Abstract
The Cpx two-component system of Gram-negative bacteria senses extracytoplasmic stresses using the histidine kinase CpxA, a membrane-bound sensor, and controls the transcription of the genes involved in stress response by the cytosolic response regulator CpxR, which is activated by the phosphorelay from CpxA. CpxP, a CpxA-associated protein, also plays an important role in the regulation of the Cpx system by inhibiting the autophosphorylation of CpxA. Although the stress signals and physiological roles of the Cpx system have been extensively studied, the lack of structural information has limited the understanding of the detailed mechanism of ligand binding and regulation of CpxA. In this study, we solved the crystal structure of the periplasmic domain of Vibrio parahaemolyticus CpxA (VpCpxA-peri) to a resolution of 2.1 Å and investigated its interaction with CpxP. VpCpxA-peri has a globular Per-ARNT-SIM (PAS) domain and a protruded C-terminal tail, which may be required for ligand sensing and CpxP binding, respectively. The direct interaction of the PAS core of VpCpxA-peri with VpCpxP was not detected by NMR, suggesting that the C-terminal tail or other factors, such as the membrane environment, are necessary for the binding of CpxA to CpxP.
Keywords: Gram-negative bacterium, PAS domain, extracytoplasmic stress response, two-component system, periplasm
Introduction
Gram-negative bacteria have developed extracytoplasmic stress response (ESR) systems to deal with environmental stress.1–7 The Cpx system is one of several bacterial ESR systems operated by a two-component signaling system that is abundantly used for bacteria to sense, respond, and adapt to environmental changes. The two-component system generally consists of a sensory histidine kinase (HK) and a response regulator (RR) as basal elements.8 In the Cpx system, CpxA is the inner membrane-spanning, sensory HK.9 Recognition of a specific envelope stress by the periplasmic domain of CpxA triggers signal transduction to the cytoplasmic domain, which results in the autophosphorylation of a conserved histidine residue (His248 in Escherichia coli CpxA). Then, the phosphate group is transferred to its cognate RR, CpxR, which controls the transcription of the genes involved in the ESR.10
Many factors seemingly unrelated to each other are responsible for the activation of the Cpx pathway in E. coli.1–3, 7 Adhesion to abiotic surfaces triggers the Cpx response via NlpE, an outer membrane lipoprotein that is overexpressed by LacZ accumulation in the periplasm and alleviates the toxicity of periplasmic LacZ in a CpxAR-dependent manner.11, 12 YafY, an inner membrane lipoprotein, is also reported to be responsible for the Cpx response.13 The Cpx pathway can also be activated in an NlpE-independent manner by various factors, such as bacterial growth,14 misfolded or aggregated pili subunits,15 alkaline pH,16 and the presence of heavy metals.17, 18 However, to date, the ligand responsible for the activation of CpxA by direct binding has not been identified. The Cpx signaling system and the Bae two-component system, another ESR system, share spheroplast formation as an envelope stress signal.19 In addition, both the Cpx and Arc two-component systems are commonly activated by antibiotic-mediated oxidative stress.20
The Cpx pathway regulates a variety of genes responsible for the envelope stress response and bacterial virulence. Approximately 100 genes were predicted as members of the cpx regulon based on bioinformatic predictions of CpxR-binding sites.21 More recently, the systemic screening of known CpxR-binding promoters revealed that more than 40 genes are under the control of CpxR.22 Alternatively, the Cpx signaling pathway is regulated by the negative feedback loop in the cells as the Cpx-regulated protein, CpxP, inhibits the Cpx signal via the periplasmic domain of CpxA.23 Tethering CpxP to the inner membrane using an MBP tag constitutively blocks the Cpx signaling, suggesting that inhibition occurs at or near the inner membrane surface where CpxA is localized.16 An in vitro reconstituted system containing recombinant CpxA, CpxP, and CpxR showed that CpxP inhibits the autophosphorylation activity of CpxA in the absence of other periplasmic proteins.24 Recent structural and functional studies demonstrated the direct interaction between CpxP and the periplasmic domain of CpxA.25, 26 Therefore, it is proposed that CpxP represses the Cpx pathway, especially the autophosphorylation of CpxA, by directly interacting with the periplasmic domain of CpxA. The feedback loop of Cpx signaling is completed by the DegP-mediated degradation of CpxP and the activation of CpxA.27 CpxP also plays a role in protein quality control by delivering the misfolded PapE or PapG to DegP.28
Structural studies of CpxA and CpxP were conducted to gain the structural implication of signal recognition of CpxA and its interaction with CpxP. In this study, we elucidated the crystal structure of the periplasmic domain of Vibrio parahaemolyticus CpxA (VpCpxA-peri) and tested its interaction with VpCpxP. V. parahaemolyticus, a Gram-negative halophilic bacterium causing gastroenteritis,29 which belongs to the same class as E. coli, a γ-proteobacteria, harbors the cpxa, cpxr, and cpxp genes in the Cpx two-component system. Although the detailed Cpx pathways of bacteria belonging to the Vibrio genes have not been studied intensively, it is expected that the overall regulation mechanisms of the Cpx pathways must be similar to those in E. coli.
From the crystal structure analysis, we found that the sensory domain of CpxA contains a Per-ARNT-SIM (PAS) fold, which is not predicted from the sequence alignment. On the basis of NMR chemical shift perturbation analysis, we also confirmed that the PAS domain of CpxA does not directly interact with CpxP. Our results will provide a foundation for additional biochemical and genetic studies aimed at dissecting the mechanism of ligand sensing by CpxA and inhibition by CpxP.
Results
The overall structure of VpCpxA-peri
The crystal of the periplasmic domain of V. parahaemolyticus CpxA (VpCpxA-peri) belongs to the R3 space group and diffracted to a 2.1 Å resolution. The structure was solved by the multi-wavelength anomalous dispersion (MAD) method using selenomethionine-derived VpCpxA-peri crystals. There were two molecules in the asymmetric unit, which are referred to as Chain A and Chain B (Fig. 1). The amino acid residues 41–172 were assigned to Chain A, whereas the amino acid residues 42–172 were assigned to Chain B. The Rwork and Rfree of the final model were 22.0% and 26.5%, respectively. Table I summarizes the data collection and refinement statistics. Chain A is used for the structural description in this study unless otherwise indicated.
