Abstract
IL-2 inducible T-cell kinase (Itk) is a Tec family non-receptor tyrosine kinase involved in signaling downstream of the T-cell receptor. Itk contains an amino-terminal Pleckstrin Homology (PH) domain that binds phosphatidylinositol (3,4,5)-trisphosphate, recruiting Itk to the plasma membrane upon T-cell receptor activation. In addition to phosphoinositide binding, accumulating data suggest that the Itk PH domain likely mediates additional interactions outside of the phosphoinositide ligand binding pocket. The structural basis for additional PH domain functions remains elusive because of the poor recombinant expression and in vitro solution behavior of the Itk PH domain. Here, we determine that the lone α-helix in the Itk PH domain is responsible for the poor solution properties and that mutation of just two residues in the Itk α-helix to the corresponding amino acids in Btk or Tec dramatically improves the soluble recombinant expression and solution behavior of the Itk PH domain. We present this double mutant as a valuable tool to characterize the structure and function of the Itk PH domain. It is also interesting to note that the precise sites of mutation identified in this study appear as somatic mutations associated with cancerous tissue. Collectively, the findings suggest that the two helical residues in the Itk PH domain may serve an important and unique structural role in wild-type Itk that differentiates this tyrosine kinase from its related family members.
Keywords: Tec family kinases, IL-2 inducible T-cell kinase (Itk), Pleckstrin Homology domain, protein engineering
Introduction
IL-2 inducible T-cell kinase (Itk) is a member of the Tec family of non-receptor tyrosine kinases that plays a role in T-cell receptor-mediated signaling.1–3 In addition to mediating numerous protein-protein interactions through its non-catalytic domains, Itk catalytic activity is responsible for phosphorylation, and thus activation, of phospholipase C γ1 (PLCγ1).4–6 A recent review coined Itk as the “rheostat” of the T-cell, in that it fine-tunes the strength of the signaling cascade and subsequent immune response.7 This makes Itk an appealing target for modulating the immune response in the context of disease states such as allergy and autoimmunity. Detailed knowledge of the structures and functions of the individual domains of Itk, as well as how the different domains work together to control Itk function, are required steps to advance toward exerting control over this T-cell signaling protein.
Itk consists of five domains; the amino-terminal Pleckstrin Homolgy (PH) domain, followed by the Tec Homology (TH) domain, Src Homology 3 (SH3) domain, Src Homology 2 (SH2) domain, and the catalytic Kinase domain at the carboxy-terminus. We have extensively characterized the structural and functional details of the SH3, SH2, and Kinase domains of Itk7–13 but have not, to date, been able to investigate the Itk PH domain in any detail because of its extremely poor solution behavior. Numerous structures of PH domains from other proteins have been solved, including that of Bruton's Tyrosine Kinase (Btk), the Tec family kinase expressed in B-cells14, 15 and all share a common fold consisting of a single α-helix adjacent to a seven-stranded β-barrel. PH domains generally bind phosphatidylinositols, and despite the conserved fold, have surprisingly low sequence homology.16 In T-cells, the Itk PH domain binds phosphatidylinositol (3,4,5)-trisphosphate (PtdIns(3,4,5)P3 or PIP3) that is transiently produced by the action of phosphoinositide 3-kinase at the plasma membrane following activation of the T-cell receptor.17
In addition to phosphoinositide binding, a subset of PH domains displays a range of additional functions.18 Touhara et al. show that nine different PH domains bind βγ-subunits of G Protein coupled receptors, suggesting that this might be a conserved feature of many PH domains.19 The Btk PH domain has also been shown to bind βγ-subunits,20 and another study suggests that both Btk and Itk can be activated by βγ-subunits of G Proteins.21 The Itk PH domain also exhibits activities beyond PIP3 binding; specifically, kinetic data point to a role for the Itk PH domain in regulating the catalytic activity of the Itk kinase domain,13, 17 perhaps in a manner similar to Protein Kinase B,22, 23 and the Itk PH domain is thought to mediate intermolecular association of multiple molecules of Itk24 forming intermolecular clusters that affect Itk catalytic activity.25 The Itk and Btk PH domains have also been shown to bind filamentous actin.26 Despite the numerous potential physiological roles of the Itk PH domain, efforts to elucidate the structural basis for these observations have been hampered by the fact that recombinant Itk PH domain is very poorly expressed in bacterial systems and exhibits poor solution behavior in vitro.
Here, we compare the expression and solution behavior of the PH domains of three Tec family kinases, Itk, Btk and Tec, and show that the poor recombinant expression and in vitro solution behavior of the Itk PH domain is unique among these Tec family kinases. Using the well-behaved Tec and Btk PH domains as a template, we deliver mutations to the Itk PH domain that remedy the poor solution behavior, without adversely affecting the binding of the Itk PH domain to PIP3. We aim to use this mutant Itk PH domain as a tool to probe the structure and function of the Itk PH domain and shed light on the multiple roles of this domain in controlling Itk function in T-cells.
Results
Wild-type Itk PH domain, unlike Tec and Btk PH, shows poor recombinant expression yields
The PH-TH domain fragments of Itk (residues 1-154), Tec (residues 1-154), and Btk (residues 1-176) were cloned into modified pET-vectors containing N-terminal tags of either His6-MBP or His6-GB1. Itk PH-TH and Btk PH-TH were also cloned into pGEX-4T vectors, containing an N-terminal GST tag [Fig. 1(A)]. Previous work in Btk suggests that the TH domain (also known as the Btk motif) is necessary for a stably folded PH domain,15, 27 and so in this work, we use constructs that include both the PH and TH domain of either Itk, Btk or Tec. Hereafter, the PH-TH region from each kinase is referred to as ‘PH’. Expression levels for all the PH domain constructs were tested in BL21(DE3) cells. After induction with IPTG and overnight expression at 15°C, cells were lysed and pellet (P) and soluble (S) fractions were subjected to gel electrophoresis on 12% SDS-PAGE gels [Fig. 1(B–D)]. For all three solubility tags, GB1, MBP, and GST, ItkPH shows almost no soluble expression, whereas BtkPH, and especially TecPH are readily expressed as soluble protein regardless of which purification tag is used.
Figure 1.
