however, localization of CLN3 in early endosomes occurred independently of lysosomal targeting. Therefore, it is possible that CLN3 contains additional targeting information, which is only recognized in specialized cells (such as neurons), and/or which regulates the rate of trafficking at the level of endosomes (Johnson et al., 2001).
Endosomes present in neuronal processes are dynamic and heterogeneous structures (Sulzer and Holtzman, 1989; Cooney et al., 2002). Among these organelles, multivesicular bodies have been suggested to be the major structures mediating transport of endocytic and autophagic material from the nerve terminals to the lysosomes in the cell body (Parton et al., 1992; Hollenbeck, 1993; Nixon and Cataldo, 1995). It is tempting to hypothesize that the function of CLN3 is to regulate the physiological properties of such organelles (including their degradative characteristics). This would be consistent with previous reports demonstrating altered intravesicular pH when CLN3 was absent or functionally inactive (Golabek et al., 2000; Holopainen et al., 2001). The function of many lysosomal proteins is known to be regulated by the proteolytic activation of their precursors in acidic environments. For example, gradual proteolytic processing of cathepsin D in acidic compartments is required to yield an active enzyme, which has been suggested to have an important role in neuronal development and/or homeostasis (Tyynelä et al., 2000). Interestingly, altered levels of CLN3 expression have been reported to affect processing of cathepsin D as well as amyloid-β protein precursor (Golabek et al., 2000). The highly specialized endosomal-lysosomal system may render neurons especially sensitive to impaired (or accelerated) processing of certain proteins and lipids and therefore, lead to the accumulation of lipofuscin typical for all NCL disorders.
Acknowledgments
We thank Sally Gray for technical assistance and Dr. Karin Römish for advice in determination of topology. The work was supported by grants from the Wellcome Trust (United Kingdom) to A.K. (Traveling Research Fellowship 061077), to J.P.L. and G.I. (057263); The Medical Research Council (United Kingdom) to J.P.L.; and The Academy of Finland (48047) and the Maud Kuistila Foundation (Finland) to A.K. M.J.S. was supported by the Royal Society (United Kingdom). The Cambridge Institute for Medical Research is in receipt of a strategic award from the Wellcome Trust.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-02-0120. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-02-0120.
Abbreviations used: CI-M6PR, cation-independent mannose 6-phosphate receptor; EEA1, early endosomal antigen-1; GST, glutathione S-transferase; lgp, lysosomal membrane glycoprotein; NCL, neuronal ceroid lipofuscinosis; NRK, normal rat kidney; PDI, protein disulfide isomerase; PBS, phosphate-buffered saline; PFA, paraformaldehyde; TMD, transmembrane domain; wt, wild type.
References
- Andersson, S., Davis, D.L., Dahlback, H., Jornvall, H., and Russell, D.W. (1989). Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J. Biol. Chem. 264, 8222-8229. [PubMed] [Google Scholar]
- Blagoveshchenskaya, A.D., Norcott, J.P., and Cutler, D.F. (1998). Lysosomal targeting of P-selectin is mediated by a novel sequence within its cytoplasmic tail. J. Biol. Chem. 273, 2729-2737. [DOI] [PubMed] [Google Scholar]
- Bonifacino, J.S., and Traub, L.M. (2003). Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72, 395-447. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay, S., Muzaffar, N.E., Sherman, F., and Pearce, D.A. (2000). The yeast model for Batten disease: mutations in btn1, btn2, and hsp30 alter pH homeostasis. J. Bacteriol. 182, 6418-6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay, S., Roberts, P.M., and Pearce, D.A. (2003). The yeast model for Batten disease: a role for Btn2p in the trafficking of the Golgi-associated vesicular targeting protein, Yif1p. Biochem. Biophys. Res. Commun. 302, 534-538. [DOI] [PubMed] [Google Scholar]