Figure 1.
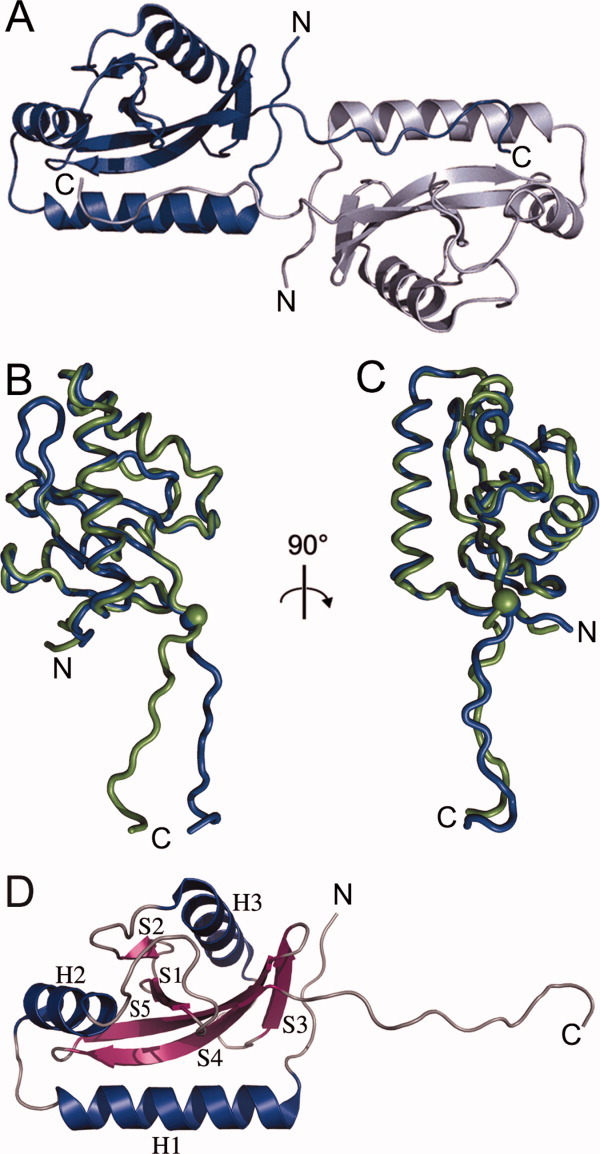
The overall structure of VpCpxA-peri. (A) Ribbon diagrams of Chain A and Chain B in an asymmetric unit are shown in blue and grey, respectively. (B) and (C) The Cα traces of Chain B of VpCpxA-peri were superimposed on Chain B in two different views. Chain A and Chain B are represented in blue and green, respectively. The Cα atom of Asn161 is drawn as a sphere. N and C represent the N- and C-termini of VpCpxA-peri, respectively. (D) The secondary structure elements of Chain A of VpCpxA-peri are indicated. α-helices (H1–H3), β-strands (S1–S5), and loops are colored blue, violet, and grey, respectively. An interactive view is available in the electronic version of the article.
Table I.
Data Collection and Refinement Statistics
| MAD data collection statistics (PAL BL 4A) | |||||
|---|---|---|---|---|---|
| Native | Peak | Edge | Remote 1 | Remote 2 | |
| Space group | R3 | ||||
| Unit cell (Å, °) | a = 148.58, b = 148.58, c = 32.31, α = 90, β = 90, γ = 120 | ||||
| Resolution (Å) | 30.0–2.1 | 30.0–2.3 | |||
| Wavelength (Å) | 0.97943 | 0.97943 | 0.97970 | 0.98751 | 0.97176 |
| Total/unique reflections | 65658/29204 | 67638/11801 | 67729/11812 | 67852/11830 | 67810/11823 |
| Completeness (%) | 94.0 (80.5)a | 100.0 (100.0) | 100.0 (100.0) | 100.0 (100.0) | 100.0 (100.0) |
| I/ó(I) | 18.1 (2.8) | 23.0 (4.0) | 22.2 (3.4) | 22.9 (3.6) | 20.4 (3.0) |
| Rsym (%) | 10.0 (34.4) | 11.5 (36.7) | 10.7 (40.3) | 10.0 (40.2) | 11.4 (48.4) |
| Refinement statistics | |||||
| Resolution (Å) | 28.9–2.1 | ||||
| No. of reflections | 15389 | ||||
| Rwork/Rfree | 22.0/26.5 | ||||
| No. of atoms protein/water | 2191/128 | ||||
| r.m.s. deviations bond lengths (Å)/bond angles (°) | 0.008/1.348 | ||||
| Ramachandran statistics(%) b | 85.0/15.0/0.0 | ||||
The highest resolution shell is shown in parenthesis.
The fraction of residues in the favored, allowed, and disallowed regions of the Ramachandran diagram calculated by PROCHECK.30
The structure of VpCpxA-peri has a globular domain with a protruded C-terminal tail (Fig. 1). The five-stranded β-sheet (S1-S5) surrounded by three α-helices (H1-H3) composes the globular domain [Fig. 1(D)]. Two α-helices (H1 and H2) are placed at the N-terminus, which are followed by two β-strands (S1 and S2). S2 is at the end of the β-sheet and connected to S3 via an α-helix, H3. The C-terminal tail may be more flexible than the globular domain [Fig. 1(B and C)]; when the Cα atoms of Chain A are superimposed on those of Chain B, the root mean square deviation (RMSD) in the presence of C-terminal tail is 2.1 Å, whereas the RMSD in the absence of the C-terminal tail (amino acids 162–172) is 0.8 Å.