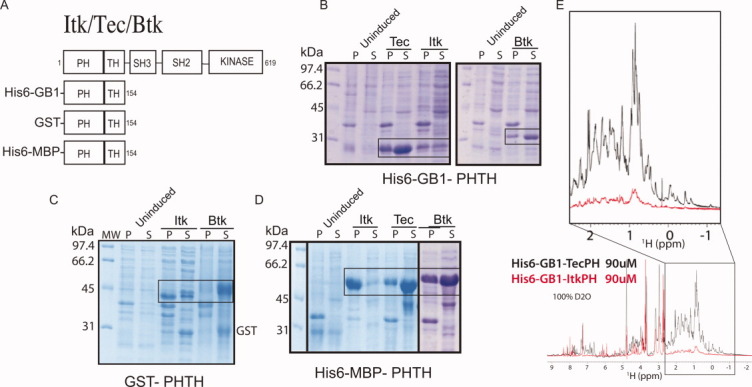
ItkPH shows poor soluble expression compared with TecPH and BtkPH. (A) Domain architecture of constructs used in this study: full-length Itk and three PH-TH domain constructs with N-terminal purification/solubility tags as indicated. Numbering refers to the mouse Itk sequence. (B-D) Itk, Tec, and Btk PH-TH domain constructs with different N-terminal purification/solubility tags were expressed in BL21(DE3) cells, and the pellet (P) and soluble (S) fractions were analyzed by SDS-PAGE; the boxes indicate the bands corresponding to the PH-TH domain on each gel. (E) Superposition of one-dimensional 1H NMR spectra for His6-GB1-TecPH (black spectrum) and His6-GB1-ItkPH (red spectrum) in 100% D2O at identical concentrations (90 μM). The non-exchangeable, aliphatic region is expanded for clarity.
Using a large culture volume (3L), we attempted to purify soluble His6-GB1-ItkPH for preliminary NMR analysis; however, ItkPH precipitates immediately on cleavage from the His6-GB1 tag. Retaining the His6-GB1 tag, we were able to purify and concentrate ItkPH for acquisition of a one-dimensional 1H NMR spectrum [Fig. 1(E)]. Purified His6-GB1-ItkPH (90 μM in 100% D2O) was compared directly with His6-GB1-TecPH under identical conditions. The signal intensity for the TecPH protein is consistent with the sample concentration whereas the Itk spectrum exhibits much lower intensity [Fig. 1(E)]. We hypothesize that the weak signal of ItkPH is due to formation of large soluble aggregates that broaden the NMR signal and the fact that only a fraction of the Itk PH domain remains soluble during the course of the NMR experiment. Indeed, the His6-GB1-ItkPH sample shows visible precipitate in the NMR tube that increases with time, consistent with protein aggregation. The poor expression characteristics of the Itk PH domain combined with the insoluble nature of the purified domain prevent detailed analysis of this domain in vitro.
The α-helix of ItkPH is responsible for its poor recombinant expression behavior
The contrast between the low soluble expression levels and poor in vitro behavior of ItkPH, and the readily over-expressed and well-behaved TecPH led us to use the Tec sequence as a template to pinpoint the region of the Itk PH domain responsible for the poor solubility. Using a chimeric approach, we swapped secondary structure elements of Itk into the Tec sequence at logical secondary-structure junctions [Fig. 2(A,B)]. We expected that systematic replacement of portions of the Tec PH domain sequence with those of Itk would diminish the soluble expression of Tec for at least one of the chimeras. The expression results for this series of constructs should pinpoint the region of Itk responsible for the observed lack of expression. To test this hypothesis, we screened for soluble expression of the Tec/Itk chimeras in BL21(DE3) cells. Chimera “Tec/ItkHα,” in which the α-helix of the Tec PH domain was swapped for the α-helix of Itk, all but eliminated soluble expression [Fig. 2(C)], whereas all other Tec/Itk chimeras yielded significant soluble expression comparable with wild-type TecPH domain [Fig. 2(C)]. On the basis of this result, we next replaced the α-helix of the ItkPH domain with that of Tec (Itk/TecHα) and tested the bacterial expression behavior. The Itk/TecHα construct yielded substantial soluble expression compared with wild-type ItkPH [Fig. 2(C)]. The data therefore show that swapping eight residues within the α-helix region of Itk PH to those of Tec [Fig. 2(D)] rescues the poor expression behavior observed for wild-type Itk.
Figure 2.
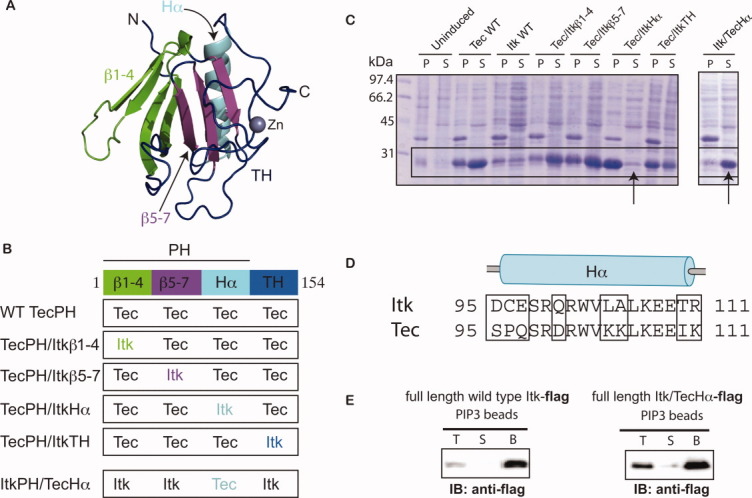
A chimeric approach based on the TecPH sequence reveals that the α-helix of ItkPH is responsible for its poor expression behavior. (A) Threaded structure of ItkPHTH onto the available BtkPHTH structures14, 15 (PDB: 1BTK and 1B55); threading was performed using I-TASSER;28 the ribbon tracing the protein backbone is colored to indicate the four regions used in construction of the Tec/Itk chimeras. β strands 1–4 are green, β strands 5–7 are purple, the α-helix is cyan, and the TH domain (or Btk motif) is dark blue. The structural model of ItkPH was visualized in PyMOL29 and is used in the subsequent figures. (B) Diagram describing the construction of the four Tec/Itk chimeras and the single Itk/Tec chimera. Coloring for each of the regions matches that described in (A). (C) Wild type (WT) and chimeric proteins were expressed in BL21(DE3) cells and analyzed for soluble expression; pellet (P) and soluble (S) fractions were analyzed by SDS-PAGE and the boxes indicate the bands corresponding to the PHTH constructs; the arrows point to the chimeras in which the α-helices of Tec and Itk have been swapped. (D) Sequence alignment of the alpha-helices of ItkPH and TecPH, showing the residues that were swapped in the helix chimeras (amino acids 95–111); boxes highlight the eight amino acid differences between Tec and Itk in this region. (E) Itk/TecHα retains canonical PIP3 binding activity in the context of full-length Itk; FLAG-tagged full-length wild-type Itk and full length Itk/TecHα are captured using PIP3-coated beads, washed, and analyzed by immunoblot (anti-FLAG): lanes correspond to total input (T), supernatant (S), and washed bead (B) samples.