- Cherqui, S., Kalatzis, V., Trugnan, G., and Antignac, C. (2001). The targeting of cystinosin to the lysosomal membrane requires a tyrosine-based signal and a novel sorting motif. J. Biol. Chem. 276, 13314-13321. [DOI] [PubMed] [Google Scholar]
- Cooney, J.R., Hurlburt, J.L., Selig, D.K., Harris, K.M., and Fiala, J.C. (2002). Endosomal compartments serve multiple hippocampal dendritic spines from a widespread rather than a local store of recycling membrane. J. Neurosci. 22, 2215-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, A.M., von Harlem, R., Feist, M., Lucke, T., and Kohlschutter, A. (2001). Altered levels of high-energy phosphate compounds in fibroblasts from different forms of neuronal ceroid lipofuscinoses: further evidence for mitochondrial involvement. Eur. J. Paediatr. Neurol. 5, 143-146. [DOI] [PubMed] [Google Scholar]
- Davies, J.P., and Ioannou, Y.A. (2000). Topological analysis of Niemann-Pick C1 protein reveals that the membrane orientation of the putative sterol-sensing domain is identical to those of 3-hydroxy-3-methylglutaryl-CoA reductase and sterol regulatory element binding protein cleavage-activating protein. J. Biol. Chem. 275, 24367-24374. [DOI] [PubMed] [Google Scholar]
- Girotti, M., and Banting, G. (1996). TGN38-green fluorescent protein hybrid proteins expressed in stably transfected eukaryotic cells provide a tool for the real-time, in vivo study of membrane traffic pathways and suggest a possible role for rat TGN38. J. Cell Sci. 109, 2915-2926. [DOI] [PubMed] [Google Scholar]
- Golabek, A.A., Kida, E., Walus, M., Kaczmarski, W., Michalewski, M., and Wisniewski, K.E. (2000). CLN3 protein regulates lysosomal pH and alters intracellular processing of Alzheimer's amyloid-beta protein precursor and cathepsin D in human cells. Mol. Genet. Metab. 70, 203-213. [DOI] [PubMed] [Google Scholar]
- Haskell, R.E., Carr, C.J., Pearce, D.A., Bennett, M.J., and Davidson, B.L. (2000). Batten disease: evaluation of CLN3 mutations on protein localization and function. Hum. Mol. Genet. 9, 735-744. [DOI] [PubMed] [Google Scholar]
- Hirokawa, T., Boon-Chieng, S., and Mitaku, S. (1998). SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14, 378-379. [DOI] [PubMed] [Google Scholar]
- Hollenbeck, P.J. (1993). Products of endocytosis and autophagy are retrieved from axons by regulated retrograde organelle transport. J. Cell Biol. 121, 305-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holopainen, J.M., Saarikoski, J., Kinnunen, P.K., and Järvelä, I.I. (2001). Elevated lysosomal pH in neuronal ceroid lipofuscinoses (NCLs). Eur. J. Biochem. 268, 5851-5856. [DOI] [PubMed] [Google Scholar]
- Höning, S., Sandoval, I.V., and von Figura, K. (1998). A di-leucine-based motif in the cytoplasmic tail of LIMP-II and tyrosinase mediates selective binding of AP-3. EMBO J. 17, 1304-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihrke, G., Gray, S.R., and Luzio, J.P. (2000). Endolyn is a mucin-like type I membrane protein targeted to lysosomes by its cytoplasmic tail. Biochem. J. 345, 287-296. [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, L., Madsen, P., Nielsen, M.S., Geraerts, W.P.M., Gliemann, J., Smit, A.B., and Petersen, C.M. (2002). The sorLA cytoplasmic domain interacts with GGA1 and -2 and defines minimum requirements for GGA binding. FEBS Lett. 511, 155-158. [DOI] [PubMed] [Google Scholar]
- Janes, R.W., Munroe, P.B., Mitchison, H.A., Gardiner, R.M., Mole, S.E., and Wallace, B.A. (1996). A model for Batten disease protein CLN 3, functional implications from homology and mutations. FEBS Lett. 399, 75-77. [DOI] [PubMed] [Google Scholar]
- Johnson, A.O., Lampson, M.A., and McGraw, T.E. (2001). A di-leucine sequence and a cluster of acidic amino acids are required for dynamic retention in the endosomal recycling compartment of fibroblasts. Mol. Biol. Cell 12, 367-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Järvelä, I., Lehtovirta, M., Tikkanen, R., Kyttälä, A., and Jalanko, A. (1999). Defective intracellular transport of CLN3 is the molecular basis of Batten disease (JNCL). Hum. Mol. Genet. 8, 1091-1098. [DOI] [PubMed] [Google Scholar]