VpCpxA-peri possesses a PAS domain
The periplasmic sensory domain of VpCpxA has no significant sequence homology with known proteins, whereas the remaining region is comprised of three conserved domains found in other two-component HKs: the HAMP domain, the ATPase domain, and the phosphoacceptor domain. However, VpCpxA-peri matches many PAS domains in various sensory HKs when the structural homologues were searched against the structure in Protein Data Bank. Five structures with z scores greater than 6.0 were selected for structural comparison to VpCpxA-peri (Table II and Fig. 2): the AbfS arabinofuranosidase two-component sensor protein of Cellvibrio japonicus (PDB ID: 2VA0),31 the methyl-accepting chemotaxis protein of V. parahaemolyticus (PDB ID: 2QHK), the KinD sporulation kinase D of Bacillus subtilis (PDB ID: 3FOS), the LuxQ autoinducer binding proteins of Vibrio cholerae (PDB ID: 3C38) and Vibrio harveyi (PDB ID: 1ZHH).32 Therefore, the fold of the globular domain of VpCpxA-peri can be classified into a classical PAS domain fold, which is found in many kinds of sensory proteins across the three kingdoms, such as serine/threonine kinases, HKs, photo receptors, and circadian clock proteins, and is involved in sensing of signals such as oxygen, redox molecules, light, or chemical compounds.33 Every PAS domain shows a low sequence identity (less than 10%) to the sequence of the VpCpxA PAS domain (Table II), so this PAS domain was not predicted from sequence-based homology search.
Table II.
The Structural Homologues of VpCpxA-Peri
| PDB ID (chain) | Name | Z score | RMSD | Amino acids | Sequence identity (%) |
|---|---|---|---|---|---|
| 2VA0 (B) | AbfS | 7.0 | 4.3 | 97a/119b | 10 |
| 2QHK (A) | MCP | 6.6 | 2.8 | 99/148 | 6 |
| 3C38 (A) | LuxQ | 6.3 | 4.3 | 109/217 | 10 |
| 3FOS (B) | KinD | 6.3 | 3.7 | 108/202 | 8 |
| 1ZHH (B) | LuxQ | 6.3 | 4.6 | 107/208 | 7 |
Number of amino acid residues that are used for the RMSD calculation.
Number of amino acid residues present in the homologous structures.
Figure 2.
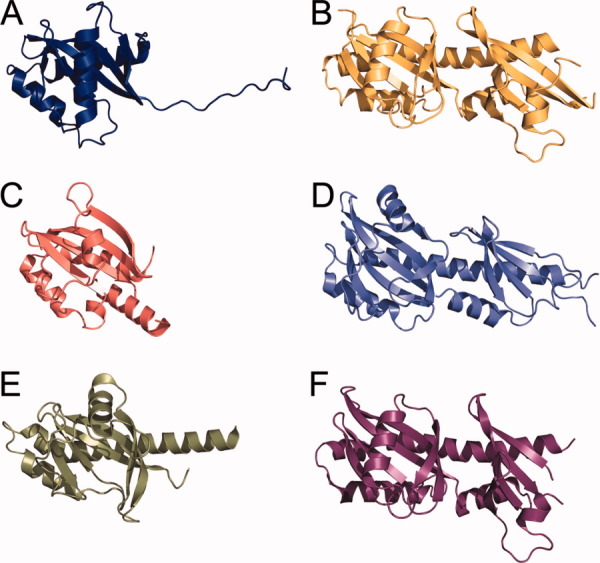
Structural similarity between the periplasmic domain of VpCpxA-peri and other HKs containing PAS domains in their periplasmic regions. VpCpxA-peri (A) is compared with the LuxQ autoinducer binding protein of V. cholerae (B) (3C38), AbfS arabinofuranosidase two-component sensor protein of Cellvibrio japonicus (C) (2VA0), KinD sporulation kinase D of Bacillus subtilis (D) (3FOS), methyl-accepting chemotaxis protein of V. parahaemolyticus (E) (2QHK), and LuxQ autoinducer 2-binding periplasmic protein of Vibrio harveyi (F) (1ZHH) in ribbon diagrams. The orientations of (B–F) were determined by overlapping (B–F) with (A). N- and C-termini were indicated by N and C, respectively.
The central β-pleated sheet consisting of five antiparallel β-strands which is the most conserved structural feature in the PAS domain is also found in VpCpxA-peri. In the classical PAS domain, the second and third β-strands are connected by a long loop or additional helices. In the crystal structure of VpCpxA-peri, the helix H3 is found between two strands, S2 and S3. Although the PAS domains share the overall fold, they must harbor structural differences to accommodate various ligands.34 Accordingly, the conformational diversity is observed in the N-terminus, the C-terminus, and the loop or helix between the second and third strands (Fig. 2). In addition, the structural elements and their arrangement are also quite different in each PAS domain (Fig. 2). The structure of AbfS is most similar to the VpCpxA-peri structure, except for the additional small helix between H2 and S1 and the orientation of H1 [Fig. 2(C)]. The methyl-accepting chemotaxis protein has an additional α-helix and the long H1 [Fig. 2(E)]. KinD and LuxQ also have a long H1 helix that extends to the second β-sheet and an additional PAS domain [Fig. 2(B, D, F)].
Surface charge distribution
The periplasmic domain of VpCpxA has 14 arginine and 8 lysine residues out of a total of 136 residues, and thus it can be categorized as a basic polypeptide because its Arg+Lys frequency (16.2%) is higher than the average value (11.4%). When the electrostatic surface potential of the VpCpxA-peri monomer was visualized (Fig. 3), most of the basic residues are distributed over four regions: a basic juxtamembrane region, a basic bulge, a basic side, and a basic hole [Fig. 3(A–C)]. Arg42, Lys43, and Arg45 in the N-terminal loop, Arg114 on H3, Arg136 on S4, and Lys159 on S5 constitute the basic juxtamembrane region [Fig. 3(D)]. The basic bulge consists of residues on S1 and loops nearby: Arg83, Lys86, and Arg90 on LH2-S1, Arg92 on S1, and Lys107 and Arg108 on LS2-H3 [Fig. 3(E)]. The basic side is distributed over H1 (Arg54 and Lys65), LH1-H2 (Lys68), H2 (Arg74 and Arg78), and LH2-S1 (Arg90) [Fig. 3(G)]. In addition to these basic surfaces, Lys130 and Arg132 on S3 form a basic hole on a rather hydrophobic surface [Fig. 3(C and F)]. Arg59 and Lys128 do not belong to these basic regions, whereas Arg169 is placed at the C-terminus. VpCpxA-peri also has two acidic regions [Fig. 3(A and B)]. Seven acidic residues, Glu69, Asp71, Asp97, Asp125, Glu124, Asp149, and Glu151, form a large acidic region [Fig. 3(H)]. Four acidic residues on H1 constitute the other small acidic region: Asp49, Glu50, Glu57, and Asp60 [Fig. 3(I)].