ItkPH/TecHα binds PIP3
We next engineered the ItkPH/TecHα mutation into full-length Itk to examine the effect of this sequence change on PH domain ligand binding. Direct comparisons between wild-type and mutant must be made in the context of the full-length Itk protein because the wild-type Itk PH domain cannot be expressed. Flag-tagged wild-type full-length Itk and full-length Itk/TecHα baculoviral constructs were created and expressed in Hi-Five cells. Purification was achieved as described previously for wild-type full-length Itk using the FLAG-affinity resin.30 The purified full-length proteins (wild-type Itk and Itk/TecHα) were subjected to a pull-down assay using immobilized PIP3 ligand. Both wild-type Itk and Itk/TecHα bind to PIP3 suggesting that mutations within the α-helix of the PH domain do not adversely affect the PIP3 ligand binding pocket [Fig. 2(E)].
Mutation of the amino acids at each end of the α-helix, C96 and T110, is sufficient to rescue ItkPH expression and solubility
Having established that the α-helix region of the Itk PH domain is the origin of the poor expression and solution behavior, we next set out to determine the minimal set of mutations necessary to generate a well-behaved Itk PH domain construct. Individual and pairs of amino acids in the α-helix of Itk PH domain were mutated to their corresponding residues in Tec and screened for soluble expression as before [Fig. 3(A,B)]. The minimal sequence change that permits soluble expression levels of Itk PH involves mutation of residues C96 and T110 to the amino acids of either Tec (C96P/T110I) or Btk (C96E/T110I) [Fig. 3(B, arrows)]. Positions 96 and 110 are located at opposite ends of the α-helix [Fig. 3(C)] suggesting that helix stability may play a role in the observed soluble expression. Moreover, the mutation of T110 to isoleucine likely improves side-chain packing within the PH domain; the modeled structure of ItkPH reveals a significant void surrounding T110, whereas the isoleucine side chain at this position fills the space [Fig. 3(D)]. Regardless of mechanism, mutation of C96 to either Glu (derived from Btk) or Pro (derived from Tec) combined with mutation of T110 to isoleucine results in robust soluble expression of ItkPH [Fig. 3(B)].
Figure 3.
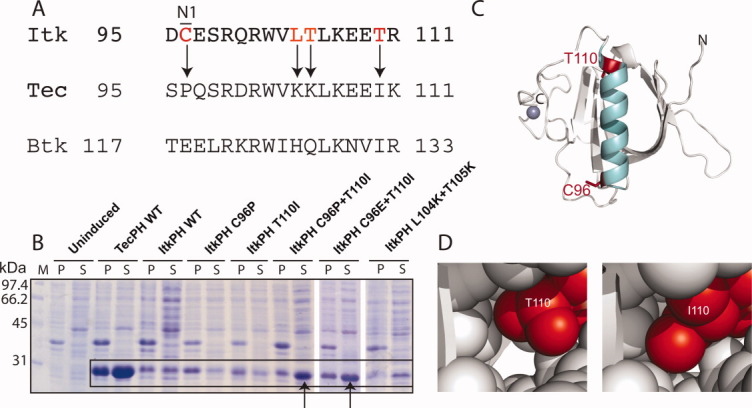
Minimal sequence changes in ItkPH required to produce soluble protein. (A) Sequence alignment of the α-helices of ItkPH, TecPH, and BtkPH, with arrows indicating the most significant amino acid differences that were targeted for mutation. It is interesting to note that position 96 is not conserved between mouse and human Itk sequences; mutation of the mouse sequence to that of human (C96R) did not alter the expression and solubility behavior of ItkPH (data not shown). (B) Soluble expression analysis of single and double helix mutations; arrows point to the C96P/T110I and C96E/T110I mutations that rescue the poor expression of wild type ItkPH. The overall expression level for wild-type ItkPH is quite low when compared with ItkPH C96P/T110I, ItkPH C96E/T110I, and TecPH. This suggests that expression of the wild-type ItkPH sequence leads to protein degradation rather than incorporation into inclusion bodies. (C) Structural model of the Itk PH-TH domain: α-helix shown in cyan, positions C96 and T110 shown in red, N- and C-termini are labeled and the Zn2+ ion coordinated by the TH domain is shown in blue. (D) Comparison of side-chain packing for threonine versus isoleucine at position 110 in the threaded model of ItkPH reveals a significant cavity for T110, whereas I110 packs tightly against adjacent side-chains.
Expression and purification of soluble ItkPH
Having created a well-behaved double mutant of ItkPH, we next set out to express and purify this protein for biophysical characterization. The panel of four PH domains, wild-type ItkPH, ItkPH/TecHα, wild-type TecPH, and ItkPH(C96P/T110I) were each expressed as His6-GB1 fusion proteins in Escherichia coli and purified by nickel affinity chromatography. Next, each protein was subjected to size exclusion chromatography before cleavage of the His6-GB1 tag [Fig. 4(A)]. The elution profile for wild-type ItkPH is consistent with the poor solution behavior and aggregation-prone nature of this protein, eluting over a range of fractions, indicating a heterogenous mixture of apparent molecular weights. In contrast, wild-type TecPH and Itk/TecHα both elute from the column as a single band at fractions consistent with monomeric protein. Interestingly, Itk (C96P/T110I) shows an elution profile that is intermediate to wild-type ItkPH and wild-type TecPH or Itk/TecHα. A significant portion of Itk(C96P/T110I) elutes as a monomer whereas a portion of the sample elutes at earlier fractions in a manner that is similar to wild-type ItkPH.
Figure 4.