- Järvelä, I., Sainio, M., Rantamäki, T., Olkkonen, V.M., Carpen, O., Peltonen, L., and Jalanko, A. (1998). Biosynthesis and intracellular targeting of the CLN3 protein defective in Batten disease. Hum. Mol. Genet. 7, 85-90. [DOI] [PubMed] [Google Scholar]
- Kaczmarski, W., Wisniewski, K.E., Golabek, A., Kaczmarski, A., Kida, E., and Michalewski, M. (1999). Studies of membrane association of CLN3 protein. Mol. Genet. Metab. 66, 261-264. [DOI] [PubMed] [Google Scholar]
- Kida, E., Kaczmarski, W., Golabek, A.A., Kaczmarski, A., Michalewski, M., and Wisniewski, K.E. (1999). Analysis of intracellular distribution and trafficking of the CLN3 protein in fusion with the green fluorescent protein in vitro. Mol. Genet. Metab. 66, 265-271. [DOI] [PubMed] [Google Scholar]
- Luiro, K., Kopra, O., Lehtovirta, M., and Jalanko, A. (2001). CLN3 protein is targeted to neuronal synapses but excluded from synaptic vesicles: new clues to Batten disease. Hum. Mol. Genet. 10, 2123-2131. [DOI] [PubMed] [Google Scholar]
- Mao, Q., Foster, B.J., Xia, H., and Davidson, B.L. (2003). Membrane topology of CLN3, the protein underlying Batten disease. FEBS Lett. 27143, 1-7. [DOI] [PubMed] [Google Scholar]
- Misra, S., Puertollano, R., Kato, Y., Bonifacino, J.S., and Hurley, J.H. (2002). Structural basis for acidic-cluster-dileucine sorting-signal recognition by VHS domains. Nature 415, 933-937. [DOI] [PubMed] [Google Scholar]
- Munro, S. (1995). An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J. 14, 4695-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe, P.B., et al. (1997). Spectrum of mutations in the Batten disease gene, CLN3. Am. J. Hum. Genet. 61, 310-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon, R.A., and Cataldo, A.M. (1995). The endosomal-lysosomal system of neurons: new roles. Trends Neurosci. 18, 489-496. [DOI] [PubMed] [Google Scholar]
- Owen, D.J., Setiadi, H., Evans, P.R., McEver, R.P., and Green, S.A. (2001). A third specificity-determining site in μ2 adaptin for sequences upstream of Yxxφ sorting motifs. Traffic 2, 105-110. [DOI] [PubMed] [Google Scholar]
- Parton, R.G., Simons, K., and Dotti, C.G. (1992). Axonal and dendritic endocytic pathways in cultured neurons. J. Cell Biol. 119, 123-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce, D.A. (2000). Localization and processing of CLN3, the protein associated to Batten disease: where is it and what does it do? J. Neurosci. Res. 59, 19-23. [PubMed] [Google Scholar]
- Pearce, D.A., Ferea, T., Nosel, S.A., Das, B., and Sherman, F. (1999). Action of BTN1, the yeast orthologue of the gene mutated in Batten disease. Nat. Genet. 22, 55-58. [DOI] [PubMed] [Google Scholar]
- Persaud-Sawin, D.A., VanDongen, A., and Boustany, R.M. (2002). Motifs within the CLN3 protein: modulation of cell growth rates and apoptosis. Hum. Mol. Genet. 11, 2129-2142. [DOI] [PubMed] [Google Scholar]
- Ponnambalam, S., Rabouille, C., Luzio, J.P., Nilsson, T., and Warren, G. (1994). The TGN38 glycoprotein contains two non-overlapping signals that mediate localization to the trans-Golgi network. J. Cell Biol. 125, 253-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond, L., Kuhn, L.A., Teyton, L., Schutze, M.P., Tainer, J.A., Jackson, M.R., and Peterson, P.A. (1995). A role for acidic residues in di-leucine motif-based targeting to the endocytic pathway. J. Biol. Chem. 270, 19989-19997. [DOI] [PubMed] [Google Scholar]
- Puranam, K.L., Guo, W.-X., Qian, W.-H., Nikbakht, K., and Boustany, R.M. (1999). CLN3 defines a novel antiapoptotic pathway operative in neurodegeneration and mediated by ceramide. Mol. Genet. Metab. 66, 294-308. [DOI] [PubMed] [Google Scholar]