Figure 3.
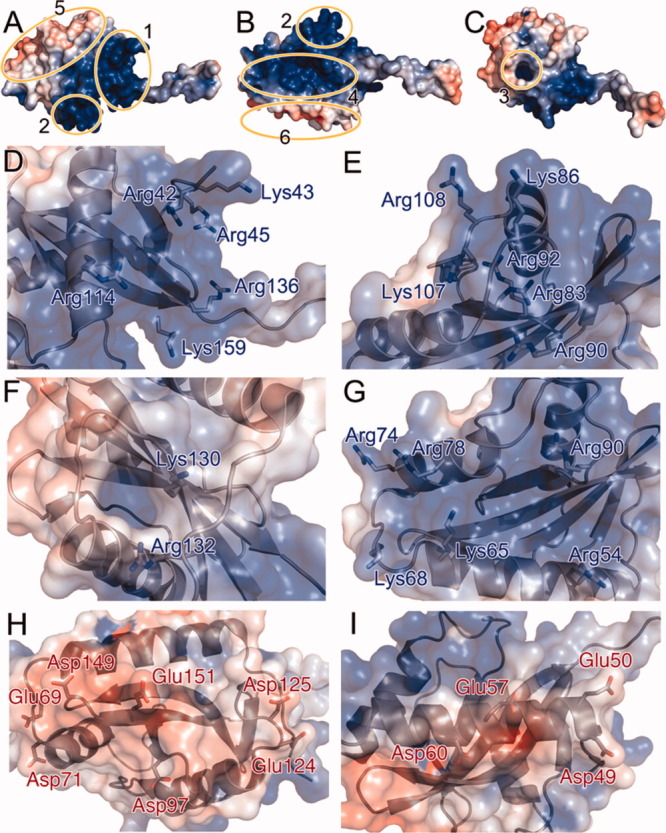
The surface charge distribution of VpCpxA-peri. (A-C) The charge of VpCpxA-peri is calculated within a range of −5 kT (red) to +5 kT (blue) and displayed on the surface diagram of VpCpxA-peri. The basic juxtamembrane region, the basic bulge, the basic hole, the basic side, the large acidic surface, and the small acidic surface are depicted with numbers 1–6, respectively. The side chains of residues constituting each basic or acidic region are drawn as a stick diagram: (D) the basic juxtamembrane region, (E) the basic bulge, (F) the basic hole, (G) the basic side, (H) the large acidic surface, and (I) the small acidic surface.
The interaction between the periplasmic domains of VpCpxA and VpCpxP
It has been suggested that CpxP represses the Cpx signal by direct association with the periplasmic domain of CpxA in E. coli.25, 26 To confirm the binding between VpCpxA and VpCPxP and investigate the residues involved in the CpxP binding, an NMR chemical shift perturbation assay was performed by titrating VpCpxA-peri with VpCpxP. Because VpCpxA-peri was not suitable for the NMR experiment due to poor spectral resolution of the 2D 15N HSQC spectrum, VpCpxA-peri lacking the C-terminal 11 residues, VpCpxA-ΔCT (amino acid residues 38–162), was introduced. The 15N HSQC spectra of VpCpxA-ΔCT were measured in the absence or presence of a five-fold molar excess of VpCpxP, and the two spectra were overlapped for comparison (Fig. 4). The peaks in both spectra are identical, which means VpCpxP does not interact with the VpCpxA periplasmic domain at least in the experimental conditions we used.
Figure 4.
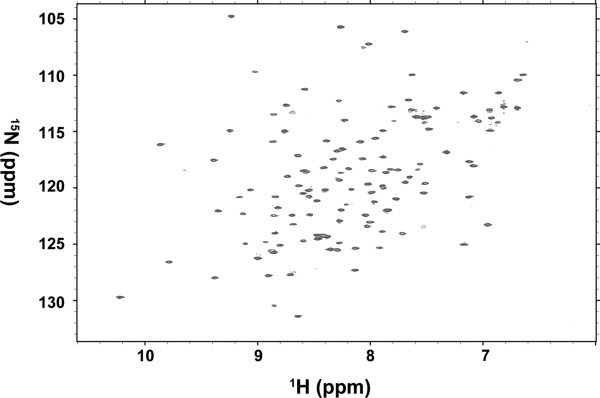
The 1H-15N HSQC spectra of 15N-labeled VpCpxA-ΔCT in the presence and absence of VpCpxP. The 1H-15N HSQC spectrum of 15N-labeled VpCpxA-ΔCT in the presence of a five-fold molar excess of VpCpxP is superimposed with the spectrum of VpCpxA-ΔCT alone, but they are indistinguishable.
Discussion
The sequence identities of the full-length CpxA proteins belonging to γ-proteobacteria vary from 23% to 44%, whereas those of the periplasmic domains of CpxA proteins are in the range of 8%–17% except the sequence identity between two Vibrio CpxA proteins (Table III). Compared with the periplasmic domain, the remaining regions, two transmembrane helices and the cytoplasmic domains, show higher sequence homology, and particularly, all the essential motifs found in HK are well conserved in the cytoplasmic domain. Thereby, it is expected that the Cpx signal is transduced in the similar way as other known two-component system in spite of low sequence identities among the CpxA proteins belonging to the different genera. Although the periplasmic domains of the CpxA proteins are not homologous in their sequences, they seem to have similar three-dimensional topology because the predicted secondary structures are well matched with those of VpCpxA-peri (Fig. 5). This may reflect that the inducing signal recognized by the sensing domain of CpxA is diverse in each species, whereas the signaling mechanisms through the PAS, transmembrane, and cytoplasmic domain are similar.