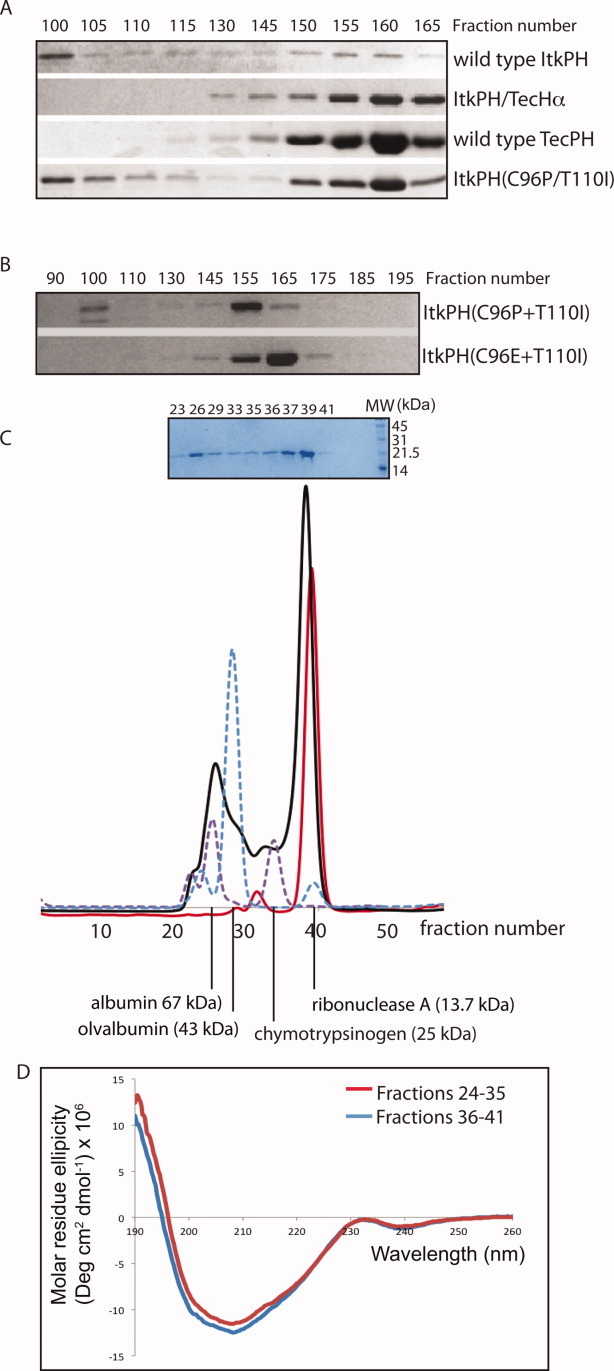
Characterization of purified PH domain proteins. (A) Four different His6-GB1 fusion proteins were purified and run through a Sephadex S300 sizing column; wild type ItkPH and ItkPH(C96P/T110I) show signs of aggregation, eluting in two peaks, whereas ItkPH/TecHα and TecPH WT each elute as a single peak corresponding to the size of the monomeric PH domain. (B) Purified His6-GB1-ItkPH(C96P/T110I) and His6-GB1-ItkPH(C96E/T110I) were run through a Sephadex S100 sizing column. Unlike ItkPH(C96P/T110I), the ItkPH(C96E/T110I) mutant elutes as a single peak. (C) After cleavage of the His6-GB1 tag, purified 13C/15N-labeled ItkPH(C96E/T110I) was passed over a Superdex-75 column on an Aktä FPLC system (solid black line). SDS-PAGE analysis of the resulting fractions is shown above the FPLC trace. Protein standards (dashed purple and blue lines) are shown and indicate the major peak for ItkPH(C96E/T110I) elutes in a manner consistent with its 19 kDa monomeric molecular weight. After completion of NMR data acquisition, the IP4-bound ItkPH(C96E/T110I) was re-injected onto the Superdex-75 column and run under identical conditions (solid red line). The slight shift in the elution profile of IP4 bound ItkPH (compare red with black trace) is consistent with previously reported findings that PH domains exhibit restricted motions on binding to IP4.31 (D) The two peaks (fractions 24–35 and 36–41) from the initial ItkPH(C96E/T110I) Superdex-75 run (black trace in (C)) were analyzed by CD spectroscopy. The two samples show no significant difference in secondary structure.
Because the double mutant ItkPH(C96E/I110I) based on the Btk amino acid sequence [Fig. 3(A)] also produced soluble protein [Fig. 3(B)], we expressed and purified this ItkPH mutant to assess its behavior on a sizing column. Figure 4(B) shows that the His6-GB1-ItkPH(C96E/T110I) protein elutes in a manner consistent with a monomeric species in contrast to the elution prolife of His6-GB1-ItkPH(C96P/T110I). We therefore proceeded with the ItkPH(C96E/T110I) mutant for further characterization.
The His6-GB1-ItkPH(C96E/T110I) construct was next expressed in 3 liters of M9 minimal media containing 15N-NH4Cl as the sole source of nitrogen and purified using nickel affinity (Ni-NTA) chromatography. The His6-GB1 purification tag was cleaved using the engineered Factor Xa site and separated from ItkPH(C96E/T110I) by a second pass through a Ni-NTA column. The ItkPH(C96E/T110I) protein was then subjected to size exclusion chromatography for the final step of purification. Unlike the uniform elution of His6-GB1-ItkPH(C96E/T110I) from the sizing column [Fig. 4(B)], the cleaved ItkPH(C96E/T110I) protein elutes from the sizing column over a range of fractions primarily clustered into two peaks [Fig. 4(C, black trace)]. SDS PAGE indicates that both elutions correspond to the ItkPH(C96E/T110I) protein [see inset, Fig. 4(C)] and circular dichroism (CD) spectra of the pooled fractions for each peak (fractions 24–35 and 36–41) are nearly identical [Fig. 4(D)]. These data suggest that the ItkPH(C96E/T110I) mutant forms higher molecular weight aggregates in solution (approximately 40–70 kDa in size) in a manner that might be similar to wild-type ItkPH.
Unlike wild-type ItkPH, the major ItkPH(C96E/T110I) species in solution corresponds to the molecular weight of the monomer [Fig. 4(C)]. Moreover, we find that we can concentrate the ItkPH(C96E/T110I) fractions corresponding to the monomeric peak and successfully acquire 2D heteronuclear single quantum coherence (HSQC) NMR data [Fig. 5(A)]. The HSQC spectrum of ItkPH(C96E/T110I) is characterized by resonances with well-dispersed chemical shifts indicative of a folded protein domain, and average 1H line-widths of 23 Hz consistent with that of a monomeric protein32 [Fig. 5(A)]. We also concentrated the higher molecular weight species that elutes in fractions 24–35 [Fig. 4(C)] and find that NMR resonances cannot be detected for this sample (not shown). Given the purity of the sample [see inset Fig. 4(C)], the fact that both peaks run identically on SDS PAGE, and the presence of 2 mM DTT in the buffer to disfavor formation of disulfide bonds, we suspect that a portion of ItkPH(C96E/T110I) aggregates to form a higher molecular weight species giving rise to broad NMR lines and that exchange processes further broaden the NMR linewidths beyond detection. The purified ItkPH(C96E/T110I) sample is stable over time without detectable formation of the larger aggregate. Evidence for the stable monomeric species comes from lack of a higher molecular weight peak after reinjection of the purified fractions (36–41) onto the same sizing column [Fig. 4(C, red trace)] and no change in the HSQC data over a period of 3 months (data not shown).
Figure 5.