- Reaves, B., and Banting, G. (1994). Overexpression of TGN38/41 leads to mislocalisation of γ-adaptin. FEBS Lett. 351, 448-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaves, B.J., Banting, G., and Luzio, J.P. (1998). Lumenal and trans-membrane domains play a role in sorting type I membrane proteins on endocytic pathways. Mol. Biol. Cell 9, 1107-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaves, B.J., Bright, N.A., Mullock, B.M., and Luzio, J.P. (1996). The effect of wortmannin on the localisation of lysosomal type I integral membrane glycoproteins suggests a role for phosphoinositide 3-kinase activity in regulating membrane traffic late in the endocytic pathway. J. Cell Sci. 109, 749-762. [DOI] [PubMed] [Google Scholar]
- Reusch, U., Bernhard, O., Koszinowski, U., and Schu, P. (2002). AP-1A and AP-3A lysosomal sorting functions. Traffic 3, 752-761. [DOI] [PubMed] [Google Scholar]
- Robinson, M.S., and Bonifacino, J.S. (2001). Adaptor-related proteins. Curr. Opin. Cell Biol. 13, 444-453. [DOI] [PubMed] [Google Scholar]
- Rodionov, D.G., Honing, S., Silye, A., Kongsvik, T.L., Von Figura, K., and Bakke, O. (2002). Structural requirements for interactions between leucine-sorting signals and clathrin-associated adaptor protein complex AP3. J. Biol. Chem. 277, 47436-47443. [DOI] [PubMed] [Google Scholar]
- Rous, B.A., Reaves, B.J., Ihrke, G., Briggs, J.A.G., Gray, S.R., Stephens, D.J., Banting, G., and Luzio, J.P. (2002). The role of the adaptor complex AP-3 in targeting wild type and mutated CD63 to lysosomes. Mol. Biol. Cell 13, 1071-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval, I.V., Martinez-Arca, S., Valdueza, J., Palacios, S., and Holman, G.D. (2000). Distinct reading of different structural determinants modulates the dileucine-mediated transport steps of the lysosomal membrane protein LIMPII and the insulin-sensitive glucose transporter GLUT4. J. Biol. Chem. 275, 39874-39885. [DOI] [PubMed] [Google Scholar]
- Santavuori, P. (1988). Neuronal ceroid-lipofuscinoses in childhood. Brain Dev. 10, 80-83. [DOI] [PubMed] [Google Scholar]
- Schell, M.J., Erneux, C., and Irvine, R.F. (2001). Inositol 1, 4, 5-trisphosphate 3-kinase A associates with F-actin and dendritic spines via its N terminus. J. Biol. Chem. 276, 37537-37546. [DOI] [PubMed] [Google Scholar]
- Shiba, T., et al. (2002). Structural basis for recognition of acidic-cluster dileucine sequence by GGA1. Nature 415, 937-941. [DOI] [PubMed] [Google Scholar]
- Sulzer, D., and Holtzman, E. (1989). Acidification and endosome-like compartments in the presynaptic terminals of frog retinal photoreceptors. J. Neurocytol. 18, 529-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Batten Disease Consortium. (1995). Isolation of a novel gene underlying Batten disease, CLN3. Cell 82, 949-957. [DOI] [PubMed] [Google Scholar]
- Tyynelä, J., Sohar, I., Sleat, D.E., Gin, R.M., Donnelly, R.J., Baumann, M., Haltia, M., and Lobel, P. (2000). A mutation in the ovine cathepsin D gene causes a congenital lysosomal storage disease with profound neurodegeneration. EMBO J. 19, 2786-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesa, J., Chin, M.H., Oelgeschlager, K., Isosomppi, J., DellAngelica, E.C., Jalanko, A., and Peltonen, L. (2002). Neuronal ceroid lipofuscinoses are connected at molecular level: interaction of CLN5 protein with CLN2 and CLN3. Mol. Biol. Cell 13, 2410-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne, G. (1992). Membrane protein structure prediction, hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225, 487-494. [DOI] [PubMed] [Google Scholar]
- Voorhees, P., Deignan, E., van Donselaar, E., Humphrey, J., Marks, M.S., Peters, P.J., and Bonifacino, J.S. (1995). An acidic sequence within the cytoplasmic domain of furin functions as a determinant of trans-Golgi network localization and internalization from the cell surface. EMBO J. 14, 4961-4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer, J.M., Kriscenski-Perry, E., Elshatory, Y., and Pearce, D.A. (2002). The neuronal ceroid lipofuscinoses: mutations in different proteins result in similar disease. Neuromol. Med. 1, 111-124. [DOI] [PubMed] [Google Scholar]