Table III.
The Sequence Identities Among CpxA Proteins
| CpxA of γ-proteobacteriaa | ||||||
|---|---|---|---|---|---|---|
| Vp | So | Lp | Ps | Vc | ||
| CpxA of γ-proteobacteria | Ec | 44b (13c) | 35 (16) | 27 (11) | 25 (8) | 44 (15) |
| Vp | — | 33 (17) | 25 (9) | 23 (7) | 68 (48) | |
| So | — | — | 32 (10) | 27 (16) | 33 (13) | |
| Lp | — | — | — | 25 (8) | 25 (10) | |
| Ps | — | — | — | — | 24 (8) | |
Ec, Escherichia coli str. K-12 sub. W3110 (geneID: 7438256); Vp, V. parahaemolyticus RIMD 2210633 (geneID: 1190422); So, Shewanella oneidensis MR-1 (geneID: 1172073); Lp, Legionella pneumophila str. Lens (geneID: 3114153); Ps, Pseudomonas syringae pv. tomato str. DC3000 (geneID: 1183444); Vc, V. cholerae O1 biovar El Tor str. N16961 (geneID: 2615521).
Sequence identity among the CpxA proteins (%).
Sequence identity of the periplasmic domain of CpxA proteins (%). Residues corresponding to 38–173 of VpCpxA-peri in Fig. 5 were used for the calculation of the sequence identity.
Figure 5.
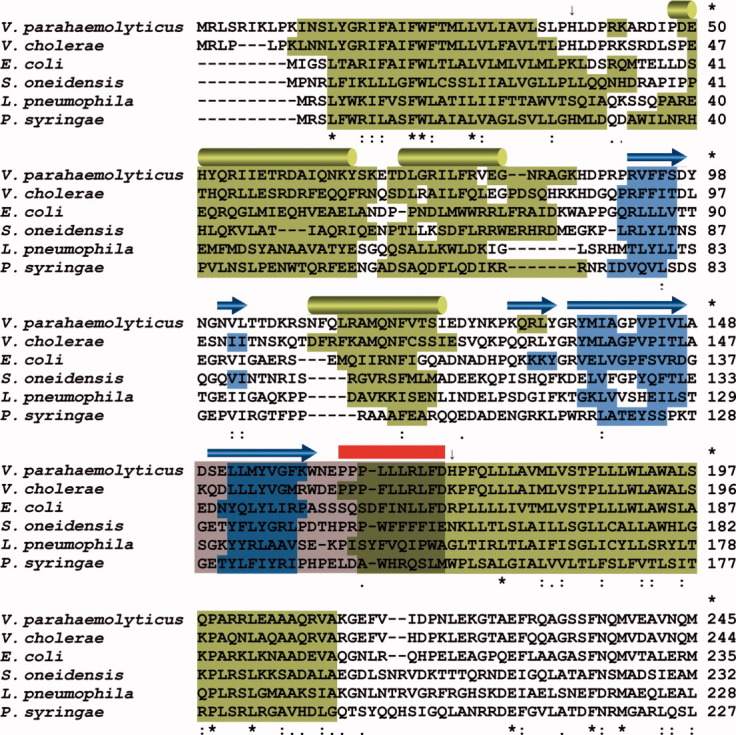
Multiple sequence alignment of CpxA proteins of γ-proteobacteria. Multiple sequence alignment of the CpxA proteins of V. parahaemolyticus, V. cholerae, E. coli, Shevanella oneidensis, Legionella pneumophila, and Pseudomonas syringae. Predicted α-helices and β-strands are highlighted on the sequence of each species in green and blue, respectively. α-helices (H1–H3) and β-strands (S1–S5) in Chain A of VpCpxA-peri are displayed in the first row as green cylinders and blue arrows, respectively. The C-terminal tail of VpCpxA-peri deleted for the NMR experiment is indicated by a red line, and the region predicted to play a role in E. coli CpxP binding is represented by a grey box.25 The N- and the C-terminal residues of VpCpxA-peri are indicated by arrows.
Some stimuli of the E. coli Cpx system are shared with those of the Vibrio Cpx system such as copper,35 but not all them. Based on the stimulation of the Vibrio Cpx pathway by chloride and magnesium ions, it was proposed that the Vibrio Cpx pathway have a role in responding to salinity.35V. parahaemolyticus and V. cholerae belong to the same genus and share a similar habitat. The high sequence homology between the CpxA periplasmic domains of these two species implies that stimuli for VpCpxA are similar to triggers for V. cholerae CpxA.
When the predicted secondary structure of VpCpxA-peri is compared with that of the crystal structure, the α-helices are in good agreement with each other, except the C-terminus that is connected to the second transmembrane helix (Fig. 5). This α-helix is also predicted to be present in the same position in other CpxA proteins despite the low sequence identities in the periplasmic domain (Fig. 5 and Table III). In addition, residues 169–173 of Chain A form a 310 helix. Based on these results, it is tempting to propose that the C-terminal end of the periplasmic domain forms a helix that continues to the second transmembrane helix but is unfolded due to interruption of the helix in the crystal structure.
The PAS domain of VpCpxA-peri has low sequence homology to other PAS domains (Table II). In addition, each PAS domain uses a different region to recognize its cognate ligand. Neither the ligand nor a ligand-binding site was observed in the crystal structure of the periplasmic domain of VpCpxA, and thus it is difficult to identify the ligand-binding region in the structure of VpCpxA-peri. However, assuming that VpCpxA also recognizes Cl− ions like V. cholera CpxA, one of the basic surfaces and especially the highly basic side between H1 and H2 may be a good candidate for the ligand recognition site [Fig. 3(B)]. If the basic side is accommodated by negatively charged Cl− ions, then the flat surface can be used for dimerization, which is a requirement for the autophosphorylation of HK in the two-component system. This possibility should be addressed in a further study.