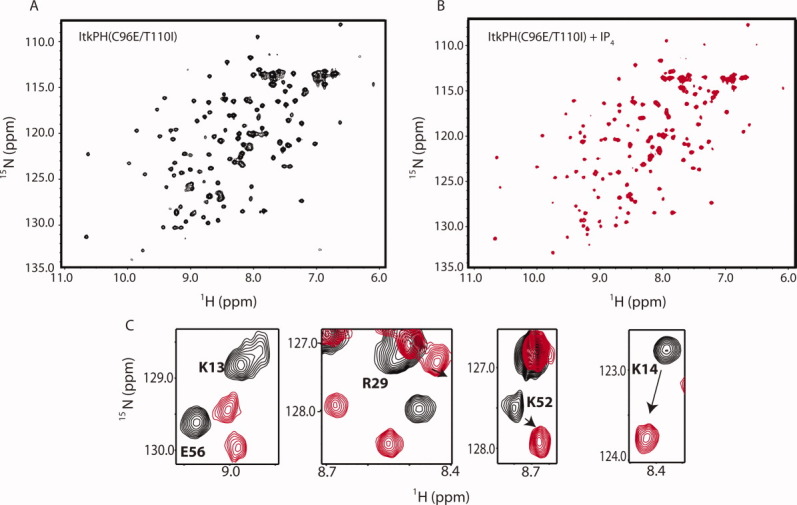
Preliminary NMR characterization of apo- and IP4-bound ItkPH(C96E/T110I). (A) Complete 1H-15N HSQC spectrum of 600 μM ItkPH(C96E/T110I). (B) 1H-15N HSQC spectrum of ItkPH(C96E/T110I) in the presence of 1.2 molar equivalent of IP4. (C) Superposition of HSQC spectra acquired in the absence (black) and presence (red) of IP4. Chemical shift changes of a subset of Itk resonances are shown.
Triple-resonance NMR datasets have been acquired and backbone resonance assignments for ItkPH(C96E/T110I) are underway. HSQC spectra for ItkPH(C96E/T110I) in the absence and presence of inositol 1,3,4,5-tetrakisphosphate (IP4), the soluble head-group of PI(3,4,5)P3, show significant chemical shift perturbations for a subset of ItkPH resonances [Fig. 5(A,B)]. The backbone amide resonances of several of the positively charged amino acids surrounding the ligand binding pocket of the PH domain have been assigned and exhibit chemical shift perturbations consistent with IP4 binding [Fig. 5(C)]. Together, the preliminary NMR data and the improved expression characteristics illustrate that introducing two amino acid sequence changes into the α-helix region of the ItkPH domain yields a significantly better behaved protein that binds the IP4 ligand [Fig. 2(E), Fig. 5] and partially retains the propensity of the wild type Itk PH domain to aggregate in solution [Fig. 4(C)].
Discussion
Two amino acids within the single α-helix of the Itk PH domain are the origin of the low expression and lack of solubility observed for this protein when expressed and purified from bacteria. The extremely poor solution behavior of the wild-type ItkPH prevents production of this protein for detailed structural and biochemical analysis. Indeed, despite evidence supporting a PIP3 binding role for the Itk PH domain in T-cells,17, 33 the failure of ItkPH to bind IP4 in previous in vitro assays34 has led to the suggestion that ItkPH does not bind inositol polyphosphates. We suggest that these previously reported in vitro results were hampered by the poor behavior of ItkPH in solution. In this work, we have identified two Itk PH domain residues, C96 and T110, that when mutated to the corresponding Tec or Btk amino acids, alleviate the poor expression and solution behavior of ItkPH, providing a tool to probe structural and mechanistic features of the Itk PH domain that are relevant to T-cell signaling.
The specific mutations, C96E and T110I, are located at the N- and C-termini of the PH domain α-helix, pointing to a role for this helix in the aggregation prone behavior of the wild-type Itk PH domain. We propose that poor side-chain packing interactions at position 110 might destabilize the PH domain [Fig. 3(D)]. We find, however, that the single point mutation of T110 to isoleucine is not sufficient to produce soluble and stable Itk PH domain [Fig. 3(B)]. The other mutation required to generate a well-behaved Itk PH domain (C96E) lies at the amino-terminal end of the same α-helix (Fig. 3).
Within the ItkPH α-helix, residue 96 occupies the N1 position, defined as the first residue with helical torsion angles.35 Position specific amino acid preferences in helices have been extensively examined over the years.36, 37 Proline and glutamate (the amino acids in the TecPH and BtkPH N1 position, respectively) show the highest propensity for the N1 position of α helices, whereas cysteine and arginine (amino acids in the N1 position of mouse and human ItkPH, respectively) are significantly less favorable in this position. Given the intractable nature of the wild-type ItkPH protein, it is not possible to measure and directly compare the stability of the α-helix in the context of the wild-type Itk PH sequence versus the C96E/T110I mutation. It is nevertheless tempting to speculate that the wild-type residues found at both positions 110 and 96 within ItkPH serve to destabilize the α-helix leading to unfolding and subsequent aggregation that results in the poor behavior observed for this domain in solution. The C96E/T110I mutation rescues this aggregation, presumably by increasing the stability of the α-helix, which likely reduces the population of unfolded species and limits aggregation and precipitation.
The aggregation prone behavior of ItkPH may be an integral part of this domain's function and not simply an artifact of recombinant expression and purification in vitro. Previous work has shown that full-length Itk and the ItkPH fragment form intermolecular clusters both in cells38 and in a co-immunoprecipitation assay from 293T-cells.24 These observations, combined with intermolecular cluster formation via the SH3 and SH2 domains of Itk,8, 39 suggest that the entire regulatory domain region of Itk promotes intermolecular self-association. Disrupting the SH3/SH2 interface results in increased catalytic activity suggesting the self-association may reflect a negative regulatory conformation of Itk.25 The extent and manner by which the Itk PH domain contributes to regulation of Itk catalytic activity (and whether the PH domain mutations described here alter clustering of full-length Itk) remain to be determined. Another regulatory model is that the overall stability of the Itk PH domain fold may be evolutionarily tuned to achieve appropriate levels of PIP3 binding after T-cell receptor activation. The identity of the amino acids at the ends of the ItkPH helix may have evolved to shift the equilibrium away from the well-folded, ligand binding competent conformation of the Itk PH domain thereby regulating the extent to which Itk associates with the membrane during T-cell signaling. Evidence for intrinsic disorder in the PH domain of SWAP-70 has been recently described40 suggesting that conformational flexibility might be a shared mechanism for regulating the function of PH domains more generally. All the questions related to the precise mechanism by which the Itk PH domain contributes to and modulates T-cell signaling can, with the ItkPH(C96E/T110I) mutant described here, be addressed in a more quantitative manner.