Recently, two groups independently reported the crystal structure of E. coli CpxP.25, 26 It shows the same fold as its homologue, E. coli Spy, which contains a basic concave surface.36 Zhou et al. suggested that E. coli CpxA likely binds CpxP via its C-terminus (amino acid residues 138–162) that corresponds to S5 of the PAS domain (residues 149–160) and the protruded C-terminal tail (residues 161–172) of VpCpxA-peri (Fig. 5).25 Assuming that the E. coli CpxA has the same structure as VpCpxA-peri, it is unlikely that the residues in S5 are involved in CpxP binding because they are buried in the PAS domain and form a secondary structure with other strands (Figs. 1 and 5). Therefore, the reported interaction between the E. coli CpxP and CpxA are not likely biologically relevant because the synthesized peptide may not reflect the structural property of the native protein. Alternatively, current results can be interpreted as that the binding between CpxP and CpxA is mediated not by the S5 region but by the C-terminal end of the periplasmic domain of CpxA (residues 163–172) because the CpxP binding peptide identified by Zhou et al. harbors the C-terminal end25 and the binding of VpCpxP to VpCpxA-ΔCT was not detected via NMR. Another possible scenario is that the lack of a helical structure on the C-terminal end of VpCpxP hinders binding to CpxP or the membrane lipids may mediate the binding between CpxP and CpxA because the proposed binding site is near the membrane. From the binding assay between E. coli CpxP and CpxA, it was proposed that their binding was mediated by the charge-charge interaction involving the positively charged patch of CpxA (Arg56, Arg60, and Arg67).25 These residues are well conserved as Arg67, Lys73, and Arg80 in VpCpxP, respectively. Consistently, the negatively charged residues of E. coli CpxA in the proposed CpxP binding site are also found in VpCpxA (Fig. 5). Therefore, the CpxA-CpxP interaction may be similar in E. coli and V. parahaemolyticus.
VpCpxA-peri exists as a dimer in an asymmetric unit [Fig. 1(A)]; however, its biological relevance is not confirmed. In the crystal structure, the C-terminal tail of one subunit is protruded to and grabbed by the other subunit [Fig. 1(A)]. Both the N-terminus and C-terminus of VpCpxA-peri have to be connected to transmembrane helices, and hence the dimer structure seems to be improper as a membrane-spanning protein. These results suggest that the dimer formation is likely caused by the crystal packing or the intervention of the native structure in the recombinant protein. Many PAS domains of the sensory HKs, such as PhoQ,37 LuxQ,32 and FixL,34 form a dimer with diverse dimeric interfaces. Furthermore, PAS domains can exist as several dimeric forms in the case of FixL from Bradyrhizobium japonicum34 and Sinorhizobium meliloti.38 Although there is no report that the CpxA periplasmic domain exists as a dimer, VpCpxA-peri is expected to form a dimer by ligand binding or the intact VpCpxA can exist as a dimer. However, predicting the dimeric interface of VpCpxA-peri from this study is not plausible because of its flexibility.
In summary, we solved the crystal structure of the periplasmic domain of V. parahaemolyticus CpxA. Although not predicted based on sequence, VpCpxA-peri contains a PAS domain that could play a role in sensing envelope stress signals. However, no direct clue necessary for identifying the ligand of CpxA was provided. We also examined the interaction between the periplasmic domains of CpxA and CpxP of V. parahaemolyticus using NMR spectroscopy, but their binding was not observed under the current experimental conditions. Our work that defined the structure of the sensor domain of CpxA may help future biochemical and genetic studies in the identification of natural ligands and the elucidation of its signaling and regulation mechanisms.
Material and Methods
Cloning and purification
The N-terminal signal sequence of CpxP and the transmembrane regions of CpxA were predicted using SignalP39 and TMHMM,40 respectively. The mature form of CpxP and the periplasmic domain of CpxA of V. parahaemolyticus were designed based on the following domain boundaries: VpCpxP amino acid residues 15–170, VpCpxA-peri amino acid residues 38–173, and VpCpxAΔCT amino acid residues 38–162. Each gene was amplified by polymerase chain reaction (PCR) from genomic DNA and cloned into the modified pET-28a(+) or pDUET vector that has a 6xHis-tag and tobacco etch virus (TEV) protease cleavage site. Every protein was expressed using Escherichia coli BL21(DE3), except selenomethionine-labeled VpCpxA-peri, which was expressed using a Escherichia coli B834(DE3) cell. A single colony of transformed cells was inoculated into 50 ml lysogeny broth (LB) with 50 μg/ml ampicilin or kanamycin at 37°C overnight. Cultured cells were transferred into 500 ml LB for the unlabeled protein, 600 ml M9 minimal medium with 1 g/L of 15NH4Cl for 15N-labeled protein or 600 ml M9 minimal medium with 50 mg/L of selenomethionine. Cells were cultured at 37°C to an optical density 0.5–0.6 at 600 nm. The recombinant proteins were expressed at 18°C in the presence 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 12 hours.
The cells were harvested by centrifugation at 3500 g at 4°C for 15 min, resuspended in buffer A (20 mM Tris-HCl pH 8.5) and disturbed by sonication. Remaining cells and aggregates were pelleted by centrifugation at 16,000 g at 4°C for 1 hour. Each protein was purified by the same method: the protein was loaded into a HisTrap column (GE Healthcare), washed with 40 mM imidazole, and subjected to a 40–500 mM imidazole gradient using FPLC (GE Healthcare). After the eluted protein was pooled, the sample was dialyzed to buffer A and treated with TEV protease to cut the N-terminal 6xHis-tag. After the cleaved tag was removed by the HisTrap column, the sample was purified by size-exclusion chromatography with a Superdex75 column (GE Healthcare). The purified VpCpxA-peri was concentrated to 10 mg/ml for crystallization purpose. The purity of VpCpxA-peri checked by SDS-gel electrophoresis was more than 95%.