In addition to the well-documented role for Itk in immune cell signaling processes, Itk mutations appear in the COSMIC database (Catalogue Of Somatic Mutations In Cancer) that records mutations in select genes sequenced from cancerous tissues.41, 42 Eight of the Itk mutations in the COSMIC database are in the Itk PH domain; and three of the eight mutations are located within the α-helix of the Itk PH domain. Remarkably, two of these mutated sites coincide exactly with the sites identified in this study: R96H and T110M,43 lending further support to the idea that these sites within the ItkPH α-helix are important for the structure and function of this domain. Although a direct link between the Itk mutations listed in the COSMIC database and disease has not been firmly established, our results suggest that specific amino acid changes at positions 96 and 110 within the Itk PH domain can overcome an intrinsically unfolded and unstable domain to stabilize the structure possibly promoting Itk activity.
Using the sequences of the related Tec family kinases, Tec and Btk, as a guide, we have solved a practical problem related to expression and purification of ItkPH. Structural, biochemical, and mechanistic studies can now proceed to elucidate what are likely numerous roles for the PH domain in controlling Itk function during T-cell signaling.
Materials and Methods
Bacterial expression constructs
We modified pET-20b expression vectors to incorporate N-terminal tags of either His6-GB1 or His6-MBP. GB1 and MBP were PCR amplified and subcloned into pET-20b at the NdeI and Xho1 restriction sites; overhangs of the primers were designed to incorporate the His6 tag, the restriction sites, as well as a BamH1 site C-terminal to the purification tag. ItkPHTH (amino acids 1–154), TecPHTH (amino acids 1–154), and BtkPHTH (1–176) were then PCR amplified and subcloned into the BamH1 and Xho1 sites of the modified pET-20b vectors. Additionally, ItkPHTH and BtkPHTH were subcloned into the BamH1 and Xho1 sites of the pGEX-4T expression vector (GE Healthcare). In all these constructs, a Factor Xa cleavage site was engineered between the purification tag and the PHTH sequence by overhang primer design. Wild-type sequences for all Itk, Tec, and Btk constructs used in this article correspond to mouse (mus musculus) cDNA. Chimeric Tec/Itk sequences were constructed by amplifying fragments of Tec and Itk that were then fused by overlap extension PCR and inserted into pET-20b-His6-GB1 at the BamH1 and XhoI restriction sites: TecPH/Itkβ1–4 consists of Itk amino acids 1–58 fused with Tec 58–154; TecPH/Itkβ5–7 consists of Tec 1–57 fused with Itk 59–90 and Tec 91–154; TecPH/ItkHα consists of Tec 1–94 fused with Itk 95–111 and Tec 112–154; TecPH/ItkTH consists of Tec 1–111 fused with Itk 112–154; and ItkPH/TecHα consists of Itk 1–94 fused with Tec 95–111 and Itk 112–154. Point mutants were generated by site-directed mutagenesis (QuikChange II kit, Stratagene). Sequences of all constructs were verified by the Iowa State University DNA Synthesis and Sequencing Facility.
Tests for soluble expression of PHTH constructs
The plasmids described above were transformed into BL21(DE3) cells. Cells were grown in LB media with 100 μg/mL ampicillin at 37°C, 250 rpm until OD600 reached approximately 0.6, at which point temperature was lowered to 15°C and expression was induced with 0.1 mM IPTG. Expression was performed overnight (12–18 hours) at 15°C. One millilitre of cells were centrifuged and cell pellets re-suspended in lysis buffer of 25 mM Tris pH 7.5, 150 mM NaCl, 20 mM imidazole, and lysozyme. Re-suspensions were stored overnight at −80 °C and then thawed at room temperature, lysing the cells, and DNAse and 1 mM phenylmethanesulfonylfluoride were added. Lysates were centrifuged for 10 minutes at 14 K rpm in a microcentrifuge at 4°C. Samples were then taken from the supernatant (S) and pellet (P) fractions, boiled for 5 minutes in SDS loading buffer and analyzed by 12% SDS-PAGE and staining with coomassie brilliant blue.
PI(3,4,5)P3 binding assay of full-length Itk
Full-length Itk/TecHα was constructed using the same internal primers as for the ItkPH/TecHα construct and inserted into the pENTR/D-TOPO vector (Invitrogen) by TOPO cloning, with a C-terminal FLAG tag. Baculovirus was then generated and used to express the protein in insect cells, which was then purified via FLAG-affinity resin as described previously for wild-type full-length Itk.13 For both wild-type and Itk/TecHα, 300 nM of purified full-length enzyme was incubated for 1 hour at 4°C in 150 μl of 20 mM HEPES pH 7.4, 150 mM NaCl, 0.1% Nonidet P-40, with 15 μl of PI(3,4,5)P3 coated agarose beads (Echelon Biosciences #P-B345a). Beads were then washed five times in the same buffer, re-suspended in 2X SDS-loading buffer and boiled for 5 minutes. Samples were run on 8% SDS-PAGE gels and transferred to PVDF membranes by semi-dry western transfer and incubated overnight in primary antibody (1:1000 anti-FLAG). Membranes were then washed three times in TBST, probed with secondary anti-Mouse-HRP, washed an additional three times in TBST and imaged by chemiluminescence.
Purification of soluble PH domains
Protein was expressed, harvested, and lysed as described above (Tests for soluble expression of PHTH constructs). Cleared lysate was passed over an Ni-NTA column equilibrated with lysis buffer (25 mM Tris pH 7.5 at 4°C, 150 mM NaCl, 20 mM Imidazole) washed and eluted with the same buffer containing 40 mM and 200 mM Imidazole, respectively. Protein was then concentrated and loaded onto Sephadex S300 or S100 size-exclusion columns, equilibrated in 25 mM Tris pH 7.5, 150 mM NaCl, 2 mM DTT, and sodium azide.
For NMR studies, His6-GB1-ItkPH(C96E/T110I) was expressed in modified M9 Minimal Media supplemented with 100 μM ZnCl2, 50 μg/mL ampicillin, 15N ammonium chloride (1 g/l, Cambridge Isotope Laboratories) and either 13C glucose (2 g/l, Cambridge Isotope Laboratories) or un-enriched glucose as sole nitrogen and carbon sources. Cells were induced with 0.1 mM IPTG at OD600 = 0.7 and expression was performed for 16 hours for at 17°C. Protein was then purified as described above with the following additions: the His6-GB1 tag was cleaved with Factor Xa for 16 hours at room temperature; cleavage was stopped with 1 mM phenylmethanesulfonylfluoride and once again passed over an Ni-NTA column, concentrated and loaded onto a Superdex-75 column equilibrated in 50 mM potassium phosphate buffer pH 7.0, 150 mM NaCl, 2 mM DTT, and sodium azide; the protein was then concentrated to 600 μM for NMR data acquisition.