Structure determination and analysis
The crystallization of selenomethinone-labeled VpCpxA-peri was performed using the microbatch method at 22°C. One microliter of protein solution at a concentration of 10 mg/ml was mixed with an equal volume of crystallization reagent containing 0.1 M Tris-HCl pH 7.0, 18–24% PEG 3000, 0.2 M Ca(OAc)2, 3% (v/v) 1,6-hexanediol, and 2 mM Tris (2-carboxyethyl) phosphine under a layer of Al's oil (Hampton). The crystals were flash-frozen in a cold nitrogen stream. The X-ray diffraction data were collected at a resolution of 2.3 Å with an ADSC Quantum 210 CCD detector at beam line 4A, Pohang Light Source, South Korea. The data was processed and integrated using HKL2000 and scaled using SCALEPACK.41 The VpCpxA-peri structure was determined by MAD using SOLVE/RESOLVE.42, 43 The model building was performed by Coot.44 Several cycles of rigid-body refinement, positional refinement, restrained refinement, simulated annealing, B-factor refinement, and model rebuilding were performed at 2.3 Å resolution using Phenix,45 Refmac,46 CNS,47 and Coot44 programs.
The quality of the structure was analyzed by PROCHECK.30 The RMSD values of the overlapped structure were calculated using LSQKAB.48 A DaliLite v. 3 server49 was used for searching the structural homologues in the Protein Data Bank.50 Figures were drawn using PyMOL,51 and the surface charge was calculated and visualized by PDB2PQR52 and APBS,53 respectively. The domain structure was searched using the Pfam database.54 The multiple sequence alignment was implicated using ClustalW2.55
Interaction test
15N HSQC spectra were recorded at 20°C on a Bruker Avance 800 MHz spectrometer outfitted with cryogenic probes, and the data were processed with nmrPipe.56 Isotopically labeled VpCpxAΔCT was made from M9 minimal media containing 15N NH4Cl as the sole nitrogen source. The sample buffer for NMR was 50 mM HEPES pH 7.5, 0.2 M NaCl, 5% glycerol, 50 mM glutamate, and 50 mM arginine. HSQC spectra were recorded on VpCpxAΔCT at a final concentration of 60 μM in the absence or presence of VpCpxP.
Acknowledgments
The authors thank laboratory members John and Carol Gross for helpful discussions and the staff of the PAL BL 4A beamline for their assistance during data collection.
References
- 1.Dorel C, Lejeune P, Rodrigue A. The Cpx system of Escherichia coli, a strategic signaling pathway for confronting adverse conditions and for settling biofilm communities? Res Microbiol. 2006;157:306–314. doi: 10.1016/j.resmic.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Raivio TL. Envelope stress responses and Gram-negative bacterial pathogenesis. Mol Microbiol. 2005;56:1119–1128. doi: 10.1111/j.1365-2958.2005.04625.x. [DOI] [PubMed] [Google Scholar]
- 3.Raivio TL, Silhavy TJ. The sigmaE and Cpx regulatory pathways: overlapping but distinct envelope stress responses. Curr Opin Microbiol. 1999;2:159–165. doi: 10.1016/S1369-5274(99)80028-9. [DOI] [PubMed] [Google Scholar]
- 4.Raivio TL, Silhavy TJ. Periplasmic stress and ECF sigma factors. Annu Rev Microbiol. 2001;55:591–624. doi: 10.1146/annurev.micro.55.1.591. [DOI] [PubMed] [Google Scholar]
- 5.Rowley G, Spector M, Kormanec J, Roberts M. Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nat Rev Microbiol. 2006;4:383–394. doi: 10.1038/nrmicro1394. [DOI] [PubMed] [Google Scholar]
- 6.Ruiz N, Silhavy TJ. Sensing external stress: watchdogs of the Escherichia coli cell envelope. Curr Opin Microbiol. 2005;8:122–126. doi: 10.1016/j.mib.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Wick LM, Egli T. Molecular components of physiological stress responses in Escherichia coli. Adv Biochem Eng Biotechnol. 2004;89:1–45. doi: 10.1007/b93957. [DOI] [PubMed] [Google Scholar]
- 8.Chang C, Stewart RC. The two-component system. Regulation of diverse signaling pathways in prokaryotes and eukaryotes. Plant Physiol. 1998;117:723–731. doi: 10.1104/pp.117.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber RF, Silverman PM. The cpx proteins of Escherichia coli K12. Structure of the cpxA polypeptide as an inner membrane component. J Mol Biol. 1988;203:467–478. doi: 10.1016/0022-2836(88)90013-7. [DOI] [PubMed] [Google Scholar]
- 10.Dong J, Iuchi S, Kwan HS, Lu Z, Lin EC. The deduced amino-acid sequence of the cloned cpxR gene suggests the protein is the cognate regulator for the membrane sensor, CpxA, in a two-component signal transduction system of Escherichia coli. Gene. 1993;136:227–230. doi: 10.1016/0378-1119(93)90469-j. [DOI] [PubMed] [Google Scholar]
- 11.Snyder WB, Davis LJ, Danese PN, Cosma CL, Silhavy TJ. Overproduction of NlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. J Bacteriol. 1995;177:4216–4223. doi: 10.1128/jb.177.15.4216-4223.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otto K, Silhavy TJ. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc Natl Acad Sci USA. 2002;99:2287–2292. doi: 10.1073/pnas.042521699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyadai H, Tanaka-Masuda K, Matsuyama S, Tokuda H. Effects of lipoprotein overproduction on the induction of DegP (HtrA) involved in quality control in the Escherichia coli periplasm. J Biol Chem. 2004;279:39807–39813. doi: 10.1074/jbc.M406390200. [DOI] [PubMed] [Google Scholar]
- 14.DiGiuseppe PA, Silhavy TJ. Signal detection and target gene induction by the CpxRA two-component system. J Bacteriol. 2003;185:2432–2440. doi: 10.1128/JB.185.8.2432-2440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee YM, DiGiuseppe PA, Silhavy TJ, Hultgren SJ. P pilus assembly motif necessary for activation of the CpxRA pathway by PapE in Escherichia coli. J Bacterial. 2004;186:4326–4337. doi: 10.1128/JB.186.13.4326-4337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danese PN, Silhavy TJ. CpxP, a stress-combative member of the Cpx regulon. J Bacteriol. 1998;180:831–839. doi: 10.1128/jb.180.4.831-839.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kershaw CJ, Brown NL, Constantinidou C, Patel MD, Hobman JL. The expression profile of Escherichia coli K-12 in response to minimal, optimal and excess copper concentrations. Microbiology. 2005;151:1187–1198. doi: 10.1099/mic.0.27650-0. [DOI] [PubMed] [Google Scholar]