CD measurements
CD measurements were performed on a Jasco J-715 spectropolarimeter (Jasco) for the far-UV region of 190–260 nm, at 25°C, as described previously.10
NMR spectroscopy
NMR spectra were collected on a Bruker AVII 700 spectrometer with a 5 mm HCN z-gradient cryoprobe operating at a 1H frequency 700.13 MHz. Backbone chemical shifts of apo-ItkPH(C96E/T110I) are assigned using Sparky44 and MARS45 software programs, from the following pairs of triple-resonance experiments: HNCA and HN(CO)CA, HNCO and HN(CA)CO, and CBCA(CO)NH and CBCANH. Spectra are referenced to DSS, directly in the 1H dimension and indirectly for the 13C and 15N dimensions, according to standard procedures. HSQC spectra were also acquired for ItkPH(96E/T110I) in the presence and absence of 1.2 molar equivalent of inositol 1,3,4,5-tetrakisphophate (IP4, A.G. Scientific). 1D 1H NMR spectra were collected for wild-type His6-GB1-ItkPH and His6-GB1-TecPH that was lyophilized and resuspended in 100% D2O. All spectra were acquired at 298 K. NMRPipe46 and NMRViewJ47 were also used for data processing, visualization, and analysis.
Acknowledgments
GB1 was amplified from the GEV-1 vector, which was kindly provided by Dr. Angela Gronenborn. The MBP sequence was generously provided by Dr. Reuben Peters. Btk and Tec cDNAs were kindly provided by Dr. Hiroyuki Mano. The authors thank Dr. Raji Joseph for valuable discussions and guidance during this work.
References
- 1.Andreotti AH, Schwartzberg PL, Joseph RE, Berg LJ. T-cell signaling regulated by the Tec family kinase, Itk. Cold Spring Harb Perspect Biol. 2010;2:a002287. doi: 10.1101/cshperspect.a002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwartzberg PL, Finkelstein LD, Readinger JA. TEC-family kinases: regulators of T-helper-cell differentiation. Nat Rev Immunol. 2005;5:284–295. doi: 10.1038/nri1591. [DOI] [PubMed] [Google Scholar]
- 3.Berg LJ, Finkelstein LD, Lucas JA, Schwartzberg PL. Tec family kinases in T lymphocyte development and function. Annu Rev Immunol. 2005;23:549–600. doi: 10.1146/annurev.immunol.22.012703.104743. [DOI] [PubMed] [Google Scholar]
- 4.Lee SB, Rhee SG. Significance of PIP2 hydrolysis and regulation of phospholipase C isozymes. Curr Opin Cell Biol. 1995;7:183–189. doi: 10.1016/0955-0674(95)80026-3. [DOI] [PubMed] [Google Scholar]
- 5.Liu KQ, Bunnell SC, Gurniak CB, Berg LJ. T cell receptor-initiated calcium release is uncoupled from capacitative calcium entry in Itk-deficient T cells. J Exp Med. 1998;187:1721–1727. doi: 10.1084/jem.187.10.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perez-Villar JJ, Kanner SB. Regulated association between the tyrosine kinase Emt/Itk/Tsk and phospholipase-C gamma 1 in human T lymphocytes. J Immunol. 1999;163:6435–6441. [PubMed] [Google Scholar]
- 7.Grasis JA, Tsoukas CD. Itk: the rheostat of the T cell response. J Signal Transduct. 2011;2011:297868. doi: 10.1155/2011/297868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brazin KN, Fulton DB, Andreotti AH. A specific intermolecular association between the regulatory domains of a Tec family kinase. J Mol Biol. 2000;302:607–623. doi: 10.1006/jmbi.2000.4091. [DOI] [PubMed] [Google Scholar]
- 9.Breheny PJ, Laederach A, Fulton DB, Andreotti AH. Ligand specificity modulated by prolyl imide bond cis/trans isomerization in the Itk SH2 domain: a quantitative NMR study. J Am Chem Soc. 2003;125:15706–15707. doi: 10.1021/ja0375380. [DOI] [PubMed] [Google Scholar]
- 10.Joseph RE, Severin A, Min L, Fulton DB, Andreotti AH. SH2-dependent autophosphorylation within the Tec Family Kinase Itk. J Mol Biol. 2009;391:164–177. doi: 10.1016/j.jmb.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joseph RE, Xie Q, Andreotti AH. Identification of an allosteric signaling network within Tec family kinases. J Mol Biol. 2010;403:231–242. doi: 10.1016/j.jmb.2010.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joseph RE, Andreotti AH. Conformational snapshots of Tec kinases during signaling. Immunol Rev. 2009;228:74–92. doi: 10.1111/j.1600-065X.2008.00740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joseph RE, Min L, Andreotti AH. The linker between SH2 and kinase domains positively regulates catalysis of the Tec family kinases. Biochemistry. 2007;46:5455–5462. doi: 10.1021/bi602512e. [DOI] [PubMed] [Google Scholar]
- 14.Baraldi E, Djinovic Carugo K, Hyvonen M, Surdo PL, Riley AM, Potter BV, O'Brien R, Ladbury JE, Saraste M. Structure of the PH domain from Bruton's tyrosine kinase in complex with inositol 1,3,4,5-tetrakisphosphate. Structure. 1999;7:449–460. doi: 10.1016/s0969-2126(99)80057-4. [DOI] [PubMed] [Google Scholar]
- 15.Hyvonen M, Saraste M. Structure of the PH domain and Btk motif from Bruton's tyrosine kinase: molecular explanations for X-linked agammaglobulinaemia. EMBO J. 1997;16:3396–3404. doi: 10.1093/emboj/16.12.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemmon MA. Pleckstrin homology(PH) domains and phosphoinositides. Biochem Soc Symp. 2007;74:81–93. doi: 10.1042/BSS0740081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ching KA, Kawakami Y, Kawakami T, Tsoukas CD. Emt/Itk associates with activated TCR complexes: role of the pleckstrin homology domain. J Immunol. 1999;163:6006–6013. [PubMed] [Google Scholar]
- 18.Blomberg N, Baraldi E, Nilges M, Saraste M. The PH superfold: a structural scaffold for multiple functions. Trends Biochem Sci. 1999;24:441–445. doi: 10.1016/s0968-0004(99)01472-3. [DOI] [PubMed] [Google Scholar]
- 19.Touhara K, Inglese J, Pitcher JA, Shaw G, Lefkowitz RJ. Binding of G protein beta gamma-subunits to pleckstrin homology domains. J Biol Chem. 1994;269:10217–10220. [PubMed] [Google Scholar]