- 18.Yamamoto K, Ishihama A. Transcriptional response of Escherichia coli to external copper. Mol Microbiol. 2005;56:215–227. doi: 10.1111/j.1365-2958.2005.04532.x. [DOI] [PubMed] [Google Scholar]
- 19.Raffa RG, Raivio TL. A third envelope stress signal transduction pathway in Escherichia coli. Mol Microbiol. 2002;45:1599–1611. doi: 10.1046/j.1365-2958.2002.03112.x. [DOI] [PubMed] [Google Scholar]
- 20.Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell. 2008;135:679–690. doi: 10.1016/j.cell.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Wulf P, McGuire AM, Liu X, Lin EC. Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli. J Biol Chem. 2002;277:26652–26661. doi: 10.1074/jbc.M203487200. [DOI] [PubMed] [Google Scholar]
- 22.Price NL, Raivio TL. Characterization of the Cpx regulon in Escherichia coli strain MC4100. J Bacteriol. 2009;191:1798–1815. doi: 10.1128/JB.00798-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raivio TL, Popkin DL, Silhavy TJ. The Cpx envelope stress response is controlled by amplification and feedback inhibition. J Bacteriol. 1999;181:5263–5272. doi: 10.1128/jb.181.17.5263-5272.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleischer R, Heermann R, Jung K, Hunke S. Purification, reconstitution, and characterization of the CpxRAP envelope stress system of Escherichia coli. J Biol Chem. 2007;282:8583–8593. doi: 10.1074/jbc.M605785200. [DOI] [PubMed] [Google Scholar]
- 25.Zhou X, Keller R, Volkmer R, Krauss N, Scheerer P, Hunke S. Structural basis for two-component system inhibition and pilus sensing by the auxiliary CpxP protein. J Biol Chem. 2011;286:9805–9814. doi: 10.1074/jbc.M110.194092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thede GL, Arthur DC, Edwards RA, Buelow DR, Wong JL, Raivio TL, Glover JN. Structure of the periplasmic stress response protein CpxP. J Bacteriol. 2011;193:2149–2157. doi: 10.1128/JB.01296-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buelow DR, Raivio TL. Cpx signal transduction is influenced by a conserved N-terminal domain in the novel inhibitor CpxP and the periplasmic protease DegP. J Bacteriol. 2005;187:6622–6630. doi: 10.1128/JB.187.19.6622-6630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Isaac DD, Pinkner JS, Hultgren SJ, Silhavy TJ. The extracytoplasmic adaptor protein CpxP is degraded with substrate by DegP. Proc Natl Acad Sci USA. 2005;102:17775–17779. doi: 10.1073/pnas.0508936102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakazaki R, Iwanami S, Fukumi H. Studies on the enteropathogenic, facultatively halophilic bacteria, Vibrio parahaemolyticus. I. Morphological, cultural and biochemical properties and its taxonomical position. Jpn J Med Sci Biol. 1963;16:161–188. doi: 10.7883/yoken1952.16.161. [DOI] [PubMed] [Google Scholar]
- 30.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:9. [Google Scholar]
- 31.Emami K, Topakas E, Nagy T, Henshaw J, Jackson KA, Nelson KE, Mongodin EF, Murray JW, Lewis RJ, Gilbert HJ. Regulation of the xylan-degrading apparatus of Cellvibrio japonicus by a novel two-component system. J Biol Chem. 2009;284:1086–1096. doi: 10.1074/jbc.M805100200. [DOI] [PubMed] [Google Scholar]
- 32.Neiditch MB, Federle MJ, Miller ST, Bassler BL, Hughson FM. Regulation of LuxPQ receptor activity by the quorum-sensing signal autoinducer-2. Mol Cell. 2005;18:507–518. doi: 10.1016/j.molcel.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 33.Ponting CP, Aravind L. PAS: a multifunctional domain family comes to light. Curr Biol. 1997;7:R674–R677. doi: 10.1016/s0960-9822(06)00352-6. [DOI] [PubMed] [Google Scholar]
- 34.Ayers RA, Moffat K. Changes in quaternary structure in the signaling mechanisms of PAS domains. Biochemistry. 2008;47:12078–12086. doi: 10.1021/bi801254c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slamti L, Waldor MK. Genetic analysis of activation of the Vibrio cholerae Cpx pathway. J Bacteriol. 2009;191:5044–5056. doi: 10.1128/JB.00406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwon E, Kim DY, Gross CA, Gross JD, Kim KK. The crystal structure Escherichia coli Spy. Protein Sci. 2010;19:2252–2259. doi: 10.1002/pro.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheung J, Bingman CA, Reyngold M, Hendrickson WA, Waldburger CD. Crystal structure of a functional dimer of the PhoQ sensor domain. J Biol Chem. 2008;283:13762–13770. doi: 10.1074/jbc.M710592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyatake H, Mukai M, Park SY, Adachi S, Tamura K, Nakamura H, Nakamura K, Tsuchiya T, Iizuka T, Shiro Y. Sensory mechanism of oxygen sensor FixL from Rhizobium meliloti: crystallographic, mutagenesis and resonance Raman spectroscopic studies. J Mol Biol. 2000;301:415–431. doi: 10.1006/jmbi.2000.3954. [DOI] [PubMed] [Google Scholar]
- 39.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 40.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 41.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:20. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 42.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Cryst. 1999;D55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Cryst. 2003;D59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 47.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 48.Kabsch W. A solution for the best rotation to relate two sets of vectors. Acta Cryst. 1976;A32:2. [Google Scholar]
- 49.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 51.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 52.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 56.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]