- 20.Tsukada S, Simon MI, Witte ON, Katz A. Binding of beta gamma subunits of heterotrimeric G proteins to the PH domain of Bruton tyrosine kinase. Proc Natl Acad Sci USA. 1994;91:11256–11260. doi: 10.1073/pnas.91.23.11256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langhans-Rajasekaran SA, Wan Y, Huang XY. Activation of Tsk and Btk tyrosine kinases by G protein beta gamma subunits. Proc Natl Acad Sci USA. 1995;92:8601–8605. doi: 10.1073/pnas.92.19.8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sable CL, Filippa N, Filloux C, Hemmings BA, Van Obberghen E. Involvement of the pleckstrin homology domain in the insulin-stimulated activation of protein kinase B. J Biol Chem. 1998;273:29600–29606. doi: 10.1074/jbc.273.45.29600. [DOI] [PubMed] [Google Scholar]
- 23.Wu WI, Voegtli WC, Sturgis HL, Dizon FP, Vigers GP, Brandhuber BJ. Crystal structure of human AKT1 with an allosteric inhibitor reveals a new mode of kinase inhibition. PLoS One. 2010;5:e12913. doi: 10.1371/journal.pone.0012913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang YH, Grasis JA, Miller AT, Xu R, Soonthornvacharin S, Andreotti AH, Tsoukas CD, Cooke MP, Sauer K. Positive regulation of Itk PH domain function by soluble IP4. Science. 2007;316:886–889. doi: 10.1126/science.1138684. [DOI] [PubMed] [Google Scholar]
- 25.Min L, Wu W, Joseph RE, Fulton DB, Berg L, Andreotti AH. Disrupting the intermolecular self-association of Itk enhances T cell signaling. J Immunol. 2010;184:4228–4235. doi: 10.4049/jimmunol.0901908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao L, Janmey P, Frigeri LG, Han W, Fujita J, Kawakami Y, Apgar JR, Kawakami T. Pleckstrin homology domains interact with filamentous actin. J Biol Chem. 1999;274:19752–19761. doi: 10.1074/jbc.274.28.19752. [DOI] [PubMed] [Google Scholar]
- 27.Vihinen M, Nore BF, Mattsson PT, Backesjo CM, Nars M, Koutaniemi S, Watanabe C, Lester T, Jones A, Ochs HD, Smith CI. Missense mutations affecting a conserved cysteine pair in the TH domain of Btk. FEBS Lett. 1997;413:205–210. doi: 10.1016/s0014-5793(97)00912-5. [DOI] [PubMed] [Google Scholar]
- 28.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeLano WL. The PyMOL molecular graphics system. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 30.Joseph RE, Min L, Xu R, Musselman ED, Andreotti AH. A remote substrate docking mechanism for the tec family tyrosine kinases. Biochemistry. 2007;46:5595–5603. doi: 10.1021/bi700127c. [DOI] [PubMed] [Google Scholar]
- 31.Gryk MR, Abseher R, Simon B, Nilges M, Oschkinat H. Heteronuclear relaxation study of the PH domain of beta-spectrin: restriction of loop motions upon binding inositol trisphosphate. J Mol Biol. 1998;280:879–896. doi: 10.1006/jmbi.1998.1731. [DOI] [PubMed] [Google Scholar]
- 32.Cavanagh JFWPAGRMSN. Protein NMR principles and practice. Burlington, MA: Elsevier; 2007. [Google Scholar]
- 33.Cruz-Orcutt N, Houtman JC. PI3 kinase function is vital for the function but not formation of LAT-mediated signaling complexes. Mol Immunol. 2009;46:2274–2283. doi: 10.1016/j.molimm.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 34.Kojima TFM, Watanabe Y, Hamazato F, Mikoshiba K. Characterization of the pleckstrin homology domain of Btk as an inositol polyphosphate and phosphoinositide binding domain. Biochem Biophys Res Commun. 1997;236:333–339. doi: 10.1006/bbrc.1997.6947. [DOI] [PubMed] [Google Scholar]
- 35.Presta LG, Rose GD. Helix signals in proteins. Science. 1988;240:1632–1641. doi: 10.1126/science.2837824. [DOI] [PubMed] [Google Scholar]
- 36.Richardson JS, Richardson DC. Amino acid preferences for specific locations at the ends of alpha helices. Science. 1988;240:1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
- 37.Kumar S, Bansal M. Dissecting alpha-helices: position-specific analysis of alpha-helices in globular proteins. Proteins. 1998;31:460–476. doi: 10.1002/(sici)1097-0134(19980601)31:4<460::aid-prot12>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 38.Qi Q, Sahu N, August A. Tec kinase Itk forms membrane clusters specifically in the vicinity of recruiting receptors. J Biol Chem. 2006;281:38529–38534. doi: 10.1074/jbc.M609180200. [DOI] [PubMed] [Google Scholar]
- 39.Severin AJR, Boyken S, Fulton DB, Andreotti AH. Proline isomerization preorganizes the Itk SH2 domain for binding to the Itk SH3 domain. J Mol Biol. 2009;387:726–743. doi: 10.1016/j.jmb.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tokuda N, Kawai K, Lee YH, Ikegami T, Yamaguchi S, Yagisawa H, Fukui Y, Tuzi S. Membrane-induced alteration of the secondary structure in the SWAP-70 pleckstrin homology domain. J Biochem. 2012;151:391–401. doi: 10.1093/jb/mvr146. [DOI] [PubMed] [Google Scholar]
- 41.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, Teague JW, Campbell PJ, Stratton MR, Futreal PA. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, Cole C, Kok CY, Jia M, Ewing R, Menzies A, Teague JW, Stratton MR, Futreal PA. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38:D652–657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, Yue P, Haverty PM, Bourgon R, Zheng J, Moorhead M, Chaudhuri S, Tomsho LP, Peters BA, Pujara K, Cordes S, Davis DP, Carlton VE, Yuan W, Li L, Wang W, Eigenbrot C, Kaminker JS, Eberhard DA, Waring P, Schuster SC, Modrusan Z, Zhang Z, Stokoe D, de Sauvage FJ, Faham M, Seshagiri S. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 44.Goddard TD, Kneller DG. Sparky 3. San Fransisco, CA: University of California; 2008. [Google Scholar]
- 45.Jung YS, Zweckstetter M. Mars—robust automatic backbone assignment of proteins. J Biomol NMR. 2004;30:11–23. doi: 10.1023/B:JNMR.0000042954.99056.ad. [DOI] [PubMed] [Google Scholar]
- 46.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 47.Johnson BA, Blevins RA. NMRView: a computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]


